Abstract
Introduction:
MicroRNAs (miRNA) are a class of small non-coding RNA that play a major role in various cellular processes by negatively regulating gene expression. In the past decade, miRNA dysregulation has been reported to be closely linked to inflammatory diseases. The immune response modulates cancer initiation and progression; miRNAs including let-7 family members have been shown to act as key regulators of the immune responses in various diseases and cancers. Notably, the let-7 miRNA has been reported to be closely associated with immunity, specifically with Toll-like receptors that mediate cytokine expression during pathogen infection and with the regulation of various other immune effectors.
Areas covered:
In this review, the authors describe the discovery of let-7 as the starting point of the RNA revolution and highlight let-7 as an efficient tool for cancer and immune therapy.
Expert opinion:
let-7 miRNA has emerged as a key player in cancer therapy and immune responses and it has potential role as a new immunotherapeutic target. However, while there are challenges regarding miRNA delivery, the exciting emergence of personalized medicine for cancer and immunotherapy could be beneficial for the development of let-7 therapeutics.
Keywords: let-7, miRNAs, RNA therapy, immune response, immunotherapy
1. Introduction
MicroRNAs (miRNA or miRs) are small non-coding RNA, which were first discovered in 1993 in Caenorhabditis elegans (C. elegans), followed 7 years later by the discovery of the first mammalian miRNA [1,2]. Their discovery opened a new paradigm for gene regulation [3]. MiRNAs are transcribed by RNA polymerase-II, are 18–25 nucleotides-long, and play a major role in various cellular processes. miRNAs negatively regulate gene expression by acting through imperfect complementary base pairing to the 3′-untranslated region (UTR) of target mRNAs, regulating their translational inhibition or decay. let-7, the second miRNA discovered in C. elegans is extremely conserved among species including humans. Since its discovery, let-7 studies have substantially expanded and paved the way for the ncRNA revolution, allowing the emergence of the concept of miRNAs acting as tumor suppressors, and later as oncomiRs.
MiRNAs, including let-7 are dysregulated in a plethora of diseases, including cancers, and let-7 is currently implicated in a variety of approaches for therapy and diagnosis. let-7 has been largely described as a tumor-suppressive miRNA because it is under-expressed in multiple cancers. The emergence of the use of small chemically modified nucleic acid compounds as miRNA mimics or antimiRs allows one to manipulate let-7 expression for therapeutic approaches. let-7 reintroduction, using mimics, in cancer has led to repressed tumor growth and metastasis in numerous tumor types. let-7 dysregulation has been reported to be closely linked to inflammatory diseases and immune response modulates cancer initiation and progression. In this review the authors will summarize how, since its discovery, the let-7 miRNA, has emerged as a key player in cancer therapy and immune responses and we highlight its potential role as a new immunotherapeutic target.
2. let-7 discovery
The first miRNA, lin-4, discovered in the C. elegans model paved the way to a new gene regulation paradigm where short non-coding RNAs of 22 nucleotides long have the ability to repress mRNA expression by specific binding to the 3′UTR [1, 4]. Reinhart et al. subsequently demonstrated that another small non-coding RNA named let-7 (for lethal-7) encoded for a 21-nucleotide RNA that is complementary to the 3′ UTR regions of lin-14, lin-28, lin-41, lin-42 and daf-12, and was a key regulator of the temporal sequence of stem cell development in C. elegans development [2,5]. An important aspect of let-7’s role during C. elegans late-stage development has been demonstrated through the use of mutant let-7 worms. The authors showed that the mutants die because some of their cells cannot successfully complete terminal differentiation. Specifically, the mutant seam stem cells fail to exit the cell cycle and fail to express determinants required for proper formation of adult cell fates [2]. This evidence bolstered the idea that a small RNA gene is able to control the temporal sequence of development in C. elegans and more generally across animal phylogeny [6]. In 2000, the same group reported the expression of let-7 RNA in human cells and that expression levels of the human let-7 are different among various tissues, indicating a possible cell-type regulation of let-7 expression [6]. In 2004, a study investigating miRNA expression level by northern blot from 159 non-small cell lung carcinoma (NSCLC) tissues and cell lines showed for the first time a reduced expression of let-7 in tumor tissues [7]. Later in 2006, by using LNA microarrays to compare and profile miRNA expression in tumor breast samples compared to normal, it was shown that high expression of miRNAs including let-7 occurs in various adult tissues but more predominantly in luminal epithelial cells than malignant cells [8,9]. Later let-7 expression level was found to be downregulated in many human cancers [10].
let-7 and many other miRNAs are extremely well conserved across animal species and have multiple orthologs in each species, including in human and mouse [6]. In humans, mature let-7 members are encoded by 13 genomic loci. The let-7 family is composed of 10 different let-7 sequences annotated from let-7-a to let-7-i that share the same seed sequence (nucleotides 2–8) for target recognition. Only the let-7-a sequence UGAGGUAGUAGGUUGUAUAGUU is fully conserved across the following species: mouse, human, nematode, fly, frog, chicken, zebrafish, and dog; whereas the other isoforms differ in some nucleotide positions [11].
Since the discovery of lin-4 and let-7, 439 mature C. elegans miRNA sequences (cel-miR) have been discovered. From that starting point, the miRNA revolution has led to the discovery of 2694 new mature sequences for human miRNAs (hsamiRNAs), 2014 mature sequences for mouse (mmu-miRNAs), and 430 in Arabidopsis thaliana (ath-mIR), the majority of which have not yet been extensively studied, used, developed, and modified for therapy (http://miRBase.org).
3. let-7: the starting point of the RNA revolution
Since the discovery of human let-7, which sparked the ncRNA revolution and led to a variety of impressive discoveries, miRNA has emerged as an innovative tool for therapy [12]. Briefly, from early 2000, by studying miRNAs as a new class of gene products in chronic lymphocytic leukemias, the dogma that miRNAs can be dysregulated in cancer and associated with a cancer signature emerged [13]. In 2003, the discovery that one miRNA can target multiple genes by its specific miRNA seed sequence revolutionized our understanding of the regulation of the mammalian transcriptome [14]. The emergence of advanced techniques and methods of analysis led to the first report of down-regulation of let-7 in lung tumor samples, paving the way to using alteration of miRNAs with high clinical potential as biomarkers [7]. Concomitantly, by analyzing the increase of mature miRNAs from the mir-17–92 locus in B-cell lymphoma samples compared to normal tissues, the new concept that miRNA could act as an ‘oncomiR’ was developed [15]. One strong piece of evidence of oncomiR addiction was demonstrated by our group in the model of miRNA-21-induced pre-B-cell lymphoma in 2010 [16]. By generating a mouse model that allows tissue-specific and doxycycline-controlled expression of miR-21 we obtained mice that develop hematological malignancies. To highlight the role of miR-21 addiction to tumor development, we switched off miR-21 expression in established tumors by feeding mice with doxycycline-impregnated food and observed tumor regression after 48 h. This strong evidence demonstrated a central role of miR-21 expression in tumor initiation and maintenance and confirmed the dogma of miRNA addiction in tumors [16]. The complete comprehension of miRNA expression in disease has allowed identification of misregulation of miRNA in other sites like tumor metastases [17,18], blood circulation [19,20] and later in tumor Cancer Cell-Derived exosomes and the microenvironment [21].
To treat oncomiR-addicted tumors, researchers have used a number of innovative tools including the first generation of an inhibitor of miRNA function in C. elegans that consists of 2′-O-methyl oligonucleotides. To test it, researchers injected this let-7-c oligonucleotide modified compound into C. elegans larvae and observed an efficient derepression of lin-41, a well-known let-7 target [22]. From that point, a variety of innovative tools for inhibition or reintroduction of miRNAs have been developed, including locked-nucleic acid (LNA)-inhibitor for various miRNA with oncogenic functions like miR-221 or miR-155 [23,24] see Rupaimoole and Slack for a review [25].
Because it is known that naked miRNA oligonucleotides are rapidly degraded in blood circulation, optimization of new drug delivery systems allowing potent and safe delivery of RNA-based therapy including miRNAs are needed for in vivo applications. One of them, first described in 2008, highlights a reduction of orthotopic KRAS G12D lung tumors after intranasal let-7 administration. Then, enhancement of anti-miRs stability using 2′-O-methyl-modified nucleotides and phospho rothioate bonds lead to significantly reduces tumor burden in another model of lung cancer [26,27]. These let-7 therapeutics has led to a generation of modified mimic formulations, including delivering the miRNA in a lipid-based delivery vehicle designed for systemic delivery of the miR-34a to lung tumor [28]. In the meantime, the first miRNA antagomir targeting miR-122 entered clinical phase I trials, followed by a miR-34a mimic in 2012. The RNA revolution continues to progress with development of various innovative engineering tools for RNA-based delivery, including use of peptide nucleic acid antimiRs with a low pH-induced transmembrane structure (pHLIP) targeting the acidic tumor microenvironment [29], and the tumor-penetrating-nanoparticles miR-21 (TPN-21) that pave the way to using miRNAs for personalized medicine in pancreatic cancer [30].
Meanwhile, deep sequencing has uncovered lncRNAs, circRNAs and various epigenetic regulations of non-coding RNA elements that are now extensively studied for further fundamental comprehension and therapeutic applications [31,32]. To date, http://Clinicaltrial.gov, a website that collects data on privately and publicly funded clinical trials, shows that there are 516 clinical trials ongoing or recruiting for miRNA research, 21 for lncRNAs and 2 for circRNAs.
Let-7 sparked the start of this RNA revolution and continues to play a central role in RNA therapeutic fields with more than 1479 publications to date. Currently, six clinical trials are recruiting patients, active, or terminated for studying let-7 in various diseases including obesity, cancer, and diabetes, where it can be detected in a variety of samples from serum to tissues. For example, in one of these trials, they are using let-7 for disease prognosis by investigating one let-7 family member (let-7i) in the patient’s serum to detect intracranial traumatic lesions [33]. A second study hopes to detect the level of various let-7 members before and after radiotherapy to study neurological complications after radiotherapy in the brain, and in that case, let-7 will be studied to predict chemoresistance [34]. For a clinical trial regarding obesity, they are measuring let-7e in the blood after a specific diet to follow diet loss and search for a biomarker for obesity.
Altogether, these examples of let-7 utilization in different clinical trials highlight that let-7, and more generally miRNAs, are extensively used to classify and identify disease/cancer tissue origin, determine prognosis and disease progression, predict chemoresistance, monitor therapy, and screen for diseases – demonstrating huge promise for human therapy.
4. Let-7 as a tool for therapy
A widely employed approach used to study miRNA gain or loss of function is the use of chemically modified oligonucleo-tide sequestering or reintroducing miRNAs. The first in vivo miRNA inhibitor was tested by microinjection of 2′-O-Me oligonucleotide complementary to let-7 in C. elegans, promoting a loss-of-function phenotype in larvae [22]. Then, to allow a better contact with the cell membrane, 2′-O-Me oligonucleo-tides were modified by conjugation of a 3′ cholesterol motif, called antagomirs and tested first by intravenous injection of miR-16 in normal mice [35]. Chemical inhibition using an anti- let-7 2′OMe oligonucleotide underlined that let-7 miRNA is able to promote the repression of cell proliferation pathways in human cells [36]. From this evidence, various therapeutic approaches have emerged for the targeting or re-introduction of let-7 for therapy.
Our group showed one of the first pieces of evidence that let-7 plays a key role in cancer. By showing that RAS is regulated by the let-7 miRNA family in C. elegans and by highlighting let-7 complementary sites in human RAS 3′UTRs, we first validated that the let-7 miRNA family negatively regulates RAS in two different C. elegans tissues and human cell lines and lung tissue. This was the first report of the mechanism of action of a miRNA acting as a tumor suppressor [37].
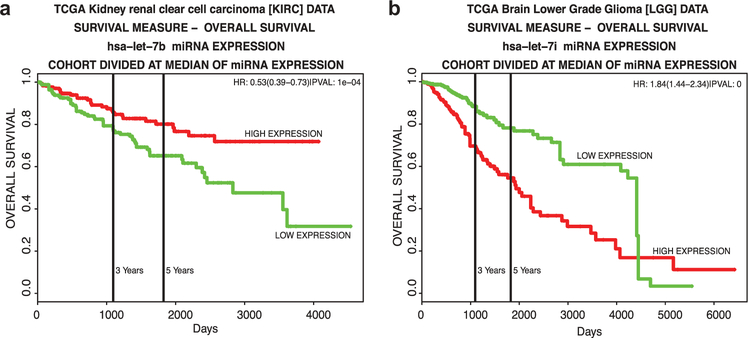
Later, let-7 was associated with lung cancer by showing that ectopic expression of let-7g in Kras driven lung cancer cells promoted both cell cycle arrest and cell death [38]. More generally, all the let-7 family members are dysregulated in cancer with the majority of them found downregulated. Loss of let-7 family members also predicts poor survival in various types of cancer [39]. For therapy, numerous innovative approaches using tumor-suppressive activity of let-7 have been used for cancer therapy. For example, systemic mimic delivery using neutral lipid emulsions to reintroduce let-7 in lung tumors. This delivery system showed that systemic delivery of let-7b promoted its accumulation into the tumor site and induced tumor-inhibitory effects in an autochthonous KRASG12D transgenic mouse model of lung cancer [40]. Table 1, highlights which specific let-7 family member expression levels (high or low) is associated with a significant low overall survival from a variety of cancers patient databases [41]. This table suggests not only which tumor types might benefit most from let-7 therapy, but also suggests which let-7 family members have to be targeted or reintroduced for therapy. This analysis suggests that Kidney renal clear cell carcinoma and Brain lower grade glioma tumor types may benefit most from let-7 therapy (respectively mimics and anti-miRs) (Figure 1).
Table 1.
Let-7 family member expression associated with poor overall survival in cancers. Table 1 has been generated from the PROGmiR tool by querying each let-7 family member’s expression level and correlating this with overall survival (p-value ≤0.1) among 33 human cancer databases. Unsignificant result (p-value ≥0.1) from Cholangiocarcinoma [CHOL] Glioblastoma multiforme [GBM], Thymoma [THYM], Breast invasive carcinoma [BRCA], Diffuse Large B-cell Lymphoma, Cervical squamous cell carcinoma, and endocervical carcinoma, Rectum adenocarcinoma [READ] are not presented in the table.
| Cancer types | Low miRNA expression associated to low survival | High miRNA expression associated to low survival | ||
|---|---|---|---|---|
| Adrenocortical carcinoma [ACC] |
let-7 family member let-7c |
p-Value 0.016948897 |
let-7 family member let-7b let-7d let-7g let-71 |
p-value 0.030754687 0.081720566 0.052540823 0.033604282 |
| Kidney renal papillary cell carcinoma [KIRP] |
let-7 family member let-7g |
p-Value 0.006148305 |
let-7 family member let-71 |
p-value 0.006714306 |
| Lung adenocarcinoma [LUAD] |
let-7 family member let-7b let-7c |
p-Value 0.053530532 0.020505798 |
let-7 family member | p-value |
| Pancreatic adenocarcinoma [PAAD] |
let-7 family member let-76 let-7g |
p-Value 0.000419855 0.007191681 |
let-7 family member let-7b |
p-value 0.000614112 |
| Sarcoma [SARC] | let-7 family member | p-Value |
let-7 family member let-7d let-71 |
p-value 0.01726064 0.019594756 |
| Thyroid carcinoma [THCA] |
let-7 family member let-7d |
p-Value 0.034278599 |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7f1 Iet-7f2 let-7g |
p-value 0.04003319 0.040378847 0.039595321 0.038909032 0.023087711 0.008662061 |
| Uveal melanoma [UVM] |
let-7 family member let-7c let-7g |
p-Value 0.000107251 0.104325918 |
let-7 family member let-7b let-7e |
p-value 0.002430699 0.04168769 |
| Bladder urothelial carcinoma [BLCA] |
let-7 family member et-7f1 let-7g |
p-Value 0.019933527 0.015729926 |
let-7 family member let-7c |
p-value 4.72E-05 |
| Colon adenocarcinoma [COAD] | let-7 family member | p-Value |
let-7 family member let-7c let-7e let-7g let-7i |
p-value 0.086099976 0.063650106 0.032521736 0.073548534 |
| Head and neck squamous cell carcinoma [HNSC] |
let-7 family member let-7c |
p-Value 0.004211464 |
let-7 family member | p-value |
| Acute myeloid leukemia [LAML] | let-7 family member | p-Value |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7b let-7c |
p-value 0.068066729 0.110382351 0.069096686 0.000228442 0.03908679 |
| Lung squamous cell carcinoma [LUSC] |
let-7 family member Iet-7f1 Iet-7f2 let-71 |
p-Value 0.028973805 0.098768735 0.099783464 |
let-7 family member | p-value |
| Pheochromocytoma and paraganglioma [PCPG] |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7b let-7c let-7e Iet-7f2 let-7g |
p-Value 0.006725665 0.006375698 0.00709643 0.090803967 0.082592342 0.032377698 0.013186697 0.109684624 |
let-7 family member | p-value |
| Skin cutaneous melanoma [SKCM] |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7b let-7f1 let-7g |
p-Value 0.00202646 0.002124937 0.001942797 0.014905228 0.088167942 0.009275654 |
let-7 family member | p-value |
| Kidney chromophobe [KICH] |
let-7 family member let-7c |
p-Value 0.001515206 |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7d let-712 let-71 |
p-value 0.038444651 0.039861791 0.037566956 0.032730936 0.086293043 0.070056101 |
| Brain lower grade glioma [LGG] | let-7 family member | p-Value |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7c let-7e let-7g let-7i |
p-value 0.037891823 0.038907961 0.039434459 0.006373482 6.17E-05 0.002189787 9.02E-07 |
| Mesothelioma [MESO] |
let-7 family member Iet-7f1 let-7i |
p-Value 0.059549538 0.101150698 |
let-7 family member | p-value |
| Prostate adenocarcinoma [PRAD] |
let-7 family member let-7c |
p-Value 0.028908849 |
let-7 family member let-7d let-7g |
p-value 0.006208984 0.05622126 |
| Stomach adenocarcinoma [STAD] |
let-7 family member Iet-7f2 |
p-Value 0.078859686 |
let-7 family member let-7c |
p-value 0.033118777 |
| Uterine corpus endometrial carcinoma [UCEC] |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7b Iet-7f2 |
p-Value 0.005929407 0.005870681 0.005808579 0.013321126 0.029634711 |
let-7 family member | p-value |
| Esophageal carcinoma [ESCA] |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7b |
p-Value 0.058000018 0.054346774 0.054789831 0.119825178 |
let-7 family member | p-value |
| Kidney renal clear cell carcinoma [KIRC] |
let-7 family member let-7a-1 let-7a-2 let-7a-3 let-7b let-7c let-7e |
p-Value 0.009771075 0.01042424 0.009462964 9.10E-05 0.001029453 0.082738752 |
let-7 family member let-7i |
p-value 0.005679393 |
| Liver hepatocellular carcinoma [LIHC] |
let-7 family member let-7c |
p-Value 0.019666702 |
let-7 family member let-7d let-7i |
p-value 0.086695391 0.083386848 |
| Testicular ggerm ccell tumors [TGCT] | let-7 family member | p-Value |
let-7 family member let-7f1 let-7i |
p-value 0.108508216 0.055108285 |
| Uterine carcinosarcoma [UCS] | let-7 family member | p-Value |
let-7 family member Iet-7f1 |
p-value 0.081338369 |
| Ovarian serous cystadenocarcinoma [OV] |
let-7 family member let-7d |
p-Value 0.091576628 |
let-7 family member let-7a-1 let-7a-2 let-7a-3 |
p-value 0.0770986 0.078229956 0.078569352 |
Figure 1.
miRNA prognostic plots from two let-7 family member that may benefit most from let-7 therapy. (a) Overall survival of hsa-let-7b miRNA expression in Kidney renal clear cell carcinoma. (b) Overall survival of hsa-let-7i miRNA expression in Brain lower grade glioma.
Inhibition of the oncogene c-MYC by the reintroduction of the same let-7b mimic has been shown to reverse multidrug resistance in gastric cancer and similarly, let-7 sensitizes cells to chemotherapy in a model of KRAS mutant cells [42,43]. Favorable outcomes in other diseases, such as preventing obesity-induced glucose intolerance or improving cardioprotection and cardiac function, using anti-let-7 strategies have also been observed [44–46]. Diverse mimics have also been used to reintroduce let-7 family members to counteract HCV, brain injury, endometrioses or inflammation [47–50].
Interestingly, a variety of approaches have also targeted key upstream effectors of let-7 for therapy. The most notable way is to target Lin28b, a well-known regulator of the let-7 family member expression/accumulation [51]. Lin28b targeting has been used for pancreatic cancer therapy where downregulation of oncofetal protein Lin28b through SIRT6 led to increased expression of let-7 family member and acted as a method to promote growth inhibition of PDAC cells models [52].
5. let-7 interacts with immune effectors during infections
MiRNAs have been reported to contribute to the formation and progression of diseases like cancer, but also to inflammatory disorders [53]. The body’s initial defense against external pathogen infection is called the innate immune response. This immune response is well organized and starts with detection of the external pathogen by macrophages or dendritic cells and their specific receptors like (Toll-like). Activation of Toll-like receptors triggers many biological processes including apoptosis and cytokine expression that help counteract infections. In 2004, a Science paper showing that the production of mature B and T lymphocytes was modified after ectopic expression of miRNAs in hematopoietic stem cells and immature lymphocytes supported the idea that miRNAs represent a class of molecules that regulate hematopoiesis and are important in the immune system [54]. One of the first pieces of evidence linking the let-7 miRNA and the immune response emerged in the model of SV40-transformed human cholangiocytes derived from the normal liver that were screened using a semi-quantitative microarray for 385 human miRNAs. In this study, using computational prediction analysis, they found that three members of the let-7 family (let-7b, let-7i, let-7g) have complementarity to the 3′-UTR of toll-like receptor 4 (TLR4) mRNA [55]. In an in vitro model, modulation of let-7i caused alterations in immune response to C. parvum infection through modulation of TLR4 expression [55]. Additionally, let-7 miRNA has been linked to the TLR-mediated inflammatory response in many other diseases such as coronary artery disease and in an oxygen-glucose deprivation model [56,57]. In dilated cardiomyopathy, levels of let-7i are negatively correlated with TLR4 protein levels. Furthermore, low levels were associated with poor clinical outcome [58]. As miRNAs take part in the immune response against viral, bacterial, fungal, and parasitic infections, studies have also highlighted a relationship between let-7 family members and the regulation of the innate immune response to pathogenic infection and stress. They all show that let-7 family members target TLR4 to regulate innate immunity and inflammation in response to P. aeruginosa or Helicobacter pylori infection [59–61]. It has also been reported that use of loss-of-function let-7 mutants impaired the colony-forming units of a type of P. aeruginosa in the nematode model. This change of let-7 expression drove increased transcriptional expression of specific antimicrobial genes that enhance the innate immune response [59].
By performing a functional anti-HCV (Hepatitis C Virus) luciferase reporter screen using mimic and inhibitors, a study demonstrated an important role for let-7b during HCV infection. They showed that deletion of the wild-type seed region of let-7b plays a crucial role in its antiviral activity via targeting of the HCV genome and host gene [62].
Another key effector of immune response and inflammation is the transcription factor NF-κB, which promotes immunity by driving expression of target genes that mediate cell proliferation and the release of antimicrobial molecules and cytokines to activate the immune response. During Mycobacterium tuberculosis infection (Mtb), let-7f has been shown to target A20, a feedback inhibitor of the NF-κB pathway that regulates the innate immune response. Moreover, downregulation of let-7f is associated with concomitant upregulation of a number of its putative targets. These results reveal a role for let-7f and its target A20 in regulating immune responses to Mtb and is directly associated with enhanced NF-κB activity [63].
In other infectious diseases, let-7 has been linked to the regulation of NF-κB. First, a significant decrease in let-7i was observed by northern blot in H69 cells after C. parvum infection. Moreover, reintroduction or inhibition of let-7i precursor in H69 cell line can modify SIRT1 protein expression and could modulate NF-κB activation [64]. Taken together, these examples highlight a strong role for let-7 in the immune response and inflammation, and suggest that let-7 may also regulate cancer cell immune evasion.
6. Let-7 as a new tool for immunotherapy
Cancer development and progression have been extensively studied and a variety of hallmarks of cancer are now extremely well-known, allowing researchers to propose innovative therapies. Among them, cancer immunity, i.e. immune cell infiltration into the tumor site and tumor microenvironment, regulates the equilibrium between pro or anti-tumoral behavior [65]. Various molecular players are known to regulate cancer immune response, including miRNAs [66]. Let-7 has been reported to control maintenance and development of immature and mature immune cells [66] but also to control differentiation and function of natural killer T cells, a subset of T cells [67,68]. The role of let-7 in the suppressive function of Tregs has also been reported [3] as well as that let-7 family members suppresses B cell activation [69].
MiRNA and specifically, let-7c has been reported to play a role in regulating macrophage plasticity. Briefly, macrophages have the ability to switch between different functional pheno-types that are called M1 or M2. M1 macrophages are critical to counteract external infections by bacteria or viruses whereas M2 macrophages are found in the inflammatory zone and participate in many remodeling biological process such as angiogenesis and tumor progression. In a fibrotic mouse model, isolated macrophages showed significantly increased let-7c levels. By loss and gain of function studies, the authors show that let-7c is a negative regulator of M1 macrophage phenotypes and that let-7c overexpression promotes the GMBMM transition to the M2 phenotype [70].
Another study reported that let-7 facilitates T-cell anergy. mTOR and Rictor, two members of MTOR, are miRNA targets in cancer. By using miRNA deficiency approaches, it has been shown that ability of CD4 T cells to decide between an activating and anergizing state requires post-transcriptional regulation of the mTOR components by let-7 and miR-16. These results show that expression of mTOR components in T cells is regulated by let-7 miRNAs [71].
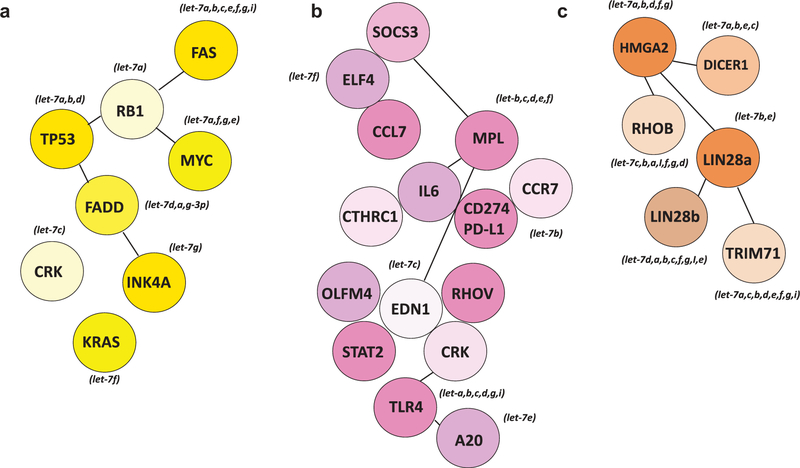
As described earlier, let-7 family members generally function as tumor suppressors by targeting multiple oncogenes, including RAS, HMGA2, and c-Myc, but also act as immune regulators (Figure 2) [72]. Multiples lines of evidence described earlier suggest that the let-7 family can distort or inhibit immune responses and promote tumor immune evasion. A study performed in 2016 in a colorectal cancer model highlighted the inverse association between let-7a expression levels and the densities of CD8+ cells and CD45RO+ T cells in the tumor site, associated with a favorable prognosis. They also highlighted a correlation between let-7a expression and colorectal cancer mortality. This work nicely supports the idea that let-7a can suppress antitumor immunity in this cancer and suggests that let-7a could inhibit the T cell immune response against colorectal cancer, highlighting let-7a as a good target for immunotherapy [73].
Figure 2.
let-7 targets with oncogene/tumor suppressor properties or immune-related properties. Validated targets gene were identified in DIANA-TarBase v8 considering the top candidates (1 ~ 40) of validated targets for each let-7 family member. Shown are key let-7 family target genes with oncogene or tumor suppressor properties (a) or immune-related properties (b) or general let-7 family targets conserved among family members (c).
CD8 T cells promote rapid clearance of cancer cells or virus-infected-cells. In a study from 2017, the authors identified a new role for let-7 in CD8 T cell differentiation and function. They demonstrated that a high level of let-7 miRNAs is required to maintain the naive phenotype of CD8 T cells. Moreover, they show that overexpression of let-7 copy number impairs the proliferation and differentiation of CD8 T cells by modulating the expression of genes involved in the regulation of the cell cycle and metabolism, such as Myc [74].
Cytokines are soluble substances of 8 to 50 kDa, synthesized and secreted by various tissues or cells, including immune cells, to regulate the functions other cells. Cytokines include different subgroups like interferons, interleukins or chemokines. Their production is mediated in response to foreign antigen presentation by the immune system. One of the first reports linking let-7 to cytokine expression was published in 2008. By computational approaches, the authors reported a putative miRNA binding site for let-7 in the interleukin 6 sequence (IL-6) [75]. In another study using RAW 264.7 cells that have been infected with the bacteria Salmonella, the authors observed a downregulation of let-7 family members. By monitoring cytokine expression using a Renilla luciferase ORF in a reporter vector that also expresses firefly luciferase in mouse embryonic fibroblast (MEF) cells, they observed that let-7 family members post-transcriptionally repress the production of cytokines interleukin-10 and 6 (IL-10, IL-6). This study was the first to provide molecular evidence of how the let-7 family was implicated in anti-bacterial defense during Salmonella infection and to show that
Repression of let-7 family alleviates the negative post-transcriptional regulation of IL-6 and IL-10, which are the key players of the immune response. This work establishes a strong link between let-7 expression and the modulation of activation of inflammatory factors during pathogen invasion [76]. Similarly, in HIV-1 infection, downregulation of let-7 miRNAs promotes IL-10 expression from CD4(+) T cells. As elevated IL-10 occurs during HIV-1 infection, and because let-7 regulates IL-10 expression in CD4 T cells, the decrease of let-7b and c expression after infection drives an immune change favorable for the virus survival [77].
It has been also reported that IL-10, an anti-inflammatory cytokine, has a well-established role in neuroprotection. One paper demonstrated that extracellular miRNA let-7, which is highly expressed in the central nervous system, induces neuronal cell death [78].
Let-7 family members have been also shown to target another interleukin (IL-13) in vivo and in vitro in a lung model of experimental asthma [79]. Interferon-γ (IFN-γ) is an important member of the interferon family that regulates various biological purposes like the antiviral response, and immunomodulatory responses. Interferon-γ is an essential player in the modulation of anti-tumor effector because it sensitizes Fas-related apoptosis in colon cancer cells. Using a luciferase expression assay, it has been reported that let-7 directly inhibits expression of Fas by binding to a defined target sequence in the 3′UTR. By combining let-7 inhibitor and FAS mABs they also determined that let-7 suppression sensitizes Fas-induced apoptosis of HT29 cells in vitro as well as in vivo [80]. As FAS and let-7 are inversely correlated in various other cancer cells these findings could notably influence new clinical approaches for tumor therapy.
In another recent study, a group has studied the relationship between let-7 expression, Fas and FasL and bone marrow-derived mesenchymal stem cells (MSCs) for the treatment of inflammatory diseases. It is already known that Fas and FasL are essential for MSCs to induce T cell apoptosis. Using an in vitro transwell system and an experimental colitis mouse model, they confirmed that knockdown of let-7a in MSCs significantly promoted MSC-induced T cell migration and apoptosis in vitro and in vivo. In addition, they observed a strong change in immune response by suppression of the inflammation reaction in MSCs from a let-7a knocked down mouse model. Altogether, by identifying let-7a as a negative modulator of FasL/Fas they highlight a nice way to improve MSC immunotherapy by enhancing FasL/Fas expression [81]. Finally, these studies show that let-7 modulation can be a tool to suppress tumor immune reactions.
7. Conclusion
miRNAs and specifically let-7 are very attractive targets for cancer therapy but have also emerged now as key players in immune responses that modulate cancer initiation and progression. As described earlier, many studies suggest that the let-7 family can regulate or inhibit immune responses and promote tumor immune evasion, and multiple strategies have emerged to develop therapeutic approaches for the targeting or re-introduction of let-7 for therapy. Taken together, this review highlights let-7 as a new target for immunotherapy.
8. Expert opinion
let-7 is the most studied miRNA in biology and the let-7 family plays a major role in the regulation of pluripotency and differentiation in many species. It plays a major role in various process including development, lifespan, cell proliferation, differentiation, signaling pathways, apoptosis and metabolism but also in aging-associated diseases, viral infections, genetic disorders and cancer [82]. Here we have collated a non-exhaustive list of examples and scientific reports which highlight that let-7 family members are an important node in the miRNA regulatory network that governs the immune response. More specifically, through regulation of Toll-like receptors, immune cell activation, T-cell anergy, and cytokine regulation, let-7 appears to be a great tool for immunotherapy. Specifically, let-7 therapy could be beneficial for some specific immune cell types including macrophages [70], lymphocytes T cells (NK, CD8+, CD4+, Treg) [67,68,73,77], lymphocytes B cells [69] and Bone Marrow-Derived Mesenchymal Stem Cells [81]. As novel systems of drug delivery for RNA medicine have emerged [25,29,30], the reintroduction of let-7 could be a strategy for the modulation of immune reactions and immunotherapy in cancer. However, as let-7 is closely linked to immune response in various cancer cell models, utilization of RNA medicine for human therapy has to be handled with care and precision.
The first-in-human miRNA therapeutic trial (MRX-34 Phase I clinical trial) demonstrated some immune abnormalities including lymphopenia, neutropenia, and thrombocytopenia and side effects leading to five immune-related serious adverse events, causing the cessation of the phase I trial [83]. Because let-7 family members can also display strong immune response upon various immune mediators (myeloid compartment, T cell, or Toll-like receptors) that we previously highlighted: utilization of let-7 family members for therapy has to carefully developed to avoid unexpected and non-specific immune response. To this end, utilization of specific drug delivery systems that will target only tumor cells and relevant stroma has to be selected for human therapy. In addition, targeting specific cell surface receptors and adoption of next-generation oligonucleotide chemistry that will not initiate unwanted immune responses have to be developed for let-7 RNA delivery. Finally, as current anti-cancer strategy has shown better efficiency by combining two or multiple therapeutic approaches (such as chemotherapy and immunotherapy), combo-therapy between the let-7 family member and any other cancer modulator has to carefully studied to avoid non-specific cancer-related immune responses.
Another interesting point is the emergence of personalized medicine for therapy with use of personalized avatar models that help guide precision medicine for individual patients that show different genetic or immune patterns. In our previous study, through the utilization of Patient-Derived-Organoids as a rapid screening platform for our RNA-based-therapeutic (TPN-21) we showed for the first time, the potential of personalized models for RNA-based precision medicine therapy in PDAC [30]. By working with patient avatars, we have shown the potential to predict clinical outcomes of patients and thus align pre-clinical work to patient response to our RNA therapy. This major change of therapeutically screening is changing the landscape of cancer therapy and immunotherapy and are a valuable approach to deepen our knowledge regarding the roles and impact of let-7 as a new target for immunotherapy.
Article Highlights.
let-7 discovered as the first mammalian miRNA
let-7 discovery was the starting point of the RNA revolution
Emergence of chemically modified oligonucleotide sequestering or reintroducing miRNAs as a novel therapeutic modality
let-7 family members are an important node in the miRNA regulatory network that governs cancer progression and the immune responses
This box summarizes key points contained in the article.
Acknowledgments
We thank Lisa Walker for critical reading of this manuscript.
Funding
This work was supported by a grant to FJ Slack by the Harvard Medical School Initiative for RNA Medicine, a grant from the Ludwig Center at Harvard ; National Cancer Institute, [P50CA196530–03S1
Footnotes
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993. December 3;75(5):843–854. [DOI] [PubMed] [Google Scholar]; • Key papers reporting let-7 discovery
- 2.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000. February 24;403(6772):901–906. [DOI] [PubMed] [Google Scholar]; • Key papers reporting let-7 discovery
- 3.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008. September;14(9):400–409.. [DOI] [PubMed] [Google Scholar]
- 4.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993. December 3;75(5):855–862. [DOI] [PubMed] [Google Scholar]
- 5.Slack FJ, Basson M, Liu Z, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000. April;5(4):659–669. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000. November 2;408(6808):86–89. [DOI] [PubMed] [Google Scholar]; • Key papers reporting let-7 discovery
- 7.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004. June 1;64(11):3753–3756. [DOI] [PubMed] [Google Scholar]
- 8.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007. December 15;67(24):11612–11620. [DOI] [PubMed] [Google Scholar]
- 9.Thomson JM, Newman M, Parker JS, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006. August 15;20(16):2202–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SM, Shell S, Radjabi AR, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007. November 1;6(21):2585–2590. [DOI] [PubMed] [Google Scholar]
- 11.Ruby JG, Jan C, Player C, et al. Large-scale sequencing reveals 21URNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006. December 15;127(6):1193–1207. [DOI] [PubMed] [Google Scholar]
- 12.Lieberman J Tapping the RNA world for therapeutics. Nat Struct Mol Biol. 2018. April 16 DOI: 10.1038/s41594-018-0054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002. November 26;99(24):15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003. December 26;115(7):787–798. [DOI] [PubMed] [Google Scholar]
- 15.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005. June 9;435(7043):828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development
- 16.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010. September 02;467(7311):86–90. [DOI] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development
- 17.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007. October 11;449(7163):682–688.. [DOI] [PubMed] [Google Scholar]
- 18.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008. January 10;451(7175):147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008. July 29;105(30):10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008. October;18(10):997–1006. [DOI] [PubMed] [Google Scholar]
- 21.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012. July 31;109(31):E2110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutvagner G, Simard MJ, Mello CC, et al. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004. April;2(4):E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Martino MT, Gulla A, Gallo Cantafio ME, et al. In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One. 2014;9(2):e89659.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012. June 26;109(26):E1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017. March;16(3):203–222.. [DOI] [PubMed] [Google Scholar]
- 26.Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008. March 15;7(6):759–764. [DOI] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development
- 27.Trang P, Medina PP, Wiggins JF, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010. March 18;29 (11):1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development
- 28.Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010. July 15;70(14):5923–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015. February 5;518(7537):107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development.
- 30.Gilles ME, Hao L, Huang L, et al. Personalized RNA-medicine for pancreatic cancer. Clin Cancer Res. 2018;24(7). Doi: 10.1158/1078-0432.CCR-17-2733. [DOI] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development.
- 31.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018. January;18(1):5–18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jegu T, Aeby E, Lee JT. The X chromosome in space. Nat Rev Genet. 2017. June;18(6):377–389.. [DOI] [PubMed] [Google Scholar]
- 33.Balakathiresan N, Bhomia M, Chandran R, et al. MicroRNA let-7i is a promising serum biomarker for blast-induced traumatic brain injury. J Neurotrauma. 2012. May 1;29(7):1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durand T, Jacob S, Lebouil L, et al. EpiBrainRad: an epidemiologic study of the neurotoxicity induced by radiotherapy in high grade glioma patients. BMC Neurol. 2015. December 18;15:261 Doi: 10.1186/s12883-015-0519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005. December 1;438(7068):685–689. [DOI] [PubMed] [Google Scholar]
- 36.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007. August 15;67(16):7713–7722. [DOI] [PubMed] [Google Scholar]
- 37.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005. March 11;120(5):635–647. [DOI] [PubMed] [Google Scholar]
- 38.Kumar MS, Erkeland SJ, Pester RE, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008. March 11;105(10):3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyerinas B, Park SM, Hau A, et al. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010. March;17(1):F19–36. [DOI] [PubMed] [Google Scholar]
- 40.Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011. June;19(6):1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Key papers reporting use of miRNAs and let-7 as a tool for therapy and starting point of RNA revolution and drug delivery development.
- 41.Goswami CP, Nakshatri H. PROGmiR: a tool for identifying prognostic miRNA biomarkers in multiple cancers using publicly available data. J Clin Bioinforma. 2012. December 28;2(1):23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Cai H, Liang Y, et al. Inhibition of c-Myc by let-7b mimic reverses mutidrug resistance in gastric cancer cells. Oncol Rep. 2015. April;33(4):1723–1730. [DOI] [PubMed] [Google Scholar]
- 43.Dai X, Jiang Y, Tan C. Let-7 Sensitizes KRAS Mutant Tumor Cells to Chemotherapy. PLoS One. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011. December 27;108(52):21075–21080.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeger T, Xu QF, Muhly-Reinholz M, et al. Inhibition of let-7 augments the recruitment of epicardial cells and improves cardiac function after myocardial infarction. J Mol Cell Cardiol. 2016. May;94:145–152. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Ren Y, Shi E, et al. Inhibition of the Let-7 family micrornas induces cardioprotection against ischemia-reperfusion injury in diabetic rats. Ann Thorac Surg. 2016. September;102(3):829–835. [DOI] [PubMed] [Google Scholar]
- 47.Li Q, Lowey B, Sodroski C, et al. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat Commun. 2017. November 27;8(1):1789 Doi: 10.1038/s41467-017-01954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv J, Zeng Y, Qian Y, et al. MicroRNA let-7c-5p improves neurological outcomes in a murine model of traumatic brain injury by suppressing neuroinflammation and regulating microglial activation. Brain Res. 2018. April;15(1685):91–104.. [DOI] [PubMed] [Google Scholar]
- 49.Cho S, Mutlu L, Zhou Y, et al. Aromatase inhibitor regulates let-7 expression and let-7f-induced cell migration in endometrial cells from women with endometriosis. Fertil Steril. 2016. September 1;106 (3):673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brennan E, Wang B, McClelland A, et al. Protective effect of let-7 miRNA family in regulating inflammation in diabetes-associated atherosclerosis. Diabetes. 2017. August;66(8):2266–2277. [DOI] [PubMed] [Google Scholar]
- 51.Okoye IS, Coomes SM, Pelly VS, et al. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity. 2014. September 18;41(3):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kugel S, Sebastian C, Fitamant J, et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. 2016. June 2;165(6):1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu G, Abraham E. MicroRNAs in immune response and macrophage polarization. Arterioscler Thromb Vasc Biol. 2013. February;33 (2):170–177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004. January 2;303(5654):83–86. [DOI] [PubMed] [Google Scholar]; ••• Key papers reporting use of let-7 as a tool immunotherapy
- 55.Chen XM, Splinter PL, O’Hara SP, et al. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007. September 28;282(39):28929–28938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satoh M, Tabuchi T, Minami Y, et al. Expression of let-7i is associated with Toll-like receptor 4 signal in coronary artery disease: effect of statins on let-7i and Toll-like receptor 4 signal. Immunobiology. 2012. May;217(5):533–539. [DOI] [PubMed] [Google Scholar]
- 57.Xiang W, Tian C, Peng S, et al. Let-7i attenuates human brain microvascular endothelial cell damage in oxygen glucose deprivation model by decreasing toll-like receptor 4 expression. Biochem Biophys Res Commun. 2017. November 4;493(1):788–793. [DOI] [PubMed] [Google Scholar]
- 58.Satoh M, Minami Y, Takahashi Y, et al. A cellular microRNA, let-7i, is a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. J Card Fail. 2011. November;17(11):923–929. [DOI] [PubMed] [Google Scholar]
- 59.Zhi L, Yu Y, Li X, et al. Molecular Control of Innate Immune Response to Pseudomonas aeruginosa Infection by Intestinal let-7 in Caenorhabditis elegans. PLoS Pathog. 2017. January;13(1):e1006152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren Z, Ambros VR. Caenorhabditis elegans microRNAs of the let-7 family act in innate immune response circuits and confer robust developmental timing against pathogen stress. Proc Natl Acad Sci U S A. 2015. May 5;112(18):E2366–75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teng GG, Wang WH, Dai Y, et al. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS One. 2013;8(2): e56709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng M, Si Y, Niu Y, et al. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013. September;87(17):9707–9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar M, Sahu SK, Kumar R, et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe. 2015. March 11;17(3):345–356. [DOI] [PubMed] [Google Scholar]
- 64.Xie H, Lei N, Gong AY, et al. Cryptosporidium parvum induces SIRT1 expression in host epithelial cells through downregulating let-7i. Hum Immunol. 2014. August;75(8):760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011. March 4;144(5):646–674.. [DOI] [PubMed] [Google Scholar]
- 66.Paladini L, Fabris L, Bottai G, et al. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. 2016. June 27;35:103 Doi: 10.1186/s13046-016-0375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••• Key papers reporting use of let-7 as a tool immunotherapy
- 67.Pobezinsky LA, Etzensperger R, Jeurling S, et al. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol. 2015. May;16(5):517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••• Key papers reporting use of let-7 as a tool immunotherapy.
- 68.Yuan J, Nguyen CK, Liu X, et al. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012. March 9;335(6073):1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••• Key papers reporting use of let-7 as a tool immunotherapy
- 69.Jiang S, Yan W, Wang SE, et al. Let-7 suppresses B cell activation through restricting the availability of necessary nutrients. Cell Metab. 2018. February 6;27(2):393–403e4. [DOI] [PubMed] [Google Scholar]
- 70.Banerjee S, Xie N, Cui H, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013. June 15;190(12):6542–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••• Key papers reporting use of let-7 as a tool immunotherapy
- 71.Marcais A, Blevins R, Graumann J, et al. microRNA-mediated regulation of mTOR complex components facilitates discrimination between activation and anergy in CD4 T cells. J Exp Med. 2014. October 20;211(11):2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018. January 4;46(D1): D239–D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dou R, Nishihara R, Cao Y, et al. MicroRNA let-7, T Cells, and Patient Survival in Colorectal Cancer. Cancer Immunol Res. 2016. November;4(11):927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••• Key papers reporting use of let-7 as a tool immunotherapy
- 74.Wells AC, Daniels KA, Angelou CC, et al. Modulation of let-7 miRNAs controls the differentiation of effector CD8 T cells. Elife. 2017. July 24;6 DOI: 10.7554/eLife.26398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asirvatham AJ, Gregorie CJ, Hu Z, et al. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008. April;45(7):1995–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schulte LN, Eulalio A, Mollenkopf HJ, et al. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. Embo J. 2011. May 18;30(10):1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swaminathan S, Suzuki K, Seddiki N, et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012. June 15;188(12):6238–6246. [DOI] [PubMed] [Google Scholar]
- 78.Mueller M, Zhou J, Yang L, et al. PreImplantation factor promotes neuroprotection by targeting microRNA let-7. Proc Natl Acad Sci U S A. 2014. September 23;111(38):13882–13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Polikepahad S, Knight JM, Naghavi AO, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010. September 24;285(39):30139–30149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geng L, Zhu B, Dai BH, et al. A let-7/Fas double-negative feedback loop regulates human colon carcinoma cells sensitivity to Fas-related apoptosis. Biochem Biophys Res Commun. 2011. May 13;408 (3):494–499. [DOI] [PubMed] [Google Scholar]
- 81.Yu Y, Liao L, Shao B, et al. Knockdown of microRNA Let-7a improves the functionality of bone marrow-derived mesenchymal stem cells in immunotherapy. Mol Ther. 2017. February 1;25(2):480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008. October;18(10):505–516.. [DOI] [PubMed] [Google Scholar]
- 83.Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017. April;35(2):180–188. Doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]