Sir,
Aminoglycosides are widely used to treat MDR Gram-negative bacterial infections with bactericidal activity mediated by binding the 16S rRNA aminoacyl-tRNA recognition site to prevent protein synthesis. Multiple aminoglycoside resistance mechanisms have been documented including 16S rRNA modification.1 NpmA, an uncommon 16S rRNA methyltransferase originally identified in a clinical Escherichia coli isolate confers pan-aminoglycoside resistance.2 In this study, routine WGS of hospital-acquired Clostridioides (Clostridium) difficile identified npmA in the genome of a clinical isolate (CD7814). C. difficile is a Gram-positive, spore-forming enteric pathogen and the cause of most hospital-acquired, antibiotic-associated diarrhoea. The epidemiology of C. difficile has changed in the past several decades with infections increasingly being reported outside of acute care settings.3 The discovery of npmA in the genome of a clinical C. difficile isolate has implications for the spread of aminoglycoside resistance.
Clinical C. difficile isolates are routinely sequenced using Illumina NextSeq500 and Nextera libraries as part of an infection prevention initiative at our hospital.4 Genomes are assembled using SPAdes, annotated with Prokka and characterized by searches of ResFinder and pubMLST databases (https://pubmlst.org/cdifficile/).5–7 This analysis identified NpmA-coding sequences in C. difficile isolate CD7814 belonging to ST11. CD7814 carries two additional aminoglycoside resistance determinants: (i) aph(3′)-III, which encodes an aminoglycoside phosphotransferase; and (ii) ant(6)-Ia, which encodes an aminoglycoside nucleotidyltransferase.
Aminoglycoside susceptibility testing was performed by Etest on CD7814 and two additional ST11 isolates from our hospital that lack npmA (CD7861 and CD7786). Cell suspensions corresponding to 0.5 McFarland were prepared and plated onto Brucella blood agar plates supplemented with vitamin K1 and haemin (Anaerobe Systems, Morgan Hill, CA, USA). Etest strips (bioMérieux, Durham, NC, USA) were applied and the plates were incubated anaerobically at 37°C for 48 h as previously described.8 Etests were read according to the manufacturer’s instructions. CD7814 demonstrated high-level resistance to gentamicin (>256 mg/L) relative to CD7861 (64 mg/L) and CD7786 (24 mg/L). High-level resistance to tobramycin and amikacin was observed in all ST11 C. difficile isolates tested. Although CLSI breakpoints for C. difficile are not defined for aminoglycosides, these data suggest that NpmA is expressed and associated with increased gentamicin resistance in CD7814.
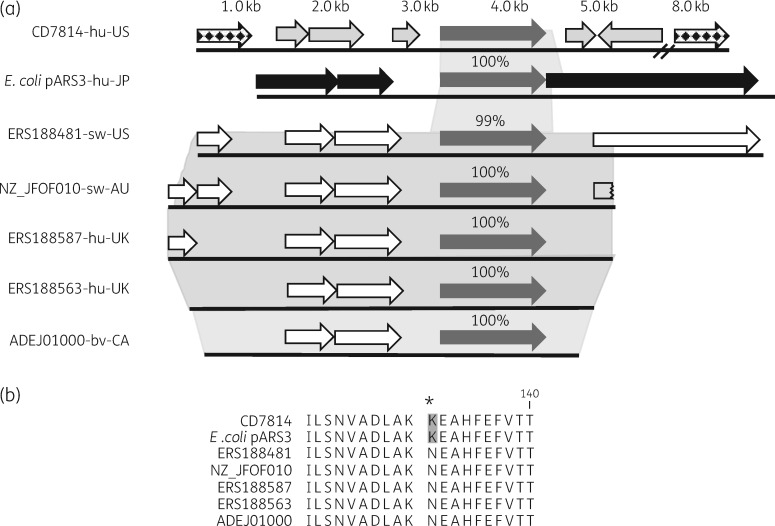
Genomic analysis of the CD7814 assembly identified a large, presumably chromosomal contig of ∼150 kb containing the npmA gene. The majority of predicted ORFs surrounding npmA in CD7814 are hypothetical proteins whereas others encode proteins involved in recombination suggesting that npmA was acquired via horizontal gene transfer, which is consistent with the mosaic structure of the C. difficile chromosome (Figure 1a).9 Five additional C. difficile genomes bearing npmA gene sequences that show 99% nucleotide identity to npmA from CD7814 and pARS3 were identified through BLAST and PubMed literature searches (Figure 1a). The DNA sequence flanking npmA in CD7814 shows little or no nucleotide identity to either E. coli pARS3 or the five C. difficile npmA flanking regions. The five C. difficile genomes are of human and animal origin and were collected from three different continents over a period of at least 10 years. Interestingly, these C. difficile isolates belong to three different STs but their genomes share 99% nucleotide identity across ∼3kb of the npmA region (Figure 1a). None of the five C. difficile genomes share any other sequence similarity to pARS3 outside of npmA. Together, these genomic data suggest that npmA is carried on a conserved element in the five C. difficile genomes and that the mechanism of npmA acquisition in CD7814 is different. In addition, all five C. difficile genomes encode a missense mutation in npmA resulting in a K131N substitution in NpmA relative to CD7814 and pARS3 sequences (Figure 1b). To maintain the established nomenclature for 16S rRNA methyltransferase genes, the CD7814 and, by default, the E. coli pARS3 npmA genes can be re-designated as npmA1 while npmA sequences containing the K131N mutation can be designated npmA2.10 CD7814 npmA1 has been deposited in GenBank under accession number MH249957.
Figure 1.
(a) Predicted ORFs surrounding the npmA gene (dark grey) in CD7814 (light grey), the E. coli pARS3 plasmid (black) and five C. difficile genomes of human and animal origin (white). Shaded areas highlight regions of sequence homology. Chequered arrows indicate ORFs encoding recombinases potentially associated with horizontal transfer of npmA into CD7814. hu, human; sw, swine; bv, bovine; US, USA, JP, Japan; AU, Australia; CA, Canada. (b) NpmA protein alignment depicting the K131N substitution in CD7814 and E. coli pARS3.
To the best of our knowledge, this is the first description of npmA and high-level aminoglycoside resistance in a hospital-acquired C. difficile isolate. Because of strict anaerobic growth conditions, Etest is the most practical method to measure antibiotic susceptibility in C. difficile. As Etests for aramycin and neomycin are unavailable, a limitation of this study is our inability to demonstrate the specificity of NpmA methyltransferase activity for the N1-A1408 16S rRNA. However, the nucleotide identity between CD7814 npmA and the original E. coli sequence and the high-level aminoglycoside resistance observed support NpmA-mediated resistance in this clinical C. difficile isolate. To the best of our knowledge, no evidence to support high-level gentamicin resistance in the presence of ant(6)-Ia and aph(3′)-III has been reported.
In conclusion, this study demonstrates the presence of npmA and high-level aminoglycoside resistance in a clinical C. difficile isolate. The ability of C. difficile to persist in the environment as a spore former may facilitate acquisition of novel antibiotic resistance determinants. These data suggest that hospital-acquired C. difficile may be a reservoir for uncommon antibiotic resistance determinants such as npmA.
Funding
This work was supported by a research grant from the National Institute of Allergy and Infectious Diseases (R01AI127472 to L. H. H.).
Transparency declarations
None to declare.
References
- 1. Doi Y, Wachino JI, Arakawa Y.. Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect Dis Clin North Am 2016; 30: 523–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wachino J, Shibayama K, Kurokawa H. et al. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 2007; 51: 4401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lessa FC, Winston LG, McDonald LC. et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372: 2369–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baym M, Kryazhimskiy S, Lieberman TD. et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 2015; 10: e0128036.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014; 30: 2068–9. [DOI] [PubMed] [Google Scholar]
- 7. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Citron DM, Ostovari MI, Karlsson A. et al. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J Clin Microbiol 1991; 29: 2197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amy J, Johanesen P, Lyras D.. Extrachromosomal and integrated genetic elements in Clostridium difficile. Plasmid 2015; 80: 97–110. [DOI] [PubMed] [Google Scholar]
- 10. Doi Y, Wachino J, Arakawa Y.. Nomenclature of plasmid-mediated 16S rRNA methylases responsible for panaminoglycoside resistance. Antimicrob Agents Chemother 2008; 52: 2287–8. [DOI] [PMC free article] [PubMed] [Google Scholar]