Abstract
Cell migration is a highly regulated event that is initiated by cell membrane protrusion and actin reorganization. Robo1, a single-pass transmembrane receptor, is crucial for neuronal guidance and cell migration. ADP-ribosylation factor (Arf)–like 4A (Arl4A), an Arf small GTPase, functions in cell morphology, cell migration, and actin cytoskeleton remodeling; however, the molecular mechanisms of Arl4A in cell migration are unclear. Here, we report that the binding of Arl4A to Robo1 modulates cell migration by promoting Cdc42 activation. We found that Arl4A interacts with Robo1 in a GTP-dependent manner and that the Robo1 amino acid residues 1394–1398 are required for this interaction. The Arl4A-Robo1 interaction is essential for Arl4A-induced cell migration and Cdc42 activation but not for the plasma membrane localization of Robo1. In addition, we show that the binding of Arl4A to Robo1 decreases the association of Robo1 with the Cdc42 GTPase-activating protein srGAP1. Furthermore, Slit2/Robo1 binding down-regulates the Arl4A-Robo1 interaction in vivo, thus attenuating Cdc42-mediated cell migration. Therefore, our study reveals a novel mechanism by which Arl4A participates in Slit2/Robo1 signaling to modulate cell motility by regulating Cdc42 activity.
INTRODUCTION
Arf-like (Arl) proteins are structurally similar to members of the Arf protein family, which belong to the Ras superfamily of small GTPases and regulate vesicular transport, membrane trafficking, organelle structure, and cytoskeletal remodeling via cyclic regulation between their GTP-bound active form and their GDP-bound inactive form (D’Souza-Schorey and Chavrier, 2006; Chavrier and Menetrey, 2010). Like other GTP-binding proteins, the GTP-GDP cycle is regulated by guanine nucleotide exchange factors (GEFs) to exchange GDP for a triphosphate nucleotide and GTPase-activating proteins (GAPs) to stimulate GTP hydrolysis. Arl4 proteins (Arl4A, Arl4C, and Arl4D) are distinct from other Arf/Arl proteins due to their unique structures, which include a nuclear localization signal peptide at the carboxy terminus and a long interswitch region between two switch domains (Pasqualato et al., 2002). Constituting the subfamily, Arl4A, Arl4C, and Arl4D have different expression patterns during fetal development (Schurmann et al., 1994; Jacobs et al., 1999; Lin et al., 2000, 2002). Our previous studies showed that Arl4C and Arl4D modulate actin cytoskeleton remodeling and cell migration by affecting their interacting partners FLNa and ARNO, respectively (Li et al., 2007; Chiang et al., 2017). Arl4A shares a 90% amino acid sequence consensus with Arl4C and Arl4D and is mainly expressed in the forebrain–midbrain and midbrain–hindbrain junctions (Lin et al., 2000). Recently, Arl4A was found to play a role in actin cytoskeleton rearrangement via a pathway that stimulates ELMO/DOCK180-induced Rac signaling (Patel et al., 2011). However, the mechanism underlying how Arl4A affects cell motility remains unclear.
Cell migration is a highly regulated event that is initiated by protrusion of the cell membrane (Le Clainche and Carlier, 2008). Among the Rho GTPase family, Cdc42 reportedly plays a major role in regulating cell polarity, cell migration, and actin reorganization (Etienne-Manneville, 2004; Raftopoulou and Hall, 2004; Ridley, 2015). The secreted proteins Slits and their transmembrane receptor Roundabout (Robo) are highly expressed in the neuronal system and are crucial for neuronal guidance and cell migration (Ballard and Hinck, 2012). Recent studies have shown that a pathway mediated by Slit2 and Robo1 also plays important roles in other physiological and pathological processes outside of the nervous system, including the vascular system and tumorigenesis (Wu et al., 2001; Hinck, 2004; Legg et al., 2008). Many studies have indicated that Slit2/Robo1 signaling regulates multiple types of signaling responses, such as cell proliferation, adhesion, and migration (Andrews et al., 2006; Legg et al., 2008). Interestingly, emerging evidence postulates that Slit2/Robo1 often function in both the promotion and prevention of cell migration in various cell types in the same tissue (Schmid et al., 2007; Prasad et al., 2008). Slit2/Robo1 showed an ability to prevent cell migration and promote cell–cell adhesion via E-cadherin and β-catenin in lung cancer and breast cancer cells, respectively, as well as to attenuate Cdc42 activity in epithelial cell lines (Prasad et al., 2008; Yiin et al., 2009; Tseng et al., 2010). By contrast, Slit2/Robo1 also reportedly promotes cell migration by chemokines or by regulating Rho GTPases in breast cancer and epithelial cells (Schmid et al., 2007; Khusial et al., 2010).
In this study, we first identified Robo1 as a novel effector of Arl4A and found that Robo1 is required for Arl4A-induced cell migration. We showed that decreased cell migration resulting from Robo1 knockdown can be rescued by the expression of wild-type Robo1 but not by a Robo1 mutant deficient in Arl4A binding. Furthermore, the Arl4A-Robo1 interaction promotes Cdc42 activation by decreasing the association of a Cdc42-GAP, srGAP1, with Robo1. The binding of Slit2 to Robo1 decreases the Arl4A-Robo1 interaction and increases the srGAP1-Robo1 association, resulting in decreased Cdc42 activation and prohibited cell migration. Our results demonstrate that Arl4A functions together with Robo1 to modulate Cdc42 activation and cell migration via regulating the Robo1-srGAP1 association.
RESULTS
Identification of Robo1 as an Arl4A GTPase interaction partner
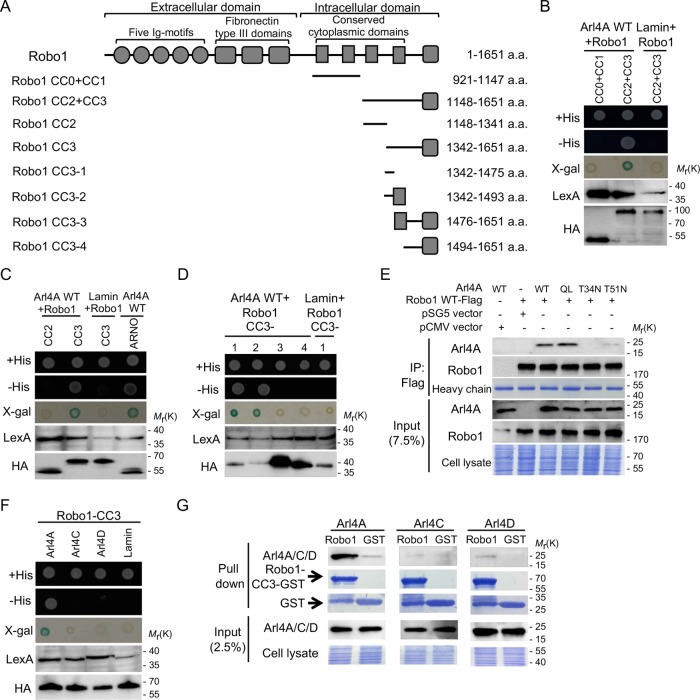
To identify Arl4A interaction proteins, a yeast two-hybrid screen with a human fetal brain cDNA library was performed using Arl4A-QL, a GTP-bound form of Arl4A, as the bait; one Arl4A-interacting protein was identified, the cytoplasmic domain of Robo1 (residues 921–1651). To identify the region within Robo1 required for its interaction with Arl4A, we constructed fragments of the intracellular region of Robo1 as illustrated in Figure 1A, and their interactions with Arl4A were tested using a yeast two-hybrid system. Only the CC2+CC3 domain of Robo1 was found to interact with Arl4A-WT (Figure 1B). To determine the specific region of Robo1 that interacts with Arl4A, the CC2+CC3 domain was divided into CC2 and CC3 fragments. Only Robo1-CC3 was accountable for its interaction with Arl4A (Figure 1C). To further investigate the specific regions within Robo1-CC3 responsible for its interaction with Arl4A, we generated three truncated forms of the Robo1-CC3 domain. Both Robo1-CC3-1 and Robo1-CC3-2 positively interacted with Arl4A in a yeast two-hybrid system, indicating that the Robo1-CC3-1 region (residues 1342–1475) is necessary for the Robo1-Arl4A interaction (Figure 1D). In addition, in vivo coimmunoprecipitation assays also suggested that the interaction between Arl4A and Robo1 is GTP-dependent (Figure 1E). Furthermore, yeast two-hybrid and pull-down assays showed that an interaction was not detected between the Robo1-CC3 region and the other two Arl4 family members, Arl4C and Arl4D (Figure 1, F and G). These results suggest that Arl4A specifically and directly interacts with Robo1 in a GTP-dependent manner.
FIGURE 1:
Arl4A interacts with Robo1 in a GTP-dependent manner. (A) Schematic diagram of Robo1 and several fragments constructed in this study. Robo1 is a single-pass transmembrane protein that contains five Ig motifs, three fibronectin type III domains, and four conserved cytoplasmic (CC) domains at its carboxy terminus. The N-terminal region of Robo1 is an extracellular region, while the four CC domains located at the carboxy terminus are in an intracellular region of Robo1. Yeast two-hybrid assays were used to verify the interactions of WT Arl4A with (B) two intracellular domains of Robo1 fragments (CC0+CC1 and CC2+CC3), (C) CC2 and CC3 Robo1 domains, and (D) different fragments of the Robo1 CC3 domain (CC3-1, CC3-2, CC3-3, and CC3-4). Lamin was used as the negative control, and ARNO was used as the positive control. The levels of proteins expressed by the transforming plasmids were confirmed by immunoblotting. After cotransformation with the indicated plasmids, interactions were verified by growth of the yeast on a synthetic His+ plate and a His− plate followed by filter assays for β-galactosidase activity. (E) In vivo interaction between Arl4A and Robo1 was confirmed by coimmunoprecipitation. HEK293T cells were transfected with the indicated plasmids. To verify the initial protein expression level, 7.5% of the total cell lysate (input) was loaded. Equal binding abilities of the magnetic beads were used in the assays as shown by the Flag signal in the IP group. (F) Yeast two-hybrid assays were used to verify the interactions of Robo1-CC3 with three members of the Arl4A family. Lamin was used as the negative control. The levels of proteins expressed by the transforming plasmids were confirmed by immunoblotting. After cotransformation with the indicated plasmids, interactions were verified by growth of the yeast on a synthetic His+ plate and a His− plate followed by filter assays for β-galactosidase activity. (G) The interactions of Robo1-CC3 with the Arl4 family members were verified by pull-down assays. His-tagged Arl4-WT was generated from E. coli, and soluble cell lysate fractions were incubated with GST and Robo1-CC3-GST immobilized on glutathione–Sepharose beads. Bound proteins were detected by Western blotting, and Coomassie Brilliant Blue staining indicated that equal amounts of His-tagged or GST-fusion proteins were used in the pull-down assays.
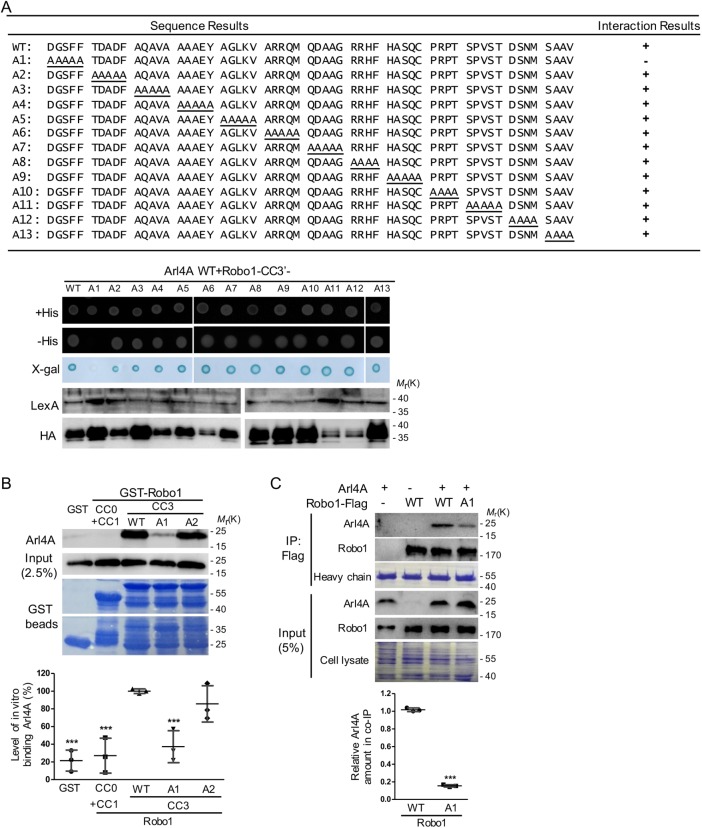
Residual Robo1 amino acids 1394–1398 are necessary for the Arl4A-Robo1 interaction
We further narrowed the amino acids responsible for the rlA4A-Robo1 interaction to Robo1 fragments 1342–1475 (Robo1 CC3-1) and 1370–1475 (Robo1-CC3′) using the yeast two-hybrid system (unpublished data). To identify which residues are important for its interaction with Arl4A, we generated Arl4A-binding defective Robo1 mutants by alanine scanning and assayed these mutants using the yeast two-hybrid system. Among these 13 mutants, only the Robo1-A1 (1394–1398 amino acids) mutant was unable to interact with Arl4A-WT in the yeast two-hybrid assay (Figure 2A). As shown in Figure 2B, Arl4A directly interacted with Robo1-CC3-1-WT fused with glutathione S-transferase (GST) beads but not with beads containing Robo1-CC3-1-A1, Robo1-CC0+CC1, or GST alone. Notably, Robo1-CC3-1-A2 fused with GST beads did not lose much interaction ability with the purified Arl4A protein, suggesting that residues 1394–1398 of Robo1 play a more critical role in the Arl4A-Robo1 interaction than other regions in this fragment. The interaction was also verified by an in vivo coimmunoprecipitation assay. HeLa cells cotransfected with Arl4A and the Robo1-Flag-WT or Robo1-Flag-A1 mutant were immunoprecipitated using magnetic beads conjugated to an anti-Flag M2 antibody. Similarly, the interaction ability of the Robo1-A1 mutant with Arl4A was decreased significantly compared with that of Robo1-WT (Figure 2C). To elucidate the effects of the Arl4A-binding defective Robo1-A1 mutant on other Robo1 partner proteins, interactions between Robo1-srGAP1 and Robo1-Nck1 were assayed by the yeast two-hybrid system. Like wild-type Robo1, the Robo1-A1 mutant was capable of interacting with both srGAP1 and Nck1, indicating that the interaction defect of Robo1-A1 was specific to Arl4A but not to the other Robo1-interacting proteins srGAP1 and Nck1 (Supplemental Figure 1).
FIGURE 2:
A specific region in Robo1 is required for its interaction with Arl4A. (A) The region of Robo1 responsible for binding Arl4A was identified by a yeast two-hybrid system using fragments comprising amino acid residues 1370–1475 and Arl4A Q79L as the bait. Alanine scanning was used to determine which Robo1 fragments (from amino acid residues 1394–1454) were required for its interaction with Arl4A using a yeast two-hybrid system. Interaction between Robo1-WT and Arl4A-WT served as the positive control, and lamin was used as the negative control. (B) Direct interactions between Arl4A-WT and different Robo1 fragments were examined by an in vitro binding assay. His-tagged Arl4-WT was generated and purified from E. coli and then incubated with GST and four truncated/mutated GST-Robo1 genes (CC0+CC1, CC3-WT, CC3-A1, and CC3-A2) immobilized on glutathione–Sepharose beads, respectively. Bound proteins were detected by Western blotting, and Coomassie Brilliant Blue staining was used to ensure that equal amounts of GST and GST-Robo1 proteins were used in the in vitro binding assay. Arl4A signals were quantified based on in vitro binding assay data obtained from three biological replicates. The solid bars represent the mean ± SD. ***, P < 0.001 (one-way ANOVA with Dunnett's post hoc multiple comparison test, GST-Robo1-WT was used as the reference). (C) Interaction between Arl4A and Robo1-WT or Robo1-A1 was verified by in vivo coimmunoprecipitation. HeLa cells transiently transfected with the indicated plasmids were lysed and immunoprecipitated with anti-Flag M2 magnetic beads. The bound proteins were separated by SDS–PAGE and subjected to immunoblotting with antibodies against Arl4A and Robo1. To confirm the initial expression level, 5% of the total cell lysate (input) was loaded. Equal amounts of magnetic beads were used in the assays as shown by Coomassie Blue staining of the heavy chain. Co-IP assay data were quantified based on three biological replicates. The solid bars represent the mean ± SD. ***, P < 0.001 (Student's t test).
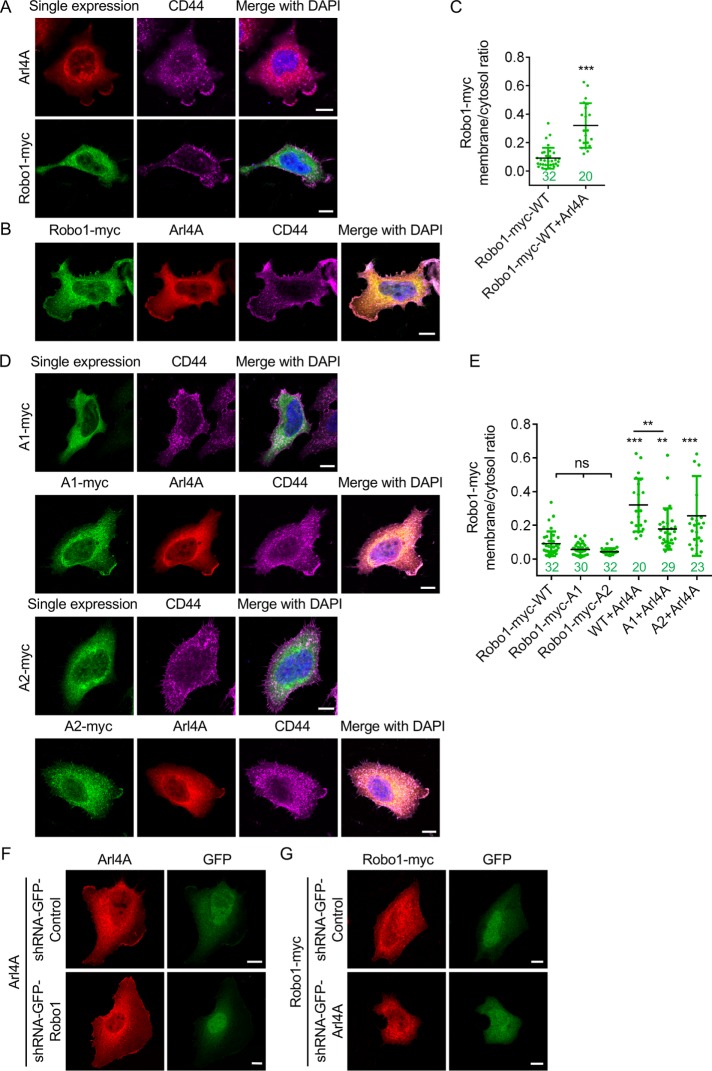
Arl4A induces Robo1 localization at the plasma membrane
Several studies have shown that the expression of Robo1 on the cell surface is regulated by factors involved in exocytosis and the endosomal system (Keleman et al., 2002; Myat et al., 2002; Philipp et al., 2012; Justice et al., 2017). We next examined whether the Arl4A-Robo1 interaction corresponds with their localization in HeLa cells by immunofluorescence staining. As previously reported, the typical pattern of Arl4A signals includes Golgi and plasma membrane signals (Lin et al., 2011). Robo1, which had a C-terminal myc tag in our experiments, localized primarily in the cytosol in diffuse or vesicle-like punctate forms in HeLa cells (Figure 3A). When Robo1 and Arl4A were coexpressed, Robo1, which appeared to be sequestered in the cytosol when expressed alone, localized partially at the plasma membrane (Figure 3B). To carefully determine the extent of Robo1 potentiation at the plasma membrane upon the expression of Arl4A, we costained HeLa cells with the plasma membrane marker CD44 to clearly demarcate the margins of the cells. With the boundaries of the HeLa cells defined by the CD44 signal, we calculated the plasma membrane-to-cytosol (PM/C) ratio (see Materials and Methods) for Robo1 and found that this ratio significantly increased upon Arl4A expression (Figure 3C). In addition, Arl4A expression significantly promoted plasma membrane localization of the Arl4A-binding defective Robo1 A1 mutant (Figure 3, D and E), although the PM/C ratios for Robo1 A1 were slightly lower than those for Robo1 WT. We further monitored the change in localization of Arl4A or Robo1 by knocking down either Robo1 or Arl4A and showed that the cellular localization of Arl4A or Robo1 did not change (Figure 3, F and G). These results suggest that although Arl4A facilitates the plasma membrane localization of Robo1, the Arl4A-Robo1 interaction is not the major factor contributing to the Arl4A-induced localization of Robo1 at the plasma membrane.
FIGURE 3:
Arl4A induces the localization of Robo1 at the plasma membrane. (A) HeLa cells were transfected with Arl4A-WT (red) or Robo-myc-WT (green) alone. (B) HeLa cells were cotransfected with Arl4A-WT and Robo1-Myc-WT. CD44 staining was used to define the plasma membrane boundary of HeLa cells. Cell nuclei were stained with DAPI. (C) PM/C ratios for Robo-myc-WT in the single-expression and Arl4A coexpression groups. (D) Cellular localization of Arl4A-binding defective mutants (A1 and A2) in the single-expression and Arl4A coexpression groups. (E) Quantitative data from A, B, and D. Scale bar = 10 µm. The numbers of analyzed cells are shown in each column. The solid bars represent the mean ± SD. **, P < 0.005; ***, P < 0.001 (C: Student's t test; E: one-way ANOVA with Dunnett's post hoc multiple comparison test).
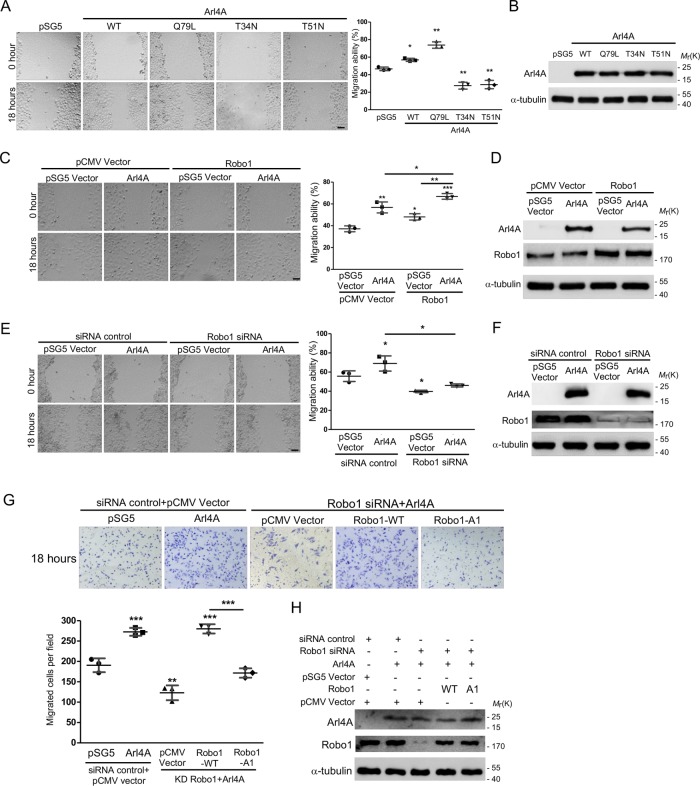
Arl4A-induced cell migration requires interaction with Robo1
Although Arl4A induces cellular protrusion and plays a role in the regulation of actin dynamics (Patel et al., 2011), its function in modulating cellular mobility remains to be established. The wound healing assay showed that HeLa cells expressing Arl4A-WT and Arl4A-Q79L had higher migration abilities than those expressing either Arl4A-T34N or Arl4-T51N (Figure 4, A and B). Knocking down Arl4A or Robo1 decreased the motility of HeLa cells (Supplemental Figure 2), although the endogenous Arl4A protein level was extremely low. Importantly, the coexpression of Arl4A and Robo1 had an additive effect on promoting cell mobility, as the increased migration abilities of Arl4A- or Robo1-expressing cells were enhanced by the coexpression of these two proteins in HeLa cells (Figure 4, C and D). The increased migration ability of Arl4A-expressing cells was abolished by Robo-1 knockdown (Figure 4, E and F). Our preliminary screening showed that HEK293T cells possess abundant endogenous Robo1; testing whether Robo1 is required for Arl4A-induced cell migration in a different cell model is thus ideal. Owing to the tendency of HEK293T cells to detach easily from the plate after the wound is made, it therefore compromises the suitability of the wound closure assay for these cells (Justus et al., 2014). Transwell migration inserts were used instead to assess the migration ability of HEK293T cells. According to the Transwell assay results, the overexpression of Arl4A did not restore the reduced motility of Robo1-knockdown cells, supporting a critical role for Robo1 in Arl4A-induced cell migration. This assumption was further supported by the observation that Arl4A and Robo1-WT coexpression but not Arl4A and Robo1-A1 coexpression rescued the cell migration ability of Robo1-knockdown cells (Figure 4, G and H). These results collectively demonstrate that the Arl4A-Robo1 interaction is necessary for Arl4A-induced cell migration.
FIGURE 4:
The Arl4A-Robo1 interaction is necessary for increased cell migration. (A, C, E) Representative images of wound healing assays. HeLa cells were transfected with the indicated plasmids or siRNA for 18 h and then subjected to wound healing migration assays. Scale bar = 45 µm. Histograms: Wound healing migration data were quantified based on three biological replicates. (B, D, F) Total protein (20 μg) was loaded onto a 10-well gel to detect proteins. Western blot analysis of lysates from HeLa cells transfected with the indicated plasmids was performed to confirm equal expression. (F) The percentages of Robo1 after siRNA treatment were 23.1 ± 0.6% and 21.7 ± 0.4%. (G) Representative images of HEK293T cells transfected with the indicated plasmids and siRNA and then subjected to Transwell assays. The number of migrated cells in a field was calculated using ImageJ software after 18 h of migration. Histogram: Migration assay data were quantified based on three biological replicates. (H) Total protein (20 μg) was loaded onto a 10-well gel to detect protein expression. Immunoblotting analyses were used to evaluate protein expression levels in cells transfected with the indicated plasmids and siRNA. The percentage of Robo1 after siRNA treatment was 17.2 ± 0.5%. The solid bars represent the mean ± SD. *, P < 0.05; **, P < 0.005; ***, P < 0.001 (A: two-tailed Student's t test; C, E, and G: one-way ANOVA with Dunnett's post hoc multiple comparison test).
The Arl4A-Robo1 interaction promotes cell migration by activating Cdc42
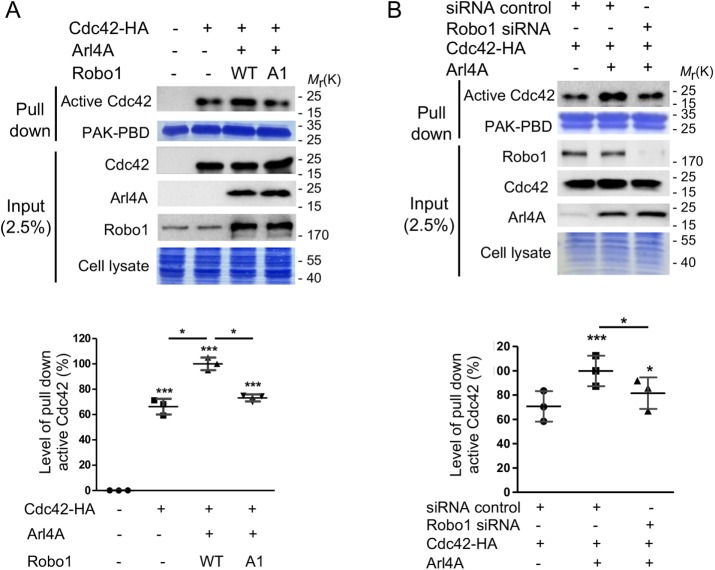
Because Cdc42 is reportedly important for regulating cell motility, we examined its role in affecting the migration of HEK293T and HeLa cells expressing Arl4A and Robo1. We tested whether the Arl4A-Robo1 interaction promotes Cdc42 activation using an activity pull-down assay with PAK1-PBD beads. No active Cdc42 was found in mock-transfected HEK293T cells, while only a low level of active Cdc42 was detected in cells expressing exogenous Cdc42. The amount of active Cdc42 increased in cells cotransfected with Cdc42, Arl4A, and Robo1-WT, suggesting that the coexpression of Arl4A and Robo1 induces Cdc42 activation. By contrast, the amount of active Cdc42 decreased when Robo1-WT was replaced with the Arl4A-binding defective Robo1-A1 mutant (Figure 5A). Similar results were also found in HeLa cells (Supplemental Figure 3). Moreover, the level of active Cdc42 decreased when Robo1 was knocked down in Arl4A-expressing HEK293T cells (Figure 5B). These results indicate that the Arl4A-Robo1 interaction is critical for promoting Cdc42 activation.
FIGURE 5:
The Arl4A-Robo1 interaction is required for Cdc42 activation. HEK293T cells were transiently transfected with (A) Cdc42-HA, Arl4A, Flag-Robo1-WT, and Flag-Robo1-A1 or (B) Cdc42-HA, Arl4A, Robo1 siRNA, and an siRNA control. The cells were lysed and Cdc42 activity pull-down assays were performed. To confirm the initial expression level, 2.5% of the total cell lysate (input) was loaded. Equal amounts of GST beads and cell lysates were used in each experiment as shown by Coomassie Blue staining. Histograms: Active Cdc42 was quantified based on three biological replicates. The solid bars represent the mean ± SD. *, P < 0.05; ***, P < 0.001 (A: one-way ANOVA with Dunnett's post hoc multiple comparison test; B: two-tailed Student's t test).
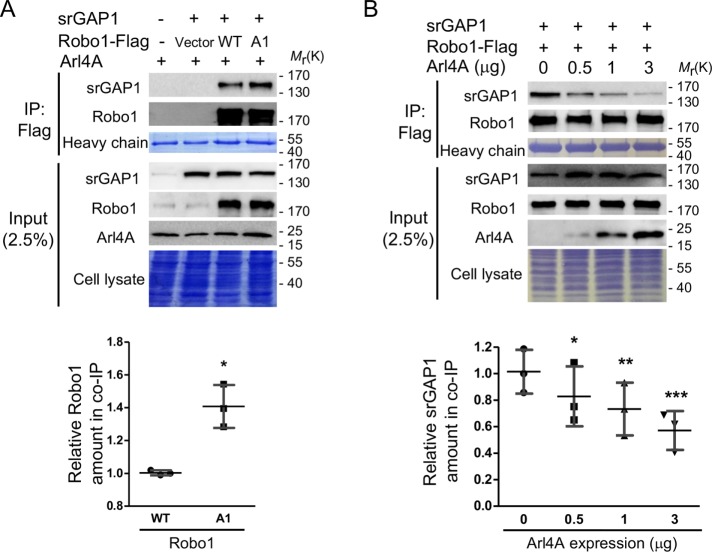
Robo1 and Arl4A binding decreases the association of Robo1 with srGAP1
Previous studies reported that the association between Robo1 and srGAP1, a Cdc42 GTPase-activating protein, regulates Cdc42 activity (Wong et al., 2001). We examined whether srGAP1 also plays a role in Arl4A-induced Cdc42 activation. The Robo1-srGAP1 association was verified by an in vivo coimmunoprecipitation assay in which HEK293T cells were transfected with Arl4A, srGAP1, Robo1-Flag-WT, and Robo1-Flag-A1. A higher level of srGAP1 coimmunoprecipitated with Robo1-A1 than with Robo1-WT (Figure 6A). To further investigate whether Arl4A affects the association between Robo1 and srGAP1, a coimmunoprecipitation assay was performed in HEK293T cells coexpressing srGAP1, Robo1-Flag, and different amounts of Arl4A. The level of immunoprecipitated srGAP1 decreased with increasing Arl4A concentrations in a dose-dependent manner (Figure 6B). Together, the Arl4A-Robo1 interaction promotes Cdc42 activation by down-regulating the Robo1-srGAP1 association.
FIGURE 6:
The Arl4A-Robo1 interaction induces Cdc42 activation via reducing the association between Robo1 and srGAP1. HEK293T cells were transiently transfected with an (A) empty vector, srGAP1, Flag-Robo1-WT, Flag-Robo1-A1, and Arl4A or (B) srGAP1, Flag-Robo1-WT, and Arl4A. The interaction was verified by in vivo coimmunoprecipitation with anti-Flag M2 magnetic beads. To confirm the initial expression level, 2.5% of the total cell lysate (input) was loaded. Equal amounts of cell lysates were used in each experiment as shown by Coomassie Blue staining. The heavy chain was used to ensure equal amounts of M2 magnetic beads were loaded in each group. SDS–PAGE analysis was used to verify srGAP1, Robo1, and Arl4A expression and total cell lysates. Histograms: Co-IP assay data were quantified based on three biological replicates. The solid bars represent the mean ± SD. *, P < 0.01; **, P < 0.005; ***, P < 0.001 (A: two-tailed Student's t test; B: one-way ANOVA with Dunnett's post hoc multiple comparison test).
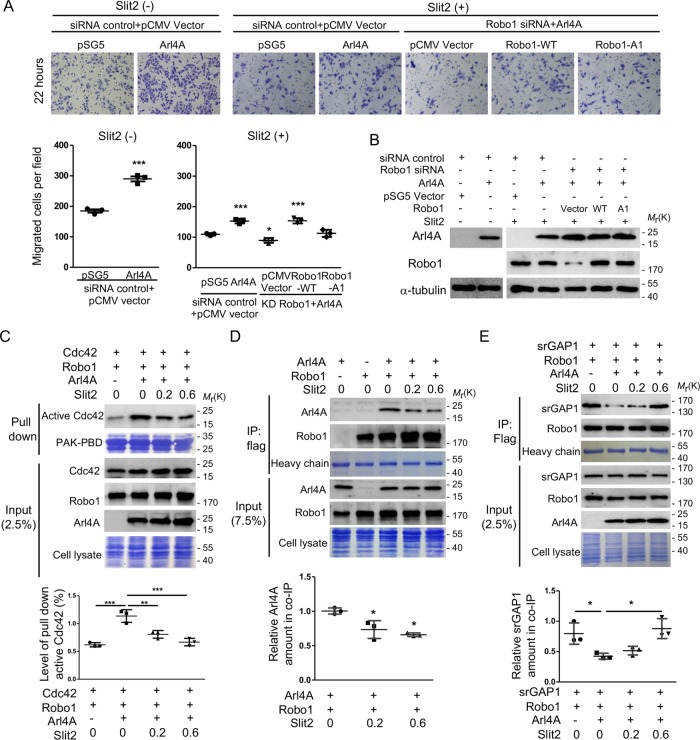
Slit2 down-regulates Cdc42 activation and cell motility by affecting the Robo1-srGAP1 and Robo1-Arl4A associations
A previous study showed that upon Slit2 stimulation, Robo1 recruits srGAP1 in order to inhibit Cdc42 activity, thus controlling neuronal migration (Wong et al., 2001). Slit2, a secreted extracellular matrix protein, binds the Robo1 receptor and controls many morphogenesis and cellular functions, including migration, proliferation, and adhesion (Zhao et al., 2016). We herein showed that Slit2 treatment reduced cell migration in a dose-dependent manner; however, this treatment did not abolish Arl4A-induced cell migration (Figure 7, A and B and Supplemental Figure 4). To test the activity of Cdc42 under Slit2 treatment, HEK293T cells were transiently transfected with Cdc42, Robo1, and Arl4A. Consistently, Slit2 treatment reduced the amount of active Cdc42 in a dose-dependent manner (Figure 7C). Notably, the quantitative level of active Cdc42 in Arl4A-overexpressing cells treated with the highest amount of Slit2 was reduced to nearly its basal level, but this reduction did not occur in cells not overexpressing Arl4A (Figure 7C), thus indicating that Slit2 fully suppresses Arl4A-induced Cdc42 activation. Next, we examined whether Slit2 modulates cellular migration by regulating the Robo1-srGAP1 and Robo1-Arl4A associations. As shown in Figure 7D, Slit2 treatment significantly reduced the association between Robo-1 and Arl4A in a dose-dependent manner. By contrast, the association between Robo1 and srGAP1 was increased when cells were treated with Slit2 (Figure 7E). These results collectively demonstrate that Slit2 plays a role in modulating the association between Robo1 and its two interacting proteins Arl4A and srGAP1 and thus contributes to the dynamic regulation of cell migration.
FIGURE 7:
Slit2 affects Arl4A-induced Cdc42 activation and cell migration via regulating the Robo1-Arl4A and Robo1-srGAP1 associations. (A) Representative images of HEK293T cells transfected with the indicated plasmids and siRNA and then subjected to the Transwell assay. Purified Slit2 (0.2 µg/ml) was added to the lower chamber of the Transwell apparatus. The number of migrated cells in a field was calculated using ImageJ software after 22 h of migration. Histograms: Migration assay data were quantified based on three biological replicates. (B) Total protein (20 μg) was loaded onto a 10-well gel to detect proteins. Immunoblotting analyses were used to evaluate protein expression levels in cells transfected with the indicated plasmids and siRNA. The percentage of Robo1 after siRNA treatment was 11.5 ± 0.75%. (C) HEK293T cells were transiently transfected with the indicated plasmid and treated with 0.2 or 0.6 µg/ml purified Slit2. The cells were lysed and subjected to Cdc42 activity pull-down assays. To confirm the initial expression level, 2.5% of the total cell lysate (input) was loaded. Equal amounts of GST-tagged beads and cell lysates were used in each experiment as shown by Coomassie Blue staining. Histogram: Active Cdc42 was quantified based on three biological replicates. (D, E) HEK293T cells were transiently transfected with the indicated plasmids and then treated with purified Slit2 (µg/ml). Interactions were verified by in vivo coimmunoprecipitation with anti-Flag M2 magnetic beads. To confirm the initial expression level, 7.5% and 2.5% of the total cell lysates (input) were loaded. The heavy chain was used to ensure that equal amounts of M2 magnetic beads were loaded in each group. SDS–PAGE analysis was used to verify srGAP1, Robo1, and Arl4A expression and total cell lysates. Histograms: Co-IP assay data were quantified based on three biological replicates. The solid bars represent the mean ± SD. *, P < 0.01; **, P < 0.005; ***, P < 0.001 (A, C, and E: one-way ANOVA with Dunnett's post hoc multiple comparison test; D: two-tailed Student's t test).
DISCUSSION
In this study, we identified Robo1 as a novel effector of Arl4A and demonstrated that the Arl4A-Robo1 interaction modulates cell migration by potentiating Cdc42 activation. Arl4A-induced Cdc42 activation was impaired in Robo1-knockdown cells (Figure 5B). Moreover, the Robo1-A1 mutant, which can no longer interact with Arl4A, failed to activate Cdc42 in either HeLa or HEK293T cells (Figure 5A and Supplemental Figure S3). The Arl4A-Robo1 interaction reduces protein–protein associations between Robo1 and the Cdc42 GAP srGAP1 (Figure 6). We further showed that the binding of Robo1 to the neuronal repulsive factor Slit2 inhibits HEK293T cell motility and Cdc42 activity by decreasing the Arl4A-Robo1 interaction and increasing the Robo1-srGAP1 association (Figure 7).
Our previous study showed that Arl4A plays a role in actin cytoskeleton rearrangement via a pathway that stimulates ELMO/DOCK180-induced Rac signaling (Patel et al., 2011). Studies have demonstrated that Arl4A and its close relatives Arl4C and Arl4D promote actin restructuring via the recruitment of ARNO, an Arf6 GEF, to the plasma membrane. Interestingly, Arf6 is positioned upstream of Rac activation in various biological processes. Arf6 activation has been proposed to recruit the DOCK180–ELMO complex to the leading edge of a cell to promote lamellipodia formation (Santy et al., 2005). However, our previous studies demonstrated that Arl4A-induced cytoskeletal remodeling occurs via an Arf6-independent pathway. This finding may signify that Arl4A can act as a central signaling node for two divergent GTPase pathways. RhoG is known to promote migration and phagocytosis when coexpressed with ELMO (Katoh et al., 2006). In contrast to the RhoG-ELMO interaction, Arl4A was incapable of synergizing with ELMO/DOCK180 in both cell migration and engulfment assays. These data suggest that Arl4A and RhoG are not located in the same membrane microdomain because their interaction with ELMO led to different biological outputs.
Robo1, originally observed in the nervous system, plays a vital role in axon guidance and cell motility during the forebrain development process. Previous studies have shown that Robo1 regulates cell proliferation and motility in both neuronal and nonneuronal cells. Apart from the central nervous system, several studies have shown that Robo1 plays dual roles in cancer cell migration, as it acts as both an oncogene and a suppressor in different types of cancers (Ballard and Hinck, 2012). Robo1 has been found to either inhibit or promote cell migration depending on its downstream interacting partners (Stella et al., 2009; Tole et al., 2009; Yiin et al., 2009). The different directions of Robo1's regulation of cell motility have something in common, as both phenomena have been reported to affect the activities of Rho GTPases, such as Cdc42, in several cell types (Wu et al., 2001; Stella et al., 2009; Tole et al., 2009; Yiin et al., 2009). Our results demonstrated that Arl4A-expressing Robo1-knockdown cells exhibited significantly decreased migration. These data are consistent with those of previous studies showing that Robo1 knockdown by small interfering RNA (siRNA) in human retinal pigmented epithelium cells significantly reduced their adhesion, proliferation, and migration capacities (Huang et al., 2010). However, cells coexpressing Arl4A and Robo1 had higher migration abilities than those expressing Arl4A or Robo1 alone. Based on our findings, we postulated that Arl4A might participate in the Robo1-regulated migration pathway, as HeLa and HEK293T cells expressing Arl4A in which Robo1 was knocked down exhibited down-regulated cell motility.
The expression of Robo1 on the cell surface is regulated by factors involved in exocytosis and the endosomal system (Keleman et al., 2002; Myat et al., 2002; Philipp et al., 2012; Justice et al., 2017). The Commissureless protein (Comm) in Drosophila can bind Robo1 in the Golgi and directly traffic Robo1 to endosomes, thus preventing it from reaching the cell surface (Keleman et al., 2002; Myat et al., 2002). In vertebrates, Robo1 was shown to be predominantly stored in Rab11-positive vesicles, and RabGDI controls axonal midline crossing by up-regulating Robo1 surface expression (Philipp et al., 2012). Consistent with this report, we observed that the majority of the Robo1 protein appeared to be located in vesicle-like puncta in HeLa cells and that Arl4A can induce Robo1 localization at the plasma membrane. Our previous study showed that Arl4A has a role in the maintenance of the Golgi structure and endosome-to-Golgi transport (Lin et al., 2011). ARL4A was partially localized at the plasma membrane and presented a punctate pattern in intracellular areas, where it partially colocalized with the trans-Golgi, early endosomes, late endosomes, and recycling endosomes. Although the Arl4A-Robo1 interaction is not required for Arl4A-induced Robo1 localization at the plasma membrane, whether Arl4A-dependent vesicle trafficking plays a role in axonal behavior remains to be investigated.
Robo1 is a transmembrane receptor with a Slit2 binding site at its amino terminus to control axonal guidance, cell motility, and active downstream signaling pathways. The intracellular region of Robo1, located at the carboxy terminus, is responsible for its association with interacting proteins, including Nck1, Dock, and srGAP1. These Robo1 interacting partners are involved in Slit2-Robo1 signaling in different ways, which are coordinated with a variety of cellular processes and result in various functions and phenotypes. Slit2 promotes Robo1 to inhibit glioma cell migration and invasion by down-regulating Cdc42 activation (Stella et al., 2009; Yiin et al., 2009). Stimulation of Robo1 by Slit2 leads to the recruitment and activation of srGAP1, consequently inhibiting Cdc42 activity. Conversely, our results showed that Arl4A binding to Robo1 decreased the srGAP1-Robo1 association in vivo following Cdc42 activation. Consistent with most Robo1 interacting proteins, Arl4A also bound to the C-terminal region of Robo1. Although the Arl4A-binding defective Robo1-A1 mutant did not lose its ability to interact with srGAP1 or Nck1 in the yeast two-hybrid assay, this mutant showed a stronger interaction ability with srGAP1 in vivo. Conversely, Arl4A binding to Robo1 reduced the Robo1-srGAP1 association (Figure 5). The Robo1-A1 mutant, which could not interact with Arl4A, failed to rescue cell migration in Robo1-knockdown HEK293T cells expressing Arl4A, suggesting that Arl4A and Robo1 function in collaboration to regulate cell motility outside the nervous system.
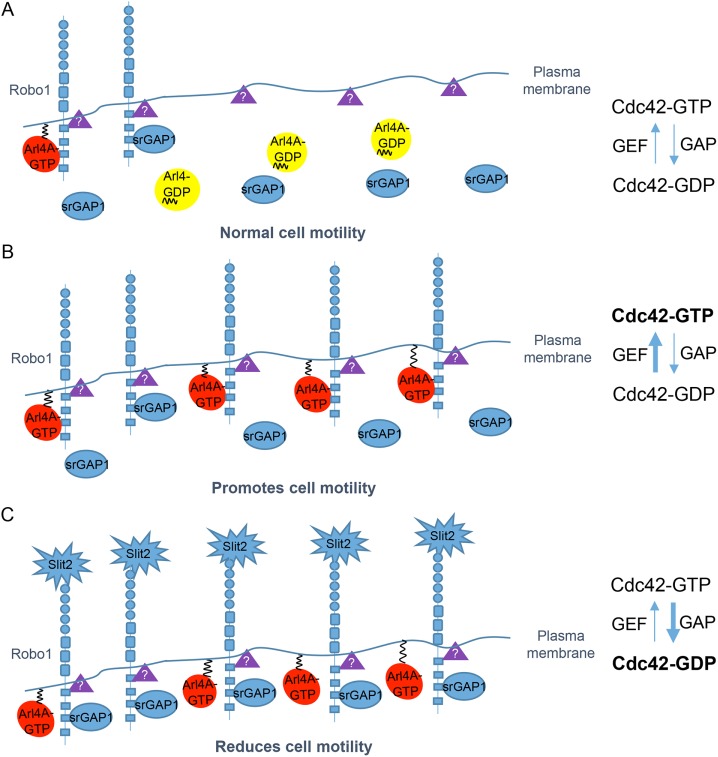
Our study suggests that the plasma membrane association of GTP-bound Arl4A can induce Robo1 to localize at the plasma membrane, and its binding to Robo1 may lead to a conformational change in Robo1, decreasing the recruitment of srGAP1 to Robo1. In addition, we speculate that an intermediate Robo1 structure associates with an unknown Cdc42 GEF, which is much more susceptible to Cdc42 activation by Arl4A stimulation. Our studies suggest that Arl4A plays a role in regulating the conformation of Robo1, which can also be modulated by other stimulators. Slit2 binds Robo1 and decreases the Arl4A-Robo1 association, leading to the recruitment and activation of srGAP1 (Figure 8).
FIGURE 8:
Model of Arl4A involvement in Robo1-mediated Cdc42 activation. Robo1 is a single-pass transmembrane receptor. GTP-bound Arl4A can interact with Robo1 via its cytoplasmic CC3 domain. (A) An intermediate Robo1 structure may be associated with an unknown Cdc42 GEF (purple triangles), which is much more susceptible to Cdc42 activation by Arl4A stimulation. (B) More activated Arl4A molecules induce the localization of Robo1 at the plasma membrane and subsequently bind its CC3 domain. The Arl4A-Robo1 interaction prevents srGAP1 from binding Robo1 and promotes Cdc42 activation. (C) Slit2 binding to the amino terminus of Robo1 decreases the Arl4A-Robo1 interaction and enhances the srGAP1-Robo1 interaction, ultimately promoting Cdc42 inactivation.
Our previous study showed that the expression of mouse Arl4A mRNA is developmentally regulated and is consistent with involvement of the protein in early events of embryogenesis of the central nervous system, somitogenesis, and spermatogenesis (Lin et al., 2000). At mouse embryonic day 14.5 (E14.5), Arl4A mRNA is expressed strongly in the cortical plate in an overlapping pattern with that of Robo1 and Slit1/Slit2 mRNA (Lin et al., 2000; Yeh et al., 2014), suggesting possible comodulation of these molecules during the development of the dorsal telencephalon. Given that Slit-Robo signaling plays a crucial role in the axon guidance of developing cortical neurons (Brose et al., 1999; Dickson, 2002) and that rsGAP1 is functionally involved in neuronal migration in response to Slit (Wong et al., 2001), our current study suggests that Arl4A might functionally participate in axonal pathfinding or neuronal migration via modulation of the Slit-Robo/rsGAP1 signal axis. Because Arl4A binds with Robo1 in a competitive manner with rsGAP1, a possible scenario for the developing migratory neurons is that Arl4A keeps the migratory-brake function of Robo1 inactivated by constitutively binding with Robo1, thus preventing Robo1 from association with the downstream signaling rsGAP1 for execution of migratory inhibition. When migratory neurons navigate through the developing cortex, contact with the guidance cue Slit releases Robo1 from Arl4A's binding, allowing for Robo1's association with rsGAP1 and subsequent Cdc42 down-regulation concomitant with inhibition of neuronal migration. It is also possible that other environmental cues or intrinsic factors, such as changes in intracellular calcium concentration or neuronal activity, could regulate the expression level or activity of Arl4A to fine-tune the Slit-Robo/rsGAP1/Cdc42 signal axis in the developing migratory neurons. Our preliminary data in cultured nonneuronal cells showed that Arl4A activity was reduced when cells were grown in serum starvation conditions. They suggest that Arl4A activity requires certain extrinsic factors, which remain to be further investigated. The proceeding dissection of possible promigratory or environmental cues will cast light on the regulation of Arl4 activation for cell migration.
In conclusion, we demonstrate that the activated GTP-bound form of Arl4A functions as a novel interacting partner of Robo1 to modulate cell motility. The Arl4A-Robo1 interaction regulates cell migration via Cdc42 activation. The cell migration enhanced by the Arl4A-Robo1 interaction is caused by the down-regulation of the srGAP1 association with Robo1. Our work addresses a long-standing knowledge gap in the effects of regulating Robo1 signaling on cell migration by defining a role for dynamic protein interactions governing Cdc42 activation.
MATERIALS AND METHODS
Antibodies
Rabbit anti-Robo1 and anti-Arl4A/C/D antibodies were generated as described previously (Li et al., 2007). The following antibodies were used: anti-Myc (1:1000, catalogue no. MMS-150R; Covance, Princeton, NJ), anti-HA (1:1000, catalogue no. SC-7392; Santa Cruz Biotechnology), anti-LexA (1:1000, catalogue no. 5397-1; Clontech, Mountain View, CA), anti-CD44 (1:200 for IF, catalogue no. MA4400; Invitrogen), anti-Cdc42 (1:500, catalogue no. 2462; Cell Signaling, Danvers, MA), anti-srGAP1 (1:1000, catalogue no. 76926; Abcam), and anti-α-tubulin (1:5000, catalogue no. T5168; Sigma-Aldrich, St. Louis, MO). A blue-fluorescent DNA stain 4′,6-diamidino-2-phenylindole (DAPI) solution was purchased from Millipore (1:5000, catalogue no. S7113). Horseradish peroxidase (HRP)–conjugated goat anti-rabbit and anti-mouse antibodies were purchased from GE Healthcare (1:5000, catalogue nos. NA934V and NA931V, respectively; Waukesha, WI). Alexa Fluor–conjugated secondary antibodies were purchased from Invitrogen (Grand Island, NY). Alexa Fluor 594/488–conjugated anti-rabbit/anti-mouse and Alexa Fluor 647–conjugated anti-rat secondary antibodies were from Invitrogen (1:1000, catalogue nos. A-11012 for Alexa Fluor 594-rabbit, A-11034 for Alexa Fluor 488-rabbit, A-11001 for Alexa Fluor 488-mouse, A-11032 for Alexa Fluor 594-mouse, and A-21247 for Alexa Fluor 647-rat).
Cell culture and transfection
HEK293T, COS-7, and HeLa cells were purchased from the American Type Culture Collection (ATCC) and cultured in high-glucose DMEM (HG-DMEM; HyClone, Logan, UT) containing 10% fetal bovine serum (HyClone). All cells were maintained in a humidified incubator supplemented with 5% CO2 at 37°C. Cells were transiently transfected with the indicated short hairpin RNA (shRNA), siRNA, and plasmids using the Lipofectamine 2000 transfection reagent (Life Technologies, Grand Island, NY) following the manufacturer's procedure. All siRNAs were purchased from Dharmacon (GE Healthcare Life Sciences). The specific siRNA and shRNA used were as follows: Arl4A: GGAACUCAUUGUCACUUUCUU and Robo1: GCAGGUACUUG-GAGGAUAU. After transfection, the transfected cells were harvested at 24 or 48 h for overexpression and knockdown, respectively.
Plasmid construction
cDNA sequences of Arl4A/C/D and its mutants were cloned as previously described (Li et al., 2007). Briefly, Arl4A was cloned into the mammalian expression vector pSG5 (Stratagene, La Jolla, CA) to generate untagged Arl4A. The cDNA sequence was subcloned into pET32a (Novagen, Madison, WI) for His-tagged Arl4A expressed in Escherichia coli or pBTM116 for LexA-tagged Arl4A expressed in yeast. Full-length Robo1 and srGAP1 cDNA sequences were amplified from a human fetal brain cDNA library (Clontech) using PCR. Robo1 and srGAP1 were cloned into the mammalian expression vectors pCMV-Tag4A and pcDNA3.1A (Invitrogen), the E. coli expression vector pGEX4T-1, or the yeast expression vector pACT2 (Clontech). Replacement of alanine was accomplished using a two-step PCR technique. All constructs were confirmed by DNA sequencing.
Total protein extraction and immunoblotting analysis
Total proteins were extracted as described elsewhere (Li et al., 2007). The proteins in the lysates were separated by 7.5% or 12.5% SDS–PAGE and transferred to Immobilon-P membranes (Millipore). The membranes were blocked in PBST buffer (0.1% Tween 20 in phosphate-buffered saline [PBS]) containing 5% skim milk powder or bovine serum albumin (BSA) and then incubated with the indicated primary antibodies. After washing, the membranes were incubated in PBST containing the appropriate HRP-conjugated secondary antibody (1:5000). α-Tubulin was used as the internal control for protein loading.
Total RNA isolation, reverse transcription, and quantitative-PCR
Total RNA isolation, reverse transcription, and quantitative-PCR (Q-PCR) were performed as previously described (Chiang et al., 2017). RNA (2 μg) was reverse transcribed into cDNA and then used for Q-PCR with the TaqMan system (Applied Biosystems [ABI], Foster City, CA). The Arl4A primer/probe (Hs01932504-s1; ABI) and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs00266705-g1; ABI) were used.
Yeast two-hybrid interaction assay
The yeast two-hybrid screen was performed as previously described (Li et al., 2007). In brief, the Saccharomyces cerevisiae strain L40 (MATα trp1 leu2 his3 LYS::[lexAop]4-HIS3 URA3::[lexAop]8-lacZ) was used. A human fetal brain cDNA library in pACT2 (Clontech) was screened using Arl4A-Q72L as the bait. Colonies exhibiting histidine auxotrophy were patched onto selective plates and assayed for β-galactosidase activity using a colony-lift filter assay.
Yeast protein extraction
Yeast cells were harvested after 5 d of culture. Yeast protein extraction was performed as described previously (Li et al., 2007), and the extracted proteins were subjected to immunoblotting analysis.
Immunofluorescence staining and quantification
To stain Robo1 and Arl4A, HeLa cells were seeded on coverslips and transfected with the indicated plasmids. After transfection, cells were fixed with 4% paraformaldehyde supplemented with 1 mM CaCl2 and 1 mM MgCl2 for 15 min. Cells were then permeabilized with PBS supplemented with 1 mM CaCl2 and 1 mM MgCl2 containing 0.1% saponin for 10 min followed by blocking in 0.02% Tween-20 PBST containing 1% BSA for 1 h at room temperature. Cells were incubated with either rabbit anti-Arl4A (1:500) or mouse anti-Myc (1:200) and anti-CD44 (1:200) antibodies. After extensive washing, cells were incubated with the corresponding Alexa Fluor–conjugated secondary antibody (1:1000) for at least 1 h. Cells were then mounted in 90% glycerol in PBS containing 1 mg/ml p-phenylenediamine and were visualized using a Carl Zeiss LSM780 system (Carl Zeiss MicroImaging, Jena, Germany).
LSM780 confocal microscope analyses of HeLa cells singly expressing or coexpressing Robo-myc and Arl4A were performed with a 63× oil immersion objective. Midplane optical sections were captured for presentation and quantification. ImageJ2 software was used for intensity and PM/C ratio calculations. We determined the border of the plasma membrane by CD44 signals, which clearly outlined the cell boundary. A 10-pixel-wide freehand Line was drawn along the boundary demarcated by the CD44 signal as a cell margin denoting the plasma membrane. The recorded coordinates were transferred to the Robo-myc detection channel to obtain the intensity of the plasma membrane signal. The freehand selection tool was used to enclose the cytosolic Region along the inner boundary of the CD44 signal inside the cell outline. Similarly, the coordinates were transferred to the Robo-myc detection channel to measure the intensity of the cytosolic signal. The PM/C ratio was calculated using the following equation:
where intensity on the line and intensity in the region are defined in the CD44 detection channel but measured in the Robo-myc detection channel.
In vitro binding assay
His-tagged Arl4A and GST-tagged Robo1 were purified using Ni-NTA resins (Qiagen, Valencia, CA) and glutathione resins (GE Healthcare), respectively. In total, 4 µg of purified His-Arl4A was incubated with 4 µg of various GST-Robo1 fragments in binding assay buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 10% glycerol, 1% Triton X-100, 1 mM NaN3, 1× protease inhibitor cocktail) for at least 2 h at 4°C. Unbound proteins were washed away with assay buffer, and proteins bound to GST beads were then eluted with protein loading buffer for immunoblotting analysis.
Coimmunoprecipitation
Immunoprecipitation was performed according to a previously described protocol with slight modifications (Chiang et al., 2017). Cells were lysed with IP buffer for 30 min on a shaker and cleared by centrifugation at 15,000 × g for 10 min at 4°C. The IP buffer for immunoprecipitation of the Robo1-srGAP1 complex comprised 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1% NP40, 10 mM NaF, 1 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail. The IP buffer for immunoprecipitation of the Robo1-Arl4A complex comprised 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP40, and 1× protease inhibitor cocktail. The cell supernatants were incubated with 20 µl of anti-Flag M2 magnetic beads (Sigma-Aldrich) for 2 h at 4°C and then extensively washed with IP buffer at least five times. The immunoprecipitated proteins were analyzed by immunoblotting.
Preparation and purification of Slit2
COS-7 cells were transiently transfected with the empty pSecTag2 vector or the pSecTag 2-human Slit2 plasmid. Purification was performed as previously described (Jones et al., 2008). Finally, ∼500 µl of the concentrated Slit2 solution was collected (Jones et al., 2008). Concentrated Slit2 was analyzed by Coomassie Blue staining and stored at 4°C for up to 1 wk. In addition, we prepared the empty pSecTag2 vector as the “mock” and used it as the negative control in all the experiments.
Cdc42 activity pull-down assay
Cells were transfected with the indicated plasmids and then lysed in Cdc42 activity pull-down assay buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1 mM DTT, 1% NP-40, 5 mM MgCl2, 5% glycerol, and 1× protease inhibitor cocktail). Then, the cells were incubated with purified PAK-PBD-GST beads for 2 h at 4°C. The beads were washed with assay buffer, and bound proteins were eluted with protein loading buffer for immunoblotting analysis.
Migration assay
Wound healing migration assays were performed as described previously (Chiang et al., 2017). Cells were observed using a time-lapse microscope (Axiovert 200M; Carl Zeiss MicroImaging) equipped with a temperature and CO2 controller. The cell migration capacity was determined as described previously using ImageJ software (Chiang et al., 2017).
In addition, Transwell cell migration assays were performed using the Boyden chamber assay method as previously described (Chiang et al., 2017). Cells were serum-starved for 16 h before being seeded into the upper chamber. For Slit2 treatment experiments, the indicated concentrations of purified Slit2 and the vector control were added to the lower chamber. After the assay, the migrated cells were stained with 1% crystal violet and images were captured using a phase-contrast microscope (Eclipse TS-100; Nikon, Melville, NY) equipped with a digital camera (DS-5M; Nikon).
Statistical analysis
All data are presented as the mean ± SD, and P values were calculated by one-way analysis of variance (ANOVA) followed by Dunnett's post hoc multiple comparison test or two-tailed Student's t test. Significant statistical differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) are indicated. For each independent in vitro experiment, three biological replicates were used.
Supplementary Material
Acknowledgments
We thank Randy Haun, Chia-Jung Yu, Ya-Wen Liu, and Pei-Hsin Huang for their critical review of this article. This work was supported by Grant no. NHRI-EX106-10601B1 from the National Health Research Institutes (NHRI) in Taiwan to F.-J.S.L.
Abbreviations used:
- Arf
ADP-ribosylation factor
- Arl
ADP-ribosylation factor–like
- GDP
guanosine-5’-diphosphate
- GTP
guanosine-5’-triphosphate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-01-0001) on November 14, 2018.
REFERENCES
- Andrews W, Liapi A, Plachez C, Camurri L, Zhang J, Mori S, Murakami F, Parnavelas JG, Sundaresan V, Richards LJ. (2006). Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development , 2243–2252. [DOI] [PubMed] [Google Scholar]
- Ballard MS, Hinck L. (2012). A roundabout way to cancer. Adv Cancer Res , 187–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. (1999). Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell , 795–806. [DOI] [PubMed] [Google Scholar]
- Chavrier P, Menetrey J. (2010). Toward a structural understanding of arf family:effector specificity. Structure , 1552–1558. [DOI] [PubMed] [Google Scholar]
- Chiang TS, Wu HF, Lee FS. (2017). ADP-ribosylation factor–like 4C binding to filamin-A modulates filopodium formation and cell migration. Mol Biol Cell , 3013–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. (2002). Molecular mechanisms of axon guidance. Science , 1959–1964. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol , 347–358. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. (2004). Cdc42—the centre of polarity. J Cell Sci , 1291–1300. [DOI] [PubMed] [Google Scholar]
- Hinck L. (2004). The versatile roles of “axon guidance” cues in tissue morphogenesis. Dev Cell , 783–793. [DOI] [PubMed] [Google Scholar]
- Huang L, Xu Y, Yu W, Li Y, Chu L, Dong J, Li X. (2010). Effect of Robo1 on retinal pigment epithelial cells and experimental proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci , 3193–3204. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Schilf C, Fliegert F, Koling S, Weber Y, Schurmann A, Joost HG. (1999). ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterization by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett , 384–388. [DOI] [PubMed] [Google Scholar]
- Jones CA, London NR, Chen H, Park KW, Sauvaget D, Stockton RA, Wythe JD, Suh W, Larrieu-Lahargue F, Mukouyama YS, et al. (2008). Robo4 stabilizes the vascular network by inhibiting pathologic angiogenesis and endothelial hyperpermeability. Nat Med , 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice ED, Barnum SJ, Kidd T. (2017). The WAGR syndrome gene PRRG4 is a functional homologue of the commissureless axon guidance gene. PLoS Genet , e1006865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justus CR, Leffler N, Ruiz-Echevarria M, Yang LV. (2014). In vitro cell migration and invasion assays. J Vis Exp , e51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H, Hiramoto K, Negishi M. (2006). Activation of Rac1 by RhoG regulates cell migration. J Cell Sci , 56–65. [DOI] [PubMed] [Google Scholar]
- Keleman K, Rajagopalan S, Cleppien D, Teis D, Paiha K, Huber LA, Technau GM, Dickson BJ. (2002). Comm sorts robo to control axon guidance at the Drosophila midline. Cell , 415–427. [DOI] [PubMed] [Google Scholar]
- Khusial PR, Vadla B, Krishnan H, Ramlall TF, Shen Y, Ichikawa H, Geng JG, Goldberg GS. (2010). Src activates Abl to augment Robo1 expression in order to promote tumor cell migration. Oncotarget , 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF. (2008). Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev , 489–513. [DOI] [PubMed] [Google Scholar]
- Legg JA, Herbert JM, Clissold P, Bicknell R. (2008). Slits and Roundabouts in cancer, tumour angiogenesis and endothelial cell migration. Angiogenesis , 13–21. [DOI] [PubMed] [Google Scholar]
- Li CC, Chiang TC, Wu TS, Pacheco-Rodriguez G, Moss J, Lee FJ. (2007). ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol Biol Cell , 4420–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Chiang TC, Liu YT, Tsai YT, Jang LT, Lee FJ. (2011). ARL4A acts with GCC185 to modulate Golgi complex organization. J Cell Sci , 4014–4026. [DOI] [PubMed] [Google Scholar]
- Lin CY, Huang PH, Liao WL, Cheng HJ, Huang CF, Kuo JC, Patton WA, Massenburg D, Moss J, Lee FJ. (2000). ARL4, an ARF-like protein that is developmentally regulated and localized to nuclei and nucleoli. J Biol Chem , 37815–37823. [DOI] [PubMed] [Google Scholar]
- Lin CY, Li CC, Huang PH, Lee FJ. (2002). A developmentally regulated ARF-like 5 protein (ARL5), localized to nuclei and nucleoli, interacts with heterochromatin protein 1. J Cell Sci , 4433–4445. [DOI] [PubMed] [Google Scholar]
- Myat A, Henry P, McCabe V, Flintoft L, Rotin D, Tear G. (2002). Drosophila Nedd4, a ubiquitin ligase, is recruited by Commissureless to control cell surface levels of the roundabout receptor. Neuron , 447–459. [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Renault L, Cherfils J. (2002). Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for “front-back” communication. EMBO Rep , 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M, Chiang T-C, Tran V, Lee F-JS, Côté J-F. (2011). The Arf family GTPase Arl4A complexes with ELMO proteins to promote actin cytoskeleton remodeling and reveals a versatile Ras-binding domain in the ELMO proteins family. J Biol Chem , 38969–38979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp M, Niederkofler V, Debrunner M, Alther T, Kunz B, Stoeckli ET. (2012). RabGDI controls axonal midline crossing by regulating Robo1 surface expression. Neural Dev , 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Paruchuri V, Preet A, Latif F, Ganju RK. (2008). Slit-2 induces a tumor-suppressive effect by regulating β-catenin in breast cancer cells. J Biol Chem , 26624–26633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. (2004). Cell migration: Rho GTPases lead the way. Dev Biol , 23–32. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. (2015). Rho GTPase signalling in cell migration. Curr Opin Cell Biol , 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy LC, Ravichandran KS, Casanova JE. (2005). The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol , 1749–1754. [DOI] [PubMed] [Google Scholar]
- Schmid BC, Rezniczek GA, Fabjani G, Yoneda T, Leodolter S, Zeillinger R. (2007). The neuronal guidance cue Slit2 induces targeted migration and may play a role in brain metastasis of breast cancer cells. Breast Cancer Res Treat , 333–342. [DOI] [PubMed] [Google Scholar]
- Schurmann A, Breiner M, Becker W, Huppertz C, Kainulainen H, Kentrup H, Joost HG. (1994). Cloning of two novel ADP-ribosylation factor-like proteins and characterization of their differential expression in 3T3-L1 cells. J Biol Chem , 15683–15688. [PubMed] [Google Scholar]
- Stella MC, Trusolino L, Comoglio PM. (2009). The Slit/Robo system suppresses hepatocyte growth factor-dependent invasion and morphogenesis. Mol Biol Cell , 642–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S, Mukovozov IM, Huang YW, Magalhaes MA, Yan M, Crow MR, Liu GY, Sun CX, Durocher Y, Glogauer M, et al. (2009). The axonal repellent, Slit2, inhibits directional migration of circulating neutrophils. J Leukoc Biol , 1403–1415. [DOI] [PubMed] [Google Scholar]
- Tseng RC, Lee SH, Hsu HS, Chen BH, Tsai WC, Tzao C, Wang YC. (2010). SLIT2 attenuation during lung cancer progression deregulates β-catenin and E-cadherin and associates with poor prognosis. Cancer Res , 543–551. [DOI] [PubMed] [Google Scholar]
- Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, et al. (2001). Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell , 209–221. [DOI] [PubMed] [Google Scholar]
- Wu JY, Feng L, Park HT, Havlioglu N, Wen L, Tang H, Bacon KB, Jiang Z, Zhang X, Rao Y. (2001). The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature , 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ML, Gonda Y, Mommersteeg MT, Barber M, Ypsilanti AR, Hanashima C, Parnavelas JG, Andrews WD. (2014). Robo1 modulates proliferation and neurogenesis in the developing neocortex. J Neurosci , 5717–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiin JJ, Hu B, Jarzynka MJ, Feng H, Liu KW, Wu JY, Ma HI, Cheng SY. (2009). Slit2 inhibits glioma cell invasion in the brain by suppression of Cdc42 activity. Neuro-oncology , 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhou FL, Li WP, Wang J, Wang LJ. (2016). Slit2Robo1 signaling promotes the adhesion, invasion and migration of tongue carcinoma cells via upregulating matrix metalloproteinases 2 and 9, and downregulating Ecadherin. Mol Med Rep , 1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.