Abstract
Senescence associated gene (SAG) 12, which encodes a cysteine protease is considered to be important in nitrogen (N) allocation to Arabidopsis thaliana seeds. A decrease in the yield and N content of the seeds was observed in the Arabidopsis SAG12 knockout mutants (sag12) relative to the wild type (Col0) under limited nitrogen nutrition. However, leaf senescence was similar in both lines. To test whether SAG12 is involved in N remobilization from organs other than the leaves, we tested whether root N could be used in N mobilization to the seeds. Root architecture, N uptake capacity and 15N partitioning were compared in the wild type and sag12 under either high nitrogen (HN) or low nitrogen (LN) conditions. No differences in root architecture or root N uptake capacity were observed between the lines under HN or LN. However, under LN conditions, there was an accumulation of 15N in the sag12 roots compared to the wild type with lower allocation of 15N to the seeds. This was accompanied by an increase in root N protein contents and a significant decrease in root cysteine protease activity. SAG12 is expressed in the root stele of the plants at the reproductive stage, particularly under conditions of LN nutrition. Taken together, these results suggest a new role for SAG12. This cysteine protease plays a crucial role in root N remobilization that ensures seed filling and sustains yields when nitrogen availability is low.
Keywords: cysteine protease activity, N remobilization, reproductive stage, roots, SAG12, N uptake
Introduction
Many field crop species are high nitrogen (N) demanding plants. In the context of switching to a sustainable agricultural model, a reduction in inorganic nitrogen inputs is required. To reach this goal, it is necessary to deepen the knowledge of the physiological mechanisms related to N management in plants. Among them, senescence metabolism is essential as it allows the redistribution of nutrients from the source organs to the sink organs (Peoples and Dalling, 1988; Masclaux et al., 2000; Gregersen, 2011). In contrast to other elements such as sulfur stored, which is stored as an inorganic form in vacuoles, N is mainly stored as proteins that require some proteolysis steps to generate peptides and amino acids for remobilization to occur during senescence (Hörtensteiner and Feller, 2002; Masclaux-Daubresse et al., 2008; Tegeder and Rentsch, 2010). Such N recycling metabolism is especially important during the reproductive stage to ensure seed N filling. Metabolic events are under the control of a large panel of transcription factors and enzymes (Kusaba et al., 2013) that are also modulated by biotic and abiotic stresses such as N deprivation (Gregersen et al., 2013; Avice and Etienne, 2014; Balazadeh et al., 2014).
Senescence, which corresponds to catabolic pathways occurring before cell death, takes place in all organs (Wojciechowska et al., 2017), but it is commonly accepted that in many plant species, leaves are the main source organs for seed filling (Masclaux-Daubresse et al., 2008). During leaf senescence, the chloroplasts, which contain more than 75% of the total N of the leaf (50% in the form of RuBisCO), are the first organelles to be degraded (Peoples and Dalling, 1988; Hörtensteiner and Feller, 2002). In contrast, the mitochondria and nucleus remain functional until cell death (Lim et al., 2007; Chrobok et al., 2016), to allow the production of energy and the expression of Senescence associated genes (SAGs) that encode proteins necessary for transport and catabolism reactions (Gan and Amasino, 1997).
Protein degradation associated with senescence requires a multitude of proteases (Guo et al., 2004). They belong to five major classes: Cysteine Proteases (CPs), Serine Proteases (SPs), Aspartate Proteases (APs), Metallo Proteases (MPs), and Threonine Proteases (TPs) (Guo et al., 2004). Their specific role in protein breakdown during leaf senescence is not well-known, and the major class overexpressed in many plant species during senescence is CPs (Guo et al., 2004; Poret et al., 2016). The SAG12 papain-like cysteine protease (Noh and Amasino, 1999a,b) is the most strongly induced CP in senescent leaves of Brassica napus L. and Arabidopsis thaliana, especially in plants cultivated under nitrogen limitation (Desclos et al., 2008; Poret et al., 2016). In addition, high SAG12 protein levels are detected in senescing leaf tissues and in fallen leaves (Desclos-Théveniau et al., 2015). For all these reasons, a major role for SAG12 in N remobilization during senescence has long been proposed. Surprisingly, no difference in the leaf senescence phenotype between Col and KO-SAG12 (sag12) plants has ever been observed (Otegui et al., 2005; James et al., 2018). Otegui et al. (2005) showed that a lack of SAG12 did not prevent the formation of senescence associated vacuoles (SAVs), nor was their proteolytic activity affected, suggesting that other cysteine proteases accumulated in SAVs in the senescent leaves of Arabidopsis thaliana (Otegui et al., 2005). More recently, James et al. (2018) confirmed the absence of a leaf senescence phenotype in sag12. The authors also showed that there was no difference in yield and seed N content between Col and sag12 when cultivated under optimal N nutrition, but there was lower seed N content and yield in sag12 when cultivated under limiting nitrogen conditions. The absence of phenotype under high nitrogen was explained by an induction of cysteine and aspartate protease activities in sag12 that could preserve N remobilization. Nevertheless, the authors did not exclude the potential that better root N uptake in sag12 could have maintained seed filling and plant productivity. This hypothesis is in agreement with the decrease in seed nitrogen content and yields observed in sag12 cultivated under LN conditions. Indeed, under this particular N limitation condition, N seed filling can only be achieved by N remobilization, in spite of a possible increase in the N uptake capacity in the roots of sag12.
The aim of the present study was to determine whether root N uptake and/or N remobilization are involved in the preservation of seed N filling in sag12. Root morphology and N uptake were monitored in Col and sag12. Experiments using 15N pulse/chase labeling were performed to investigate the distribution of 15N and estimate the source and rate of remobilized nitrogen dedicated to seed filling.
Materials and Methods
Plant Growth Conditions
Arabidopsis thaliana Columbia (Col) and sag12 (SALK_124030) T-DNA mutants were used in this study. The KO-SAG12 SALK_124030 (sag12) was chosen because it was the only germplasm which was available in ABRC stock and recently characterized for some study focused on protease activities (Pružinská et al., 2017; James et al., 2018). Seeds were stratified for 48 h in 0.1% agar (Select agar, Sigma, L’Isle d’Abeau Chesnes, France) at 4°C in the dark and then sown into Eppendorf tubes (0.5 ml) filled with 0.8% agar (w/v) that had their bottoms removed. Plants were placed in a glasshouse on a tank containing 10 L of 3.75 mM nutrient solution during 44 days. The solutions contained 3.75 mM KNO3, 0.5 mM MgSO4, 0.25 mM KH2PO4, 0.2 mM EDTA.NaFe.3H2O, 1.25 mM CaCl2, 2H2O 14 μM H3BO3, 5 μM MnSO4, 3 μM ZnSO4, 0.7 μM CuSO4, 0.1 μM CoCl2, and 0.1 μM Na2MoO4, and was renewed every week. A batch of plants was cultivated under the same conditions except that a long term 15N labeling was performed (5% of atom excess, 3.75 mM K15NO3 during 44 days) in order to obtain homogeneous 15N-labeled plants and to further appreciate the 15N partitioning at harvest time (125 DAS). At 44 days after sowing (DAS) all plants were transferred to two contrasting N conditions: High Nitrogen with 3.75 mM N [HN; 1.25 mM Ca(NO3)2.4H2O, 1.25 mM KNO3, 0.7 μM (NH4)6Mo7O24] and Low Nitrogen with 4.2 μM N [LN; 0 mM Ca(NO3)2.4H2O, 0 mM KNO3, 0.7 μM (NH4)6Mo7O24]. Photosynthetic photon flux density was 110 mmol m-2 s-1 and day and night temperatures were 21 and 18°C, respectively. During the first 64 DAS, plants were cultivated with 8 h light/16 h dark photoperiod and then the reproductive stage was induced with a 16 h light/8 h dark photoperiod. Plants were harvested at the vegetative stage (64 DAS) and at the reproductive stage: 85 DAS corresponding to seed filling and 125 DAS corresponding to mature seed stage (Supplementary Figure S1). At each harvest time, the plant compartments present were separated, weighed and stored at -80°C for further analysis. In addition, the root and tip density was performed at 64 DAS with a flat scan (Epson expression 10000XL scanner, Suwa, Japan) coupled with Winrizho software (Regent, Sainte-Anne, QC, Canada).
Root N Uptake Capacity Analysis
Col and sag12 plants were cultivated in HN conditions as previously described but without any 15N labeling. The root N uptake analysis was performed at vegetative (64 DAS) and reproductive stages (85 DAS) as previously described by (Lainé et al., 1993). Briefly, roots were washed twice for 1 min in a solution of CaSO4 (1 mM) before immersion for 5 min in a solution of 250 μM or 2 mM K15NO3 (99% of atom excess) to study N uptake by the high affinity transport system (HATS) or the low affinity transport system (LATS), respectively. Then roots were rinsed twice in a solution of CaSO4 (1 mM) at 4°C for 1 min to stop the N uptake. Roots and shoots were separated and weighed before 15N analysis by an isotope mass ratio spectrometer (IRMS, IsoPrime GV Instruments, Manchester, United Kingdom). Uptake capacity was expressed as the total amount of 15N in a whole plant per gram of dry roots per hour.
Isotopic Nitrogen Analysis
N and 15N contents were quantified in the different plant compartments with an elemental analyzer (EA3000, EuroVector, Milan, Italy) coupled with an isotope mass ratio spectrometer (IsoPrime IRMS, GV Instruments, Manchester, United Kingdom).
The nitrogen quantity (NQ) in the sample was obtained with the following formula:
where MS represents the dry matter of the sample.
The isotopic abundance (A%) was determined with the formula:
With 15N and 14N representing the amount of 15N and 14N isotopes, respectively.
The isotopic excess (E%) corresponds to the difference between the isotopic abundance of sample (A%) and the N natural abundance (0.3660%): E% = A% - 0.3660%
Finally, the isotopic excess was used to estimate the quantity of 15N (μg) (Q15N): Q15N = (E% × QN) × 1000.
Determination of Amino Acid Content
Ten mg of lyophilized roots were added to 400 μl of MeOH containing 0.625 nmol/μL of norvaline used as internal standard (Sigma, L’Isle d’Abeau Chesnes, France). The mix was stirred for 15 min and then 200 μl of chloroform and 400 μl of ddH2O were added. After centrifugation (12000 rpm, 10°C, 5 min), the supernatant was recovered, evaporated and resuspended in 100 μl of ddH2O and then filtered on a 0.2 μm membrane before derivatization using an AccQ-Tag Ultra Derivatization Kit (Waters, Guyancourt, France) following the manufacturer’s protocol (Waters, Guyancourt, France). Amino acids were separated and quantified using a UPLC/PDA H-Class system (Waters, Guyancourt, France) with a BEH C18 100 mm × 2.1 mm column.
Extraction and Quantification of Soluble Proteins
Two hundred mg of frozen root leaf tissue were ground in a mortar with 250 μL of citrate-phosphate buffer (20 mM citrate, 160 mM phosphate, pH 6.8 containing 50 mg of PVPP). After centrifugation (1 h, 12,000 g, 4°C), the concentration of the soluble protein extract was determined in the supernatant by protein staining (Bradford, 1976) using bovine serum albumin (BSA) as standard.
Western Blot of SAG12 Protein
Twenty μg of soluble proteins were denatured by heating at 90°C for 10 min in 4x Laemmli sample buffer with β-mercaptoethanol (Laemmli, 1970). Proteins were separated on an SDS-PAGE Stainfree precast gel (4–15% acrylamide gradient; Mini-PROTEAN®, Bio-Rad, Marnes-la-Coquette, France) and transferred onto a polyvinylidene difluoride (PVDF) membrane as previously described by Desclos et al. (2008). The PVDF membrane was incubated overnight in Tris buffer saline – Tween 20 [TBST; Tris 10 mM, NaCl 150 mM, pH 8, Tween 20 0.15% (v/v)] with 3% (v/v) skimmed milk to avoid non-specific hybridization. Immunodetection of SAG12 was performed using an anti-SAG12 specific polyclonal antibody from rabbit provided by Agrisera® (AS14 2771; 1/2000 in TBST) as primary antibody and a second antibody coupled with peroxidase (1/10000 diluted in TBST, Bio-Rad®). The quantification of SAG12 was performed by the measurement of the chemiluminescence revealed with an ECL Kit (Bio-Rad®, Marnes-la-Coquette, France) using a ProXPRESS 2D proteomic imaging system (PerkinElmer, Courtaboeuf, France).
Proteolytic Activities
Proteolytic activities of cysteine proteases were determined by in vitro protein degradation analysis as previously described in James et al. (2018). Twenty μg of soluble proteins were incubated in a 200 μL reaction volume containing Na-acetate buffer (50 mM, pH 5.5) and 10 μg of BSA (exogenous protein used as loading control). Cysteine protease activities (CPact) were obtained by the addition of 50 μM of E-64, a cysteine protease inhibitor dissolved in dimethylsufoxide (DMSO). Furthermore, 2 mM dithiothreitol (DTT) was added to this mixture and total protease activity (TPact) was obtained by substituting inhibitors with an equal volume of DMSO. Then, proteins were precipitated with 1 mL of ice-cold acetone either immediately (t0), or after incubation for 300 min (t300) at 37°C under gentle agitation. After centrifugation (15 min, 16,000 g, 4°C), the pellet was dissolved in 2X SDS-PAGE gel loading buffer (140 mM sodium dodecyl sulfate, 200 mM Tris, 20% glycerol, 5% β-mercaptoethanol, 0.3 mM Bromophenol Blue) and heated at 90°C for 10 min. Then the soluble protein extracts were separated on a 4–15% gradient in SDS-PAGE Stainfree precast gels (Mini-PROTEAN® TGXTM Stain Free, Bio-Rad, Marnes-la-Coquette, France) and scanned under UV light with a Gel DocTM EZ scanner (Bio-Rad, Marnes-la-Coquette, France). The proteolytic activities were quantified by monitoring degradation of four bands (95, 76, 50, and 37 kDa) corresponding to endogenous proteins (EP) as targets of proteolysis at pH 5.5. Proteolytic activities are calculated as follows:
(1) Total protease activity (TPact expressed in %):
(2) Cysteine protease activity (CPact expressed in %):
where the amount of EP (Q(EP)) at t0 and t300, with (Inhib.) or without inhibitor, as well as the total amount of soluble proteins (Qtot) at t0 were quantified by using ImageLabTM software (Bio-Rad, Marnes-la-Coquette, France).
Extraction and Quantification of RNAs, Reverse Transcription, and PCR Analysis
Total RNAs were extracted from 200 mg of frozen leaf tissue previously ground in a mortar containing liquid nitrogen. The powder was suspended in 750 μL of extraction buffer (100 mM LiCl, 100 mM TRIS, 10 mM EDTA, 1% SDS (w/v), pH 8) and 750 μL of hot phenol (80°C, pH 4). After vortexing for 40 s and after addition of 750 μl of chloroform:isoamylalcohol (24/1, v/v), the homogenate was centrifuged (15,000 g, 5 min, 4°C). The supernatant was added to 750 μl of 4 M LiCl solution (w/v) and incubated overnight at 4°C. After centrifugation at 15,000 g for 20 min at 4°C, the pellet containing total RNAs was resuspended with 100 μL of sterile water. Then, total RNAs were purified with an RNeasy Mini Kit according to the manufacturer’s protocol (Qiagen, Courtaboeuf, France). Quantification of total RNA was performed by spectrophotometry at 260 nm (BioPhotometer, Eppendorf, Le Pecq, France) before reverse transcription (RT). For RT, 1 μg of total RNAs was converted to cDNA with an iScript cDNA synthesis kit according to the manufacturer’s protocol (Bio-Rad, Marnes-la-Coquette, France) before polymerase chain reaction (PCR) and quantitative PCR (qPCR) analyses.
SAG12 and 18S rRNA gene expressions were monitored by PCR using 1 μL of cDNA added to 10 μl of a PCR mix containing 250 μM dNTPs, 0.65 μM of forward and reverse primers and 0.5 μM (5 U μL-1) Qbiogene Taq polymerase (MP Biomedicals, Illkirch-Graffenstaden, France). The primers were designed with primer3+ software. SAG12 (At5g45890): forward: 5′-GGCAGTGGCACACCAMCCGGTTAG-3′; reverse 5′-AGAAGCMTTCATGGCAAGACCAC-3′ and 18S rRNA (NR_141642): forward: 5′-CGGATAACCGTAGTAATTCTAG-3′; reverse: 5′-GTACTCATTCCAATTACCAGAC-3′. PCRs were performed in a thermocycler (Applied Biosystems, Courtaboeuf, France) using the following program: 1 cycle at 95°C for 5 min, 25 and 18 cycles for Sag12 and 18S rRNA including a denaturing step at 95°C for 30 s, a primer hybridization step at 58°C for 45 s and an amplification step at 72°C for 1 min. Each PCR reaction was finished with one cycle at 72°C for 10 min. The identity of each amplicon was checked by sequencing and BLAST analysis. PCR products were separated by electrophoresis on agarose gels (1.2% in TAE 1X with 5 μg mL-1 of ethidium bromide) and revealed by illumination with UV light using a Gel-Doc TM EZ Scanner (Bio-Rad, Marnes-la-Coquette, France).
Additionally, SAG12 gene expression was monitored using RT-qPCRs analysis with 4 μl of 100× diluted RT product, 500 nM of forward and reverse primers and 1X SYBR Green PCR Master Mix (Bio-Rad, Marnes-la-Coquette, France) in a real-time thermocycler (CFX96 Real Time System, Bio-Rad, Marnes-la-Coquette, France). The program was: 95°C for 3 min followed by 40 cycles (95°C for 15 s followed by 40 s at 60°C). PCR amplifications were performed using specific primers for each housekeeping gene (EF1-α : forward: 5′-GCCTGGTATGGTTGTGACCT-3′; reverse: 5′-GAAGTTAGCAGCACCCTTGG-3′; 18S rRNA : forward: 5′-CGGATAACCGTAGTAATTCTAG-3′; reverse 5′-GTACTCATTCCAATTACCAGAC-3′) and target gene SAG12 (Forward: 5′-AAAGGAGCTGTGACCCCTATCAA-3′; reverse: 5′-CCAACAACATCCGCAGCTG-3′) as described by Kim et al. (2018). For each sample, the subsequent RT-qPCRs were performed in triplicate. The expression of the target gene in each sample was compared to the control sample (Col at the vegetative stage for each N condition) and was calculated with the delta delta Ct (ΔΔCt) method (Livak and Schmittgen, 2001).
Transgenic promoterSAG12::UIDA Construct
The SAG12 promoter (promoterSAG12), containing the 1–2180 bp sequence found in GenBank under the Accession No. U37336, was cloned from the pSG499 plasmid (Gan and Amasino, 1995) kindly provided by Pr. R. Amasino (University of Wisconsin-Madison, Madison, WI, United States) into the pMDC32 vector between the PmeI and AscI restriction enzyme sites, in place of the 35S promoter, giving the pMAZ01 plasmid. The pMAZ01 plasmid was fully sequenced from the RB to the Nos terminator to verify the absence of any modification especially in the SAG12 sequence. The full-length coding sequence of UIDA (GUS-coding gene; from 86 to 1897 of the AJ298139 accession number) was then cloned successively in the pENTRTM vector (Invitrogen, Carlsbad, CA, United States), then into the pMAZ01 vector by Gateway recombination giving rise to the pMAZ02 vector containing the promoterSAG12::UIDA fusion. The correct GUS sequence was verified by sequencing.
Transgenic Arabidopsis plants carrying the SAG12 promoter fused to the UIDA reporter gene were obtained by floral dipping (Clough and Bent, 1998) and eight homozygous lines of several primary transformants were selected on the basis of their hygromycin resistance and single insertion segregation rate.
GUS Staining and Observations
Based on the method of Jefferson et al. (1987), tissues from two independent promoterSAG12::UIDA homozygous lines were stained overnight at 37°C in 50 mM Na3PO4 pH 7.0, 5 mM ferricyanide, 5 mM ferrocyanide, 0.05% Triton X-100 and 5-bromo-4-chloro-3-indolyl glucuronide (1 mg mL-1). Then after destaining by successive incubations in 50, 75, and 96% (v/v) ethanol, samples were kept in glycerol at 4°C before observation.
The tissues were included in low melting point agarose (5%; w/v) and cut with a vibratome (Microm 650v; Thermo Scientific; United States) before observation with a light microscope (AX70 Olympus, and Olympus SC30 camera, Japan) with the help of cellSens software.
Statistical Analysis
For all parameters, at least three biological repeats were measured (n ≥ 3). All the data are presented as the mean ± standard error (SE). To compare Col with sag12 data, Student’s t-tests were performed after verifying compliance of normality with R software. Statistical significance was postulated at p ≤ 0.05.
Results
Root Architecture and N Uptake Capacity Are Not Affected in sag12
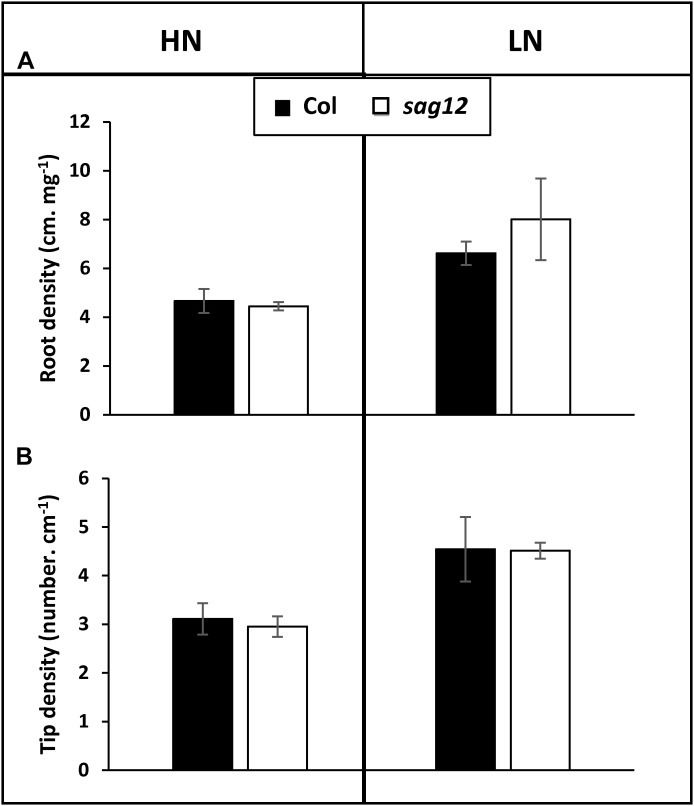
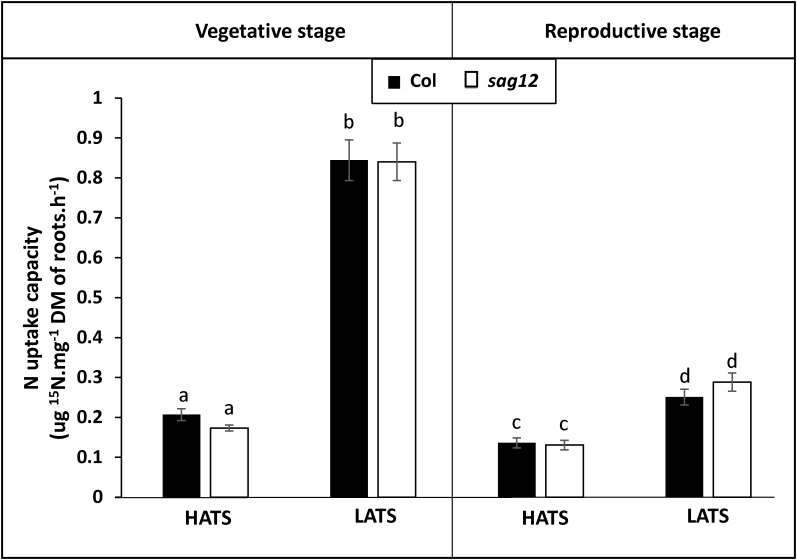
Irrespective of the N conditions (HN or LN), no significant differences were observed between Col and sag12 for root (Figure 1A) and root tip (Figure 1B) densities. N uptake capacity was monitored by measuring the amount of 15N absorbed per h and mg of root DW. In both the vegetative and reproductive stages, HATS and LATS N-uptake capacities were not different between the genotypes (Figure 2). LATS uptake was higher than HATS in both the vegetative and reproductive stages. Moreover, it could be noted that the N-uptake capacity related to LATS but even more to HATS was significantly lower in the reproductive than the vegetative stage.
FIGURE 1.
Root architecture is similar in Col and sag12. Root density (A) and tip density (B) of Col (black bars) and sag12 (white bars) plants cultivated under HN and LN conditions were determined at the vegetative stage (64 DAS). Results are presented as means ± SE (n = 3).
FIGURE 2.
Nitrogen uptake capacities by HATS and LATS are similar in Col and sag12. Col (black bars) and sag12 (white bars) plants were harvested at the vegetative (n = 12) and reproductive stages (n = 8). N uptake capacities by high (HATS) and low (LATS) affinity transport systems are presented as means ± SE. Different letters indicate statistically significant differences according to Student’s t-test (p ≤ 0.05).
The SAG12 Defect Affects N Allocation Under LN Conditions
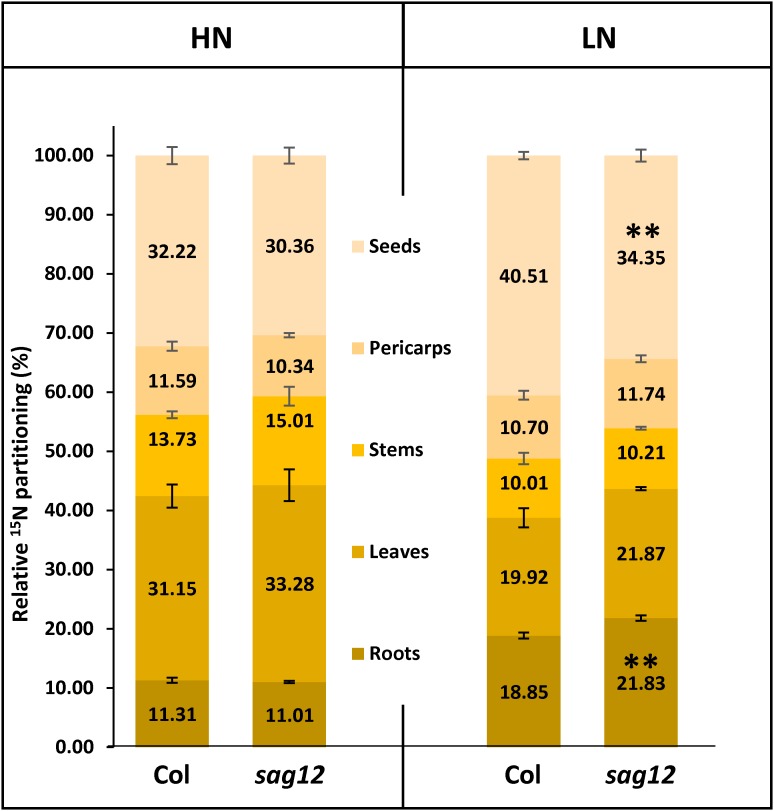
The plants were labeled with 15N for 44 DAS and then grown without 15N until harvest at 125 DAS. The similar total 15N amounts found in Col and sag12 at the end of the 15N labeling (not shown), confirmed the absence of differences between the genotypes in LATS and HATS uptake capacities at the vegetative stage. Plants were dissected into five compartments (roots, leaves, stems, pericarps, and seeds). Under HN conditions, 15N was mainly found in the leaves (Col: 31.15 ± 1.96% and sag12: 33.28 ± 2.68%) and in the seeds (Col: 32.22 ± 1.46% and sag12: 30.26 ± 1.34%) of the two genotypes (Figure 3) and the partitioning of 15N was not significantly different between Col and sag12. Under LN conditions, around 78% of the total 15N of the plant was distributed in the roots, leaves, and seeds in both genotypes. However, the 15N partitioning was significantly different in the seeds and roots of sag12 and Col. Partitioning of 15N in the seeds of sag12 (34.35 ± 1.02%) was decreased by 6.16% compared to Col (40.51 ± 0.64%) and partitioning of 15N in the roots of sag12 (21.83 ± 0.45%) was conversely increased by 2.97% compared to Col (18.85 ± 0.50%). This then suggests a defect in 15N remobilization from the roots to the seeds in sag12.
FIGURE 3.
N allocation in roots and seeds is affected in sag12 under LN conditions. Partitioning of 15N in the different plant compartments (roots, leaves, stems, pericarps, and seeds) was calculated from data obtained from plants harvested at seed maturity (125 DAS). Results are presented as means ± SE (n = 4). Significant differences between Col and sag12 are indicated by (∗∗p ≤ 0.01; n = 4).
Protein Content Is Higher in Roots of sag12 Cultivated Under LN Condition
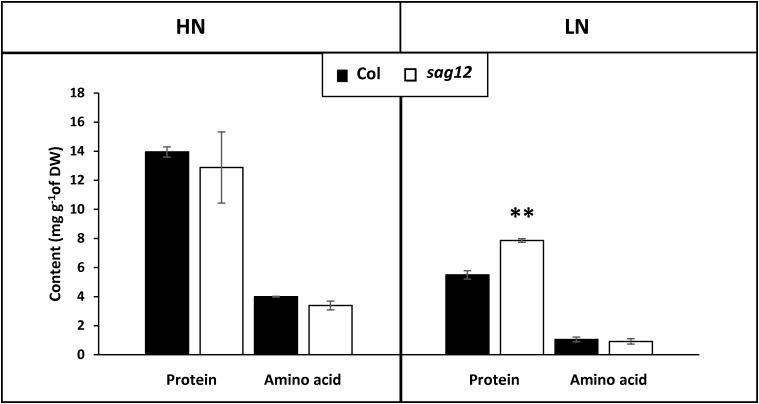
As expected, protein and amino acid contents were higher in both genotypes when cultivated under HN compared to LN conditions (Figure 4). Protein content in the roots of sag12 and Col were similar (around 13 mg g-1 of DW) when plants were cultivated under HN conditions. Under LN conditions, the protein concentration was higher in the roots of sag12 (7.87 ± 0.12 mg g-1 DW) than in Col (5.49 ± 0.28 mg g-1 DW). Interestingly irrespective of the N conditions, the amino acid contents were not significantly different between the two genotypes (Figure 4).
FIGURE 4.
Protein content is higher in roots of sag12 at the reproductive stage. The roots of Col (black bars) and sag12 (white bars) plants cultivated under high (HN) and low (LN) nitrogen conditions were harvested at the reproductive stage (85 DAS). Values are means ± SE, n = 4. Significant difference between Col and sag12 (∗∗p ≤ 0.01; n = 4).
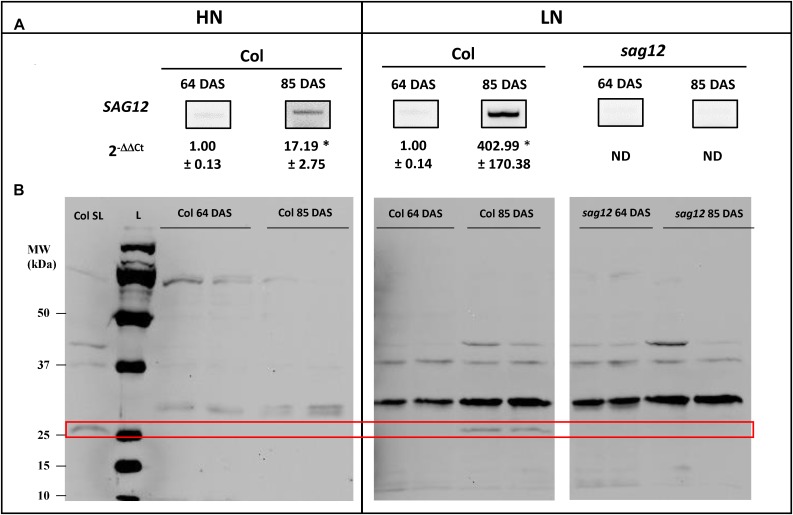
SAG12 Is Expressed in Roots at the Reproductive Stage and the Expression Is Higher Under Low Nitrogen Conditions
Low SAG12 transcripts (Figure 5A) and no protein (Figure 5B) could be detected using RT-qPCR and Western blots in the roots of Col at the vegetative stage under low and high nitrogen conditions. In contrast, high SAG12 transcripts levels (Figure 5A) were detected in roots of Col at the reproductive stage and especially under low nitrogen conditions. Accordingly, SAG12 protein was also detected in the roots of Col under LN conditions (Figure 5B). As a control, we checked that SAG12 protein and transcripts were undetectable in the roots of sag12 especially when SAG12 was detected in Col (i.e., reproductive stage and LN).
FIGURE 5.
SAG12 is expressed in roots at reproductive stage and especially under LN condition. (A) SAG12 gene expression was monitored using RT-PCR and quantified using RT-qPCR for each condition (HN and LN), the expression of the SAG12 gene at the reproductive stage (85 DAS) has been relativized with respect to the level of the SAG12 expression in Col for the corresponding N treatment at the vegetative stage. ND, not detected. For a given nitrogen condition (HN and LN), significant difference between Col at vegetative and reproductive stage (∗p ≤ 0.05; n = 3). (B) Detection of SAG12 by Western blot using an anti-SAG12 specific polyclonal antibody provided by Agrisera® (AS142771). The red frame highlight the location of SAG12 protein. The mRNAs and soluble proteins were extracted from roots of Col and sag12 cultivated under HN or LN conditions and harvested at vegetative (64 DAS) and reproductive (85 DAS) stages. Soluble proteins extracted from senescing leaves of Col (Col LS) were used as positive control for SAG12 expression. The molecular weights (MW; kDa) of the Precision Plus Protein Dual color Standards ladder (Bio-Rad) were indicated.
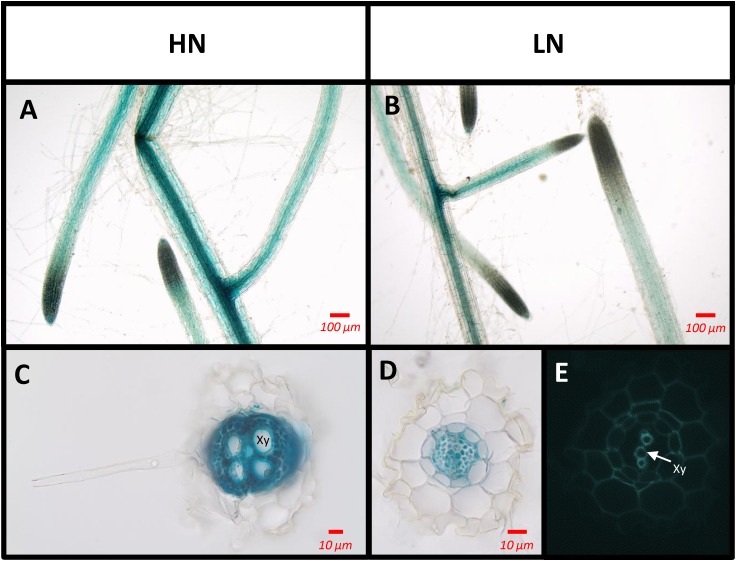
SAG12 Is Expressed in the Root Stele
In order to determine the location of SAG12 expression in the root tissues, GUS staining was performed on Arabidopsis plants transformed by the promoterSAG12::UIDA reporter fusion. Staining was performed at the reproductive stage, in which SAG12 genewas previously detected, on plants cultivated under low and high N conditions. Irrespective of the N conditions, GUS staining was observed along the entire length of the root but it was located exclusively in the stele (Figure 6). Surprisingly, although the SAG12 expression level was higher in the roots of LN cultivated plants (Figure 5), we observed that GUS staining was lower in the LN cultivated roots than in the roots of plants cultivated under HN.
FIGURE 6.
SAG12 is expressed in the root vascular tissues. The tissues where the SAG12 promoter was active were identified using the promoterSAG12::UIDA lines and GUS staining. Roots were observed using a light microscope. Transgenic lines were cultivated under HN (A,C) and LN (B,D,E) conditions and harvested at the reproductive stage. Representative pictures of the results obtained for whole root (A,B) and transverse root sections (C,D) are shown. Root section under UV excitation (E). Xy, xylem tissue.
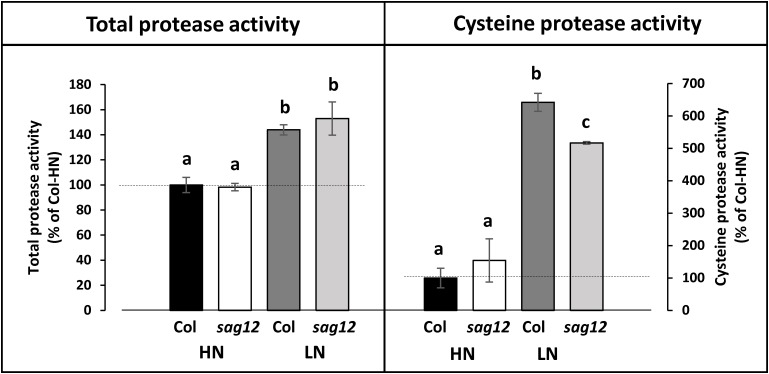
Cysteine Protease Activity Is Lower in the sag12 Root Under LN Conditions
The total protease activity measured at the reproductive stage (at the optimum pH for SAG12 activity: pH 5.5) did not reveal any difference between Col and sag12, regardless of the N conditions (Figure 7). Similar to the total protease activity, a strong increase in cysteine protease activity was observed when plants were cultivated under LN in comparison to HN conditions (Figure 7). While no difference in cysteine protease activity was observed between genotypes when plants were cultivated under HN conditions, the defect in SAG12 led to a significant decrease in cysteine protease activity relative to Col when plants were cultivated under LN conditions (642.11 ± 28.04 vs. 516.88 ± 3.99% in Col and sag12, respectively; Figure 7). Such a discrepancy could be attributed to the lack of SAG12 activity.
FIGURE 7.
Cysteine protease activity is decreased in roots of sag12 under LN conditions. Soluble proteins were extracted from roots harvested at the reproductive stage (85 DAS) from Col and sag12 plants cultivated under high (HN) or low (LN) nitrogen conditions. The cysteine and total protease activities were determined by monitoring the degradation of four endogenous proteins (95, 76, 50, 37 kDa) in the presence or the absence of the cysteine protease inhibitor E64. The protease activities are expressed as a percentage relative to the activity in the Col roots under HN conditions (100%; dot line). Values are means ± SE, n = 3. For a given protease activity (total or cysteine), significant differences (p ≤ 0.05, n = 3) are indicated by different lower case letters.
Discussion
In a recent study, James et al. (2018) have demonstrated that the absence of SAG12 in plants leads to a decrease in the production of seeds and to a lower N content in Arabidopsis seeds when cultivated under LN conditions. The absence of such a phenotype under HN conditions was explained by the increase in cysteine and aspartate protease activities, which may compensate for the SAG12 defect and sustain N remobilization during seed filling (James et al., 2018). However, although N remobilization is often considered as the major process providing N to the seeds (Tegeder and Masclaux-Daubresse, 2017), the assumption that a better N uptake in sag12 could supplement the N allocation to the seeds, especially under low N, could not be excluded. Indeed, both uptake and root architecture are known to be regulated depending on the internal N status of the plant and stimulated under LN conditions (Crawford and Forde, 2002; Nacry et al., 2013; Bellegarde et al., 2017; Gent and Forde, 2017). We here show that whatever the N conditions (LN and HN), Col and sag12 have the same root architecture (Figure 1). In addition, no matter which developmental stage was investigated (vegetative or reproductive), the N uptake by high (HATS) and low (LATS) affinity transporters was similar in Col and sag12 (Figure 2). Taken together, these results invalidated the hypotheses that an increase in N uptake (i) supplemented N allocation to the seeds in the sag12 plants cultivated under HN conditions or that (ii) N content was lower in the seeds of sag12 compared to Col when cultivated under LN condition (James et al., 2018). In order to monitor the N remobilization for seed filling, at the final stage of plant development (mature seed 125 DAS) we analyzed the distribution of the 15N provided in a pulse/chase experiment to Col and sag12 plants cultivated under HN and LN conditions. Under HN, a similar 15N distribution was observed in Col and sag12 (Figure 3), thus suggesting that in sag12 (SALK_124030), SAG12 depletion did not alter N remobilisation for seed filling when nitrogen was available. These results are in good agreement with the report of James et al. (2018) showing that the cysteine and aspartate proteases increased in sag12 might support N remobilization during seed filling. Interestingly, growing plants under low N conditions indicated that the proportion of 15N was significantly lower in the seeds of sag12 in comparison to Col and conversely, significantly higher in the roots of sag12 compared to the roots of Col (Figure 3). No significant difference between Col and sag12 was found for 15N partitioning in any organs other than the roots and seeds. This then revealed a defect in N remobilization from the roots to the seeds in sag12 and strongly suggested that the low N content previously observed by James et al. (2018) in the seeds of sag12 under LN condition was mainly due to the sequestration of N in its roots. Although N remobilization from the root to the seeds is poorly documented, a previous study performed in Brassica napus by Rossato et al (2001) showed that more than 11% of the N in seeds came from the remobilization of root N. Moreover, Girondé et al. (2015a) showed that Brassica napus genotypes with higher nitrogen remobilization efficiency had a higher contribution of N remobilised from the roots to seeds. Likewise, we observed that the protein concentration was higher in the roots of sag12 compared to Col, when cultivated under LN, while the amino acid concentrations were unchanged (Figure 4). This work performed in sag12 (SALK_124030) emphasized the role of SAG12 protease in the proteolysis of root proteins that could serve as a nitrogen source for remobilization under low nitrogen conditions.
From this finding, we then decided to investigate whether SAG12 could be expressed in the root tissue, at least under LN conditions, which has never been described before. Indeed, although the expression of SAG12 in leaves during natural and induced senescence was clearly demonstrated by numerous studies performed in various plant species (Lohman et al., 1994; Gan and Amasino, 1997; Desclos et al., 2009; Parrott et al., 2010; Carrión et al., 2013; Singh et al., 2013; Poret et al., 2016; Curci et al., 2017), to our knowledge there was no evidence for SAG12 expression in plant root tissue. In the present work the SAG12 transcripts and proteins were detected in the roots of Arabidopsis thaliana especially at the reproductive stage and were much higher in roots of plants cultivated under LN than under HN conditions (Figure 5). This makes sense because sink strength is known to be stronger at the reproductive stage due to the maturation of seeds and also stronger under LN conditions. The GUS staining of promoterSAG12::UIDA lines at the reproductive stage confirmed that SAG12 was expressed in the root and especially localized in the stele (Figure 6). We noticed that despite higher SAG12 expression in roots under LN than under HN, GUS staining was weaker in roots of LN plants. This was possibly due to the fact that the stronger protease activity measured under LN may have led to degradation of the b-glucuronidase enzyme, as previously shown in senescent leaf tissues by Noh and Amasino (1999a). Taken together, the expression of SAG12 in roots, and the increase in total protein content in the roots of sag12 suggests that SAG12 is involved in the proteolysis associated with the root N remobilization, particularly when plants are facing N limitation.
This assumption was verified by measuring the total- and cysteine-protease activities in the roots of Col and sag12 at the reproductive stage. While the depletion of SAG12 in sag12 (SALK_124030) did not affect the total protease activity in roots, it decreased significantly the cysteine protease activity of the roots of plants cultivated under LN conditions.
Altogether, the results suggest that the SAG12 protease could plays a major role in the breakdown of root proteins when plants are facing N limitation and is required for efficient N remobilization to the seeds.
Conclusion
For the first time, this work shows a clear expression of SAG12 in the stele of the roots. This expression pattern associated with results obtained in sag12 (SALK_124030) suggest a role of SAG12 for N remobilization from roots to support seed production and seed N content under N limitation. This study open new ways for improving N use efficiency in crops. For example, in Brassica napus, the level of SAG12 root expression could have partly explained the contrasting N use efficiency highlighted by Girondé et al. (2015a,b) in different genotypes cultivated under N limiting conditions.
Author Contributions
MJ, CM-D, PE, and JT conducted all the experiments, analyzed the data, and wrote the manuscript. PL performed N uptake capacity experiments and wrote the manuscript. DG performed microscopic analysis. MA and AM realized plasmid constructions, transformation and selection of transgenic lines.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Nathalie Nési (INRA, Institut de Génétique, Environnement et Protection des Plantes, Agrocampus Ouest, UMR 1349, Université de Rennes) and Prof. Jean-Christophe Avice (INRA, Ecophysiologie Végétale, Agronomie & Nutritions N.C.S., UCBN 950, UMR, Université de Caen Normandie) for the management of the RAPSODYN ANR program. They are most grateful to the PLATIN’ (Plateau d’Isotopie de Normandie) core facility for elemental analysis, to the “Centre Mondial de l’Innovation” (Groupe Roullier) for amino acid analysis and to Dr. Laurence Cantrill for proofreading and English correction.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01998/full#supplementary-material
References
- Avice J.-C., Etienne P. (2014). Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J. Exp. Bot. 65 3813–3824. 10.1093/jxb/eru177 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Schildhauer J., Araújo W. L., Munné-Bosch S., Fernie A. R., Proost S., et al. (2014). Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: transcriptomic and metabolomic consequences. J. Exp. Bot. 65 3975–3992. 10.1093/jxb/eru119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellegarde F., Gojon A., Martin A. (2017). Signals and players in the transcriptional regulation of root responses by local and systemic N signaling in Arabidopsis thaliana. J. Exp. Bot. 68 2553–2565. 10.1093/jxb/erx062 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Carrión C. A., Costa M. L., Martínez D. E., Mohr C., Humbeck K., Guiamet J. J. (2013). In vivo inhibition of cysteine proteases provides evidence for the involvement of ‘senescence-associated vacuoles’ in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J. Exp. Bot. 64 4967–4980. 10.1093/jxb/ert285 [DOI] [PubMed] [Google Scholar]
- Chrobok D., Law S. R., Brouwer B., Lindén P., Ziolkowska A., Liebsch D., et al. (2016). Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiol. 172 2132–2153. 10.1104/pp.16.01463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Crawford N. M., Forde B. G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1:e0011. 10.1199/tab.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curci P. L., Aiese Cigliano R., Zuluaga D. L., Janni M., Sanseverino W., Sonnante G. (2017). Transcriptomic response of durum wheat to nitrogen starvation. Sci. Rep. 7:1176. 10.1038/s41598-017-01377-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclos M., Dubousset L., Etienne P., Le Caherec F., Satoh H., Bonnefoy J., et al. (2008). A proteomic profiling approach to reveal a novel role of Brassica napus drought 22 kD/water-soluble chlorophyll-binding protein in young leaves during nitrogen remobilization induced by stressful conditions. Plant Physiol. 147 1830–1844. 10.1104/pp.108.116905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclos M., Etienne P., Coquet L., Jouenne T., Bonnefoy J., Segura R., et al. (2009). A combined 15N tracing/proteomics study in Brassica napus reveals the chronology of proteomics events associated with N remobilisation during leaf senescence induced by nitrate limitation or starvation. Proteomics 9 3580–3608. 10.1002/pmic.200800984 [DOI] [PubMed] [Google Scholar]
- Desclos-Théveniau M., Coquet L., Jouenne T., Etienne P. (2015). Proteomic analysis of residual proteins in blades and petioles of fallen leaves of Brassica napus. Plant Biol. 17 408–418. 10.1111/plb.12241 [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R. M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270 1986–1988. 10.1126/science.270.5244.1986 [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R. M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113 313–319. 10.1104/pp.113.2.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent L., Forde B. G. (2017). How do plants sense their nitrogen status? J. Exp. Bot. 68 2531–2539. 10.1093/jxb/erx013 [DOI] [PubMed] [Google Scholar]
- Girondé A., Etienne P., Trouverie J., Bouchereau A., Le Cahérec F., Leport L., et al. (2015a). The contrasting N management of two oilseed rape genotypes reveals the mechanisms of proteolysis associated with leaf N remobilization and the respective contributions of leaves and stems to N storage and remobilization during seed filling. BMC Plant Biol. 15:59. 10.1186/s12870-015-0437-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girondé A., Poret M., Etienne P., Trouverie J., Bouchereau A., Le Cahérec F., et al. (2015b). A profiling approach of the natural variability of foliar N remobilization at the rosette stage gives clues to understand the limiting processes involved in the low N use efficiency of winter oilseed rape. J. Exp. Bot. 66 2461–2473. 10.1093/jxb/erv031 [DOI] [PubMed] [Google Scholar]
- Gregersen P. L. (2011). “Senescence and nutrient remobilization in crop plants,” in The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops, eds Hawkesford M. J., Barraclough P. (Hoboken, NJ: Wiley-Blackwell; ), 83–102. 10.1002/9780470960707.ch5 [DOI] [Google Scholar]
- Gregersen P. L., Culetic A., Boschian L., Krupinska K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82 603–622. 10.1007/s11103-013-0013-8 [DOI] [PubMed] [Google Scholar]
- Guo Y., Cai Z., Gan S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27 521–549. 10.1111/j.1365-3040.2003.01158.x [DOI] [Google Scholar]
- Hörtensteiner S., Feller U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53 927–937. 10.1093/jexbot/53.370.927 [DOI] [PubMed] [Google Scholar]
- James M., Poret M., Masclaux-Daubresse C., Marmagne A., Coquet L., Jouenne T., et al. (2018). SAG12, a major cysteine protease involved in nitrogen mobilization during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol. 59 2052–2063. 10.1093/pcp/pcy125 [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. 10.1002/j.1460-2075.1987.tb02730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim H. J., Vu Q. T., Jung S., Robertson McClung C., Hong S., et al. (2018). Circadian control of ORE1 by PRR9 positively regulates leaf senescence in Arabidopsis. PNAS 115 8448–8453. 10.1073/pnas.1722407115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Tanaka A., Tanaka R. (2013). Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 117 221–234. 10.1007/s11120-013-9862-x [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lainé P., Ourry A., Macduff J., Boucaud J., Salette J. (1993). Kinetic parameters of nitrate uptake by different catch crop species: effects of low temperatures or previous nitrate starvation. Physiol. Plant. 88 85–92. 10.1111/j.1399-3054.1993.tb01764.x [DOI] [Google Scholar]
- Lim P. O., Kim H. J., Nam H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58 115–136. 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lohman K. N., Gan S., John M. C., Amasino R. M. (1994). Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 92 322–328. 10.1111/j.1399-3054.1994.tb05343.x [DOI] [Google Scholar]
- Masclaux C., Valadier M.-H., Brugière N., Morot-Gaudry J.-F., Hirel B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta. Planta 211 510–518. 10.1007/s004250000310 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Orsel M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. 10(Suppl. 1), 23–36. 10.1111/j.1438-8677.2008.00097.x [DOI] [PubMed] [Google Scholar]
- Nacry P., Bouguyon E., Gojon A. (2013). Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370 1–29. 10.1007/s11104-013-1645-9 [DOI] [Google Scholar]
- Noh Y.-S., Amasino R. M. (1999a). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41 181–194. 10.1023/A:1006342412688 [DOI] [PubMed] [Google Scholar]
- Noh Y. S., Amasino R. M. (1999b). Regulation of developmental senescence is conserved between Arabidopsis and Brassica napus. Plant Mol. Biol. 41 195–206. [DOI] [PubMed] [Google Scholar]
- Otegui M. S., Noh Y.-S., Martínez D. E., Vila Petroff M. G., Andrew Staehelin L., Amasino R. M., et al. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41 831–844. 10.1111/j.1365-313X.2005.02346.x [DOI] [PubMed] [Google Scholar]
- Parrott D. L., Martin J. M., Fischer A. M. (2010). Analysis of barley (Hordeum vulgare) leaf senescence and protease gene expression: a family C1A cysteine protease is specifically induced under conditions characterized by high carbohydrate, but low to moderate nitrogen levels. New Phytol. 187 313–331. 10.1111/j.1469-8137.2010.03278.x [DOI] [PubMed] [Google Scholar]
- Peoples M. B., Dalling M. J. (1988). “The interplay between proteolysis and amino acid metabolism during senescence and nitrogen reallocation,” in Senescence and Aging in Plants, eds Nooden L. D., Leopold A. C. (San Diego, CA: Academic Press; ), 181–217. [Google Scholar]
- Poret M., Chandrasekar B., van der Hoorn R. A. L., Avice J.-C. (2016). Characterization of senescence-associated protease activities involved in the efficient protein remobilization during leaf senescence of winter oilseed rape. Plant Sci. 246 139–153. 10.1016/j.plantsci.2016.02.011 [DOI] [PubMed] [Google Scholar]
- Pružinská A., Shindo T., Niessen S., Kaschani F., Tóth R., Millar A. H., et al. (2017). Major Cys protease activities are not essential for senescence in individually darkened Arabidopsis leaves. BMC Plant Biol. 17:4. 10.1186/s12870-016-0955-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossato L., Lainé P., Ourry A. (2001). Nitrogen storage and remobilization in Brassica napus L. during the growth cycle: nitrogen fluxes within the plant and changes in soluble protein patterns. J. Exp. Bot. 52 1655–1663. 10.1093/jexbot/52.361.1655 [DOI] [PubMed] [Google Scholar]
- Singh S., Giri M. K., Singh P. K., Siddiqui A., Nandi A. K. (2013). Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J. Biosci. 38 583–592. 10.1007/s12038-013-9334-7 [DOI] [PubMed] [Google Scholar]
- Tegeder M., Masclaux-Daubresse C. (2017). Source and sink mechanisms of nitrogen transport and use. New Phytol. 217 35–53. 10.1111/nph.14876 [DOI] [PubMed] [Google Scholar]
- Tegeder M., Rentsch D. (2010). Uptake and partitioning of amino acids and peptides. Mol. Plant 3 997–1011. 10.1093/mp/ssq047 [DOI] [PubMed] [Google Scholar]
- Wojciechowska N., Sobieszczuk-Nowicka E., Bagniewska-Zadworna A. (2017). Plant organ senescence - regulation by manifold pathways. Plant Biol. 20 167–181. 10.1111/plb.12672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.