Abstract
Background
Non-alcoholic fatty liver disease has been associated with increased mRNA expression of FADS2 in the liver and estimated activity of delta-6 desaturase in serum, encoded by the FADS2 gene. Since DNA methylation in the FADS1/2/3 gene cluster has been previously linked with genetic variants and desaturase activities, we now aimed to discover factors regulating DNA methylation of the CpG sites annotated to FADS1/2 genes.
Methods
DNA methylation levels in the CpG sites annotated to FADS2 and FADS1 were analyzed from liver samples of 95 obese participants of the Kuopio Obesity Surgery Study (34 men and 61 women, age 49.5 ± 7.7 years, BMI 43.0 ± 5.7 kg/m2) using the Infinium HumanMethylation450 BeadChip (Illumina). Associations between DNA methylation levels and estimated delta-6 and delta-5 desaturase enzyme activities, liver histology, hepatic mRNA expression, FADS1/2 genotypes, and erythrocyte folate levels were analyzed.
Results
We found a negative correlation between DNA methylation levels of cg06781209 and cg07999042 and hepatic FADS2 mRNA expression (both p < 0.05), and with estimated delta-6 desaturase activity based on both liver and serum fatty acids (all p < 0.05). Interestingly, the methylation level of cg07999042 (p = 0.001) but not of cg06781209 (p = 0.874) was associated with FADS2 variant rs174616.
Conclusions
Genetic variants of FADS2 may contribute to the pathogenesis of non-alcoholic fatty liver disease by modifying DNA methylation.
Electronic supplementary material
The online version of this article (10.1186/s13148-019-0609-1) contains supplementary material, which is available to authorized users.
Keywords: Non-alcoholic fatty liver disease, FADS2, DNA methylation, Epigenetics, mRNA expression, Liver, Delta-6 desaturase
Background
Non-alcoholic fatty liver disease (NAFLD) refers to a wide spectrum of liver damage that ranges from simple steatosis to liver cirrhosis, affecting approximately 24% of global population [1]. Non-alcoholic steatohepatitis (NASH) is characterized by hepatocyte damage and inflammation in addition to steatosis. In up to 40% of NASH patients, the disease progresses to liver fibrosis, and the patients have an increased risk of end-stage liver disease and liver-related mortality [1–3]. Due to high prevalence of NAFLD and its association with insulin resistance, type II diabetes, metabolic syndrome, dyslipidemia, and cardiovascular diseases, it is also a major contributor to the cardiovascular risk in the population [1–4].
The risk of NAFLD and NASH is affected by genetic and lifestyle factors that associate with lipid metabolism [5–8]. In fact, multiple alterations in fatty acid composition of the liver are linked with NAFLD and NASH, and changes in the transcription of genes regulating fatty acid metabolism may play a role in the pathogenesis of NAFLD [7, 9–14]. Recently, the progression of NAFLD has also been linked with changes in epigenetic mechanisms, including DNA methylation [15–23]. Because changes in DNA methylation affect gene expression and are known to be regulated by both genetic variants and nutritional factors, such as folate [17, 24–30], DNA methylation is a potential step in gene-diet interaction.
We have previously shown that NASH associates with increased mRNA expression of fatty acid desaturase (FADS)2 gene in the liver and the activity of delta-6 desaturase, encoded by FADS2 [12]. Moreover, Howard et al. suggested that gene variants in the FADS1/2/3 gene cluster affect desaturase activities through altered DNA methylation [28]. Thus, we investigated the associations between liver DNA methylation levels in the FADS2 and FADS1 loci with delta-6 desaturase and delta-5 desaturase activities, FADS1 and FADS2 mRNA expression, FADS1/2 genotypes, and the levels of erythrocyte folate.
Methods
Subjects in the Kuopio Obesity Surgery Study
All patients undergoing obesity surgery in Kuopio University Hospital are recruited into an ongoing study investigating metabolic consequences of obesity surgery [31]. The present analysis includes baseline data from 95 people (34 men and 61 women, age 49.5 ± 7.7 years, Table 1), who were accepted for obesity surgery. Criteria for surgery were (1) body mass index (BMI) greater than 40 kg/m2, or greater than 35 kg/m2 with a significant comorbidity (e.g., type II diabetes); (2) failure of dietary and drug treatments to reduce weight; and (3) no other contraindication for the operation. Blood samples were drawn after an overnight of fasting. People with alcohol consumption of > 2 doses per day or more or people with previously diagnosed liver diseases not related to obesity were excluded from the study. As part of the surgery protocol, subjects were instructed to follow a preoperative very low-calorie diet for an average of 4 weeks. Patients consumed special products designed for a very low-calorie diet, and the daily energy intake was aimed to be 600–800 kcal. The study protocol was approved by the Ethics Committee of the Northern Savo Hospital District, and it was performed in accordance with the Helsinki Declaration. Written informed consent was obtained from all the participants.
Table 1.
Clinical characteristics of the study subjects (n = 95)
| Mean ± SD | |
|---|---|
| Sex (m/f) | 34/61 |
| Age (y) | 49.5 ± 7.7 |
| BMI (kg/m2) | 43.0 ± 5.7 |
| ALT (U/l) | 45.6 ± 34.3 |
| Fasting glucose (mmol/l) | 6.5 ± 2.1 |
| Fasting insulin (mU/l) | 18.7 ± 13.5 |
| Total cholesterol (mmol/l) | 4.2 ± 0.9 |
| HDL cholesterol (mmol/l) | 1.0 ± 0.2 |
| LDL cholesterol (mmol/l) | 2.4 ± 0.8 |
| Triglycerides (mmol/l) | 1.6 ± 0.7 |
Data presented as mean ± SD
Clinical measurements and laboratory determinations
BMI was calculated as weight (in kilograms) divided by height (in meters) squared. The serum glucose concentration was measured by enzymatic hexokinase photometric assay (Konelab Systems Reagents; Thermo Fischer Scientific, Vantaa, Finland). Serum insulin concentration was determined by immunoassay (ADVIA Centaur Insulin IRI, no. 02230141; Siemens Medical Solutions Diagnostics, Tarrytown, NY). Cholesterol, high-density lipoprotein (HDL) cholesterol, and triglyceride concentrations from the whole serum were assayed by standard automated enzymatic methods (Roche Diagnostics, Mannheim, Germany). Plasma alanine aminotransferase (ALT) concentration was determined using kinetic International Federation of Clinical Chemistry methods (Roche Diagnostics, Mannheim, Germany). Erythrocyte folate levels were measured in the fasted state using an electrochemiluminescence immunoassay (Roche Diagnostics).
Genome-wide DNA methylation analysis in human liver
DNA methylation was assessed according to previously described methods [32] using the Illumina Infinium HumanMethylation450 BeadChip, from which CpG sites annotated to FADS1 and FADS2 genes, including 18 sites annotated to FADS2 and 29 sites annotated to FADS1, were measured. The genome-wide DNA methylation data from this study population has previously been published [22, 32]. As described earlier, the raw methylation data in β values were converted to M values {M = log2[β/(1-β)]}, which were then used for statistical tests, and to easier interpret the results, M values were reconverted to β values which were used for tables and creating figures [32].
Assessment of serum and adipose tissue fatty acid composition and estimation of enzyme activities
In Kuopio Obesity Surgery Study (KOBS), the serum samples were extracted with chloroform–methanol (2:1) and the different lipid fractions, triglycerides (TG), cholesteryl esters (CE), and phospholipids (PL) were separated by solid-phase extraction with an aminopropyl column. Fatty acids were analyzed according to previously described methods [33, 34]. We also measured liver fatty acid composition in CE and TG using the same gas chromatograph method in samples (n = 19) that were previously extracted for liver NMR analysis [35].
The results of FA analysis are expressed as molar percentages (mol/mol of all fatty acids). The enzyme activities in serum CE, TG, and PL were estimated as product-to-precursor ratios of individual FAs in all lipid fractions. Delta-5-desaturase was estimated as the ratio of 20:4 n-6/20:3 n-6. Delta-6-desaturase, the ratio of 18:3 n-6/18:2 n-6, was calculated only in CE and TG due to a naturally low proportion of 18:3 n-6 in PL.
Genotype analyses
The variants rs174547 (FADS1) and rs174616 (FADS2) were genotyped from DNA samples of the KOBS study using the TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA) according to their protocol. These SNPs were chosen for the current analysis because of their previous association with PUFA levels and desaturase activities [36–39].
TruSeq targeted RNA expression
Custom gene panel (Illumina, San Diego, CA, USA) was used for measuring the mRNA expression levels of FADS1 and FADS2 in human liver according to instructions provided by the manufacturer using MiSeq system (Illumina, San Diego, CA, USA), as described previously [40]. The expression levels for each gene per sample in the custom gene panel were normalized based on the total number of aligned reads of the corresponding sample and the results are shown as percentage of total transcript reads.
Liver histology
Liver biopsies in the KOBS study were obtained during the obesity surgery using a Trucut needle (Radiplast AB, Uppsala, Sweden) or as a wedge biopsy. Overall histological assessment of liver biopsy samples was performed by one pathologist according to the standard criteria [41, 42]. Histological diagnosis was classified into three categories: (1) normal liver without any steatosis, inflammation, ballooning or fibrosis; (2) simple steatosis (steatosis > 5%) without evidence of hepatocellular ballooning, inflammation or fibrosis; and (3) NASH. Chronic hepatitis B and C were excluded using serology if ALT levels were elevated prior to the surgery. Hemochromatosis was excluded by histological analysis of liver biopsies, and by normal serum ferritin levels in subjects that had elevated serum ALT level.
LC–MS analysis
Plasma methionine and glycine betaine levels were determined by UHPLC-qTOF-MS system (Agilent Technologies, Waldbronn, Karlsruhe, Germany), using hydrophilic interaction chromatography and positive (+) electrospray ionization. The detailed analytical parameters have been reported earlier [43]. The data acquisition software was MassHunter Acquisition B.04.00 (Agilent Technologies), and the data were evaluated with MassHunter Qualitative Analysis B.05.00 (Agilent Technologies, USA). Methionine and glycine betaine were identified based on authentic chemical standards.
Statistical analysis
Data are presented as mean ± SD, unless otherwise stated. The final number of subjects in each statistical model and figure ranged between 95 and 19 depending on availability of data. To assess differences in clinical characteristics and DNA methylation between the study groups, one-way ANOVA or a one-way Welch ANOVA with Bonferroni post-hoc test was used. A logarithmic or inverse transformation was performed for skewed variables after assessing normality graphically. Spearman correlation coefficient was used for correlation analyses, and the statistical significance was corrected for multiple comparisons using the Benjamini–Hochberg procedure with a false discovery rate (FDR) of 0.25. The IBM SPSS Statistics for Windows software, Version 24 (IBM Corp., Armonk, NY, USA), was used for statistical analyses.
Results
FADS2 DNA methylation associates with estimated desaturase enzyme activities
The characteristics of the study cohort are presented in Table 1. First, we selected the CpG sites annotated to FADS1 and FADS2 gene regions from the genome-wide DNA methylation data published before [22, 32]. Using this information, we investigated in which of these CpG sites DNA methylation correlated with estimated delta-6 desaturase and delta-5 desaturase enzyme activities based on liver and serum fatty acids analysis (data for individual CpG sites shown in Fig. 1 for FADS2 and Additional file 1 for FADS1). As a result, we discovered that methylation at two CpG sites in the FADS2, cg06781209 and cg07999042, correlated significantly with estimated activity of delta-6 desaturase in both liver and serum in all lipid fractions (all p < 0.05); therefore, they were selected for further analysis. As for DNA methylation in FADS1, some significant correlations were found (Additional file 1), but because no CpG site correlated with delta-5 desaturase activity in all lipid fractions, we focused analyses on FADS2 methylation.
Fig. 1.
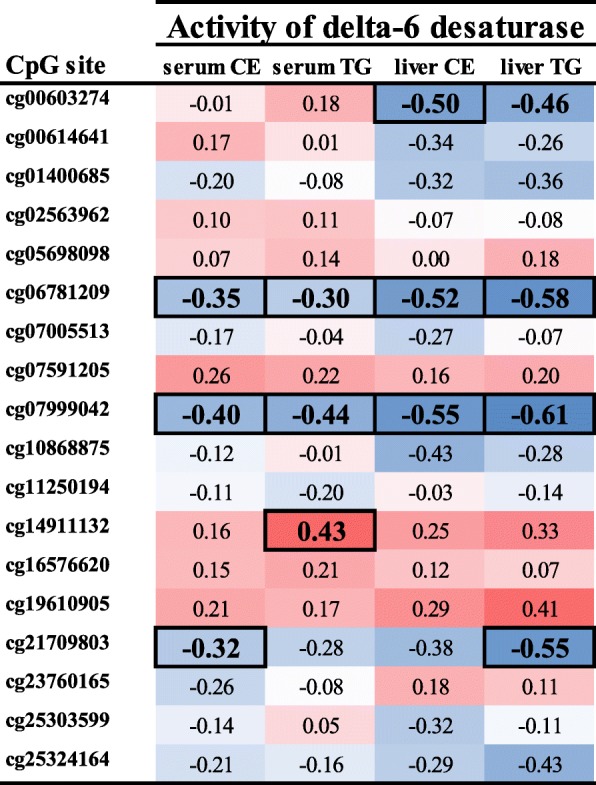
Association between estimated delta-6 desaturase activity in serum and liver and DNA methylation of CpG sites in FADS2. A Spearman’s correlation was run to assess the relationship between methylation levels and delta-6 desaturase activity (n = 49 for serum and n = 19 for liver). Data is presented as Spearman correlation coefficient, and associations with a nominal p value < 0.05, which remained significant after correction for multiple testing using the Benjamini-Hochberg procedure with FDR 0.25, are indicated by boxes around the correlation coefficient. Positive correlations indicated with red and negative correlations with blue. Activity of delta-6 desaturase was estimated by a ratio of 18:3 n-6/18:2 n-6 in cholesteryl esters (CE) and triglycerides (TG)
FADS2 DNA methylation associates with FADS2 mRNA expression in the liver
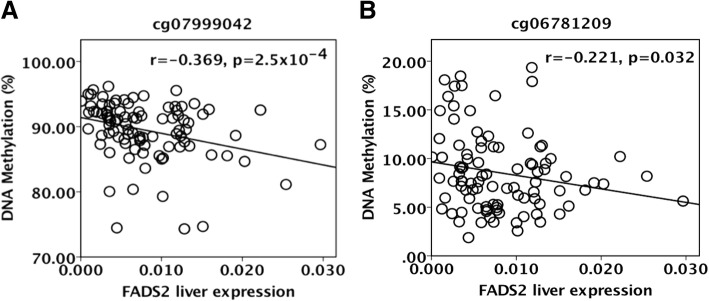
For a DNA methylation at a certain CpG site to regulate transcription, the assumption is that methylation and transcription correlate negatively. In fact, FADS2 mRNA expression in the liver had a negative correlation with the methylation level of both cg07999042 (r = − 0.369, p = 2.5 × 10−4, Fig. 2) and cg06781209 (r = − 0.221, p = 0.032) suggesting that these CpG sites could be regulatory.
Fig. 2.
FADS2 DNA methylation associates with hepatic mRNA expression of FADS2. Scatterplots demonstrating correlations between methylation levels of cg06781209 (a) and cg07999042 (b) and the mRNA expression of FADS2 in liver. Spearman correlation coefficient was used for correlation analyses, and the statistical significance was corrected for multiple comparisons using the Benjamini–Hochberg procedure with a false discovery rate (FDR) of 0.25. Nominal p values are presented
Genotype is associated with FADS2 DNA methylation levels at some CpG sites annotated to FADS2
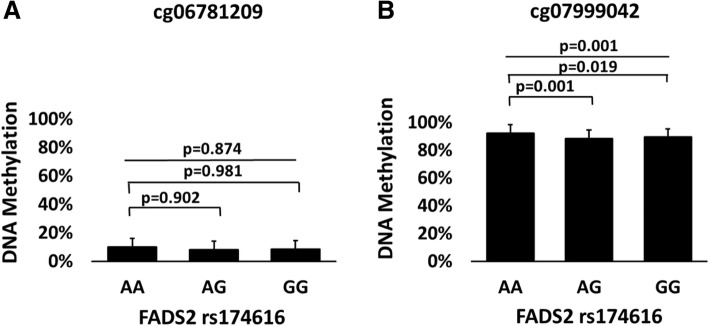
We have previously linked genetic variation of FADS2 and FADS1 to estimated delta-6 and delta-5 desaturase activities in adipose tissue [40], and Howard et al. suggested that the association between genetic variation in the FADS gene cluster and altered fatty acid metabolism is a result of altered DNA methylation [28]. Thus, we investigated if SNPs in FADS2 and FADS1 associated with methylation levels of CpG sites in this gene cluster. Interestingly, we discovered that the FADS2 variant rs174616 had a significant association with the methylation level of cg07999042 (p = 0.001 in ANOVA), but not with the methylation level of cg06781209 (p = 0.874 in Welch ANOVA, Fig. 3a, b, Additional file 2). Bonferroni post-hoc test revealed that subjects who had the major allele G of the FADS2 polymorphism had significantly lower methylation levels of cg07999042 than those with the AA genotype (88.3 ± 4.97% vs. 92.3 ± 2.40%, p = 0.001 for AG vs. AA, respectively, and 89.4 ± 4.05% vs. 92.3 ± 2.40%, p = 0.019 for GG vs. AA, Additional file 2). There were no significant differences in clinical and metabolic parameters between subjects with different FADS2 genotypes (Additional file 3). FADS1 variant rs174547 associated with methylation levels at some CpG sites annotated to the FADS1 gene (Additional file 4). Subjects with the TT genotype had higher HDL cholesterol levels compared to those with CC genotype (p = 0.041), but otherwise there were no significant differences between subjects with different FADS1 genotypes. However, we concentrated on the FADS2 methylation because of its association with mRNA expression and estimated enzyme activities (see above).
Fig. 3.
FADS2 DNA methylation levels according to groups based on FADS2 genotype. Panels (a) and (b) show DNA methylation levels in cg06781209 and cg07999042 according to FADS2 genotype (n = 88). Statistical significance was calculated with one-way ANOVA or a one-way Welch ANOVA with Bonferroni post-hoc test and nominal p values are presented
Association between DNA methylation of FADS2 and liver steatosis
Since we have previously shown in this study population that NASH associates with altered delta-6 desaturase activity [12], we wanted to investigate if the histological characteristics of NAFLD correlate also with FADS1/2 DNA methylation levels in liver. Spearman correlation analysis showed that both cg06781209 and cg07999042 associated negatively with the grade of steatosis in liver (n = 72, nominal p < 0.05 for both, Additional file 5), but the correlation did not reach statistical significance after correcting for multiple testing (FDR 0.25). Other histological features of NASH, such as lobular inflammation or hepatocellular ballooning, did not show association with the methylation levels of cg06781209 or cg07999042. The Spearman correlation coefficients for all CpG sites annotated to FADS1/2 methylation and liver histology are presented in Additional file 6.
Association between DNA methylation levels and erythrocyte folate level
Since our earlier results suggests that hypomethylation in the liver of diabetic subjects could be explained by reduced folate levels in this study cohort [32], we investigated if the erythrocyte folate levels associate with DNA methylation levels in the FADS2. We observed a nominally significant correlation between methylation levels of cg06781209 and levels of erythrocyte folate (nominal p value of 0.025, Spearman correlation, Additional file 5), but the association did not remain significant after correcting for multiple testing. Further, we divided the subjects into three groups according to erythrocyte folate levels (1st tertile 1076.0; 613.0–1279.0, (median; min-max), 2nd tertile 1417.0; 1280.0–1573.0, 3rd tertile 1939.0;1593.0–2986.0). The groups did not differ in clinical characteristics (Additional file 7). In this preliminary analysis, we observed that there was a borderline significant association between methylation levels of cg06781209 and erythrocyte folate levels (nominal p value of 0.050 in ANOVA, Additional file 8). Other nutrients potentially regulating the methylation pathway, such as plasma glycine betaine or l-methionine, did not significantly correlate with DNA methylation levels in the FADS2 (Additional file 5).
Discussion
In this study, we aimed to identify factors behind altered fatty acid desaturase activities in NAFLD, following the findings that estimated desaturase enzyme activities in serum and FADS2 mRNA expression in the liver are higher in individuals with NASH [12]. We found an association of higher hepatic FADS2 mRNA expression and serum delta-6 desaturase activity with lower levels of DNA methylation in CpG sites annotated to FADS2. Moreover, DNA methylation levels of FADS2 were linked with the FADS2 genotype, suggesting a novel mechanism for the genetic contribution of FADS2 genotypes in the pathogenesis of NAFLD.
An important finding of this study was that high estimated delta-6 desaturase activity, observed in subjects with NASH [12], associated with low DNA methylation of two CpG sites annotated to FADS2, cg06781209 and cg07999042. The first CpG site, cg06781209, is located in a CpG-rich region (CpG island) within the first 1500 bp upstream from the transcription start site, in a potential gene enhancer region indicated by monomethylation of histone H3 lysine4 (H3K4me1) and acetylation of H3K27 (H3K27Ac) (the ENCODE tracks, Additional file 9) [44, 45]. Moreover, cg06781209 is in an area with several transcription factor binding sites: POLR2A, YY1, CHD2, RELA, CHD1, TAF1, CTCF, SIN3A, and TCF7L2. High peaks of H3K27Ac indicate activation of transcription in this area. The second significant CpG site, cg07999042, is located in the body of FADS2, and it is in the same area as transcription factor binding site for POLR2A. The cg07999042 has previously been associated with the risk of type II diabetes in men with low birth weight [29]. Moreover, altered DNA methylation of several genes has been associated with obesity, type II diabetes, and NASH [15–17, 32, 46, 47], but the role of FADS2 DNA methylation in NAFLD is unknown.
In this study, we observed that lower DNA methylation levels of both cg06781209 and cg07999042 correlated with not only higher delta-6 desaturase activity in serum but also with higher FADS2 mRNA expression in the liver. These associations between FADS2 DNA methylation, mRNA expression, and desaturase activity suggest that alterations in the methylation level of these CpG sites annotated to FADS2 may affect delta-6 desaturase activities through transcriptional changes, especially considering their location in the area with several transcription binding sites (Additional file 9). Similar findings have been previously demonstrated for other genes involved in fatty acid metabolism in subjects with NAFLD [18]. However, since we were able to study only associations between DNA methylation, mRNA expression and desaturase activities, the mechanisms need to be further examined in future studies.
We discovered that altered FADS2 DNA methylation was associated with the FADS2 variant rs174616. This variant is located in intron 7 of the FADS2 gene (Additional file 9) and has been associated with alterations in fatty acid metabolism, inflammation, and type II diabetes [39, 40, 48]. In this study, subjects with the major allele G had lower DNA methylation levels of cg07999042 in FADS2 compared to homozygotes for the minor allele A. Accordingly, we have shown that the subjects with the major allele G have also higher delta-6 desaturase activity, similarly to patients with NASH [12, 40]. Thus, it is plausible that FADS2 genotype may regulate delta-6 desaturase activity at least partly through DNA methylation. Similar results have been published by Rahbar et al., who reported associations between genotype and DNA methylation in the putative enhancer region within the FADS gene cluster [49], and by Howard et al., who suggested that the association between genetic variation in the FADS gene cluster and altered fatty acid metabolism is a result of altered DNA methylation [28].
Hepatic steatosis is associated with disturbances in the methylation pathway [23, 50], and altered DNA methylation has been linked with different stages of NAFLD, possibly promoting the disease progression [15–22]. Thus, we wanted to study the associations between DNA methylation and histological features of NAFLD. After correcting for multiple testing, only one CpG site annotated to FADS1 showed significant correlation to steatosis grade and fibrosis stage (cg16213375). No significant correlations were found for FADS2, but both cg06781209 and cg07999042 had a nominally significant correlation with steatosis grade. Based on these results, it would be of interest to study the possible role of DNA methylation of genes regulating fatty acid metabolism in the future. Although altered DNA methylation in several other genes has been associated with NASH and liver fibrosis [15, 16, 18, 20–22], DNA methylation in neither cg06781209 nor cg07999042 in FADS2 correlated with fibrosis or inflammation in the liver. This supports our suggestion that altered DNA methylation in FADS2 contributes primarily to altered desaturase activity and possibly liver steatosis in NAFLD, and not to liver inflammation or fibrosis.
We wanted to investigate if DNA methylation levels in FADS2 associate with erythrocyte folate levels. Folate is a coenzyme in methylation pathway in the liver, affecting the synthesis of the universal methyl donor in DNA methylation, S-adenosyl-methionine [19, 25]. Since folate modulates the availability of methyl donors in DNA methylation, it can affect gene expression and thus influence cell function. Moreover, folate deficiency and changes in S-adenosyl-methionine have been related to hepatic accumulation of triglycerides and alterations in expression of genes involved in fatty acid metabolism [50]. Although reduced folate levels are often associated with low DNA methylation, as we have also published for genes related to type II diabetes [32], in this preliminary analysis we observed only a nominally significant negative correlation between folate levels and DNA methylation of cg06781209. Similar findings have been observed in mice, as Tryndyak et al. reported hypermethylation in response to folate-deficient diet in mice with severe NASH-like liver injury [20]. The discrepancy in these findings and previous studies warrants that the association between erythrocyte folate levels and DNA methylation in liver needs to be investigated in experimental models and in larger study populations. Finally, other nutrients in the methylation pathway, and potentially the amount and quality of fat in the diet [50], may regulate DNA methylation in the liver. For example, amino acids, such as methionine and betaine, are involved in the synthesis of S-adenosyl-methionine and serve as methyl donors [19, 25, 26]. Despite their important role in the methylation pathway and association with NAFLD [51], we found no significant correlations between serum methionine, glycine betaine, and the methylation levels of cg06781209 and cg07999042.
We recognize the following limitations in our study. The sample size was modest; thus, we may not have recognized all relevant associations. As the study was cross-sectional, we cannot conclude that there is a causal relationship between DNA methylation, FADS2 mRNA expression, and delta-6 desaturase activities. Actual enzyme activities could not be measured with human liver samples. Instead, we used estimated enzyme activities calculated from product-to-precursor ratios as surrogate measures, similarly to previous studies [28, 52, 53]. We acknowledge that because this study was performed in obese subjects, the results might not reflect the DNA methylation in normal-weighed individuals with NAFLD. Moreover, due to strong linkage disequilibrium in the FADS1/2 locus, we acknowledge that the FADS2 variant rs174616 most likely is a marker for other uncharacterized functional variants.
Conclusions
In conclusion, we observed a novel association of FADS2 DNA methylation at selected CpG sites with the FADS2 variant rs174616. Moreover, DNA methylation levels of these CpG sites correlated with FADS2 mRNA expression and estimated delta-6 desaturase activity. Thus, we suggest that genetic variants of FADS2 may contribute to the pathogenesis of NAFLD by modifying fatty acid metabolism through DNA methylation.
Additional files
Association between estimated delta-5 desaturase activity in serum and liver and DNA methylation of CpG sites in FADS1. (DOCX 26 kb)
DNA methylation levels in CpG-sites annotated to FADS2 in groups based on FADS2 variant rs174616. (DOCX 27 kb)
Characteristics of the groups based on FADS2 genotype. (DOCX 24 kb)
DNA methylation levels in CpG-sites annotated to FADS1 in groups based on FADS1 genotype. (DOCX 30 kb)
Correlation of DNA methylation levels of cg06781209 and cg07999042 with clinical parameters and liver histology. (DOCX 25 kb)
Association between liver histology and DNA methylation of CpG sites annotated to FADS2 and FADS1. (DOCX 29 kb)
Characteristics of the groups based on erythrocyte folate. (DOCX 27 kb)
DNA methylation of FADS2 in groups based on erythrocyte folate. (DOCX 26 kb)
Association between cg06781209, cg07999042 and rs174616 in the FADS gene cluster. (DOCX 135 kb)
Acknowledgements
We thank Jenna Jokkala, Sirkku Karhunen, Päivi Turunen, Erja Kinnunen, and Matti Laitinen for their assistance.
Funding
This study was supported by the University of Eastern Finland Doctoral School and The Finnish Medical Foundation grant (to PW). This study was also supported by the Finnish Diabetes Research Foundation (to JP), Kuopio University Hospital Project grant (NUDROBE, JP), the Academy of Finland grant (JP, Contract no. 120,979;138,006), the Finnish Cultural Foundation (JP), and the University of Eastern Finland Spearhead Funding (JP) as well as The Swedish Research Council (CL) and ALF (CL).
Availability of data and materials
The datasets analyzed during the current study are not publicly available, because the written informed consent from the participants does not cover publication of raw individual array data.
Abbreviations
- ALT
Alanine aminotransferase
- BMI
Body mass index
- CE
Cholesteryl esters
- FADS
Fatty acid desaturase
- HDL
High-density lipoprotein
- KOBS
Kuopio Obesity Surgery Study
- NAFLD
Non-alcoholic fatty liver disease
- NASH
Non-alcoholic steatohepatitis
- PL
Phospholipids
- TG
Triglycerides
Authors’ contributions
PW researched data and wrote the manuscript with the help of VM and VdM. MV was responsible for molecular analysis of the tissue samples. AP participated on the analysis of the DNA methylation data and reviewed the manuscript. KH was responsible for the LC-MS analysis and reviewed the manuscript. CL was responsible for the study design of DNA methylation analyses and reviewed the manuscript. JP is the guarantor of this work and, as such, was responsible for the clinical and molecular studies, and had full access to all the data to take responsibility for the integrity and the accuracy of the analyses. All authors have approved the final article.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Northern Savo Hospital District (54/2005,104/2008 and 27/2010), and it was performed in accordance with the Helsinki Declaration. Written informed consent was obtained from all the participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Paula Walle, Email: paula.walle@uef.fi.
Ville Männistö, Email: ville.mannisto@kuh.fi.
Vanessa Derenji de Mello, Email: vanessa.laaksonen@uef.fi.
Maija Vaittinen, Email: maija.vaittinen@uef.fi.
Alexander Perfilyev, Email: alexander.perfilyev@med.lu.se.
Kati Hanhineva, Email: kati.hanhineva@uef.fi.
Charlotte Ling, Email: charlotte.ling@med.lu.se.
Jussi Pihlajamäki, Phone: +358-50-3440187, Email: jussi.pihlajamaki@uef.fi.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Ekstedt M, Franzen LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, Kechagias S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44(4):865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 3.Rafiq N, Bai C, Fang Y, Srishord M, McCullough A, Gramlich T, Younossi ZM. Long-term follow-up of patients with nonalcoholic fatty liver. Clin Gastroenterol Hepatol. 2009;7(2):234–238. doi: 10.1016/j.cgh.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 4.van den Berg EH, Amini M, Schreuder TC, Dullaart RP, Faber KN, Alizadeh BZ, Blokzijl H. Prevalence and determinants of non-alcoholic fatty liver disease in lifelines: a large Dutch population cohort. PLoS One. 2017;12(2):e0171502. doi: 10.1371/journal.pone.0171502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, Nguyen P, Hernandez C, Richards L, Salotti J, Lin S, Seki E, Nelson KE, Sirlin CB, Brenner D. Genetics of NAFLD in twins consortium. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149(7):1784–1793. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelber-Sagi S, Lotan R, Shlomai A, Webb M, Harrari G, Buch A, Nitzan Kaluski D, Halpern Z, Oren R. Predictors for incidence and remission of NAFLD in the general population during a seven-year prospective follow-up. J Hepatol. 2012;56(5):1145–1151. doi: 10.1016/j.jhep.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Arendt BM, Comelli EM, Ma DW, Lou W, Teterina A, Kim T, Fung SK, Wong DK, McGilvray I, Fischer SE, Allard JP. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology. 2015;61(5):1565–1578. doi: 10.1002/hep.27695. [DOI] [PubMed] [Google Scholar]
- 8.Yki-Jarvinen H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. 2014;2(11):901–910. doi: 10.1016/S2213-8587(14)70032-4. [DOI] [PubMed] [Google Scholar]
- 9.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, Contos MJ, Sterling RK, Fuchs M, Zhou H, Watkins SM, Sanyal AJ. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50(6):1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li ZZ, Berk M, McIntyre TM, Feldstein AE. Hepatic lipid partitioning and liver damage in nonalcoholic fatty liver disease: role of stearoyl-CoA desaturase. J Biol Chem. 2009;284(9):5637–5644. doi: 10.1074/jbc.M807616200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jump DB, Tripathy S, Depner CM. Fatty acid-regulated transcription factors in the liver. Annu Rev Nutr. 2013;33:249–269. doi: 10.1146/annurev-nutr-071812-161139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walle P, Takkunen M, Mannisto V, Vaittinen M, Lankinen M, Karja V, Kakela P, Agren J, Tiainen M, Schwab U, Kuusisto J, Laakso M, Pihlajamaki J. Fatty acid metabolism is altered in non-alcoholic steatohepatitis independent of obesity. Metabolism. 2016;65(5):655–666. doi: 10.1016/j.metabol.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Yamada K, Mizukoshi E, Sunagozaka H, Arai K, Yamashita T, Takeshita Y, Misu H, Takamura T, Kitamura S, Zen Y, Nakanuma Y, Honda M, Kaneko S. Characteristics of hepatic fatty acid compositions in patients with nonalcoholic steatohepatitis. Liver Int. 2015;35(2):582–590. doi: 10.1111/liv.12685. [DOI] [PubMed] [Google Scholar]
- 14.Anjani K, Lhomme M, Sokolovska N, Poitou C, Aron-Wisnewsky J, Bouillot JL, Lesnik P, Bedossa P, Kontush A, Clement K, Dugail I, Tordjman J. Circulating phospholipid profiling identifies portal contribution to NASH signature in obesity. J Hepatol. 2015;62(4):905–912. doi: 10.1016/j.jhep.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Ahrens M, Ammerpohl O, von Schonfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H, Erhart W, Egberts J, Sipos B, Schreiber S, Hasler R, Stickel F, Becker T, Krawczak M, Rocken C, Siebert R, Schafmayer C, Hampe J. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Zeybel M, Hardy T, Robinson SM, Fox C, Anstee QM, Ness T, Masson S, Mathers JC, French J, White S, Mann J. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin. Epigenetics. 2015;7:25. doi: 10.1186/s13148-015-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145(5):1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mwinyi J, Bostrom AE, Pisanu C, Murphy SK, Erhart W, Schafmayer C, Hampe J, Moylan C, Schioth HB. NAFLD is associated with methylation shifts with relevance for the expression of genes involved in lipoprotein particle composition. Biochim Biophys Acta. 2017;1862(3):314–323. doi: 10.1016/j.bbalip.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Lee JH, Friso S, Choi SW. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients. 2014;6(8):3303–3325. doi: 10.3390/nu6083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tryndyak VP, Han T, Fuscoe JC, Ross SA, Beland FA, Pogribny IP. Status of hepatic DNA methylome predetermines and modulates the severity of non-alcoholic fatty liver injury in mice. BMC Genomics. 2016;17:298. doi: 10.1186/s12864-016-2617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitamoto T, Kitamoto A, Ogawa Y, Honda Y, Imajo K, Saito S, Yoneda M, Nakamura T, Nakajima A, Hotta K. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol. 2015;63(2):494–502. doi: 10.1016/j.jhep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 22.de Mello VD, Matte A, Perfilyev A, Mannisto V, Ronn T, Nilsson E, Kakela P, Ling C, Pihlajamaki J. Human liver epigenetic alterations in non-alcoholic steatohepatitis are related to insulin action. Epigenetics. 2017;12(4):287–295. doi: 10.1080/15592294.2017.1294305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, Kim Y, Friso S, Choi SW. Epigenetics in non-alcoholic fatty liver disease. Mol Asp Med. 2017;54:78–88. doi: 10.1016/j.mam.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 25.Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Adv Nutr. 2012;3(1):21–38. doi: 10.3945/an.111.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park LK, Friso S, Choi SW. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc. 2012;71(1):75–83. doi: 10.1017/S0029665111003302. [DOI] [PubMed] [Google Scholar]
- 27.Volkov P, Olsson AH, Gillberg L, Jorgensen SW, Brons C, Eriksson KF, Groop L, Jansson PA, Nilsson E, Ronn T, Vaag A, Ling C. A genome-wide mQTL analysis in human adipose tissue identifies genetic variants associated with DNA methylation, gene expression and metabolic traits. PLoS One. 2016;11(6):e0157776. doi: 10.1371/journal.pone.0157776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard TD, Mathias RA, Seeds MC, Herrington DM, Hixson JE, Shimmin LC, Hawkins GA, Sellers M, Ainsworth HC, Sergeant S, Miller LR, Chilton FH. DNA methylation in an enhancer region of the FADS cluster is associated with FADS activity in human liver. PLoS One. 2014;9(5):e97510. doi: 10.1371/journal.pone.0097510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillberg L, Perfilyev A, Brons C, Thomasen M, Grunnet LG, Volkov P, Rosqvist F, Iggman D, Dahlman I, Riserus U, Ronn T, Nilsson E, Vaag A, Ling C. Adipose tissue transcriptomics and epigenomics in low birthweight men and controls: role of high-fat overfeeding. Diabetologia. 2016;59(4):799–812. doi: 10.1007/s00125-015-3852-9. [DOI] [PubMed] [Google Scholar]
- 30.Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, Nilsson E, Riserus U, Ling C. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105(4):991–1000. doi: 10.3945/ajcn.116.143164. [DOI] [PubMed] [Google Scholar]
- 31.Pihlajamaki J, Gronlund S, Simonen M, Kakela P, Moilanen L, Paakkonen M, Pirinen E, Kolehmainen M, Karja V, Kainulainen S, Uusitupa M, Alhava E, Miettinen TA, Gylling H. Cholesterol absorption decreases after Roux-en-Y gastric bypass but not after gastric banding. Metabolism. 2010;59(6):866–872. doi: 10.1016/j.metabol.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson E, Matte A, Perfilyev A, de Mello VD, Kakela P, Pihlajamaki J, Ling C. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J Clin Endocrinol Metab. 2015;100(11):E1491–E1501. doi: 10.1210/jc.2015-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agren JJ, Julkunen A, Penttila I. Rapid separation of serum lipids for fatty acid analysis by a single aminopropyl column. J Lipid Res. 1992;33(12):1871–1876. [PubMed] [Google Scholar]
- 34.Venalainen T, Schwab U, Agren J, de Mello V, Lindi V, Eloranta AM, Kiiskinen S, Laaksonen D, Lakka TA. Cross-sectional associations of food consumption with plasma fatty acid composition and estimated desaturase activities in Finnish children. Lipids. 2014;49(5):467–479. doi: 10.1007/s11745-014-3894-7. [DOI] [PubMed] [Google Scholar]
- 35.Mannisto VT, Simonen M, Soininen P, Tiainen M, Kangas AJ, Kaminska D, Venesmaa S, Kakela P, Karja V, Gylling H, Ala-Korpela M, Pihlajamaki J. Lipoprotein subclass metabolism in nonalcoholic steatohepatitis. J Lipid Res. 2014;55(12):2676–2684. doi: 10.1194/jlr.P054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellstrand S, Sonestedt E, Ericson U, Gullberg B, Wirfalt E, Hedblad B, Orho-Melander M. Intake levels of dietary long-chain PUFAs modify the association between genetic variation in FADS and LDL-C. J Lipid Res. 2012;53(6):1183–1189. doi: 10.1194/jlr.P023721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merino DM, Ma DW, Mutch DM. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 2010;9:63. doi: 10.1186/1476-511X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merino DM, Johnston H, Clarke S, Roke K, Nielsen D, Badawi A, El-Sohemy A, Ma DW, Mutch DM. Polymorphisms in FADS1 and FADS2 alter desaturase activity in young Caucasian and Asian adults. Mol Genet Metab. 2011;103(2):171–178. doi: 10.1016/j.ymgme.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, Widhalm K, Moschonis G, Stehle P, Amouyel P, De Henauw S, Molnar D, Moreno LA, Meirhaeghe A, Dallongeville J. HELENA study group. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J. Lipid Res. 2010;51(8):2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaittinen M, Walle P, Kuosmanen E, Mannisto V, Kakela P, Agren J, Schwab U, Pihlajamaki J. FADS2 genotype regulates delta-6 desaturase activity and inflammation in human adipose tissue. J Lipid Res. 2016;57(1):56–65. doi: 10.1194/jlr.M059113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94(9):2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 42.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ, Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 43.Hanhineva K, Lankinen MA, Pedret A, Schwab U, Kolehmainen M, Paananen J, de Mello V, Sola R, Lehtonen M, Poutanen K, Uusitupa M, Mykkanen H. Nontargeted metabolite profiling discriminates diet-specific biomarkers for consumption of whole grains, fatty fish, and bilberries in a randomized controlled trial. J Nutr. 2015;145(1):7–17. doi: 10.3945/jn.114.196840. [DOI] [PubMed] [Google Scholar]
- 44.Kim TK, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162(5):948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pradeepa MM. Causal role of histone acetylations in enhancer function. Transcription. 2017;8(1):40–47. doi: 10.1080/21541264.2016.1253529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD, Deloukas P, Siebert R, Sipos B, Becker T, Rocken C, Schafmayer C, Hampe J. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirola CJ, Gianotti TF, Burgueno AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castano GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62(9):1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 48.Yao M, Li J, Xie T, He T, Fang L, Shi Y, Hou L, Lian K, Wang R, Jiang L. Polymorphisms of rs174616 in the FADS1-FADS2 gene cluster is associated with a reduced risk of type 2 diabetes mellitus in northern Han Chinese people. Diabetes Res Clin Pract. 2015;109(1):206–212. doi: 10.1016/j.diabres.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Rahbar E, Ainsworth HC, Howard TD, Hawkins GA, Ruczinski I, Mathias R, Seeds MC, Sergeant S, Hixson JE, Herrington DM, Langefeld CD, Chilton FH. Uncovering the DNA methylation landscape in key regulatory regions within the FADS cluster. PLoS One. 2017;12(9):e0180903. doi: 10.1371/journal.pone.0180903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.da Silva RP, Kelly KB, Al Rajabi A, Jacobs RL. Novel insights on interactions between folate and lipid metabolism. Biofactors. 2014;40(3):277–283. doi: 10.1002/biof.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sookoian S, Puri P, Castano GO, Scian R, Mirshahi F, Sanyal AJ, Pirola CJ. Nonalcoholic steatohepatitis is associated with a state of betaine-insufficiency. Liver Int. 2017;37(4):611–619. doi: 10.1111/liv.13249. [DOI] [PubMed] [Google Scholar]
- 52.Peter A, Cegan A, Wagner S, Lehmann R, Stefan N, Konigsrainer A, Konigsrainer I, Haring HU, Schleicher E. Hepatic lipid composition and stearoyl-coenzyme A desaturase 1 mRNA expression can be estimated from plasma VLDL fatty acid ratios. Clin Chem. 2009;55(12):2113–2120. doi: 10.1373/clinchem.2009.127274. [DOI] [PubMed] [Google Scholar]
- 53.Warensjo E, Ohrvall M, Vessby B. Fatty acid composition and estimated desaturase activities are associated with obesity and lifestyle variables in men and women. Nutr Metab Cardiovasc Dis. 2006;16(2):128–136. doi: 10.1016/j.numecd.2005.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association between estimated delta-5 desaturase activity in serum and liver and DNA methylation of CpG sites in FADS1. (DOCX 26 kb)
DNA methylation levels in CpG-sites annotated to FADS2 in groups based on FADS2 variant rs174616. (DOCX 27 kb)
Characteristics of the groups based on FADS2 genotype. (DOCX 24 kb)
DNA methylation levels in CpG-sites annotated to FADS1 in groups based on FADS1 genotype. (DOCX 30 kb)
Correlation of DNA methylation levels of cg06781209 and cg07999042 with clinical parameters and liver histology. (DOCX 25 kb)
Association between liver histology and DNA methylation of CpG sites annotated to FADS2 and FADS1. (DOCX 29 kb)
Characteristics of the groups based on erythrocyte folate. (DOCX 27 kb)
DNA methylation of FADS2 in groups based on erythrocyte folate. (DOCX 26 kb)
Association between cg06781209, cg07999042 and rs174616 in the FADS gene cluster. (DOCX 135 kb)
Data Availability Statement
The datasets analyzed during the current study are not publicly available, because the written informed consent from the participants does not cover publication of raw individual array data.