Abstract
Thousands of unique non-coding RNA (ncRNA) sequences exist within cells. Work from the past decade has altered our perception of ncRNAs from ‘junk’ transcriptional products to functional regulatory molecules that mediate cellular processes including chromatin remodelling, transcription, post-transcriptional modifications and signal transduction. The networks in which ncRNAs engage can influence numerous molecular targets to drive specific cell biological responses and fates. Consequently, ncRNAs act as key regulators of physiological programmes in developmental and disease contexts. Particularly relevant in cancer, ncRNAs have been identified as oncogenic drivers and tumour suppressors in every major cancer type. Thus, a deeper understanding of the complex networks of interactions that ncRNAs coordinate would provide a unique opportunity to design better therapeutic interventions.
Six decades ago, Francis Crick’s ‘central dogma’ asserted that genetic information travels from DNA through RNA towards protein synthesis1,2. With few exceptions, research in the following years characterized RNAs mainly as intermediaries in the process of protein production, principally as temporary copies of genetic information (mRNA), components of the ribosome (ribosomal RNAs (rRNAs)) or translators of codon sequence (tRNAs)3. For many years, proteins represented the primary functional end product of genetic information, though the genes that encode them account for less than 2% of the genome.
The discovery of the first small temporal RNAs, lineage defective 4 (lin‑4)4 and lethal 7 (let‑7)5, identified 23 and 16 years ago, respectively, in Caenorhabditis elegans, demonstrated that some RNAs, despite lacking protein-coding regions, are conserved functional molecules required for development. Since these early discoveries, studies have shown that functional products encoded by the genome are not limited to proteins but include a variety of unique RNAs6,7. Advances in sequencing technologies have led to the discovery of a multitude of non-coding RNA (ncRNA) species, some highly conserved, such as microRNAs (miRNAs), transcribed ultra-conserved regions6 and circular RNAs (circRNAs), and others generally lacking conservation across species, such as long ncRNAs (lncRNAs)7. Constituting almost 60% of the transcriptional output in human cells8,9, ncRNAs have been shown to regulate cellular processes and pathways in developmental and pathological contexts.
It has become increasingly difficult to view ncRNA function in isolation. Some ncRNAs, like miRNAs, target the mRNAs of many other different genes (and the mRNA of each gene can be targeted by multiple miRNAs); thus, these ncRNAs naturally link associated genes into regulatory networks10,11. To add more complexity, miRNAs also functionally interact with other species of ncRNAs, such as circRNAs and lncRNAs, to regulate their stability. In turn, lncRNAs and circRNAs regulate the abundance of available miRNAs through mechanisms including sequestration. The highly complex nature of some ncRNA interactions supports their roles as key regulators in important cellular programmes. Perturbations to these interactions have widespread consequences affecting cell fate and are common in cancer. Though complex, the networks in which ncRNAs participate are far from inscrutable. Many ncRNA interactions conform to characteristic patterns, or motifs, which are found in complex networks of all types, from biological to social12,13.
In this Review, we examine the nature of ncRNAs within cancer networks. We also highlight examples of ncRNAs participating in network motifs and other recurrent patterns of interactions and how these contribute to malignant phenotypes of cancer cells. Finally, we discuss some of the common disruptions to ncRNA interaction networks that are found in cancer. As RNA-based therapeutics have emerged in an increasing number of clinical applications14, illuminating the complex networks of ncRNAs is fundamental to applying such technologies to cancer diagnosis and treatment.
ncRNAs in network motifs
As ncRNA genes represent a growing list of therapeutic targets in cancer15, the design of effective RNA-based anticancer strategies requires a much better understanding of the diverse and context-dependent nature of ncRNA interactions. The circuitous architecture of the molecular interactions that transduce signals in cells can be illuminated through the lens of network theory. Networks are composed of nodes, the participatory members, and edges, the links or interactions between those members12,13 (FIG. 1a). In this Review, we examine networks in which nodes are represented by products of genes, both non-coding and coding, connected through the interactions that occur between them to accomplish cellular functions. In all networks, the patterns in which the nodes connect determine not only the output of the system but also the robustness or vulnerability of that system to perturbations.
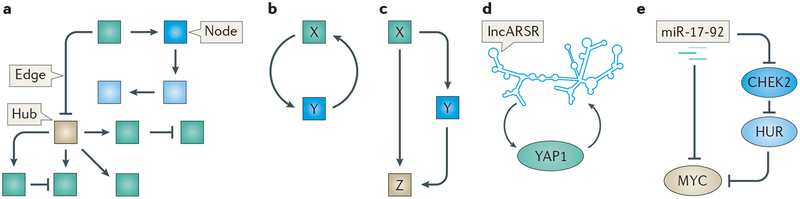
Figure 1 |. Motifs in RNA networks.
a | Schematic representation of a simplified network. Examples of nodes, edges and hubs are noted. b | Representation of a feedback loop. c | Representation of a feedforward loop. d | Example of a feedback loop between the long non-coding RNA activated in renal cell cancer with sunitinib resistance (lncARSR) and the transcriptional co-activator YAP1. e | Example of a feedforward loop between miR-17–92, serine/threonine-protein kinase Chk2 (CHEK2) and MYC in lymphoma. HUR, hu antigen R.
Biological networks have been shown to be scale-free and not random. In random networks, each node is connected to other nodes through roughly the same number of connections. In scale-free networks, by contrast, no node is representative of all other nodes, as the number of connections varies for each node. Nodes with a high number of connections, such as miRNAs and transcription factors, represent network hubs. In most real-world scale-free networks, certain patterns of interactions are enriched in frequency. These recurring clusters of interactions are known as network motifs. In this section, we examine the participation of ncRNAs in well-defined network motifs (FIG. 1a–c). These patterns of interactions are common to all complex networks, from biological to social, and provide information about the structure of networks as well as their vulnerabilities13.
Feedback loops
In any complex system, mechanisms serve to protect the integrity of that system by adapting the system to various inputs and by maintaining outputs within an appropriate range. One such mechanism found in complex networks is the feedback loop motif (FIG. 1b). In cancer, some of these feedback loops can be co-opted to promote tumori genesis rather than normal cell behaviour. Below we highlight some examples of ncRNA functions within feedback loops in cancer cells.
Throughout cell signalling networks, certain nodes function indirectly to promote their own expression. For example, a transcriptional target of the Hippo signalling pathway is the lncRNA lncARSR (a lncRNA activated in renal cell carcinoma (RCC) with sunitinib resistance). When the pathway is active its transcriptional effector, the transcriptional co-activator YAP1, translocates from the cytoplasm to the nucleus, where it engages with other transcription factors to initiate target gene transcription. Once expressed, the lncARSR transcript provides positive feedback (FIG. 1d) by binding to YAP1 at a site that sterically inhibits its phosphorylation by its negative regulator, the large tumour suppressor kinase 1 (LATS1)16. Phosphorylated YAP1 is unable to translocate into the nucleus. Thus, by binding to YAP1, lncARSR not only promotes its own expression16 but also serves to lock Hippo pathway activity in a steady state, a common characteristic of positive feedback loops13. High expression of lncARSR has been associated with poor prognosis in patients with RCC, likely because lncARSR is required for the maintenance of tumour-initiating cells in renal tumours. The activity of lncARSR to promote the tumour-initiating abilities of renal cancer cells is dependent on YAP1 signalling, indicating that the positive feedback by lncARSR also promotes transcription of other oncogenic factors by YAP116.
Feedforward loops
ncRNAs frequently participate in another canonical network motif, the feedforward loop, in which a node X simultaneously directly and indirectly regulates another node Z. The indirect regulation occurs through regulation of a third node Y, which is regulated by X and regulates Z13 (FIG. 1c). A common example of this phenomenon in cell biology is when a transcription factor promotes the expression of both another protein-coding gene and a miRNA that post-transcriptionally inhibits the target protein-coding gene. However, various combinations of miRNAs, proteins and transcription factors can exist within feedforward loops. In lymphoma cells, miR-17-92 helps to sensitively calibrate the activity of the MYC oncogene by participating in a feedforward loop with MYC, serine/threonine-protein kinase Chk2 (CHEK2) and hu antigen R (HUR; also known as ELAV1)17 (FIG. 1e). miR-17-92 inhibits the activity of MYC through direct targeting of MYC transcripts as well as through inhibition of CHEK2. This allows the RNA-binding protein HUR, normally repressed by CHEK2, to bind MYC transcripts and prevent their translation17. Without this mechanism to balance expression of MYC, which also participates in a positive feedback loop with the transcription factor E2F, it would rise unchecked to levels that promote apoptosis17.
ncRNA genes can occupy multiple nodes
One of the great challenges in understanding the function of ncRNAs in cancer is that ncRNAs do not always have a singular role in the process of tumorigenesis. An ncRNA may function as a tumour suppressor in one type of cancer but promote disease progression in another type18. In other cases, multiple species of gene products from the same ncRNA gene locus may operate synergistically or in opposition within a given pathway. The multifaceted functions of many ncRNA genes provide further evidence that the genome is not organized to contain individual genes with individual functions but instead is often highly multiplexed at particular loci to produce manifold gene products that allow for an array of possible fine-tuned responses to complex pathway stimuli.
ncRNAs with context-dependent roles
Even with the ability to computationally predict potential targets of certain species of ncRNAs, like miRNAs (BOX 1), it can be difficult to accurately predict their oncogenic or tumour-suppressive role in cancer. Part of the reason lies in the vast quantity and diversity of targets a given miRNA may have. Without knowing the function of each of the downstream targets, whether each is expressed and to what degree the miRNA inhibits the expression of each, it is difficult to predict the overall effect of the loss-of-function or gain-of-function of a miRNA on tumour growth. One reason for this challenge is that ncRNA interactions can be context- dependent or cell-type-dependent (reviewed in REF. 18). Such is the case for miR-125b, which acts as a tumour suppressor in some tumour types and as an oncogene in others18. Whereas the basic mechanisms by which miR-125b functions remain the same across all tissues, the pool of miR-125b targets that are expressed varies by cell type, allowing miR-125b to target tumour-promoting transcripts in cells of solid tumours and tumour-suppressive transcripts in cells of haematological cancers18.
Box 1 |. Tools for researching ncRNA networks in cancer.
Database tools.
Multiple bioinformatic tools are available and have been reviewed elsewhere136,137. In addition, The Cancer Genome Atlas (TCGA)138,139 database integrates DNA, RNA, protein and clinical data across numerous cancers. PROGmiR140 identifies microRNAs (miRNAs) as biomarkers in cancer prognosis. The SomamiR DB 2.0 algorithm141 analyses somatic mutations that may alter miRNA and competitive endogenous RNA (ceRNA) interactions. The database doRiNA 2.0 (REF. 142) gathers interactions between miRNAs, RNA-binding proteins and mRNAs. DIANA-mirExTra v2.0 (REF. 143) helps investigators integrate RNA sequencing data sets in order to identify miRNAs and transcription factors with central regulatory roles. The database circRNADb144 provides genomic information about circular RNAs (circRNAs) as well as internal ribosome entry sites, open reading frames and references. Additionally, circBase provides scripts to identify circRNAs in sequencing data145.
Effective analysis of non-coding RNA (ncRNA)-interacting networks in cancer with such algorithms should be validated with on-bench experiments.
Tools for the bench.
Identification of ncRNA signatures as prognostic and diagnostic biomarkers in human tumour samples is made through next-generation sequencing146,147 of ncRNAs. In addition, the photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) technique identifies interactions of ncRNA and coding RNA with proteins, such as the protein Argonaute 2 (AGO2); however, low-abundance targets may not be captured by this technique.
Another tool in the ncRNA field is a genomic editing tool called the CRISPR–Cas9 system64,148. This system allows targeted editing of the genome, for example, to cleave part of the genome that carries mutations and successfully repair them. The CRISPR–Cas9 system is also used for ncRNA genomic deletions to study the functional roles of ncRNA in cancer.
In order to study phenotypic changes in human tumour cell lines and in mouse models, complementary antisense RNA or DNA oligonucleotides149 are used that block the interaction between ncRNAs and their gene target as well as artificial decoying molecules77 that inhibit miRNAs through multiple complementary artificial binding sites cloned into and expressed from DNA plasmids150.
Tools for in vivo manipulation of ncRNA networks.
Systemic delivery of synthetic miRNA151 or long non-coding RNA inhibitors152 in vivo without toxic collateral effects is still a major challenge. A paradigm for successful delivery of antisense RNA is the example involving the oncomiR miR-155. Anti-miR-155 was placed in nanoparticles composed of poly(lactic-co-glycolic acid) (PLGA)153 as well as conjugated to a peptide with a low-pH-induced transmembrane structure (pHLIP)154 and was successfully delivered, reduced the tumour growth in mice and did not induce toxicity in mice. Furthermore, antisense DNA oligonucleotides against metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) inhibited human lung cancer metastasis in mice xenografts155.
Members of the miR-29 family of miRNAs, consisting of three isoforms (miR-29a, miR-29b and miR-29c), have been reported to play dual roles: they can act as tumour-suppressor miRNAs in mantle-cell lymphoma, acute myeloid leukaemia (AML), lung cancer, diffuse large B cell lymphoma and Burkitt lymphoma19,20 and as oncogenic miRNAs in indolent human B cell chronic lymphocytic leukaemia, AML and metastatic breast cancer19. A context-dependent dual role of another miRNA, miR-375, has been observed in prostate cancer21. miR-375 is highly expressed in an androgen- dependent human prostate cancer cell line, 22Rv1. Inhibition of miR-375 decreased the viability of 22Rv1 cells and derepressed the expression of the tumour-suppressor gene RB1. Very low expression of the same miRNA has been detected in the malignant androgen-independent prostate cancer cell line PC3 (REF. 21). Overexpression of miR-375 in PC3 cells had anti-proliferative effects and blocked the expression of one of its known targets, the gene cyclin D2 (CCND2)21. Additionally, the effects of miR-191/425 cluster expression in breast cancer cell lines were found to be dependent on oestrogen receptor α (ERα; also known as ESR1) expression22. ERα-positive cells express high levels of the miR-191/425 cluster, leading to the suppression of the tumour-suppressor gene early growth response 1 (EGR1). ERα-negative cells express low levels of the miR-191/425 cluster. When both miR-191 and miR-425 were overexpressed in ERα-negative cells, tumour growth and metastasis were impaired22. The examples reported here indicate that the same miRNA can act as an oncogene or a tumour suppressor not only in different types of cancer, like miR-125b and miR-29b, but also within the same type of cancer, like miR-375 and the miR-191/425 cluster.
Supergenes
The ability of ncRNA genes to occupy multiple nodes in cancer networks can be manifested by the context-dependent function of an individual gene product (as discussed above) as well as by the generation of diverse products from a single gene locus. The numerous products may be the result of alternative splicing, a common phenomenon during transcription in which certain exons are retained or lost, resulting in different end-product proteins or ncRNAs23. In addition, some genetic loci concurrently encode multiple products, including miRNAs, lncRNAs, circRNAs and proteins24. A ‘supergene’ yields more than one species of gene product either through the production of distinct transcripts for each product (type I supergenes) or through the production of a single transcript that is post-transcriptionally processed to generate multiple functional effectors (type II supergenes). Type III supergene loci are those that combine characteristics of both type I and type II supergenes. By supplying numerous products that occupy multiple nodes within a signalling pathway, the high density of genetic information encoded at supergenes may provide built-in measures to buffer or fine-tune the effects downstream of transcription at the locus.
Type I supergenes.
The activating signal cointegrator 1 complex subunit 3 (ASCC3) locus generates either a protein or a lncRNA to regulate cellular activity in response to ultraviolet radiation25. After DNA damage by ultra-violet radiation or chemotherapeutic agents, it is crucial for the survival of cells not only to enact mechanisms to repair the damage but also to arrest processes that may exacerbate genome instability, such as transcription26. ASCC3 is expressed under normal conditions and functions to maintain but not initiate global repression of transcription after DNA damage25. However, once DNA damage occurs and RNA synthesis is widely inhibited, the ASCC3 locus stops producing protein-coding transcripts and instead generates a shorter ncRNA transcript25. This ncRNA form of ASCC3 functions to oppose the actions of the ASCC3 protein and aids in the recovery of global RNA synthesis25. By producing multiple gene products, the ASCC3 locus can participate in the signalling pathways that control cellular responses after DNA damage in a context-specific and temporally appropriate way.
Type II supergenes.
Small RNAs, such as miRNAs, endogenous small interfering RNAs (endo-siRNAs) and tRNA-derived RNA fragments (tRFs), are not transcribed directly in their mature form. It is possible that cells are incapable of specifically transcribing very short regions of the genome. Alternatively, the transcription of long RNA transcripts may provide evolutionary advantages in the form of distinct opportunities for fine- tuning the expression of shorter mature products through the regulation of processing steps. A third possibility is that the longer transcript possesses inherent functions distinct from its role as a precursor RNA and that both the longer and shorter RNA moieties are required for cellular functions. Such genes that encode multiple functional gene products within a single transcript can be classified as type II supergenes.
Some ncRNAs are produced from the transcripts of protein-coding genes. Though the host genes of some miRNAs produce transcripts with no known function apart from being a miRNA precursor, many host genes for miRNAs are protein-coding and produce products other than miRNAs from the primary transcript. One of the first examples of this phenomenon was the discovery that the gene encoding miR-208a is located in the intron of the gene myosin heavy chain 6 (MYH6). The transcript produced from this locus generates both miR-208a and the MYH6 protein, both of which help to coordinate cardiac contractility27. In cancer, the oncogenic miRNA (oncomiR) miR-483 is produced from the intron of the oncoprotein-coding gene insulin-like growth factor 2 (IGF2)28. Other examples of ncRNAs produced from protein-coding transcripts include mirtrons29 and circRNAs30.
Endo-siRNAs can also be generated from pseudogene transcripts. For example, endo-siRNAs cleaved from naturally occurring double-stranded RNA (dsRNA) have been identified in plants, C. elegans and mouse oocytes31. The first observation of endo-siRNAs in human cancer cells was the discovery of cleaved inhibitory products from a transcribed pseudogene antisense to the gene protein phosphatase, Mg2+/Mn2+-dependent 1K (PPM1K) in human hepatocellular carcinoma (HCC)32. The double-stranded precursor of the cleaved endosiRNA is derived from either binding of the antisense pseudogene transcript to the sense transcript of PPM1K or the formation of internal hairpin structures within the pseudogene transcript32. The endo-siRNAs were downregulated in HCC tumours compared with normal tissue, and functional testing showed that the PPM1K pseudogene–endo-siRNA axis influenced the growth of tumour cells32. Taken together, these results suggest that loss of PPM1K-pseudogene-derived endo-siRNAs confers a growth advantage on cancer cells in HCC. Whether additional endo-siRNAs generated from human pseudogenes exist and contribute to tumori-genesis remains to be seen. Either result would have interesting implications not only for the functional relevance of human pseudogenes but also for the evolution of cellular mechanisms to control gene expression.
The noncanonical 3’ end processing of a larger lncRNA to generate smaller tRNA-like molecules is also involved in tumorigenesis. One example is the highly abundant lncRNA metastasis-associated lung adeno-carcinoma transcript 1 (MALAT1) initially transcribed by RNA polymerase II (RNA Pol II) as an approximately 7 kb poly-adenylated RNA transcript33. The mature MALAT1 transcript, however, is not poly-adenylated but contains a triple-helix structure at its 3’ end to protect the transcript from endonuclease activity34,35. The construction of the triple-helix structure requires the cleavage of 62 bp at the 3’ end by RNase P34,35. Further processing of the cleaved small RNA moiety produces a tRNA-like molecule termed MALAT1-associated small cytoplasmic RNA (mascRNA), which is efficiently shuttled to the cytoplasm, while mature MALAT1 is retained in the nucleus33. MALAT1 is known to be required for the retention of some poly-adenylated RNA transcripts in the nucleus and proper localization of splicing factors to nuclear paraspeckles in the nucleus36. mascRNA is more highly conserved than MALAT1, and loss-of- function or gain-of-function experiments suggest that it plays an important role in monocyte–macrophage functions by influencing tumour necrosis factor (TNF) receptor superfamily member 6 (TNFRSF6; also known as FAS), TNF ligand superfamily member 6 (TNFSF6; also known as FASLG), TNF, interleukin-6 (IL-6) and other chemokines33,37. However, its molecular mechanism of action is not yet known.
Mature tRNA molecules themselves serve as precursors for an abundant class of miRNA-like small ncRNA molecules, tRFs38 (FIG. 2). The 14-to-30-nucleotide single- stranded RNA moieties are cleaved from mature tRNAs through a not yet fully elucidated process (FIG. 2h; reviewed by Kumar et al.39). Both the endoribonuclease Dicer40,41 and angiogenin have been shown to play a role in tRF biogenesis42,43. Unlike miRNAs, tRFs are restricted to the cytoplasm; however, tRFs bind to Argonaute proteins and may regulate the abundance and translation of long RNAs in a miRNA-like manner38,44,45. The function of this class of small RNAs has been implicated in the pathogenesis of lymphoma41,45, prostate cancer46 and breast cancer47. Functioning through a competitive binding mechanism (discussed below) rather than an RNA-induced silencing complex (RISC)-mediated mechanism, tRFs act as tumour suppressors in breast cancer by sequestering the RNA-binding protein nuclease-sensitive element-binding protein 1 (YBX1) and thus permitting the destabilization of oncogenic transcripts that would be stabilized by YBX1 binding47.
Figure 2 |. Recurrent patterns of ncRNA function in cancer networks.
a | The H19, imprinted maternally expressed transcript (non-protein coding) (H19) locus encodes genes in both directions — the long non-coding RNA (lncRNA) H19, the microRNA (miRNA) miR-675, the protein H19 opposite tumour suppressor (HOTS) and the lncRNA 91H — which are not only of different gene types but also have opposing roles in tumorigenesis. Thus, the entire locus has a dual and possibly context-dependent role. b | The transcription of H19 is a tumour-suppressive event because it competes for transcription factor binding with a nearby insulin-like growth factor 2 (IGF2) locus, thereby inhibiting oncogenic IGF2 expression. c | The lncRNA H19 is processed to produce miR-675, a miRNA that inhibits a number of oncogenic transcripts. d | The lncRNA H19 acts as a competitive endogenous RNA (ceRNA) that competitively binds tumour-suppressive miRNAs. e | The lncRNA H19 binds to adenosylhomocysteinase (SAHH), which prevents it from facilitating DNA methylation through the action of DNA methyltransferase 3B (DMNT3B). The lack of methylation is associated with spurious transcription in cancer. f | The lncRNA transcribed antisense to H19, 91H, also promotes oncogenesis. g | HOTS functions as a tumour suppressor through unknown mechanisms. h | tRNAs serve as precursors for miRNA-like small RNAs (tRNA-derived RNA fragments (tRFs)). i | The lncRNA second chromosome locus associated with prostate 1 (SChLAP1) inhibits the activity of the switch/sucrose non-fermentable (SWI/SNF) complex in producing repressive chromatin modifications. j | The BRAF pseudogene 1 (BRAFP1) acts as a sponge to sequester miRNAs that target BRAF. k | A lncRNA transcribed antisense to the gene encoding paxillin (PXN), PXN-antisense1 (PXN-AS1), binds the PXN transcript and inhibits miRNA binding. 3’UTR, 3’ untranslated region; ERH, enhancer of rudimentary homologue; ME, methylation; ncRNA, non-coding RNA; RNA Pol II, RNA polymerase II.
Type III supergenes.
Some genetic loci produce multiple gene products from distinct transcripts and multiple gene products from the same transcript. Such is the case for the H19, imprinted maternally expressed transcript (non-protein coding) (H19) locus (FIG. 2a–g). The H19 locus encodes the lncRNA H19 and miR-675 (REF. 48) in one direction and the protein H19 opposite tumour suppressor (HOTS) and the lncRNA 91H in the opposite direction. Initially, H19 was thought to be a tumour suppressor because transcription of H19 competes for transcription factor binding with a nearby oncogene, IGF2 (REF. 49). However, further study revealed that though the act of H19 transcription is indeed tumour-suppressive through reduction of IGF2 transcription, the lncRNA H19 itself promotes oncogenesis. Full-length H19 regulates the epigenetic state of several genetic loci, including that of other lncRNAs50. Direct binding and inhibition of the protein adenosylhomocysteinase (AHCY; also known as SAHH) by H19 indirectly inhibits DNA methyltransferase 3B (DNMT3B)-dependent DNA methylation at these genetic loci, allowing spurious transcription in endometrial cancer cells50. H19, like other ncRNAs, tends to be expressed specifically in certain cancer types51, and different aspects of its function also seem to be restricted among certain cells within a heterogeneous tumour cell population52. The lncRNA H19 acts as a competitive endogenous RNA (ceRNA), through sequestration and competitive binding of several tumour-suppressive miRNAs, including let-7, miR-200b and miR-200c. In doing so, H19 facilitates epithelial–mesenchymal transition and mesenchymal–epithelial transition during different stages of breast cancer metastasis52,53. The other lncRNA encoded at this locus, 91H, is transcribed in the antisense direction to H19 and has also been shown to function in an oncogenic manner by promoting proliferation, migration and invasion in colorectal cancer (CRC) cells, though its exact mechanism of action is not known54. The H19 locus also encodes a tumour suppressor, HOTS, which has been shown to inhibit the growth of rhabdomyosarcoma and choriocarcinoma cells55 and suppress growth of cervical cancer tumours in mice55. It is not yet clear how the expression and functions of the various tumour suppressors and oncogenes encoded at the H19 locus are integrated to affect cancer cell behaviour.
The fact that the functional H19 transcript encodes miR-675 makes the H19 gene locus more complex than either a type I or type II supergene. miR-675 (REF. 48), encoded in the first exon of H19 (FIG. 2c), is a developmental regulator and tumour suppressor in prostate cancer56, non-small-cell lung cancer (NSCLC)57, breast cancer58 and liver cancer59. Excision and maturation of miR-675 from H19 occurs through a typical process mediated by ribonuclease 3 (Drosha) and Dicer48. During development, cells employ the RNA-binding protein HUR to inhibit processing of H19 by Drosha and allow cell proliferation60. Cancer cells may use the same process to promote cell proliferation in tumour types where HUR expression is high.
We speculate that as our understanding of the extent of multiple transcripts and functional RNAs produced from the same genetic locus improves, we may find that almost every gene has ‘supergene’ capabilities.
Other patterns of ncRNA networks
Beyond canonical network motifs, other recurrent patterns of interactions have been identified in cancer networks involving ncRNA nodes. These patterns may represent novel motifs or interactions that overlap with canonical network motifs (FIG. 2i–j).
Regulation of protein complexes
Protein coupling is a major means by which ncRNAs function in gene regulatory and cell signalling networks. ncRNAs can bind to individual proteins as well as complexes of proteins and regulate aspects of their function61. The direct binding of ncRNAs to proteins in some cases facilitates their ability to target specific proteins and in other cases provides scaffolding for protein complexes to assemble.
Riboproteins.
In eukaryotes, RNA cooperates with proteins in functional units often termed riboproteins or ribonucleoproteins. A large number of cellular housekeeping mechanisms rely on the interaction between RNAs and protein complexes. For example, the maintenance of telomeres is facilitated through the action of telomerase reverse transcriptase (TERT) and dyskerin pseudouridine synthase 1 (DKC1) using telomerase RNA as a template62,63. The two subunits of eukaryotic ribosomes are composed of four highly structured ncRNAs (18S, 5S, 28S and 5.8S) and over 75 proteins that work together to manufacture the protein products of the cell63.
ncRNA guides.
Some protein complexes draw upon ncRNAs as a source of specificity. For example, the prokaryotic defence mechanism against DNA-based viruses, CRISPR, which has been co-opted as a powerful research tool, uses lncRNAs to guide Cas9 to specific DNA regions64. In addition, the proteins in RISC interact with various combinations of small ncRNAs to inhibit the expression of mRNAs. The sequence of the small ncRNA, usually a miRNA, determines which transcripts will be targeted by RISC. RISC represents a prominent example of protein complexes recruited by one RNA species to influence another. The detailed mechanisms of miRNA-mediated silencing of mRNA transcripts have recently been reviewed elsewhere65.
Recruitment of protein complexes.
The discovery that the lncRNA HOX transcript antisense RNA (HOTAIR) alters targeting of the Polycomb repressive complex 2 (PRC2)66 first showed that ncRNAs can recruit chromatin- modifying complexes to affect gene transcription. Here, lncRNAs act as address codes to direct PRC2 to specific sites for epigenetic silencing, redirecting its occupancy and leading to genome-wide modification in DNA histone H3 lysine 27 methylation (H3K27me) that resemble epigenetic states found in early development66. Changes in the epigenetic state such as these in cancer cells lead to gene expression patterns that support migration and invasion66. In addition to HOTAIR, other lncRNAs, such as neuroblastoma-associated transcript 1 (NBAT1) and MIR31 host gene (MIR31HG), have been shown to interact with PRC2 to influence the epigenetic state of cancer cells67,68. How lncRNAs initiate interaction with PRC2, what motifs are required for binding and whether these interactions are specific have not yet been resolved. Recent studies indicate that RNA recruitment of PRC2 may simultaneously be specific, as seen with RepA RNA, and promiscuous, as seen with non- relevant bacterial mRNA, but the in vivo relevance of this observation requires further investigation69.
Targeting of other chromatin-modifying complexes often requires the function of ncRNAs. For example, the switch/sucrose non-fermentable (SWI/SNF) complex, also known as BRG1- or HBRM-associated factors (BAF), is also regulated by ncRNAs. In prostate cancer, the lncRNA second chromosome locus associated with prostate 1 (SChLAP1) impairs the ability of the SWI/SNF chromatin remodelling complex subunit SNF5 to bind to the genome, resulting in genome-wide aberrant gene expression and tumorigenesis70 (FIG. 2i). In addition, the expression of the Wnt-pathway-responsive transcription factor 7 (TCF7) is partially upregulated in HCC through recruitment of the SWI/SNF complex by the lncRNA lncTCF7 (REF. 71). In addition, the function of DNA methylases sometimes requires ncRNAs. The lncRNA TCF21 antisense RNA inducing demethylation (TARID) enables transcription of the tumour-suppressor gene transcription factor 21 (TCF21) through recruitment of the DNA demethylation regulator growth arrest and DNA damage-inducible protein GADD45α72.
Control via competitive interactions
Another method by which ncRNAs contribute to gene regulatory networks is through physical interaction and sequestration of a target molecule, such as a miRNA or protein. The phenomenon, initially proposed in plant biology as the concept of ‘target mimicry’ (REF. 73) and frequently referred to in mammalian biology as ‘sponging’, prevents the target molecule from interacting with other binding partners74. Even though synthetic molecules to inhibit specific RNAs had been widely used before as research tools75–77, the discovery that endogenous mammalian RNAs could act as sponges showed that competitive binding by RNAs is a physiological control mechanism in mammalian regulatory networks74. Since then, cell fate decisions in both developmental as well as pathological cancer contexts have been shown to be influenced by regulation of cellular machinery through ceRNAs74,75,78–81.
Multiple species of RNAs have been proposed as possible ceRNAs, including mRNAs, lncRNAs, lncRNA pseudogenes, circRNAs and tRFs. Modulation of gene regulatory network interactions through competitive binding would require precise adjustment of molecule abundance to obtain the appropriate stoichiometry for effective competition82,83. Using computational strategies to integrate gene co-expression data, the presence and abundance of miRNA target sites, data from crosslinking immunoprecipitation followed by sequencing (CLIP-seq) and the level of complementarity between RNAs, many researchers have developed models to predict ceRNA function in cells and databases to annotate possible ceRNA interactions84–89. In this section, we provide a few examples of how individual ncRNAs may participate as ceRNAs in regulatory networks in cancer. The ceRNA hypothesis has been thoughtfully and thoroughly reviewed elsewhere74,75,90 along with an extending hypothesis that groups of RNAs with similar miRNA binding sites may prove more physiologically relevant as competitive effectors in cell biology than individual ceRNAs75.
Competing lncRNAs.
Because of shared sequence homology with their parental genes, lncRNA pseudo-genes represent a potential pool of ceRNA effectors. If abundant enough, pseudogenes that contain miRNA binding sites also shared with parental transcripts can increase the stability of the parental transcript by competing for inhibitory miRNAs. Experiments in mice have shown the BRAF pseudogene (Braf transforming gene, related sequence 1 (Braf‑rs1) in mice; BRAF pseudogene 1 (BRAFP1) in humans) is capable of sequestering miRNAs that target both the BRAF pseudogene as well as BRAF (FIG. 2j). Overexpression of Braf‑rs1 in mice led to an increase in BRAF abundance, activation of the ERK pathway and the development of lymphoma91. Conversely, depletion of Braf‑rs1 resulted in decreased expression of BRAF and reduced tumorigenesis91. It remains to be seen whether a ceRNA network in human cancers activates BRAF. However, BRAFP1 expression has been found in human cancer cell lines, and copy number amplification of the genomic region containing BRAFP1 has been observed in colon cancer patient tumour data sets91.
Decoy role of circRNAs.
A handful of circRNAs are deregulated in cancer (TABLE 1), and some are capable of sequestering miRNAs (FIG. 3),though their functional role is still under investigation. The first circRNA discovered to harbour complementary sequences for miRNAs is the sex-determining region of Chr Y (Sry) gene expressed in the mouse testis, which contains 16 binding sites for miR-138 (REFS 92,93). In addition, the circular isoform of the coding gene itchy E3 ubiquitin protein ligase (ITCH), circ-ITCH, functions as a sponge for miR-7, miR-17 and miR-214 in oesophageal squamous cell carcinoma (ESCC). Circ-ITCH expression functions as a tumour suppressor in ESCC by sponging miRNAs that would inhibit the production of the ITCH protein that functions to inhibit the Wnt–β-catenin pathway94.
Table 1 |.
Circular RNAs in cancer
| circRNA in cancer | Function | Refs |
|---|---|---|
| ciRS-7 | Blocks the tumour-suppressive effects of miR-7 in human colorectal cancer cell lines | 98 |
| circ-ITCH | Has a sponging effect on miR-7, miR-17 and miR-214in oesophageal squamous cell carcinoma cell lines | 94 |
| circ-Foxo3 | Forms a ternary complex with two cell cycle regulatory proteins, p21 and CDK2, and blocks cell cycle progression in mouse mammary tumour cell lines | 99 |
| f-circM9 | Increases tumorigenicity together with the MLL-AF9 fusion protein in mice | 109 |
| circHIPK3 | Identified as the most abundant circRNA in various types of cancer. It binds and inhibitsthetumour-suppressor microRNAmiR-124 in human cell lines | 161 |
| cZNF292 | This circRNA is regulated by hypoxia, and its depletion by small interfering RNA inhibits angiogenic sprouting of endothelial cells. This suggests that cZNF292 has proangiogenic activities | 162 |
circRNAs, circular RNAs; circ-Foxo3, circRNA of the transcription factor forkhead box protein O3; circHIPK3, circRNA of homeodomain-interacting protein kinase 3; circ-ITCH, circRNA of itchy E3 ubiquitin protein ligase; ciRS-7, circular RNA sponge for miR-7; f-circM9, fusion circular RNA of the oncogenes MLL and AF9 in acute myeloid leukaemia; cZNF292, circular RNA from the gene zinc-finger protein 292; p21, cyclin-dependent kinase inhibitor.
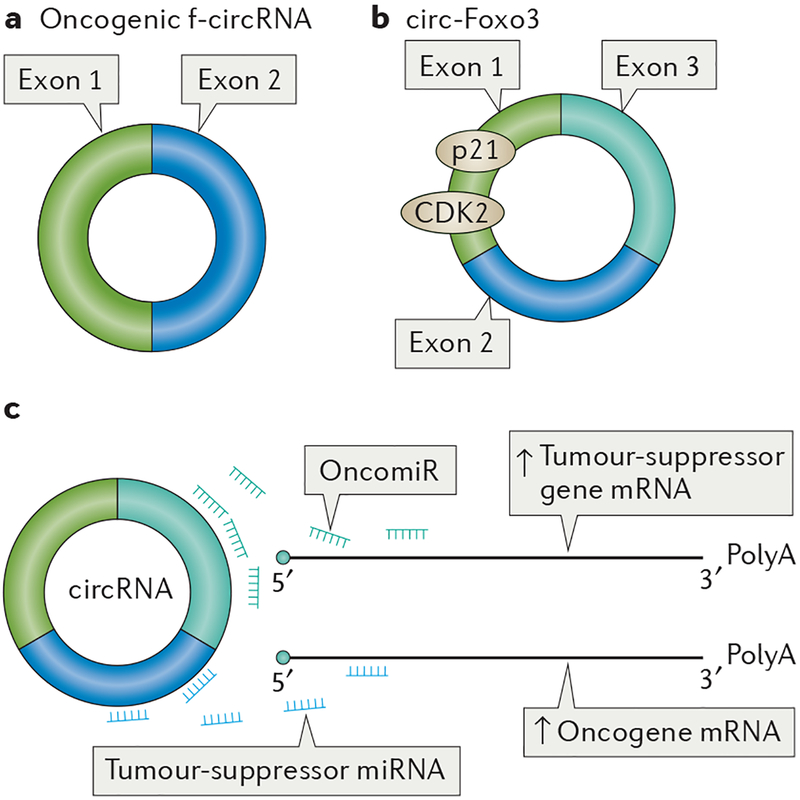
Figure 3 |. Putative functions of circular RNAs in cancer.
a | In the cytoplasm, the fusion of two oncogenes brought together upon chromosomal translocation creates a fusion circular RNA (f-circRNA) that is oncogenic in vitro and in vivo. b | The decoying function of circular RNA (circRNA) is illustrated, for example, by the circRNA of the transcription factor forkhead box protein O3 (circ-Foxo3), which forms a ternary inhibitory complex with the cell cycle proteins p21 (a cyclin-dependent kinase inhibitor) and cyclin-dependent kinase 2 (CDK2) to block cell cycle progression. c | circRNA can promote tumour progression by sequestering tumour-suppressor microRNAs (miRNAs) (for example, ciRS-7 sponges the tumour suppressor miR-7) because it alleviates expression of oncogenic targets. In an opposite manner, circRNA may exert an antitumoural effect when it sponges miRNAs that suppress tumour-suppressor genes (oncogenic miRNAs, oncomiRs); for example, circ-ITCH decoys miR-7, miR-17 and miR-214 and liberates the tumour-suppressor gene itchy E3 ubiquitin protein ligase (ITCH).
The antisense cerebellar degeneration-related protein 1 (CDR1as) contains 74 binding sites for miR-7 and may negatively regulate miR-7. Thus, CDR1as is also known as ciRS-7. Direct interaction of ciRS-7 and miR-7 was experimentally demonstrated in human cell lines, as was their colocalization in human cell lines and in the mouse embryonic brain93,95. The functional role of the sponging effect of ciRS-7 on miR-7 in cancer is now starting to be clarified. It is known that expression of the tumour suppressor miR-7 is reduced in cancer stem-like cells96 and furthermore inhibits the expression of onco-genes, such as epidermal growth factor receptor (EGFR), insulin receptor substrate 1 (IRS1) and IRS2, involved in glioblastoma97. miR-7 also targets the oncogenes EGFR and CRAF in CRC. ciRS-7 expression levels are higher in CRC compared with normal tissues, and it is associated with poor patient survival. In CRC cell lines, ciRS-7 sequesters miR-7, and consequently the expression of its target oncogenes EGFR and CRAF increases. This finding implies that ciRS-7 could be used as a therapeutic target in CRC98.
The circRNA of the transcription factor forkhead box protein O3 (circ-Foxo3) forms a ternary complex with two cell cycle regulatory proteins, the cyclin- dependent kinase inhibitor p21 and cyclin-dependent kinase 2 (CDK2)99, and blocks cell cycle progression. The discovery of the circ-Foxo3–p21–CDK2 complex99 is the first indication that circRNAs can bind and perhaps sequester proteins in addition to miRNAs. These unique properties of circRNAs suggest their promising application as therapeutic targets in cancer100.
Interactions between long RNAs
Another mechanism by which ncRNAs participate in gene regulatory networks is through direct binding of lncRNAs to other RNA molecules to regulate their stability or translation. These interactions rely on binding of the lncRNA to its RNA target and either creating a substrate for protein function or removing access for miRNA or inhibitory protein effectors.
A tumour-suppressor lncRNA that is directly induced by the tumour suppressor p53 (REF. 101), LincRNA-p21, binds directly to at least two mRNAs, including JUNB, which encodes an AP-1 transcription factor subunit and CTNNB1, which encodes β-catenin, to repress their translation102. This binding appears to be mediated by base pair interactions between complementary sequences within LincRNA-p21 and its target mRNAs102 and leads to recruitment of negative regulators of translation such as the proteins probable ATP-dependent RNA helicase DDX6 and synaptic functional regulator FMR1102. It is not known how many other mRNAs can be targeted by LincRNA-p21. Also unknown is whether this mechanism of translational inhibition is widely relevant to the binding of other ncRNAs with mRNA transcripts. However, given the important tumour-suppressive role of LincRNA-p21, further investigation into this mechanism may illuminate its role in lung cancer.
Staufen1-mediated mRNA decay (SMD) is a mechanism for protein-mediated degradation of mRNA, recognition of which occurs through lncRNAs. Specifically, most but not all mRNAs containing an Alu element in their 3’ untranslated region (3’UTR) are targeted by SMD103. A subset of lncRNAs, categorized as half-Staufen1-binding site RNAs (1/2-sbsRNAs), bind to Alu-containing mRNAs at their 3’UTR. This binding occurs through imperfect base pairing between areas adjacent to and including the Alu element, thereby forming dsRNA-binding protein Staufen homologue 1 (STAU1)-binding sites. STAU1 then binds and mediates degradation of the newly formed duplex104.
Other ncRNAs have been found to inhibit the function of STAU1. For example, the small nucleolar RNA host gene 5 (SNHG5),which serves as a precursor for the small nucleolar RNAs U50 and U50’, is localized in the cytoplasm and binds to the 3’UTRs of over 100 different Alu-containing transcripts. The SNHG5 protein competes with the mRNA-destabilizing STAU1 protein for binding to the 3’UTR of spermatogenesis associated serine rich 2 (SPATS2) transcripts, for example, thereby promoting stability of the transcipts105. The mRNAs protected by SNHG5 produce proteins that engage in cell survival pathways and promote tumour growth in CRC105.
In addition to creating recognition sites for degradation proteins, lncRNAs can protect transcripts by making miRNA binding sites on mRNAs inaccessible. The lncRNA produced in antisense to the gene encoding paxillin (PXN) performs this function specifically for its antisense mRNA106. Alternative splicing in HCC by the splicing factor muscleblind-like protein 3 (MBNL3) leads to the inclusion of an additional exon in the lncRNA. This exon contains a sequence complementary to the protein-coding mRNA for PXN. PXN-antisense1 (PXN-AS1) was shown to bind to the PXN mRNA and increase its stability by protecting it from miR-24 and protein Argonaute 2 (AGO2) mediated degradation (FIG. 2k). The resulting increase in PXN facilitates tumorigenesis in liver cells106.
Disruption of ncRNA networks in cancer
The networks in which ncRNAs perform their functions serve to provide checks and balances on important regulatory processes. Imbalances in the multiple layers of control of genes that allow the cell to repair DNA damage, organize chromatin, perform cell division, respond to mitogenic signals and interact with other cell types could have detrimental consequences. Changes in these highly regulated pathways may lead to cell death or propel the cell towards uncontrolled cell growth and malignancy. In this section, we explore some of the known mechanisms by which ncRNA nodes of gene regulatory networks are corrupted in cancer (BOX 2). Chemical modifications of RNA and epitranscriptomics have been recently reviewed107,108 and are not discussed in further detail here.
Box 2 |. Epigenetic changes in cancer alter the balance of ncRNA networks.
Non-coding RNAs (ncRNAs) routinely interact with proteins to coordinate the epigenetic marks to restrict transcription to a specific set of genes. Indeed, alterations to RNAs that participate in this process will transform the transcriptome and contribute to pathology. In addition, modifications to epigenetic marks of ncRNAs govern the actions of other nodes in ncRNA pathways, such as microRNA (miRNA) targeting of transcripts or long ncRNAs (lncRNAs) participating in competitive endogenous RNA (ceRNA) networks, have the potential to alter expression of these ncRNA effectors and disrupt their downstream targets, leading to dramatic changes in cell fate.
Numerous examples of aberrantly high or low expression of ncRNAs contributing to diseases have been studied. For example, hypermethylation of the tumour suppressor TP53-induced lncRNA TP53TG1 in cancer leads to decreased expression of TP53TG1 and promotes chemoresistance156. TP53TG1 regulates the subcellular localization of nuclease-sensitive element-binding protein 1 (YBX1). Loss of TP53TG1 in cancer allows YBX1 to accumulate in the nucleus and facilitate the expression of genes promoting cell growth156. A profile of the CpG island methylation status of transcribed ultra-conserved regions (T-UCRs) in eight different types of cancer cell lines demonstrated that T-UCRs are highly methylated157. When one of these T-UCRs, Uc.283+A, was overexpressed in a hypermethylated colon cancer cell line, the number of dead cells was increased, suggesting that T-UCRs have a growth inhibitory effect in colon cancer. In human colon cancer, melanoma and head and neck cancer, the miRNAs miR-34b, miR-34c, miR-148 and miR-9 were found to be hypermethylated, resulting in increased expression of their targets MYC and cyclin-dependent kinase 6 (CDK6)158. Epigenetic loss of miR-124a has been reported in different human cell lines and primary tumours. Treatment of a human colon cancer cell line with a DNA-demethylating drug restored expression of miR-124a, leading to suppression of its target, the oncogene CDK6, and induction of the activity of the tumour suppressor RB1159. The finding that the methylation status of miRNA loci correlates with chemosensitivity suggests that epigenetic profiling of ncRNA loci could serve to identify biomarkers160 and that DNA-demethylating drugs could be promising in ncRNA-based cancer therapies.
Remodelling of ncRNA networks
Tumorigenesis involves genomic alterations, such as chromosomal translocations, that cause a fusion between two genes that are otherwise located far from each other in the genome, resulting in a chimeric oncogene. This phenomenon may result in the fusion between two oncogenes. A recent study109 investigated the impact that these genomic alterations may have on ncRNAs and specifically on circRNAs. Upon chromosomal trans-location, two genes can bring complementary repetitive intronic sequences (Alu elements) together, favouring back- splicing events that generate aberrant circRNAs, called fusion (f)-circRNAs. Formation of f-circRNAs derived from recurrent chromosomal translocations of oncogenes may occur in liquid tumours such as in acute promyelocytic leukaemia (APL)110 and in AML109. The fusion of the two oncogenes promyelocytic leukaemia (PML) and retinoic acid receptor alpha (RARA) in APL gives rise to the f-circPR, while the fusion of lysine methyltransferase 2A (KMT2A; also known as MLL) and the super elongation complex subunit AF9 (also known as MLLT3) in AML produces f-circM9, and both circRNAs had pro-proliferative and proto-oncogenic activities in an immortalized mouse cell line109. The tumour-promoting property of f-circM9 was increased by the presence of the MLL–AF9 fusion protein in vitro and in vivo109. The formation of f-circRNAs was also examined in solid tumours such as in Ewing sarcoma and in lung cancer109,111. The fusion between the genes encoding EWS RNA-binding protein 1 (EWSR1) and the ETS transcription factor FLI1 in Ewing sarcoma and echinoderm microtubule associated protein like 4 (EML4) and activin A receptor like type 1 (ALK1; also known as ACVRL1) in lung cancer produced f-circEF1 and f-circEA1, respectively109. The relevance of these findings may be considered important, as it was estimated that almost 50% of aberrant translocations observed in various types of cancer may generate f-circRNAs. Nevertheless, the oncogenic potential of other f-circRNAs needs further investigation.
Altered targeting by ncRNA
Altered adenosine-to-inosine editing reprogrammes miRNA targeting.
A major cellular housekeeping process is the post-transcriptional converting or editing of adenosine to inosine (A-to-I) in transcripts, catalysed by three enzymes belonging to the family of dsRNA-specific adenosine deaminases (ADAR1, ADAR2 and ADAR3). These three enzymes have common functional domains but they differ in their A-to-I editing activity112. This process may occur in coding regions, although it is more frequent in non-coding regions such as introns and 3’UTRs where inverted Alu repeats form dsRNA structures113,114. By allowing the formation of new inosine–cytosine base pairings115, it has been hypothesized that A-to-I editing can alter the structure of transcripts116 (FIG. 4).
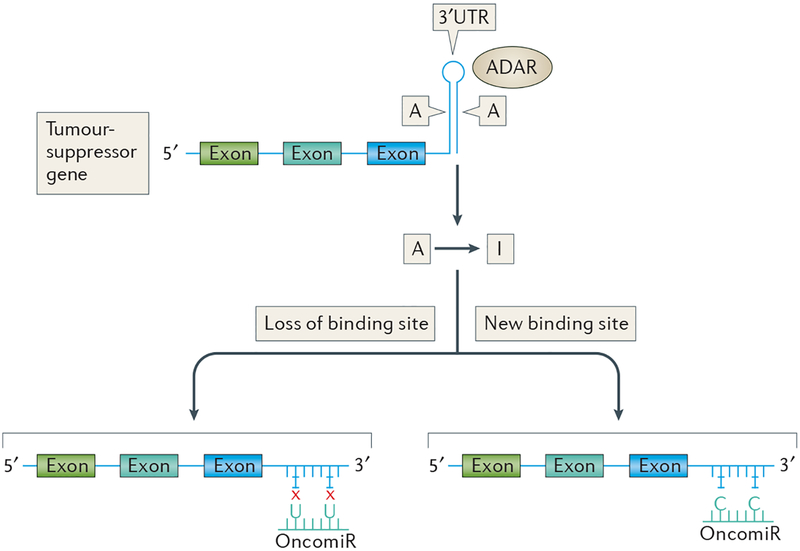
Figure 4 |. Adenosine‑to‑inosine editing of a tumour‑suppressor gene or an oncogene.
The adenosine-to-inosine (A-to-I) RNA editing mechanism may generate transcriptome diversity by influencing both tumour suppressors and oncogenes. As shown here, double-stranded RNA-specific adenosine deaminases (ADARs) may change adenosine bases in Alu sequences to inosine in the 3’ untranslated region (3’UTR) of a tumour-suppressor gene. This process sometimes results in a loss or gain of microRNA (miRNA) binding sites. By contrast, ADARs may also positively regulate oncogene expression through A-to-I editing, thus drastically reshaping miRNA nodes in non-coding RNA networks by changing the pool of possible miRNA targets genes involved in carcinogenesis. The editing dysregulation by aberrant expression of ADARs is also a frequent characteristic of cancers. The involvement of this RNA editing mechanism in cancer development seems important for tumour progression. OncomiR, oncogenic miRNA.
Recent bioinformatic comparison of A-to-I editing in tumours to normal tissue across 14 types of cancer found that whereas overall A-to-I editing remains largely unchanged in cancerous tissues, there was an increase or decrease in editing sites in the 3’UTRs of cancer-associated genes, particularly in miRNA binding sites117, which could either lead to the loss or gain of miRNA binding sites and was found to be highly context-specific. Upregulation or downregulation of A-to-I editing was dependent on the tumour type and the type of tumorigenic drivers117. Results from this study117 suggest that changes to the A-to-I editing process could drastically reshape miRNA nodes in ncRNA networks by changing the pool of possible miRNA targets such as oncogenes or tumour-suppressor genes, thereby presenting a potential selective advantage for cancer cells.
A-to-I RNA editing enzymes regulate not only miRNA targets but also miRNA precursors. ADAR enzymes form a heterodimer complex with Dicer to promote miRNA processing118 or inhibit the microprocessor complex cleavage of primary miRNAs to suppress miRNA maturation112. In cancer, loss or gain of ADAR activity influences the expression of oncogenic and tumour-suppressive miRNAs. In glioblastomas, A-to-I editing within miRNAs is decreased owing to impaired activity of ADAR2 compared with normal brain tissue119. In melanoma, low expression of ADAR1 leads to decreased A-to-I editing of miR-455-5p, resulting in a change in specificity and the targeting of different transcripts compared with normally edited miR-455-5p120. This change contributes to the metastatic ability of melanoma120. Amplification of ADAR1 in NSCLC cell lines leads to increased levels of defective transcripts of nei-like DNA glycosylase 1 (NEIL1), a DNA repair gene, and also increased levels of A-to-I editing of miR-138, an miRNA involved in chemoresistance and in cancer-related pathways. The increased editing of NEIL1 and miR‑138 transcripts led to increased growth of the NSCLC cell line A459. The high copy number of ADAR1 was also associated with poor prognosis in patients with early-stage lung cancer121.
Moreover, increased catalytic activity of ADAR1 in leukaemic stem cells inhibits biogenesis of the tumour suppressor let-7 family of miRNAs and promotes leukaemia stem cell self renewal122. Small molecule targeting of ADAR editase activity appears to restore let-7 bio genesis and impede leukaemia stem cell renewal123. Given the role of ADAR enzymes in rewiring the circuitry of miRNA networks in cancer, the ADAR enzymes themselves may provide useful targets for therapeutic intervention.
3’UTR variations in cancer alter miRNA targeting in ncRNA networks.
The 3’UTR of RNA, defined as the sequence between the stop codon and the poly(A) tail, contains a multitude of regulatory sequences for the maturation, localization, stability and translation of the RNA. More than 70% of mammalian genes contain alternative polyadenylation signals in the 3’UTR, which allow the formation of isoforms with multiple 3’UTRs derived from a single gene124. Although the ability to produce alternative ‘switches’ for the same transcript may allow fine-tuned differential expression of certain genes in a tissue-specific manner125, this system can be co-opted in cancer cells to promote expression of oncogenic signals. Indeed, one study found that cancer cells exhibit global shortening of 3’UTRs compared with normal cells, leading to the loss of miRNA and protein binding sites in 3’UTRs of oncogenic transcripts126.
Cis‑regulatory sequences in the 3’UTR are also susceptible to single nucleotide polymorphisms (SNPs), which occur frequently in the human genome in approximately every hundred base pairs127. Many silent polymorphisms, which do not alter coding sequences, may not in fact be silent. Those located in non-coding genomic regions have the potential to disrupt RNA–RNA or RNA–protein interactions in gene regulatory networks in cancer. A genome-wide DNA sequencing analysis of tumour tissues uncovered 12 SNPs in miRNA- binding sites associated with human cancer128. In lung cancer, a germline SNP in the 3’UTR of KRAS abrogated the binding site of let-7, thus increasing KRAS levels to promote tumour progression129. Moreover, a somatic SNP was found in the 3’UTR of the oncogene Mdm4, which inhibits the tumour suppressor p53, that created a new binding site for miR-191 (REF. 130). Recently, a study dissecting the KRAS 3’UTR in HeLa cells identified two 49-nucleotide cis‑regulatory sequences131 containing a miR-185 binding site and a stabilizing element. Knockdown of HUR that miRNAs and HUR cooperatively bind the same sequence in the 3’UTR of this oncogene and repress its expression. Moreover, the same repressive 49-nucleotide sequence contains a SNP at the first nucleotide of a predicted miR-185 target site, but its role has yet to be determined in cancer progression131.
Variations in non-coding sequences also have the potential to create novel regulatory sequences that may be selected in cancer cells. One such example is a SNP in LINC00673, which establishes a binding site for miR-1231. Subsequent downregulation of LINC00673 releases its target tyrosine-protein phosphatase non-receptor type 11 (PTPN11) from ubiquitin-mediated degradation. Increased PTPN11 leads to increased SRC–ERK oncogenic signalling and has been shown to be a risk factor for pancreatic cancer132.
Perspectives
What has become apparent, even from our incomplete understanding of the molecular processes through which ncRNAs contribute to cellular activity, is that ncRNAs participate extensively in gene regulatory networks. Rather than acting as a single link in a chain of cellular events, many but not all ncRNAs operate as a branch point with broad-reaching outputs capable of transforming the transcriptome and proteome of cells to define cell fates. That a substantial proportion of these pleiotropic ncRNAs in turn regulate other pleiotropic ncRNAs adds exponential depth to the complexity of regulatory networks encompassing the genome.
The framework of network science provides some level of clarity to the complex nature of ncRNAs in cellular pathways by facilitating the identification of recurrent patterns of interactions between ncRNA nodes and other pathway members. The study of transcriptional networks in bacteria has shown that the structure of network motifs also correlates with the kinetics, sensitivity and robustness of network responses to perturbations13. For example, certain network motif configurations allow for either immediate or delayed response to the application of a certain stimulus. Whereas many of these temporal aspects of network motifs were deciphered using protein networks, it is not yet clear if ncRNA nodes in biological networks would yield similar kinetics. These aspects of ncRNA network biology remain to be studied.
In this new era, when assays to investigate the DNA sequence, chromatin modifications, gene expression and protein abundance in cells at a genome-wide scale have become routine practice at the bench, the opportunity to deconvolute such complex networks is within reach. Particularly important in the design of effective cancer treatments, where single-agent targeted therapy has sometimes proved insufficient133–135, is a comprehensive understanding of the possible feedback loops, off-target effects and modes of resistance to therapeutic manipulation. A great deal of work remains to characterize the complex ncRNA networks that contribute to tumori-genesis, but continued effort has the potential to better inform the selection of therapeutic targets. Technical tools to identify and experimentally validate many bioinformatically predicted interactions within a ncRNA–coding RNA circuitry exist, are improving and have the potential to transform our understanding and treatment of diseases like cancer.
Acknowledgements
The authors thank A. L. Jiao and P. Trivedi for helpful comments on this manuscript. The authors acknowledge support from the Ludwig Center at Harvard, Boston, Massachusetts, USA, and grants from the US National Institutes of Health (R01 CA157749; P50 CA177444).
Glossary
- Network motifs
Patterns of interactions between nodes in a network that occur more often than by chance.
- Supergene
A locus that produces multiple functional RNAs.
- Endogenous small interfering RNAs
(Endo-siRNAs). Small RNAs that, unlike microRNAs, are derived from perfectly complementary sense–antisense RNA hybrids (double-stranded RNA).
- Mirtrons
MicroRNAs derived from short hairpin introns and processed by the splicing machinery but not by ribonuclease 3 (Drosha).
- Pseudogene
A nucleotide sequence that resembles a gene but does not lead to any protein expression.
- Nuclear paraspeckles
Nuclear domains within interchromatin spaces, enriched in RNA processing factors.
- Crosslinking immunoprecipitation followed by sequencing
(CLIP-seq). A method to study RNA–protein interactions by ultraviolet crosslinking followed by immunoprecipitation.
- Staufen1-mediated mRNA decay
(SMD). A process that degrades mRNA through binding of double-stranded RNA-binding protein Staufen homologue 1 (STAU1) to its binding site in the 3’ untranslated regions of target mRNA.
- Alu element
A short interspersed element, 300 bp in length, that is repeated in the human genome and forms a characteristic double-stranded RNA embedded in the precursor mRNA.
- Alternative polyadenylation signals
When more than one site within a gene locus codes for the signal that allows a string of adenosine bases (poly(A) tail) to be added to the end of the transcript.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Crick F Central dogma of molecular biology. Nature 227, 561–563 (1970). [DOI] [PubMed] [Google Scholar]
- 2.Crick FH On protein synthesis. Symp. Soc. Exp. Biol 12, 138–163 (1958). [PubMed] [Google Scholar]
- 3.Eddy SR Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet 2, 919–929 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL & Ambros V The C. elegans heterochronic gene lin‑4 encodes small RNAs with antisense complementarity to lin‑14. Cell 75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Bejerano G et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Johnsson P, Lipovich L, Grandér D & Morris KV Evolutionary conservation of long noncoding RNAs; sequence, structure, function. Biochim. Biophys. Acta 1840, 1063–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djebali S et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) project. Science 306, 636 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Ebert MS & Sharp PA Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamura S, Imai-Sumida M, Tanaka Y & Dahiya R Interaction and cross-talk between non-coding RNAs. Cell. Mol. Life Sci 10.1007/s00018-017-2626-6 (2017). [DOI] [PMC free article] [PubMed]
- 12.Barabási AL & Oltvai ZN Network biology: understanding the cell’s functional organization. Nat. Rev. Genet 5, 101–113 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Alon U Network motifs: theory and experimental approaches. Nat. Rev. Genet 8, 450–461 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Kasinski AL & Slack FJ MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 11, 849–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupaimoole R & Slack FJ MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov 16, 203–222 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Qu L et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nature Commun. 7, 12692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mihailovich M et al. miR-17–92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth. Nature Commun. 6, 8725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svoronos AA, Engelman DM & Slack FJ OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 76, 3666–3670 (2016).This is an excellent review of the dual role of miRNAs in cancer.
- 19.Pekarsky Y & Croce CM Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget 1, 224–227 (2010).This article represents one of the first examples of the dualistic function of a miRNA in the same or in different types of cancer.
- 20.Anastasiadou E et al. Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene 29, 1316–1328 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Costa-Pinheiro P et al. MicroRNA-375 plays a dual role in prostate carcinogenesis. Clin. Epigenet 7, 42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Leva G et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 9, e1003311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 20, 1639–1650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beermann J, Piccoli MT, Viereck J & Thum T Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev 96, 1297–1325 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Williamson L et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell 168, 843–855.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayne LV & Lehmann AR Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 42, 1473–1478 (1982). [PubMed] [Google Scholar]
- 27.van Rooij E et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Veronese A et al. Oncogenic role of miR-483–3p at the IGF2/483 locus. Cancer Res. 70, 3140–3149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladewig E, Okamura K, Flynt AS, Westholm JO & Lai EC Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 22, 1634–1645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol 22, 256–264 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Chan WL et al. Transcribed pseudogene psiPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 41, 3734–3747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilusz JE, Freier SM & Spector DL 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilusz JE et al. A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 26, 2392–2407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JA, Valenstein ML, Yario TA, Tycowski KT & Steitz JA Formation of triple-helical structures by the 3’-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc. Natl Acad. Sci. USA 109, 19202–19207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tripathi V et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gast M et al. Long noncoding RNA MALAT1-derived mascRNA is involved in cardiovascular innate immunity. J. Mol. Cell Biol 8, 178–181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YS, Shibata Y, Malhotra A & Dutta A A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar P, Kuscu C & Dutta A Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci 41, 679–689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole C et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maute RL et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl Acad. Sci. USA 110, 1404–1409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov P, Emara MM, Villen J, Gygi SP & Anderson P Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q et al. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther 21, 368–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haussecker D et al. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pekarsky Y et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl Acad. Sci. USA 113, 5071–5076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olvedy M et al. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget 7, 24766–24777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodarzi H et al. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai X & Cullen BR The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13, 313–316 (2007).This is one of three articles that demonstrate the various gene products made from the single H19 locus.
- 49.Yoshimizu T et al. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl Acad. Sci. USA 105, 12417–12422 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun 6, 10221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du Z et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol 20, 908–913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou W et al. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci. Signal 10, eaak9557 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Kallen AN et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52, 101–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng Q et al. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS ONE 9, e103022 (2014).This is one of three articles that demonstrate the various gene products made from the single H19 locus.
- 55.Onyango P & Feinberg AP A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript. Proc. Natl Acad. Sci. USA 108, 16759–16764 (2011).This is one of three articles that demonstrate the various gene products made from the single H19 locus.
- 56.Zhu M et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 281, 3766–3775 (2014). [DOI] [PubMed] [Google Scholar]
- 57.He D et al. Down-regulation of miR-675–5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol. Cancer 14, 73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vennin C et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 6, 29209–29223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H et al. miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget 6, 31958–31984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keniry A et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol 14, 659–665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferre F, Colantoni A & Helmer-Citterich M Revealing protein-lncRNA interaction. Brief. Bioinform 17, 106–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng J et al. The RNA component of human telomerase. Science 269, 1236–1241 (1995). [DOI] [PubMed] [Google Scholar]
- 63.Alberts B et al. Molecular Biology of the Cell 4 edn (Garland Science, 2002). [Google Scholar]
- 64.Sánchez-Rivera FJ & Jacks T Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer 15, 387–395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonas S & Izaurralde E Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet 16, 421–433 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Gupta RA et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).This study provides an example of a lncRNA that alters the occupancy of a protein complex on chromatin.
- 67.Hu P et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget 6, 32410–32425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montes M et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat. Commun 6, 6967 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Davidovich C et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 57, 552–558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prensner JR et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet 45, 1392–1398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Stem Cell 16, 413–425 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Arab K et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A Mol. Cell 55, 604–614 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Franco-Zorrilla JM et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet 39, 1033–1037 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Salmena L, Poliseno L, Tay Y, Kats L & Pandolfi PPA ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tay Y, Rinn J & Pandolfi PP The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown BD et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol 25, 1457–1467 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Ebert MS, Neilson JR & Sharp PA MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cesana M et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tay Y et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147, 344–357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sumazin P et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147, 370–381 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karreth FA et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 147, 382–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Denzler R, Agarwal V, Stefano J, Bartel DP & Stoffel M Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bosson AD, Zamudio JR & Sharp PA Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 56, 347–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J et al. Computational prediction of microRNA networks incorporating environmental toxicity and disease etiology. Sci. Rep 4, 5576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarver AL & Subramanian S Competing endogenous RNA database. Bioinformation 8, 731–733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Z et al. dbDEMC: a database of differentially expressed miRNAs in human cancers. BMC Genomics 11 (Suppl. 4), S5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chiu HS et al. Cupid: simultaneous reconstruction of microRNA-target and ceRNA networks. Genome Res. 25, 257–267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Das S, Ghosal S, Sen R & Chakrabarti J lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS ONE 9, e98965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohtsuka M, Ling H, Doki Y, Mori M & Calin GA MicroRNA processing and human cancer. J. Clin. Med 4, 1651–1667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thomson DW & Dinger ME Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet 17, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Karreth FA et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 161, 319–332 (2015).This study provides an example of a lncRNA that acts as sponge to sequester miRNAs.
- 92.Capel B et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030 (1993). [DOI] [PubMed] [Google Scholar]
- 93.Hansen TB et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Li F et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 6, 6001–6013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Memczak S et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- 96.Okuda H et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 73, 1434–1444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hansen TB, Kjems J & Damgaard CK Circular RNA and miR-7 in cancer. Cancer Res. 73, 5609–5612 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Weng W et al. Circular RNA ciRS-7 - A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 10.1158/1078-0432.ccr-16-2541 (2017).This study unravels the functional role of the ciRS‑7 sponging effect on the tumour suppressor miR‑7 in colorectal cancer.
- 99.Du WW et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rybak-Wolf A et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Huarte M et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoon JH et al. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655 (2012).This study provides an example of a lncRNA that binds to and affects the translation of mRNAs.
- 103.Ryu S et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell 23, 63–76 (2013). [DOI] [PubMed] [Google Scholar]
- 104.Gong C & Maquat LE lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470, 284–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Damas ND et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat. Commun 7, 13875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yuan JH et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol 19, 820–832 (2017).This study provides an example of a lncRNA binding to an mRNA and blocking access to a miRNA.
- 107.Roundtree IA, Evans ME, Pan T & He C Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Esteller M & Pandolfi PP The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 7, 359–368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guarnerio J et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 166, 1055–1056 (2016).This is an important study that reveals that aberrant translocations observed in various types of cancer may generate fusion circRNAs.
- 110.dos Santos GA, Kats L & Pandolfi PP Synergy against PML-RARa: targeting transcription, proteolysis, differentiation, and self-renewal in acute promyelocytic leukemia. J. Exp. Med 210, 2793–2802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martelli MP et al. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am. J. Pathol 174, 661–670 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nishikura K A-To-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol 17, 83–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim DD et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 14, 1719–1725 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Daniel C, Lagergren J & Ohman M RNA editing of non-coding RNA and its role in gene regulation. Biochimie 117, 22–27 (2015). [DOI] [PubMed] [Google Scholar]
- 115.Zinshteyn B & Nishikura K Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med 1, 202–209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liddicoat BJ et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang L, Yang CS, Varelas X & Monti S Altered RNA editing in 3’ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep 6, 23226 (2016).This interesting article suggests that changes to the editing process of miRNA target genes might be selected for as advantageous to cancer cells.
- 118.Ota H et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153, 575–589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tomaselli S et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 16, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shoshan E et al. Reduced adenosine-to-inosine miR-455–5p editing promotes melanoma growth and metastasis. Nat. Cell Biol 17, 311–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Anadon C et al. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene 35, 4407–4413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zipeto MA et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell 19, 177–191 (2016).This article describes the inhibitory mechanism of let‑7 biogenesis by ADAR1 editase activity.
- 123.Zhang WC & Slack FJ ADARs edit microRNAs to promote leukemic stem cell activity. Cell Stem Cell 19, 141–142 (2016). [DOI] [PubMed] [Google Scholar]
- 124.Elkon R, Ugalde AP & Agami R Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet 14, 496–506 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Lianoglou S, Garg V, Yang JL, Leslie CS & Mayr C Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mayr C & Bartel DP Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009).This is an important study about the 3’UTR shortening process in cancer cells as a mechanism to avoid inhibition of oncogenes by miRNAs.
- 127.Preskill C & Weidhaas JB SNPs in microRNA binding sites as prognostic and predictive cancer biomarkers. Crit. Rev. Oncog 18, 327–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu Z et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 35, 4535–4541 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chin LJ et al. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Cancer Res. 68, 8535–8540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wynendaele J et al. An illegitimate microRNA target site within the 3’ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 70, 9641–9649 (2010). [DOI] [PubMed] [Google Scholar]
- 131.Kim M, Kogan N & Slack FJ Cis-acting elements in its 3’ UTR mediate post-transcriptional regulation of KRAS. Oncotarget 7, 11770–11784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zheng J et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet 48, 747–757 (2016).This paper provides an example of a SNP in a lncRNA that creates a binding site for a miRNA.
- 133.Bozic I, Allen B & Nowak MA Dynamics of targeted cancer therapy. Trends Mol. Med 18, 311–316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bozic I et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife 2, e00747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bozic I & Nowak MA Timing and heterogeneity of mutations associated with drug resistance in metastatic cancers. Proc. Natl Acad. Sci. USA 111, 15964–15968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hamberg M et al. miRTargetLink—miRNAs, genes and interaction networks. Int. J. Mol. Sci 17, 564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang P et al. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 43, 3478–3489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.[No authors listed.] The future of cancer genomics. Nat. Med 21, 99–99 (2015). [DOI] [PubMed] [Google Scholar]
- 139.Tomczak K, Czerwin´ska P & Wiznerowicz M The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol 19, A68–A77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goswami CP & Nakshatri H PROGmiR: a tool for identifying prognostic miRNA biomarkers in multiple cancers using publicly available data. J. Clin. Bioinforma 2, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bhattacharya A & Cui Y SomamiR 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 44, D1005–D1010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blin K et al. DoRiNA 2.0—upgrading the doRiNA database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 43, D160–D167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vlachos IS et al. DIANA-mirExTra v2.0: uncovering microRNAs and transcription factors with crucial roles in NGS expression data. Nucleic Acids Res. 44, W128–W134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen X et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep 6, 34985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Glazar P, Papavasileiou P & Rajewsky N circBase: a database for circular RNAs. RNA 20, 1666–1670 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lopez JP et al. Biomarker discovery: quantification of microRNAs and other small non-coding RNAs using next generation sequencing. BMC Med. Genomics 8, 35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yarmishyn AA & Kurochkin IV Long noncoding RNAs: a potential novel class of cancer biomarkers. Front. Genet 6, 145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ho T-T et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 43, e17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Matsui M & Corey DR Non-coding RNAs as drug targets. Nat. Rev. Drug Discov 16, 167–179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bak RO, Hollensen AK & Mikkelsen JG Managing microRNAs with vector-encoded decoy-type inhibitors. Mol. Ther 21, 1478–1485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen Y, Gao D-Y & Huang L In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev 81, 128–141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fatima R, Akhade VS, Pal D & Rao SMR Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol. Cell. Therap 3, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Babar IA et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc. Natl Acad. Sci. USA 109, E1695–E1704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cheng CJ et al. MicroRNA silencing for cancer therapy targeted to the tumor microenvironment. Nature 518, 107–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gutschner T et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Diaz-Lagares A et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl Acad. Sci. USA 113, E7535–E7544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lujambio A et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 29, 6390–6401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lujambio A et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl Acad. Sci. USA 105, 13556–13561 (2008). (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lujambio A et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 67, 1424–1429 (2007). [DOI] [PubMed] [Google Scholar]
- 160.He DX et al. Genome-wide profiles of methylation, microRNAs, and gene expression in chemoresistant breast cancer. Sci. Rep 6, 24706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng Q et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun 7, 11215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boeckel JN et al. Identification and characterization of hypoxia-regulated endothelial circular RNA. Circ. Res 117, 884–890 (2015). [DOI] [PubMed] [Google Scholar]