Abstract
Reorganization of the actin cytoskeleton is crucial for cellular processes, including cytokinesis and cell–cell junction remodeling. Formins are conserved processive actin-polymerizing machines that regulate actin dynamics by nucleating, elongating, and bundling linear actin filaments. Because the formin family is large, with at least 15 members in vertebrates, there have not been any comprehensive studies examining formin localization and function within a common cell type. Here, we characterized the localization of all 15 formins in epithelial cells of Xenopus laevis gastrula-stage embryos. Dia1 and Dia2 localized to tight junctions, while Fhod1 and Fhod3 localized to adherens junctions. Only Dia3 strongly localized at the cytokinetic contractile ring. The Diaphanous inhibitory domain–dimerization domain (DID-DD) region of Dia1 was sufficient for Dia1 localization, and overexpression of a Dia1 DID-DD fragment competitively removed Dia1 and Dia2 from cell–cell junctions. In Dia1 DID-DD–overexpressing cells, Dia1 and Dia2 were mislocalized to the contractile ring, and cells exhibited increased cytokinesis failure. This work provides a comprehensive analysis of the localization of all 15 vertebrate formins in epithelial cells and suggests that misregulated formin localization results in epithelial cytokinesis failure.
INTRODUCTION
Epithelial cells cover the external and internal surface of the vertebrate body and are instrumental in maintaining homeostasis by separating distinct compartments of the body. Apical cell–cell junctions consist of tight junctions (TJs), adherens junctions (AJs), and desmosomes. AJs and desmosomes mechanically connect adjacent epithelial cells and contribute to maintenance of cell shape and tissue integrity (Hartsock and Nelson, 2008; Nekrasova and Green, 2013; Takeichi, 2014; Lecuit and Yap, 2015). TJs regulate the passage of fluids and solutes via the paracellular pathway and serve as a barrier (Hartsock and Nelson, 2008; Krug et al., 2014; Van Itallie and Anderson, 2014; Zihni et al., 2016).
Because epithelial tissues are continuously renewed, new cells must be generated by cell division, which is especially evident in the developing epithelium. Despite the drastic cell shape changes that occur during cytokinesis, cell–cell junctions must maintain cell–cell adhesion and barrier function during cell division. Although an understanding of how cell–cell junctions are maintained during cytokinesis is beginning to emerge (Higashi et al., 2016), how epithelial cells distinguish and coordinate the signaling mechanisms regulating contractile actomyosin arrays at cell–cell junctions and the cytokinetic contractile ring remains unclear.
Both cell–cell junctions and cytokinetic contractile rings are regulated by Rho GTPases (Kishi et al., 1993; Mabuchi et al., 1993; Nusrat et al., 1995; Braga et al., 1997; Miller, 2011; Arnold et al., 2017). RhoA switches between an active GTP-bound state and an inactive GDP-bound state. When RhoA is in the active GTP-bound state, it binds to and activates its effectors, including ROCKs/Rho kinases (Ishizaki et al., 1996; Matsui et al., 1996) and formins (Kohno et al., 1996; Watanabe et al., 1997; Alberts et al., 1998). It is not clear how RhoA effectors differentially regulate formation and maintenance of both RhoA-dependent junctional actomyosin bundles and cytokinetic actomyosin rings within the same cells. To address this question, we investigated the localization and functional roles of formins at epithelial cell–cell junctions and cytokinetic contractile rings in a developing vertebrate model system, the gastrula-stage Xenopus laevis embryo.
Formins constitute a family of actin regulators that is conserved among eukaryotes (Higgs and Peterson, 2005; Rivero et al., 2005; Chalkia et al., 2008). Formins mediate linear actin assembly through their formin homology (FH) 1 and FH2 domains. The FH1 domain recruits profilin-bound actin monomers and passes them to the FH2 domain. The FH2 domain directly binds to and caps the barbed end of actin filaments and simultaneously adds new actin monomers to the barbed end, which results in continuous elongation of F-actin at the barbed end (Pruyne, Evangelista, et al., 2002; Kovar et al., 2003). Vertebrate genomes have at least 15 formins (Higgs and Peterson, 2005; Rivero et al., 2005; Chalkia et al., 2008) (see Supplemental Figure S2). Among them, 10 formins (Dia1/2/3, Daam1/2, Fmnl1/2/3, Fhod1/3) are classified as Diaphanous-related formins (DRFs) (Alberts, 2002; Kuhn and Geyer, 2014). DRFs share several important regulatory domains in addition to the FH1 and FH2 domains. Binding of the Diaphanous inhibitory domain (DID) (Li and Higgs, 2005), which is located on the N-terminal (NT) side of the FH1/FH2 domains, to the Diaphanous autoinhibitory domain (DAD) (Alberts, 2001), which is located on the C-terminal (CT) side of the FH1/FH2 domains, keeps the actin-assembly activity of FH1-FH2 domains suppressed (Watanabe et al., 1999; Li and Higgs, 2003). At their NT end, DRFs have a GTPase-binding domain (GBD) (Watanabe et al., 1997; Otomo et al., 2005a; Rose, Weyand, Lammers, et al., 2005). Binding of active Rho GTPases to the GBD (and part of the DID) releases the DID-DAD autoinhibitory interaction (Watanabe et al., 1999; Lammers et al., 2005; Nezami, Poy, and Eck, 2006). Additional factors can cooperate to release DID-DAD interactions, including Anillin binding to DID for mDia2 (Dia2, also known as DIAPH3, Diap3, or DRF3) (Watanabe et al., 2010), Flightless-I binding to DAD for mDia1 and Daam1 (Higashi, Ikeda, et al., 2010), and phosphorylation of DAD by ROCK for Fhod1 (Takeya et al., 2008) and mDia2 (Staus et al., 2011). Unleashing DID-DAD autoinhibition opens up DRF molecules, making the FH1-FH2 domains accessible to bind actin.
Formins have been implicated in the regulation of cell–cell junctions (for a review, see Grikscheit and Grosse, 2016). For example, mammalian Dia1 (mDia1, also known as DIAPH1 or DRF1) has been shown to localize at AJs and regulate the stability and contractility of AJs in many cell types (Sahai and Marshall, 2002; Carramusa et al., 2007; Ryu et al., 2009; Rao and Zaidel-Bar, 2016; Acharya et al., 2017). Fmnl3 is implicated in the regulation of AJs through F-actin polymerization and stabilization of E-cadherin in migrating mouse mammary epithelial EpH4 cells (Rao and Zaidel-Bar, 2016). A three-dimensional culture model of human breast epithelial MCF10A cells showed that Fmnl2 was involved in the formation of new cell–cell junctions between daughter cells downstream of Rac1 (Grikscheit et al., 2015). Finally, several studies indicate a role for formins in regulating cell–cell adhesion downstream of Rho during developmental processes. Drosophila Diaphanous regulates junctional Myosin II levels and activity and is required for properly regulated junctional stability and cell movements during morphogenesis (Homem and Peifer, 2008). Drosophila Diaphanous can also control E-cadherin endocytosis downstream of Rho, thus regulating the level of E-cadherin at the cell–cell junction (Levayer et al., 2011). Additionally, actin-based pushing controlled by Fmn1 acting downstream of RhoA drives apical emergence of new multiciliated epithelial cells in developing X. laevis embryos (Sedzinski, Hannezo, et al., 2016); however, how this specialized actin network is linked to junctions and whether Fmn1 regulates cell–cell junctions in this setting is not clear.
Formins are also known regulators of cytokinesis (for a review, see Bohnert et al., 2013). Fission yeast formin Cdc12 is concentrated at medial nodes and mediates formation and maintenance of contractile rings (Chang et al., 1997; Kovar et al., 2003; Wu et al., 2006). In budding yeast, two formins, Bni1p and Bnr1p, are required for successful cytokinesis (Imamura et al., 1997; Tolliday et al., 2002). Caenorhabditis elegans CYK-1 and Drosophila Diaphanous are required for early embryonic divisions (Castrillon and Wasserman, 1994; Severson et al., 2002). Although mDia1, 2, and 3 are all orthologues of CYK-1 and Diaphanous, only one vertebrate formin, mDia2, has been shown to control cytokinesis. mDia2 is localized at contractile rings in mouse NIH 3T3 fibroblasts, and knockdown of mDia2 caused cytokinesis failure in NIH 3T3 cells (Watanabe et al., 2008). Additionally, mDia2 knockout mice are embryonic lethal due to cytokinesis failure in fetal erythroblasts, which results in severe anemia (Watanabe et al., 2013). Because the nomenclature of Dia group formins is frequently confused between human and mouse genes (e.g., the human orthologue of mouse mDia2 [DIAPH3] is called hDIA3, DRF3, and DIAPH3), we consistently use Dia1 (mDia1 in mice, DIAPH1 in humans), Dia2 (mDia2 in mice, DIAPH3 in humans), and Dia3 (mDia3 in mice, DIAPH2 in humans) for X. laevis genes in this paper.
To date, there has been no comprehensive study of all 15 vertebrate formins in the same model system. Furthermore, it is unclear whether any formin(s) are involved in the regulation of both cell–cell junctions and cytokinetic contractile rings, or whether these two actomyosin-based structures actively influence each other through the regulation of formin proteins. Here, we cloned the 15 formins from X. laevis and characterized their localization in epithelial cells. We identified Dia1 and Dia2 as cell–cell junction localizing formins and found that perturbing the junctional localization of Dia1 and Dia2 resulted in a cytokinesis defect.
RESULTS
Xenopus laevis has 15 formins conserved among vertebrates
To characterize which formin(s) are involved in the regulation of cell–cell junctions and contractile ring formation, we cloned all X. laevis formins. Each of the 15 formins identified in mouse and human (Higgs and Peterson, 2005; Rivero et al., 2005) was conserved in X. laevis (Supplemental Figures S1 and S2). We examined the expression level of each formin transcript using cDNA libraries from embryos at multiple X. laevis developmental stages (Supplemental Figure S3). Each formin showed a different expression pattern. In gastrula-stage embryos, which are covered with a proliferating polarized epithelial cell sheet that serves as a model for intact epithelial tissue, at least 10 formins, including Dia1, Dia2, Dia3, Daam1, Fmnl3, Inf1, Inf2, Fmn2, Fhod1, and Fhod3, are expressed.
Dia3 is localized at cytokinetic contractile rings
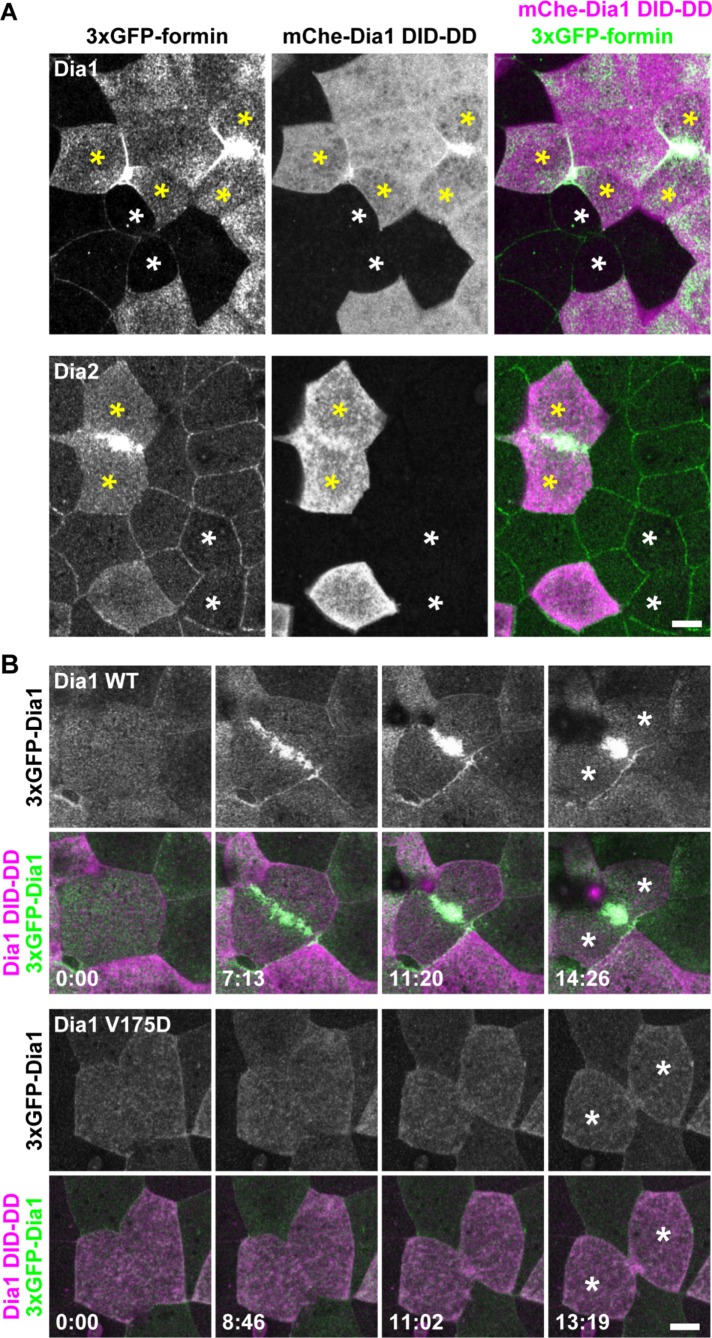
To characterize the localization of the formins, we used three green fluorescent protein (3×GFP) tags on the NT end of each formin. The expression of the tagged formins was examined by Western blot of gastrula-stage embryos (Supplemental Figure S4), and all tagged formins were detected at the expected size. Next, we coexpressed the 3×GFP-tagged formins with monomeric red fluorescent protein- (mRFP-)-ZO-1 (TJ probe) and examined the localization of the formins in gastrula-stage X. laevis embryos by confocal microscopy (Figure 1A). Among the 15 formins, only 3×GFP-Dia3 (also known as DIAPH2 or DRF2) exhibited strong localization at cytokinetic contractile rings. Dia1 and Dia2 showed very weak signal at contractile rings, and the other formins exhibited no specific signal at the division site (unpublished data). Because the contractile ring is templated by a Rho activity zone (Miller, 2011) and Dia3 can bind Rho via its NT GBD domain (Yasuda et al., 2004), we tested whether the localization of Dia3 at the cytokinetic contractile ring is dependent on Rho binding. Rho binding–deficient mutant V187D (analogous to the V161D mutation in Dia1 [Otomo et al., 2005a]) could not localize at the contractile ring during cytokinesis (Figure 1B), indicating that Dia3 is recruited to the contractile ring by active Rho GTPase. These data demonstrate that, in the X. laevis gastrula epithelium, Dia3 is the only formin strongly localized at the contractile ring.
FIGURE 1:
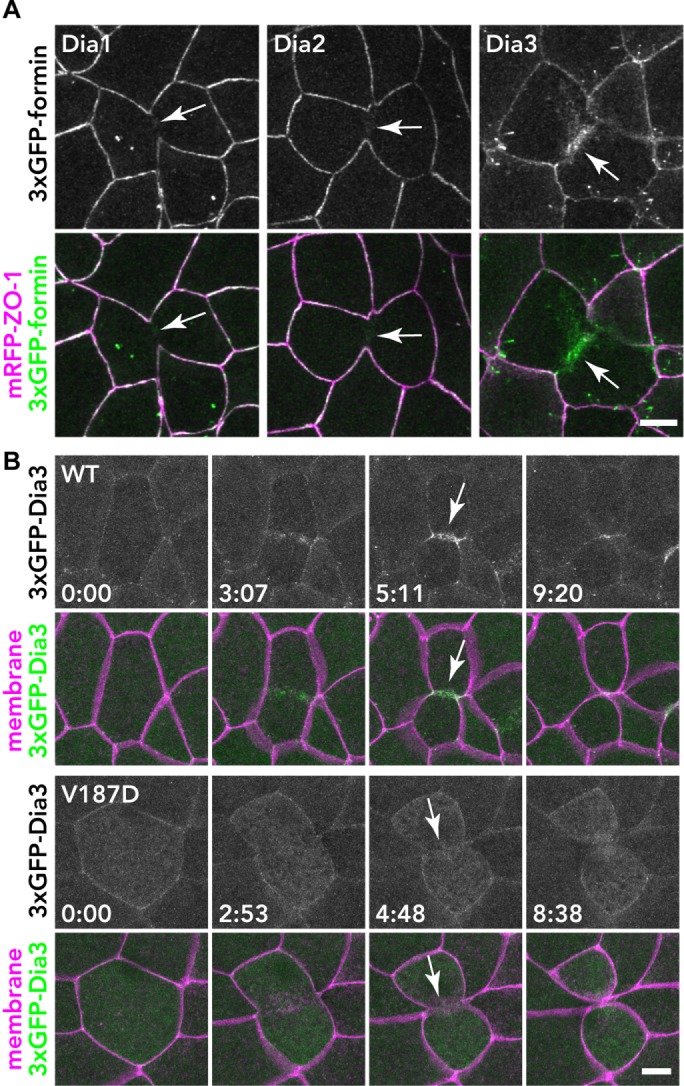
Localization of 3×GFP-tagged Dia1, Dia2, and Dia3 in the X. laevis gastrula epithelium. (A) Embryos expressing 3×GFP-tagged Dia1, Dia2, or Dia3 (green) and mRFP-ZO-1 (TJ marker; magenta) were live imaged using confocal microscopy; z-stack images of formin alone (top panels) and merged with mRFP-ZO-1 (bottom panels) are shown. Note that Dia3 is strongly localized at the contractile ring of the dividing cell. (B) The localization of Dia3 at the contractile ring is dependent on Rho binding. Embryos expressing 3×GFP-Dia1 WT or V187D (Rho-binding mutant; green) and mCherry-farnesyl (membrane probe; magenta) were imaged. Because the expression of Dia3 causes membrane deformation phenotypes (see A and Supplemental Figure S6), Dia3 was expressed at a lower level in these images. Note that the Dia3 V187D mutant cannot localize at the contractile ring. Scale bars: 10 µm.
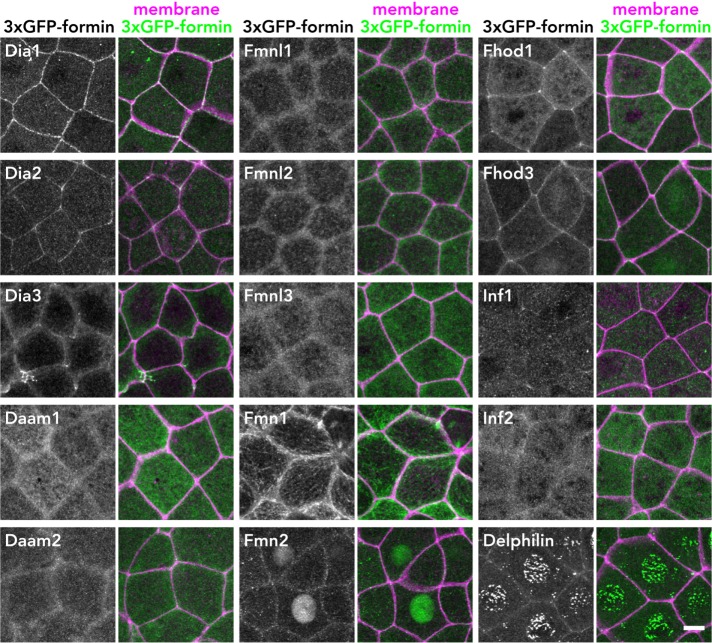
Dia1, Dia2, Fhod1, and Fhod3 are localized at cell–cell junctions
We next examined the localization of the tagged formins in the X. laevis gastrula-stage epithelium during interphase (Figure 2). Among the 15 formins, Dia1 and Dia2 exhibited strong localization at cell–cell junctions, consistent with previous reports (Sahai and Marshall, 2002; Carramusa et al., 2007; Ryu et al., 2009; Acharya et al., 2017). In addition, Fhod1 and Fhod3 also showed strong junctional localization, while Dia3, Fmn2, and Delphilin were weakly localized at cell–cell junctions. The other formins exhibited diffuse or no junctional localization (Figure 2). We quantified the extent of formin junction confinement by measuring fluorescence intensity at junctions along the x- and z-axes (Supplemental Figure S5). We conclude that Dia1, Dia2, Fhod1, and Fhod3 are localized at cell–cell junctions in epithelial cells.
FIGURE 2:
Localization of 3×GFP-tagged formin proteins in X. laevis gastrula-stage epithelium. Embryos expressing 3×GFP-tagged formin (green) and mCherry-farnesyl (membrane marker; magenta) were live imaged using confocal microscopy. Stacked images of formin alone (left panels) and merged with membrane (right panels) are shown. Note that Dia1, Dia2, Fhod1, and Fhod3 are strongly localized at cell–cell junctions. Scale bar: 10 µm.
Some formins showed interesting localizations to specific nonjunctional structures (Figure 2). For example, Fmn1 (also known as Formin-1) appeared to be localized on microtubules, which is consistent with the report that Fmn1 has a microtubule-binding sequence (Zhou et al., 2006). Fmn2 was localized primarily at the nucleus, and Delphilin also exhibited nuclear localization with a punctate pattern. Of note, exogenous expression of Dia3 induced apical, actin-rich structures at cell–cell junctions, which resemble filopodia-like or lamellipodia-like protrusions (Figure 2 and Supplemental Figure S6). The formation of these structures was not observed in Dia3 V187D (Rho binding–deficient mutant)-expressing cells or Dia3 I715A (analogous to F-actin binding– and actin assembly–deficient mutation in yeast Bni1p [Xu et al., 2004; Otomo et al., 2005b]) (Supplemental Figure S6), suggesting that these structures are formed through activation of Dia3 by a Rho GTPase and are dependent on Dia3's actin-assembly activity.
Dia1 and Dia2 are localized at TJs, and Fhod1 and Fhod3 are localized at AJs
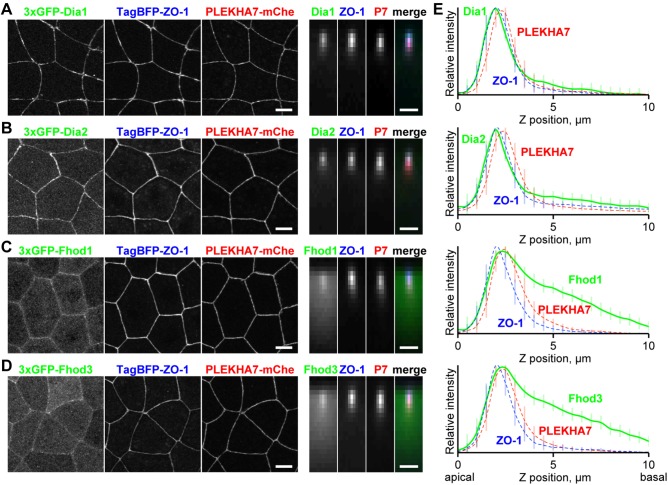
To further examine the precise localization of the junctional formins, we performed triple labeling of the 3×GFP-tagged formins with TJ- and AJ-localizing protein probes and quantified the relative fluorescence intensity along the z-axis. We used monomeric blue fluorescent protein- (TagBFP-)-ZO-1 (Anderson et al., 1988) and PLEKHA7-mCherry (Meng et al., 2008; Pulimeno et al., 2010) as TJ and AJ probes, respectively. First, we confirmed that the selected probes faithfully label the TJ and AJ and can be separated along the z-axis using our quantification approach by coexpressing TagBFP-ZO-1 and PLEKHA7-mCherry with well-characterized TJ (Claudin-GFP) and AJ (E-cadherin-3×GFP) components (Supplemental Figure S7A). Next, we expressed 3×GFP-Dia1 with the TJ and AJ probes and found that the 3×GFP-Dia1 peak overlapped with the TJ peak (Figure 3, A and E). Similarly, 3×GFP-Dia2 also exhibited its highest intensity at the TJ (Figure 3, B and E). In contrast, the Fhod1 and Fhod3 peaks overlapped with the AJ peak and exhibited more basolateral signal than Dia1 or Dia2 (Figure 3, C–E). These data indicate that Dia1 and Dia2 are localized at TJs, while Fhod1 and Fhod3 are localized at AJs in the X. laevis gastrula epithelium.
FIGURE 3:
Comparison of the localization of junctional formins with TJ and AJ proteins. (A–D) 3×GFP-tagged Dia1 (A), Dia2 (B), Fhod1 (C), and Fhod3 (D) (green) were expressed together with TagBFP-ZO-1 (TJ marker; blue) and PLEKHA7-mCherry (AJ marker; red) in gastrula-stage X. laevis embryos and imaged by confocal microscopy. Top views (left) and averaged side views (right) of cell–cell junctions (50 pixels × 16 junctions) are shown. Scale bars: 10 µm (left); 2 µm (right). (E) Graphs indicate intensity profiles of formins (green solid line), ZO-1 (blue dotted line), and PLEKHA7 (red dotted line). Note that the intensity profiles of Dia1 and Dia2 are very similar to that of ZO-1 and that Fhod1 and Fhod3 have a peak at AJs as well as signal basal to the AJ. Error bars (vertical lines) indicate SD.
Both TJs and AJs are dependent on circumferential F-actin bundles. We applied our quantification approach to examine which type of junction—TJ or AJ—is more enriched with F-actin in the X. laevis embryonic epithelium. We compared the localization of Lifeact-GFP (F-actin probe) with TagBFP-ZO-1 (TJ probe) and PLEKHA7-mCherry (AJ probe). Surprisingly, Lifeact-GFP exhibited a peak at the ZO-1 peak and not the PLEKHA7 peak, suggesting that TJ-associated F-actin is more abundant than AJ-associated F-actin (Supplemental Figure S7B). We confirmed this observation by staining fixed embryos with phalloidin (F-actin probe) as well as anti-ZO-1 (TJ protein) and anti–β-catenin (AJ protein) antibodies. The phalloidin peak overlapped with the ZO-1 peak, whereas the phalloidin signal was declining at the Z-position of the β-catenin peak (Supplemental Figure S7C), further demonstrating that TJ-associated F-actin is more prominent in the X. laevis gastrula epithelium.
The DID-DD region is necessary and sufficient for the localization of Dia1 at cell–cell junctions
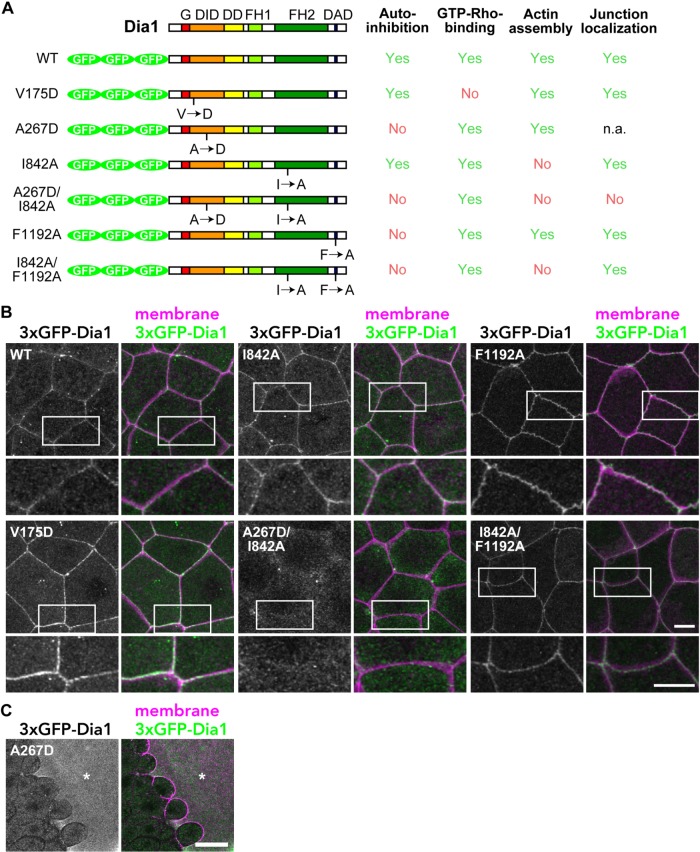
To dissect the domains responsible for junction localization of Dia1, we made several mutants of Dia1 and examined their localization (Figure 4). Because the TJ is known to be the site of a Rho activity zone (Terry et al., 2011; Ratheesh, Gomez, et al., 2012; Arnold et al., 2017) and Dia1 can bind to active Rho via the GBD (Watanabe et al., 1997, 1999), we first asked whether binding to active Rho is required for junctional localization of Dia1. A V161D mutation in the DID domain of mouse mDia1 was shown to completely abolish binding to active RhoA but did not affect DID-DAD–mediated autoinhibition (Otomo et al., 2005a). Because all of the residues at the surface of the Rho-Dia1 interface are conserved between mouse and X. laevis (Supplemental Figure S8), we introduced a corresponding mutation, V175D, into X. laevis Dia1 (Figure 4A) and examined its localization. The 3×GFP-Dia1 V175D was still localized at cell–cell junctions (Figure 4B) similar to wild-type (WT) Dia1 (Figure 4B), suggesting that active Rho binding is not required for the junctional localization of Dia1.
FIGURE 4:
Ala-267 is necessary for junctional localization of Dia1. (A) Domain structure and mutants of Dia1. G, GTPase-binding domain; DID, Diaphanous inhibitory domain; DD, dimerization domain; FH, formin homology; DAD, Diaphanous autoregulatory domain. (B) Embryos expressing Dia1 WT or mutants (green) and mCherry-farnesyl (membrane probe; magenta) were observed. Note that the A267D/I842A mutant cannot localize at cell–cell junctions, but the I842A mutant can. Enlargements of cell–cell junctions (white boxes) are shown below. (C) Embryo expressing Dia1 A267D (green) and mCherry-farnesyl (magenta). Note that the cell expressing Dia1 A267D at high level (asterisk) is enlarged, probably due to cytokinesis failure. Scale bars: 10 µm (B); 40 µm (C).
Next, we asked whether the autoinhibited closed state is required for Dia1's junctional localization. An A256D mutation in the DID domain of mouse mDia1 completely disrupts DID-DAD interaction and keeps the mDia1 molecule constitutively open, while it does not affect the binding to active RhoA (Otomo et al., 2005a). We made an analogous A267D mutant of X. laevis Dia1 (Figure 4A and Supplemental Figure S8). Expression of 3×GFP-Dia1 A267D resulted in abnormally large cell size (Figure 4C), which is likely a result of cytokinesis failure. We hypothesized that this phenotype was due to aberrant actin assembly by the constitutively active Dia1 A267D mutant. To test this possibility, we introduced another mutation in the FH2 domain, I842A (Figure 4A), which is equivalent to the I1431A mutation, an F-actin binding– and actin assembly–deficient mutation in yeast Bni1p (Xu et al., 2004; Otomo et al., 2005b). Indeed, Dia1 A267D/I842A–expressing cells exhibit normal cell size (Figure 4B), demonstrating that the large cell size of Dia1 A267D–expressing cells is due to actin assembly by constitutively open Dia1. Because it was difficult to assess the localization of Dia1 A267D at cell–cell junctions due to compromised cell shape and size, we used Dia1 A267D/I842A to test the effect of the A267D mutation on the localization of Dia1. Dia1 A267D/I842A was not localized at cell–cell junctions (Figure 4B), suggesting that the closed form of Dia1 or the Ala-267 residue itself is important for the localization. Of note, the I842A mutation alone does not affect the localization of Dia1 (Figure 4B), suggesting that actin-assembly activity is not required for junctional localization of Dia1.
To tease apart the two possibilities—that the closed-form conformation of Dia1 or the Ala-267 residue itself is required for Dia1 junctional localization—we introduced a mutation in the DAD domain as an alternate way to disrupt the DID-DAD interaction (instead of the A267D mutation). An F1195A mutation in the DAD domain of mouse mDia1 completely abolishes the affinity to DID (Lammers et al., 2005). A corresponding F1192A mutant of X. laevis Dia1 did not cause large cells and was localized at cell–cell junctions (Figure 4B and Supplemental Figure S8), suggesting that the closed form is not required for the localization of Dia1 at cell–cell junctions. Because Dia1 F1192A remained localized at cell–cell junctions, its aberrant actin-assembly activity might be restricted to the vicinity junctions, thus still allowing the actin polymerization required for cytokinesis. This might explain why Dia1 F1192A did not cause cytokinesis defects, whereas Dia1 A267A, which also disrupts the DID-DAD interaction, did. Instead, the cell–cell junctions of Dia1 F1192A–expressing cells were abnormally rippled compared with Dia1 WT–expressing cells, supporting the view that Dia1 F1192A–dependent actin assembly occurs near cell–cell junctions. Furthermore, when we added the I842A actin assembly–deficient mutation to make a Dia1 I842A/F1192A double mutant, this mutant was also localized at junctions, but did not cause the rippled junction phenotype (Figure 4B). Taken together, these results lead us to conclude that the Ala-267 residue in the DID domain is necessary for Dia1's localization at cell–cell junctions and that Dia1's localization is not affected by active Rho binding, actin-assembly activity, or the open/closed state of Dia1.
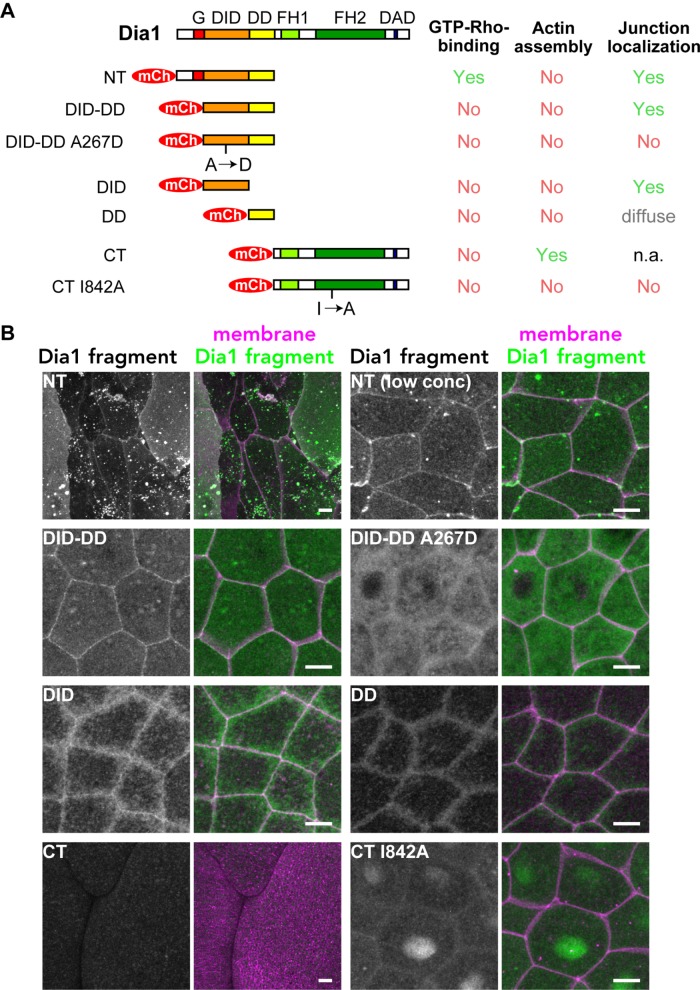
To test which region of Dia1 is sufficient for the localization at cell–cell junctions, we made fragments of Dia1 (Figure 5A). First, we split the molecule into NT and CT halves, which contain G-DID-DD and FH1-FH2-DAD, respectively. When mCherry-Dia1 NT was expressed in epithelial cells, it induced an abnormally large cell size phenotype (Figure 5B), which is likely caused by cytokinesis failure. Because altered cell size and shape prevented us from examining the localization of the NT construct, we reduced the expression level of NT to suppress the cell size phenotype. When mCherry-Dia1 NT was expressed at a low level, it was clearly localized at cell–cell junctions (Figure 5B), suggesting that NT is sufficient to recruit Dia1 to junctions. mCherry-Dia1 CT also caused abnormally large cells. This mutant was very potent, even at reduced expression levels. Therefore, we introduced the actin assembly–deficient I842A mutation, which suppressed the cell size phenotype, but mCherry-Dia1 CT I842A was not localized at junctions (Figure 5B), indicating that the CT does not have the junction-localization signal. Similar to Dia1, we found that, for both Fhod1 and Fhod3, an NT fragment was localized at cell–cell junctions, while a CT fragment was not (Supplemental Figure S9). To narrow down the Dia1 region responsible for junction localization, we made an mCherry-Dia1 DID-DD fragment. mCherry-Dia1 DID-DD was clearly localized at cell–cell junctions (Figure 5B), which indicates that Rho binding is dispensable for junctional localization. In contrast, mCherry-Dia1 DID-DD A267D was not localized at cell–cell junctions (Figure 5B). Finally, we tested mCherry-Dia1 DID and mCherry-Dia1 DD. mCherry-Dia1 DID was localized in a sharp apical line at cell–cell junctions, while mCherry-Dia1 DD was more diffusely localized around the junctions (Figure 5B). Taken together, these results lead us to conclude that Dia1's DID-DD region is necessary and sufficient for Dia1 localization at cell–cell junctions, and the Ala-267 residue in the DID domain plays an essential role in the localization.
FIGURE 5:
The DID-DD region is sufficient for junctional localization of Dia1. (A) Fragments of Dia1. The localization of CT was not directly assessed, because CT expression resulted in very large cell size (likely due to a cytokinesis defect) and compromised cell–cell junctions. (B) Embryos expressing mCherry-tagged Dia1 fragments (pseudocolored green) and GFP-farnesyl (membrane probe; pseudocolored magenta) were observed. Note that DID-DD is localized at cell–cell junctions, but DID-DD A267D is not. Both NT and CT cause abnormally large cell size. Scale bars: 10 µm.
Overexpression of Dia1 DID-DD displaces Dia1 and Dia2 from cell–cell junctions
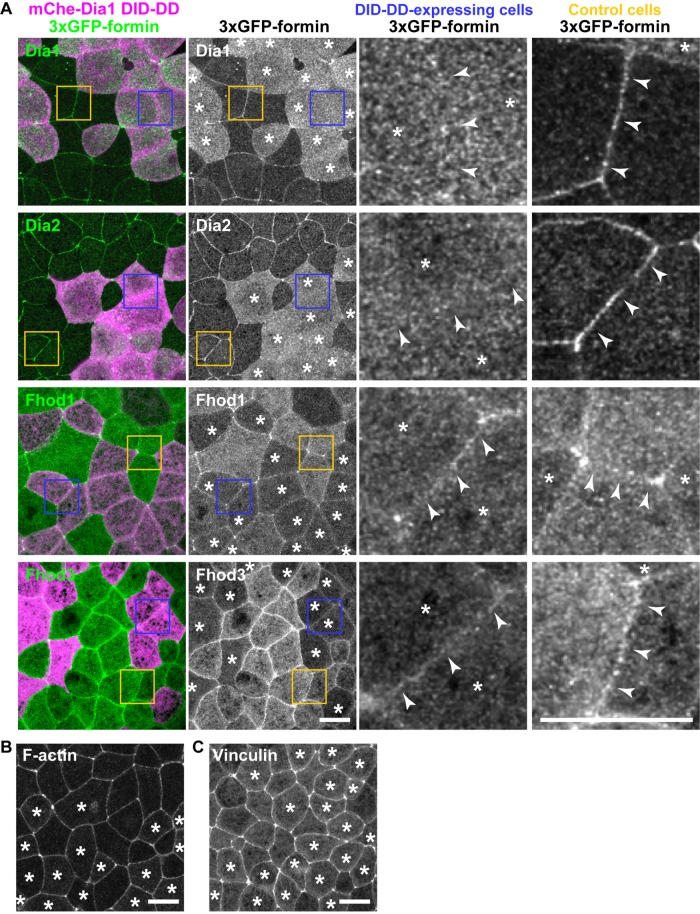
Because Dia1's DID-DD region is necessary and sufficient for junctional localization of Dia1, we hypothesized that overexpression of DID-DD could competitively remove full-length Dia1 from cell–cell junctions. To test this hypothesis, we expressed 3×GFP-Dia1 full length in all cells of the embryo and mosaically expressed mCherry-Dia1 DID-DD at a high level in part of the embryo. We found that 3×GFP-Dia1 was not localized at cell–cell junctions of adjoining Dia1 DID-DD–overexpressing cells, whereas 3×GFP-Dia1 was clearly localized at junctions in regions where cells were not expressing Dia1 DID-DD (Figure 6A), indicating that Dia1 DID-DD overexpression effectively removed full-length Dia1 from cell–cell junctions. Of interest, 3×GFP-Dia2 also lost its junctional localization in Dia1 DID-DD–overexpressing cells (Figure 6A), probably because Dia1 and Dia2 employ the same mechanism to localize at cell–cell junctions, and Dia1 DID-DD can compete with the DID of Dia2, as was previously implicated (Acharya et al., 2017). The NT halves of Fhod1 and Fhod3 are also responsible for their junctional localization (Supplemental Figure S9). However, 3×GFP-Fhod1 or 3×GFP- Fhod3 was not displaced by overexpression of Dia1 DID-DD, suggesting that Fhod1 and Fhod3 are localized at cell–cell junctions through a mechanism distinct from that of Dia1 and Dia2.
FIGURE 6:
Overexpressed Dia1 DID-DD displaces full-length Dia1 and Dia2 from cell–cell junctions but does not displace Fhod1 and Fhod3. (A) Dia1 DID-DD was mosaically expressed in the 3×GFP-tagged Dia1-, Dia2-, Fhod1-, or Fhod3-expressing embryos. Note that Dia1 and Dia2 were removed from cell–cell junctions (arrowheads) between the DID-DD–expressing cells (see enlarged blue boxes) but not from that of control cells (see enlarged yellow boxes), whereas Fhod1 and Fhod3 were not affected by Dia1 DID-DD expression. (B, C) Dia1 DID-DD was mosaically expressed in Lifeact-GFP–expressing (B) or Vinculin-3×GFP–expressing (C) embryos. Note that localization and intensity of F-actin and Vinculin are not altered in Dia1 DID-DD–expressing cells (asterisks). Scale bars: 20 µm.
To examine the effects of Dia1 and Dia2 removal from junctions on the F-actin organization at junctions, we compared the localization of Lifeact-GFP (F-actin probe) in the embryos mosaically expressing Dia1 DID-DD. The F-actin organization was indistinguishable between control cells and Dia1 DID-DD–overexpressing cells (Figure 6B). We also examined the tension at cell–cell junctions using Vinculin-3×GFP as a probe (Hara et al., 2016; Higashi et al., 2016). Again, the localization of Vinculin was not altered in Dia1 DID-DD–overexpressing cells (Figure 6C). These data imply that, although Dia1 and Dia2 are localized at junctions, they may not play a crucial role in maintenance of the F-actin bundle and junctional tension at cell–cell junctions in the X. laevis epithelium.
Dia1 and Dia2 are localized at contractile rings in dividing Dia1 DID-DD–overexpressing cells
Dia1 and Dia2 are not normally localized at the contractile ring of dividing cells in the X. laevis gastrula epithelium (Figure 1). However, when we examined dividing cells expressing Dia1 DID-DD, we noticed that full-length Dia1 was strongly localized at contractile rings (Figure 7A). Likewise, full-length Dia2 strongly accumulated at contractile rings in Dia1 DID-DD–overexpressing cells (Figure 7A). Of note, 3×GFP-Dia1 V175D did not accumulate at the contractile ring in Dia1 DID-DD–overexpressing cells, suggesting that the localization of Dia1 at the contractile ring was dependent on active Rho binding (Figure 7B).
FIGURE 7:
Dia1 and Dia2 are localized at the contractile ring in dividing Dia1 DID-DD–overexpressing cells. (A) Embryos expressing 3×GFP-Dia1 (top panels) or 3×GFP-Dia2 (bottom panels) in all cells and mCherry-Dia1 DID-DD mosaically were live imaged using confocal microscopy. Note that Dia1 and Dia2 are strongly localized at the contractile ring in the Dia1 DID-DD–overexpressing cells (yellow asterisks) but are not localized at the contractile ring in the nonexpressing cells (white asterisks). (B) Embryos expressing 3×GFP-Dia1 WT (top panels) or 3×GFP-Dia1 V175D (bottom panels) in all cells and mCherry-Dia1 DID-DD mosaically were live imaged. Note that Dia1 V175D (Rho-binding mutant) cannot localize at the contractile ring in Dia1 DID-DD–overexpressing cells. Asterisks, daughter cells. Scale bars: 10 µm.
Dia1 DID-DD–overexpressing cells exhibit cytokinesis defects
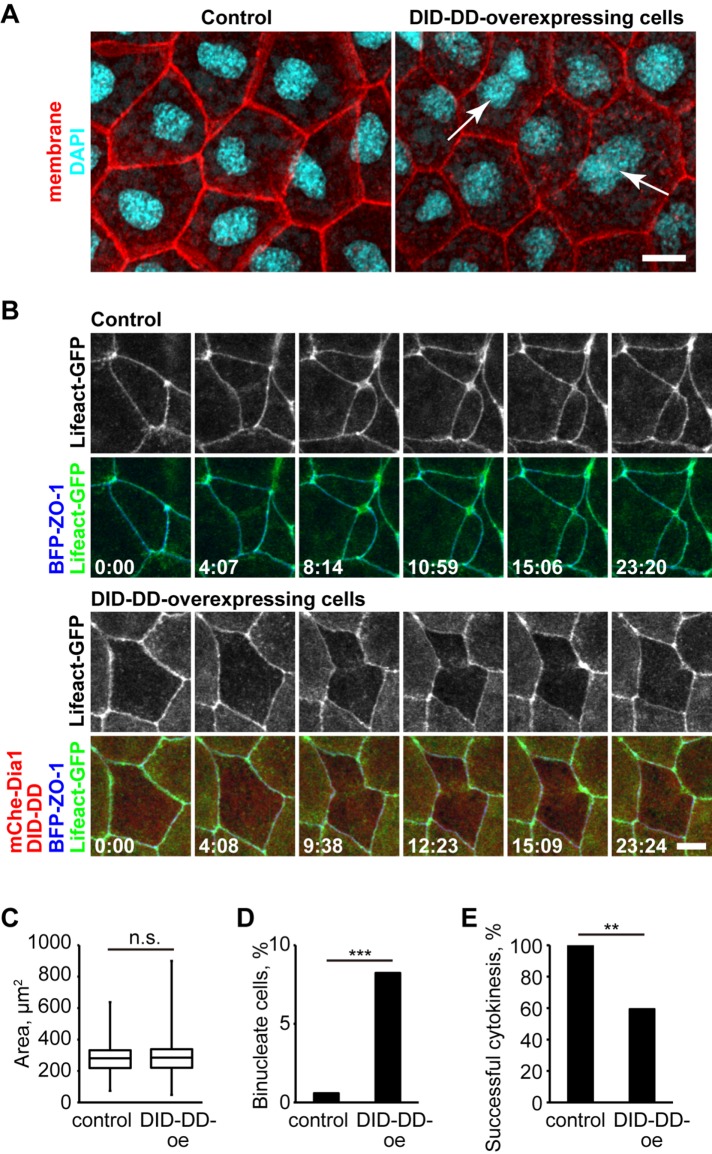
We next tested whether ectopic localization of Dia1 and Dia2 at contractile rings affects cytokinesis. We first examined fixed embryos and found that there were significantly more binucleate cells in Dia1 DID-DD–overexpressing embryos compared with the control embryos (Figure 8, A and D). The cell-surface area was not significantly larger in Dia1 DID-DD–overexpressing embryos (Figure 8C), probably because the ratio of binucleate cells was relatively small. Next, we conducted live imaging of embryos mosaically expressing Dia1 DID-DD. We observed 13 cytokinesis failures out of 32 cell divisions in the Dia1 DID-DD–overexpressing region, whereas no cytokinesis failure was observed in the WT region (Figure 8E). We also examined F-actin accumulation during cytokinesis in Dia1 DID-DD–overexpressing cells (Figure 8B). Surprisingly, in the cells that failed cytokinesis, some F-actin accumulated broadly at the division site and ingression initiated, but eventually the F-actin became discontinuous and ingression halted. Finally, the contractile ring regressed, resulting in a binucleate cell. These results suggest that competitive removal of Dia1 and Dia2 from cell–cell junctions results in cytokinesis defects.
FIGURE 8:
Dia1 DID-DD–overexpressing cells exhibit cytokinesis defects. (A) Control (left) or mCherry-Dia1 DID-DD (right) embryos expressing GFP-farnesyl (membrane probe) were fixed and stained with anti-GFP (pseudocolored red) and DAPI (cyan). Note that there are binucleate cells (arrows) in Dia1 DID-DD–overexpressing embryos. (B) Live imaging of dividing cells in control (top panels) and Dia1 DID-DD–overexpressing (bottom panels) embryos using Lifeact-GFP (F-actin probe; green) and BFP-ZO-1 (TJ marker; blue). Note that in the Dia1 DID-DD–overexpressing cells, the contractile ring is formed and regresses over time. (C, D) Apical cell-surface area (C) and percentage of binucleate cells (D) in the fixed embryos (n = 171 cells from four embryos [control] and 170 cells from four embryos [Dia1 DID-DD]). (E) Success rate of cytokinesis in live imaging (n = 12 cells from three embryos [control] and 32 cells from three embryos [Dia1 DID-DD]). p values are 0.57 (C; t test), 0.00035 (D; Fisher's exact test), 0.0067 (E; Fisher's exact test). Scale bars: 10 µm.
DISCUSSION
Both cell–cell junctions and cytokinetic contractile rings are regulated by Rho GTPases and F-actin. DRFs regulate F-actin downstream of Rho GTPases, making them good candidates to regulate linear actin assembly at both types of contractile actin arrays. In this study, we provide a comprehensive characterization of the localization of all 15 vertebrate formins in epithelial cells. We found that the DRFs Dia1, Dia2, Fhod1, and Fhod3 are localized at cell–cell junctions, and Dia3 is localized at cytokinetic contractile rings. When the localization of Dia1 and Dia2 at cell–cell junctions is perturbed by Dia1 DID-DD overexpression, Dia1 and Dia2 are mislocalized at contractile rings in dividing cells, resulting in increased cytokinesis failure, suggesting that proper localization of specific formins to distinct subcellular locations is important for successful cytokinesis.
Are specific formins sequestered at cell–cell junctions?
Because Dia1 DID-DD overexpression caused relocalization of Dia1 and Dia2 from cell–cell junctions to the contractile rings, it would be an attractive hypothesis that cell–cell junctions sequester these formins to prevent them from accumulating at contractile rings in excess amounts. An analogous idea has been proposed for Cingulin (TJ plaque protein) and Paracingulin (TJ and AJ plaque protein), which sequester GEF-H1 and Tiam1 at cell–cell junctions (Aijaz et al., 2005; Guillemot et al., 2008). It is also possible that Dia1 and Dia2 have a functional role at cell–cell junctions that we could not detect in this study.
Formins in cytokinesis
Formins have been shown to regulate cytokinesis in yeasts, worms, and flies. In vertebrates, however, the reports are quite limited. Dia2 (mDia2 in mice) is localized at the contractile ring and cleavage furrow of mouse fibroblast NIH 3T3 cells in vitro (Watanabe et al., 2008, 2010) and mouse erythroblasts in vivo (Watanabe et al., 2013) and is required for successful cytokinesis in these cells. Because these cells are not epithelial cells and do not have mature cell–cell junctions, we predict that Dia2 is not sequestered at another cellular structure and can be recruited to the contractile ring in the dividing cells. Whether Dia1, Dia2, and Dia3 have redundant roles in cytokinesis and whether they are used differentially in epithelial and nonepithelial cells are interesting questions for future study.
The possibility that formins could be sequestered by cell–cell junctions to attenuate formin activity at cytokinetic contractile rings could have physiological significance. Cytokinesis failure results in binuclear and multinuclear cells, which causes genome instability and may contribute to tumorigenesis (Fujiwara, Bandi, et al., 2005). Thus, it would be interesting to test in the future whether the mislocalization of formins induces cancer in vivo. Additionally, some specific types of epithelial cells, including hepatocytes and placental trophoblast cells, are normally tetraploid or multiploid (Alfert and Geschwind, 1958; Hoffman and Wooding, 1993; Margall-Ducos et al., 2007). Whether formin-based regulation of the contractile ring is involved in the generation of these cell types awaits future study.
Formins at cell–cell junctions
Formins have been implicated in the regulation of cell–cell junctions. Among the 15 formins, Dia1 has repeatedly been implicated in the regulation of AJs (Sahai and Marshall, 2002; Carramusa et al., 2007; Ryu et al., 2009; Rao and Zaidel-Bar, 2016; Acharya et al., 2017). In accordance with previous studies, Dia1 is also localized at cell–cell junctions in the X. laevis epithelium, but by using a quantitative imaging approach to separate the AJ and TJ, we showed that Dia1 is localized specifically at TJs. Because localization along the z-axis has not been closely assessed previously, it is possible that Dia1 is also localized at TJs in other experimental models. When we removed Dia1 and Dia2 from cell–cell junctions by Dia1 DID-DD overexpression, there was no change in junctional F-actin organization or junctional tension. This may be because the contribution of Dia1 and Dia2 to junctional organization differs among cell types or organisms, and Dia1 and Dia2 might not play major functional roles in the regulation of junctions in the X. laevis epithelium. We also identified Fhod1 and Fhod3 as AJ-localized formins. Because these formins are known to be abundantly expressed in cardiac muscle cells, the localization and function of Fhod1 and Fhod3 in epithelial cells has not been extensively assessed before. In cardiomyocytes, Fhod1 is localized at the intercalated disk, which is an AJ-like cell–cell junction structure (Al Haj et al., 2015). In the future, it will be interesting to investigate potential functional roles for Fhod1 and Fhod3 in regulating junctional actin organization.
TJ-associated actin bundles
Although both TJs and AJs are regulated by the actin cytoskeleton, AJ-associated actin bundles have gained more attention. In this study, we discovered that TJ-associated F-actin is more abundant than AJ-associated F-actin in live and fixed X. laevis gastrula-stage embryos (Supplemental Figure S7, B and C). Supporting this idea, while the AJ has been recognized as a site of actomyosin-mediated mechanotransduction, recent work shows that an actomyosin-mediated conformational change of the TJ protein ZO-1 controls junctional recruitment and signaling of its interactors (Spadaro et al., 2017). In the future, it would be interesting to examine whether the abundant TJ-associated distribution of F-actin observed here is commonly observed in other cell types or model organisms.
Mechanism of Dia1/Dia2 recruitment to cell–cell junctions
Our data strongly suggest that the Ala-267 residue in Dia1's DID domain is essential for localization of Dia1 at cell–cell junctions. This is consistent with previous reports, which showed that partial or complete deletion of the NT region, including the DID domain, abolishes the localization of mDia1 at cell–cell junctions (Carramusa et al., 2007) and that Drosophila Diaphanous loses its cortical recruitment when the DID domain is removed (Homem and Peifer, 2009). An important future question will be to determine the precise mechanism by which Dia1 and Dia2 are recruited to the cell–cell junctions. On the basis of our data that Dia1 and Dia2 are localized at TJs, we predict that the DID domain of Dia1 and Dia2 binds to some TJ protein through the Ala-267 residue. Although the Ala-267 residue is also conserved in Dia3 (Ala280), Dia3 exhibited weaker junctional localization compared with Dia1 and Dia2 (Figure 2 and Supplemental Figures S5 and S6). A better understanding of this difference awaits future study. Loss-of-function studies to reveal functional roles for Dia1, Dia2, and Dia3 and domain-swapping studies to reveal what drives their distinct localizations would be fascinating questions to pursue in future studies.
Formins and microtubules
In addition to the formins’ well-accepted roles in the regulation of the actin cytoskeleton, in vivo observations have long suggested that formins also regulate the organization and dynamics of microtubules (for a review, see Bartolini and Gundersen, 2010; Breitsprecher and Goode, 2013). Furthermore, it has recently been reported that formins are localized at microtubule plus ends and accelerate actin assembly (Henty-Ridilla et al., 2016). In this study, we found that Fmn1 is localized on microtubules (Figure 2), indicating that this formin is a good candidate for a microtubule-associated regulator of actin assembly. Additionally, we found that mDia3 is localized at cytokinetic contractile rings in epithelial cells. Of note, mDia3 has been shown to mediate microtubule attachment to kinetochores independent of its actin polymerization activity (Yasuda et al., 2004; Cheng, Zhang, et al., 2011). Future studies are required to further examine whether Fmn1 and mDia3 regulate the organization and dynamics of microtubules in epithelial cells.
Concluding remarks
In conclusion, this study provides a comprehensive analysis of the localization of the 15 vertebrate formins in epithelial cells of an intact vertebrate embryo. Our data reveal that, in epithelial cells, Dia1, Dia2, Fhod1, and Fhod3 are localized at cell–cell junctions, while Dia3 is localized at the contractile ring. Furthermore, we propose that cell–cell junctions may sequester Dia1 and Dia2, thus limiting the amount of formins localized at the contractile ring. These data suggest that cell–cell junctions may influence the cytokinetic contractile ring by calibrating the amount of localized formin activity available at the division site to ensure successful cytokinesis. It will be interesting to explore whether cells coordinately regulate these two actomyosin-based structures in other situations. Intriguingly, experimental removal of Dia1 and Dia2 from cell–cell junctions did not cause obvious defects in cell–cell junctions, suggesting that Fhod1 and/or Fhod3 may play key functional roles at cell–cell junctions.
MATERIALS AND METHODS
cDNA cloning and constructs
cDNAs encoding 15 X. laevis formins were amplified by PCR using Herculase II DNA polymerase (600675; Agilent, Santa Clara, CA) from a cDNA library that was generated from late tailbud–stage embryos using TRIzol (15596026; Thermo Fisher Scientific, Waltham, MA) and Superscript III First-Strand Synthesis System (18080-051; Thermo Fisher Scientific). Point mutants and fragments of Dia1, Dia3, Fhod1, and Fhod3 were generated by PCR using the corresponding full-length cDNAs as templates. cDNA encoding PLEKHA7 was amplified by PCR using Herculase II DNA polymerase from a cDNA library generated from gastrula-stage embryos. cDNAs encoding human ZO-1, X. laevis E-cadherin, X. laevis Claudin-6, and X. laevis Vinculin were described previously (Higashi et al., 2016). cDNAs were cloned into pCS2+ with GFP, 3×GFP, mCherry, mRFP, or mTagBFP tags. pCS2+/GFP-farnesyl, pCS2+/mCherry-farnesyl, and pCS2+/Lifeact-GFP were described previously (Reyes et al., 2014; Higashi et al., 2016). All DNA constructs were verified by sequencing (GENEWIZ, South Plainfield, NJ).
Semiquantitative reverse transcriptase-PCR
cDNA libraries were synthesized from fertilized eggs (stage 1), morula (stage 5), blastula (stage 8), gastrula (stage 10.5), neurula (stage 17), early tailbud (stage 25), late tailbud (stage 35), and tadpole (stage 45) using TRIzol and the Superscript III First-Strand Synthesis System according to the manufacturer's instructions. Fragments (∼300 base pairs) in the FH2 domain-coding sequences were amplified for each formin gene using specific primers and the stage 35 cDNA library as a template. The expression levels of each formin cDNA in the cDNA libraries from various developmental stages were assessed by PCR using GoTaq polymerase (M3005; Promega, Fitchburg, WI) with the purified fragments as standards.
Xenopus embryos and microinjection
All studies conducted using X. laevis embryos strictly adhere to the compliance standards of the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and were approved by the University of Michigan's Institutional Animal Care and Use Committee. X. laevis embryos were collected, in vitro fertilized, dejellied, and microinjected with mRNAs for fluorescent probes using methods described previously (Reyes et al., 2014; Higashi et al., 2016). Embryos were injected at either the 2-cell or the 4-cell stage and allowed to develop to the gastrula stage (Nieuwkoop and Faber stage 10–11). For mosaic expression of mCherry-Dia1 DID-DD: 3×GFP-Dia1, 3×GFP-Dia2, 3×GFP-Fhod1, 3×GFP-Fhod3, Lifeact-GFP, or Vinculin-3×GFP was injected into both cells at the 2-cell stage, and mCherry-Dia1 DID-DD was injected into two cells at the 4-cell stage such that mCherry-Dia1 DID-DD–expressing cells could be compared with neighboring internal control cells. mRNA (5 nl) was injected into each cell at the following concentrations: 3×GFP-formins (5 µg/ml), 3×GFP-Dia3 (Figure 1B) (2.5 µg/ml), mRFP-ZO-1 (5-10 µg/ml), GFP-farnesyl (4 µg/ml), mCherry-farnesyl (4 µg/ml), TagBFP-ZO-1 (5 µg/ml), PLEKHA7-mCherry (5 µg/ml), 3×GFP (5 µg/ml), Lifeact-GFP (4 µg/ml), Lifeact-RFP (5 µg/ml), Claudin-6-GFP (2 µg/ml), E-cadherin-3×GFP (5 µg/ml), Vinculin-3×GFP (5 µg/ml), mCherry-Dia1 fragments (5 µg/ml), mCherry-Dia1 NT (low conc. in Figure 5B) (1.25 µg/ml), mCherry-Dia1 DID-DD overexpression (50 µg/ml), and mCherry-Fhod1 and Fhod3 fragments (5 µg/ml).
Immunofluorescence microscopy
For fixed staining of ZO-1, β-catenin, and phalloidin, albino embryos were fixed at the gastrula stage (Nieuwkoop and Faber stage 10.5) with 1.5% formaldehyde in fixative buffer (80 mM K-PIPES [pH 6.8], 1 mM MgCl2, 5 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid [EGTA], 0.2 mM Triton X-100) overnight at room temperature. Embryos were washed three times with Tris-buffered saline (pH 7.4) containing 0.1% Nonidet P-40 (TBSN) and hemisected with a sharp scalpel, and animal hemispheres were subjected to immunostaining. Samples were blocked with TBSN containing 10% fetal bovine serum for 30 min and incubated with mouse anti–ZO-1 mAb (T8-754; generous gift from Masahiko Itoh, Dokkyo Medical University, Mibu, Japan) and rabbit anti–β-catenin pAb (ab2365; Abcam, Cambridge, MA) for 12 h. Samples were then washed three times with TBSN, incubated with Alexa Fluor 488–conjugated goat anti-mouse immunoglobulin G (IgG) (1:500, A11001; Invitrogen/Thermo Fisher Scientific), Alexa Fluor 568–conjugated goat anti-rabbit IgG (1:500, A11011; Invitrogen/Thermo Fisher Scientific), and Alexa Fluor 647–conjugated phalloidin (2.4 units/ml, A22287; Invitrogen/Thermo Fisher Scientific) in TBSN for 1 h and washed three times with TBSN.
For fixed staining of GFP-farnesyl–expressing embryos, albino embryos were injected with GFP-farnesyl with or without mCherry-Dia1 DID-DD at the 4-cell stage. At the gastrula stage (Nieuwkoop and Faber stage 10.5), the embryos were fixed with 4% formaldehyde in fixative buffer, hemisected, blocked, and stained with mouse anti-GFP mAb (JL-8, 632381; Clontech/Takara, Mountain View, CA), rabbit anti-mCherry pAb (ab167453; Abcam), and 4′,6-diamidino-2-phenylindole (DAPI; D1306; Invitrogen/Thermo Fisher Scientific). Secondary antibodies used were Alexa Fluor 488–conjugated goat anti-mouse IgG and Alexa Fluor 568–conjugated goat anti-rabbit IgG.
Live and fixed confocal microscopy
Fluorescence confocal images were collected on an inverted Olympus Fluoview 1000 microscope equipped with a 60× supercorrected PLAPON 60 × OSC objective (NA = 1.4, working distance = 0.12 mm) and FV10-ASW software. Live and fixed imaging was carried out as described previously (Reyes et al., 2014; Higashi et al., 2016).
Embryo lysates and immunoblotting
Gastrula-stage embryos (Nieuwkoop and Faber stages 10–11) were lysed as previously described (Reyes et al., 2014), with the following modification: PHEME lysis buffer did not contain phosphatase inhibitor cocktail. Samples were separated on 6% polyacrylamide gels for 3×GFP-formins and 10% polyacrylamide gels for β-tubulin and transferred onto nitrocellulose membranes. Membranes were blocked with 5% nonfat dry milk and probed with anti-GFP (JL-8, 632381; Clontech/Takara) or anti–α-tubulin (DM1A, T9026; Sigma-Aldrich, St. Louis, MO) overnight at 4°C in 1X phosphate-buffered saline/0.1% Tween-20. Horseradish peroxidase–conjugated anti-mouse IgG (W4021; Promega) was used as a secondary antibody. Membranes were developed using an ECL detection kit (32209; Pierce/Thermo Fisher, Grand Island, NY) on x-ray film.
Quantification of the localization of formins at cell–cell junctions
Gastrula-stage embryos (Nieuwkoop and Faber stages 10–11) expressing 3×GFP-tagged formins together with mCherry-farnesyl (membrane probe) were live imaged. Z-stack images of 30–40 optical slices with 0.5-µm thickness were taken from at least four embryos. Square images (50 pixels × 50 pixels) of straight junctions were excised from the original images, and 50 z-slices across the junctions were averaged using ImageJ (Schneider et al., 2012). Fluorescence intensity profiles along x- and z-axes were determined by multiple line scans using ImageJ (four line scans from each embryo) (also see Supplemental Figure S5). To determine whether formins were localized at TJs or AJs, we applied the same analysis used for the live images of embryos expressing 3×GFP-tagged formins or Lifeact-GFP (F-actin probe) together with TagBFP-ZO-1 (TJ probe) and PLEKHA7-mCherry (AJ probe) and fixed embryos stained with anti–ZO-1, anti–β-catenin, and phalloidin.
Bioinformatics analysis
The amino acid sequences of mouse and X. laevis formins and Saccharomyces cerevisiae Bni1p were aligned, and a phylogenic tree was generated by the neighbor-joining method using the ClustalX (www.clustal.org) and NJplot software packages (http://doua.prabi.fr/software/njprot) (Saitou and Nei, 1987; Perriere and Gouy, 1996; Larkin et al., 2007). The bootstrap values were calculated with the ClustalX package using the default setting.
Statistical analysis
A two-tailed unpaired t test was used for Figure 8C, and Fisher's exact test was used for Figure 8, D and E.
Supplementary Material
Acknowledgments
We thank W. M. Bement (University of Wisconsin-Madison, Madison, WI) for pCS2+/Lifeact-GFP and pCS2+/Lifeact-RFP; P.D. McCrea (University of Texas MD Anderson Cancer Center, Houston, TX) for pCS2+/xE-cadherin-3HA; E. M. De Robertis (University of California, Los Angeles, Los Angeles, CA) for pCS2+/claudin-6 (XCla)-HA; M. Itoh (Dokkyo Medical University, Mibu, Japan) for anti-ZO-1 mAb; members of our lab for helpful discussions; Torey Arnold for critical reading of the article. This work was supported by National Institutes of Health grant R01 GM112794 to A.L.M. and a postdoctoral fellowship from the Japanese Society for the Promotion of Science to T.H.
Abbreviations used:
- AJ
adherens junction
- CT
C-terminal
- DAD
Diaphanous autoinhibitory domain
- DAPI
4′,6-diamidino-2-phenylindole
- DD
dimerization domain
- DID
Diaphanous inhibitory domain
- DRF
Diaphanous-related formin
- FH
formin homology
- GBD
GTPase-binding domain
- GFP
green fluorescent protein
- NT
N-terminal
- TBSN
Tris-buffered saline containing 0.1% Nonidet P-40
- TJ
tight junction
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-02-0133) on October 31, 2018.
REFERENCES
- Acharya BR, Wu SK, Lieu ZZ, Parton RG, Grill SW, Bershadsky AD, Gomez GA, Yap AS. (2017). Mammalian Diaphanous 1 mediates a pathway for E-cadherin to stabilize epithelial barriers through junctional contractility. Cell Rep , 2854–2867. [DOI] [PubMed] [Google Scholar]
- Aijaz S, D’Atri F, Citi S, Balda MS, Matter K. (2005). Binding of GEF-H1 to the tight junction-associated adaptor cingulin results in inhibition of Rho signaling and G1/S phase transition. Dev Cell , 777–786. [DOI] [PubMed] [Google Scholar]
- Alberts AS. (2001). Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem , 2824–2830. [DOI] [PubMed] [Google Scholar]
- Alberts AS. (2002). Diaphanous-related Formin homology proteins. Curr Biol , R796. [DOI] [PubMed] [Google Scholar]
- Alberts AS, Bouquin N, Johnston LH, Treisman R. (1998). Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J Biol Chem , 8616–8622. [DOI] [PubMed] [Google Scholar]
- Alfert M, Geschwind II. (1958). The development of polysomaty in rat liver. Exp Cell Res , 230–232. [DOI] [PubMed] [Google Scholar]
- Al Haj A, Mazur AJ, Radaszkiewicz K, Radaszkiewicz T, Makowiecka A, Stopschinski BE, Schonichen A, Geyer M, Mannherz HG. (2015). Distribution of formins in cardiac muscle: FHOD1 is a component of intercalated discs and costameres. Eur J Cell Biol , 101–113. [DOI] [PubMed] [Google Scholar]
- Anderson JM, Stevenson BR, Jesaitis LA, Goodenough DA, Mooseker MS. (1988). Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin-Darby canine kidney cells. J Cell Biol , 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold TR, Stephenson RE, Miller AL. (2017). Rho GTPases and actomyosin: partners in regulating epithelial cell-cell junction structure and function. Exp Cell Res , 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. (2010). Formins and microtubules. Biochim Biophys Acta , 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Willet AH, Kovar DR, Gould KL. (2013). Formin-based control of the actin cytoskeleton during cytokinesis. Biochem Soc Trans , 1750–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga VM, Machesky LM, Hall A, Hotchin NA. (1997). The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell-cell contacts. J Cell Biol , 1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. (2013). Formins at a glance. J Cell Sci , 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carramusa L, Ballestrem C, Zilberman Y, Bershadsky AD. (2007). Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell-cell junctions. J Cell Sci , 3870–3882. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. (1994). Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development , 3367–3377. [DOI] [PubMed] [Google Scholar]
- Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M. (2008). Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol Biol Evol , 2717–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol , 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. (2011). Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell , 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. (2005). Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature , 1043–1047. [DOI] [PubMed] [Google Scholar]
- Grikscheit K, Frank T, Wang Y, Grosse R. (2015). Junctional actin assembly is mediated by Formin-like 2 downstream of Rac1. J Cell Biol , 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grikscheit K, Grosse R. (2016). Formins at the junction. Trends Biochem Sci , 148–159. [DOI] [PubMed] [Google Scholar]
- Guillemot L, Paschoud S, Jond L, Foglia A, Citi S. (2008). Paracingulin regulates the activity of Rac1 and RhoA GTPases by recruiting Tiam1 and GEF-H1 to epithelial junctions. Mol Biol Cell , 4442–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Shagirov M, Toyama Y. (2016). Cell boundary elongation by non-autonomous contractility in cell oscillation. Curr Biol , 2388–2396. [DOI] [PubMed] [Google Scholar]
- Hartsock A, Nelson WJ. (2008). Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta , 660–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Rankova A, Eskin JA, Kenny K, Goode BL. (2016). Accelerated actin filament polymerization from microtubule plus ends. Science , 1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Arnold TR, Stephenson RE, Dinshaw KM, Miller AL. (2016). Maintenance of the epithelial barrier and remodeling of cell-cell junctions during cytokinesis. Curr Biol , 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Ikeda T, Murakami T, Shirakawa R, Kawato M, Okawa K, Furuse M, Kimura T, Kita T, Horiuchi H. (2010). Flightless-I (Fli-I) regulates the actin assembly activity of diaphanous-related formins (DRFs) Daam1 and mDia1 in cooperation with active Rho GTPase. J Biol Chem , 16231–16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. (2005). Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell , 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LH, Wooding FB. (1993). Giant and binucleate trophoblast cells of mammals. J Exp Zool , 559–577. [DOI] [PubMed] [Google Scholar]
- Homem CC, Peifer M. (2008). Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development , 1005–1018. [DOI] [PubMed] [Google Scholar]
- Homem CC, Peifer M. (2009). Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell , 5138–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. (1997). Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J , 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. (1996). The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J , 1885–1893. [PMC free article] [PubMed] [Google Scholar]
- Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. (1993). Regulation of cytoplasmic division of Xenopus embryo by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI). J Cell Biol , 1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, et al. (1996). Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J , 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol , 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug SM, Schulzke JD, Fromm M. (2014). Tight junction, selective permeability, and related diseases. Semin Cell Dev Biol , 166–176. [DOI] [PubMed] [Google Scholar]
- Kuhn S, Geyer M. (2014). Formins as effector proteins of Rho GTPases. Small GTPases , e29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Rose R, Scrima A, Wittinghofer A. (2005). The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J , 4176–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics , 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Yap AS. (2015). E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol , 533–539. [DOI] [PubMed] [Google Scholar]
- Levayer R, Pelissier-Monier A, Lecuit T. (2011). Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat Cell Biol , 529–540. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. (2003). The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol , 1335–1340. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. (2005). Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem , 6986–6992. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Hamaguchi Y, Fujimoto H, Morii N, Mishima M, Narumiya S. (1993). A rho-like protein is involved in the organisation of the contractile ring in dividing sand dollar eggs. Zygote , 325–331. [DOI] [PubMed] [Google Scholar]
- Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. (2007). Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci , 3633–3639. [DOI] [PubMed] [Google Scholar]
- Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. (1996). Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J , 2208–2216. [PMC free article] [PubMed] [Google Scholar]
- Meng W, Mushika Y, Ichii T, Takeichi M. (2008). Anchorage of microtubule minus ends to adherens junctions regulates epithelial cell-cell contacts. Cell , 948–959. [DOI] [PubMed] [Google Scholar]
- Miller AL. (2011). The contractile ring. Curr Biol , R976–R978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova O, Green KJ. (2013). Desmosome assembly and dynamics. Trends Cell Biol , 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami AG, Poy F, Eck MJ. (2006). Structure of the autoinhibitory switch in formin mDia1. Structure , 257–263. [DOI] [PubMed] [Google Scholar]
- Nusrat A, Giry M, Turner JR, Colgan SP, Parkos CA, Carnes D, Lemichez E, Boquet P, Madara JL. (1995). Rho protein regulates tight junctions and perijunctional actin organization in polarized epithelia. Proc Natl Acad Sci USA , 10629–10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. (2005a). Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell , 273–281. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. (2005b). Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature , 488–494. [DOI] [PubMed] [Google Scholar]
- Perriere G, Gouy M. (1996). WWW-query: an on-line retrieval system for biological sequence banks. Biochimie , 364–369. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science , 612–615. [DOI] [PubMed] [Google Scholar]
- Pulimeno P, Bauer C, Stutz J, Citi S. (2010). PLEKHA7 is an adherens junction protein with a tissue distribution and subcellular localization distinct from ZO-1 and E-cadherin. PLoS One , e12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Zaidel-Bar R. (2016). Formin-mediated actin polymerization at cell-cell junctions stabilizes E-cadherin and maintains monolayer integrity during wound repair. Mol Biol Cell , 2844–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A, Gomez GA, Priya R, Verma S, Kovacs EM, Jiang K, Brown NH, Akhmanova A, Stehbens SJ, Yap AS. (2012). Centralspindlin and alpha-catenin regulate Rho signalling at the epithelial zonula adherens. Nat Cell Biol , 818–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes CC, Jin M, Breznau EB, Espino R, Delgado-Gonzalo R, Goryachev AB, Miller AL. (2014). Anillin regulates cell-cell junction integrity by organizing junctional accumulation of Rho-GTP and actomyosin. Curr Biol , 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F, Muramoto T, Meyer AK, Urushihara H, Uyeda TQ, Kitayama C. (2005). A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics , 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. (2005). Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature , 513–518. [DOI] [PubMed] [Google Scholar]
- Ryu JR, Echarri A, Li R, Pendergast AM. (2009). Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol , 1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ. (2002). ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat Cell Biol , 408–415. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol , 406–425. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods , 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB. (2016). Emergence of an apical epithelial cell surface in vivo. Dev Cell , 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. (2002). A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol , 2066–2075. [DOI] [PubMed] [Google Scholar]
- Spadaro D, Le S, Laroche T, Mean I, Jond L, Yan J, Citi S. (2017). Tension-dependent stretching activates ZO-1 to control the junctional localization of its interactors. Curr Biol , 3783–3795 e3788. [DOI] [PubMed] [Google Scholar]
- Staus DP, Taylor JM, Mack CP. (2011). Enhancement of mDia2 activity by Rho-kinase-dependent phosphorylation of the diaphanous autoregulatory domain. Biochem J , 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. (2014). Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol , 397–410. [DOI] [PubMed] [Google Scholar]
- Takeya R, Taniguchi K, Narumiya S, Sumimoto H. (2008). The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J , 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry SJ, Zihni C, Elbediwy A, Vitiello E, Leefa Chong San IV, Balda MS, Matter K. (2011). Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nat Cell Biol , 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday N, VerPlank L, Li R. (2002). Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol , 1864–1870. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. (2014). Architecture of tight junctions and principles of molecular composition. Semin Cell Dev Biol , 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. (1999). Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol , 136–143. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. (1997). p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J , 3044–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. (2008). mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell , 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, De Zan T, Ishizaki T, Yasuda S, Kamijo H, Yamada D, Aoki T, Kiyonari H, Kaneko H, Shimizu R, et al. (2013). Loss of a Rho-regulated actin nucleator, mDia2, impairs cytokinesis during mouse fetal erythropoiesis. Cell Rep , 926–932. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. (2010). Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell , 3193–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. (2006). Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol , 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. (2004). Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell , 711–723. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, Ishizaki T, Narumiya S. (2004). Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature , 767–771. [DOI] [PubMed] [Google Scholar]
- Zhou F, Leder P, Martin SS. (2006). Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp Cell Res , 1119–1126. [DOI] [PubMed] [Google Scholar]
- Zihni C, Mills C, Matter K, Balda MS. (2016). Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol , 564–580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.