Abstract
Using Drosophila melanogaster, we created the first animal models for myosin-based Freeman–Sheldon syndrome (FSS), a dominant form of distal arthrogryposis defined by congenital facial and distal skeletal muscle contractures. Electron microscopy of homozygous mutant indirect flight muscles showed normal (Y583S) or altered (T178I, R672C) myofibril assembly followed by progressive disruption of the myofilament lattice. In contrast, all alleles permitted normal myofibril assembly in the heterozygous state but caused myofibrillar disruption during aging. The severity of myofibril defects in heterozygotes correlated with the level of flight impairment. Thus our Drosophila models mimic the human condition in that FSS mutations are dominant and display varied degrees of phenotypic severity. Molecular modeling indicates that the mutations disrupt communication between the nucleotide-binding site of myosin and its lever arm that drives force production. Each mutant myosin showed reduced in vitro actin sliding velocity, with the two more severe alleles significantly decreasing the catalytic efficiency of actin-activated ATP hydrolysis. The observed reductions in actin motility and catalytic efficiency may serve as the mechanistic basis of the progressive myofibrillar disarray observed in the Drosophila models as well as the prolonged contractile activity responsible for skeletal muscle contractures in FSS patients.
INTRODUCTION
Patients with distal arthrogryposis (DA) display congenital contractures of two or more body parts (mainly feet and hands) without a primary neurological or muscle disease (Bamshad et al., 2009). At least 10 forms of DA have been characterized, many of which are triggered by mutations in contractile proteins (Bamshad et al., 2009; Kowalczyk and Felus, 2016). Mutations in myosin heavy chain (MYH) isoforms are major causes of DA1 (MYH3), DA2 (MYH3), DA5 (MYH2, MYH13), and DA7 (MYH8) (reviewed by Bamshad et al. [2009] and Tajsharghi and Oldfors [2013]). While muscle contractures might be expected to arise from defects in the myosin molecular motor that powers muscle contraction, it is not clear how these mutations lead to the diverse phenotypes of DA, which can include skeletal and cognitive disorders (Kowalczyk and Felus, 2016).
Dominant mutations in the embryonic form of myosin heavy chain, MYH3, appear to be the most common cause of DA2, in which patients present with contractures in hands and feet (Bamshad et al., 2009). DA2 is subdivided into more severe DA2A (Freeman–Sheldon syndrome, FSS) and less severe DA2B (Sheldon–Hall syndrome, SHS). FSS patients display facial abnormalities absent in SHS, such as H-shaped dimpling of the chin, puckered lips, as well as a small jaw, tongue, and mouth (Freeman and Sheldon, 1938). Affected individuals can have deeply set eyes, scoliosis, and other abnormalities (Wettstein et al., 1980).
Most FSS myosin mutations (R672H, R672C, E498G, Y583S, and T178I) lie in the motor domain of myosin. These alleles are predicted to affect either ATP binding or hydrolysis, thereby disrupting the ATP-dependent myosin cross-bridge cycle that is requisite for power generation (Toydemir et al., 2006; Tajsharghi et al., 2008). Prolonged binding of the myosin cross-bridge to actin could cause contractures that are observed in patients. Typically, excessive cross-bridge binding would be expected to arise from enhanced ADP affinity of the mutant myosin, since ATP that is required for cross-bridge release would be unable to readily enter the nucleotide-binding pocket. However, transient kinetic analysis of three FSS myosins (R672H, R672C, and T178I) showed that only R672H enhances ADP affinity (Walklate et al., 2016). Importantly, however, each of these proteins showed increased actin binding under saturating ADP conditions, suggesting that cross-bridge stability might be enhanced (Walklate et al., 2016).
Progress in understanding the ultrastructural and physiological defects associated with FSS is hampered by disease origination in utero and the paucity of human biopsies. Further, the defects directly engendered by the mutations can be masked or enhanced by compensatory adjustments that might be made by human muscle in down-regulating MYH3 and/or up-regulating other MYH genes. One study of a 15-mo-old FSS patient showed tibialis anterior muscle with minor structural and protein expression abnormalities, with numerous small type 1 fibers expressing perinatal MYH8 (Tajsharghi et al., 2008). MYH3 did not accumulate in mutant or control tissue at this age. Mechanical studies on isolated fibers of two adult R672C patients showed a prolonged time to relaxation as well as incomplete relaxation, that is, impaired cross-bridge detachment (Racca et al., 2015). Surprisingly, despite relatively normal muscle structure, this study found that adult muscle contains MYH3, suggesting that FSS myosin alleles can continue to influence muscle contraction into adulthood (Racca et al., 2015).
Here we focus on modeling three FSS myosin mutations in the Drosophila system to unravel the molecular, cell biological, and functional defects engendered by the mutant molecules, so that the mechanistic basis of the disease can be examined. Two of the residues assessed (R672 and T178) represent the most common mutated sites in FSS (Toydemir et al., 2006) and the third (Y583) is found in the same general area of the myosin motor domain (Figure 1A). The modeled Drosophila motor domain (Figure 1A) shows strong structural homology to the only solved human muscle myosin structure (MYH7: PDB 4DB1) and the FSS residues are embedded in sequences that are well conserved with human MYH3 (85% [R672], 84% [T178], and 61% [Y583] conservation for each residue along with its 10 N- and C-terminal residues). An advantage of the Drosophila model is that a single muscle myosin II heavy chain gene, Mhc, gives rise to all isoforms by alternative RNA splicing (George et al., 1989). Further, expression of the endogenous gene can be selectively eliminated in the indirect flight muscles (IFMs) by the Mhc10 allele (Collier et al., 1990). Hence the in vivo defects engendered by a mutant myosin allele can be studied without interference from other isoforms of the protein. This is particularly beneficial for teasing out the myofibrillar abnormalities arising from the FSS mutant alleles, for which human embryonic biopsies expressing mostly MYH3 have not been accessible.
FIGURE 1:
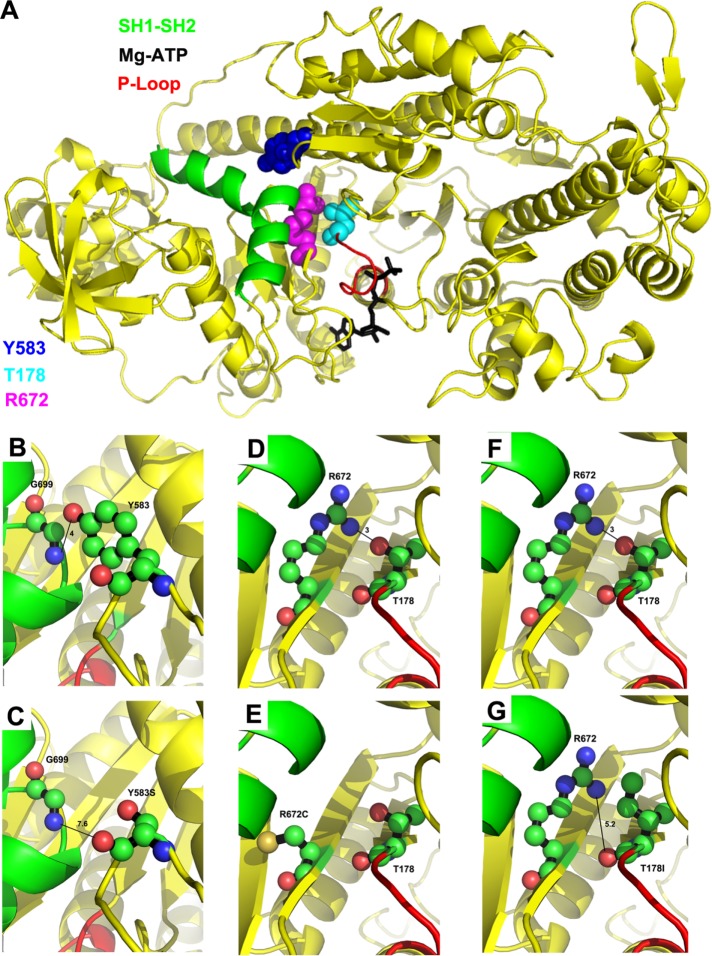
Location of T178, Y583, and R672 on the myosin molecule and interactions disrupted by the FSS mutations. (A) Human myosin FSS residues Y583 (blue), R672 (magenta), and T178 (cyan) in the Drosophila IFM myosin sequence are modeled on to the Dictyostelium discoideum myosin II motor domain structure in the presence of the Mg.ATP complex (PDB #1FMW). Functional domains of interest are illustrated: P-loop (red), SH1–SH2 helix (green), and Mg.ATP complex (black). T178 is located at the N-terminal end of the P-loop. Y583 and R672 are located near the SH1–SH2 helix. (B) Potential hydrogen bonding interaction between Y583–OH and G699–NH, with a contact distance of 4.0 Å. (C) Disruption of potential hydrogen bonding interaction with G699 by the FSS mutation Y583S yielding a new contact distance of 7.6 Å. (D) Potential hydrogen bonding interaction between R672–NH and T178–OH with a contact distance of 3.0 Å. (E) Disruption of potential hydrogen bonding interaction with T178 by the FSS mutation R672C. (F) Potential hydrogen bonding interaction between T178–OH and R672–NH with a contact distance of 3.0 Å. (G) Disruption of potential hydrogen bonding interaction with R672 by the FSS mutation T178I yielding a new contact distance of 5.2 Å. For B–G, carbon, oxygen, and nitrogen atoms are shown in green, salmon, and blue, respectively.
RESULTS
Molecular modeling of FSS mutations T178I, Y583S, and R672C maps these residues within the myosin motor domain and predicts disruptions in ionic interactions
To analyze the potential interactions of FSS T178, Y583, and R672 residues with nearby amino acids, we modeled the Drosophila IFM myosin isoform (IFI) onto a Dictyostelium discoideum myosin II motor domain crystal structure (PDB #1FMW). Since the Dictyostelium structure was solved in the presence of the Mg-ATP complex (prehydrolysis state), our analysis permits examination of FSS residue locations relative to the nucleotide. In addition to inspecting wild-type residues, we modeled the effects of FSS residue alterations to predict disrupted or aberrant interactions arising from the disease mutations.
Human residues Y583 (Y582 in Drosophila) and R672 flank the SH1–SH2 helix (colored green in Figure 1A). This helix plays a prominent role in converting nucleotide-induced conformational changes of the myosin head into myosin lever arm movement, which results in force generation when myosin is bound to actin. The SH1–SH2 helix helps define the orientation of the relay helix, which interacts with the converter to swing the lever arm (Fischer et al., 2005). The nonhelical region that separates the SH1 and SH2 helical domains contains residue G699, which acts as the pivot point for lever arm rotation (Kinose et al., 1996). The SH1–SH2 helix unwinds following the power stroke (Houdusse et al., 1999), and reformation of the helix may be important to recocking the lever arm so that it can again generate force. Interestingly, FSS residue Y583 interacts directly with G699 at the lever arm pivot point (Figure 1B). The hydroxyl group in the aromatic ring of Y583 forms a 4-Å hydrogen bond with the backbone –NH of G699. The R672 FSS residue can also interact with the SH1–SH2 helix, forming a hydrogen bond with N697 (Walklate et al., 2016). Interestingly, the terminal amino group of R672 interacts with the side-chain oxygen moiety of FSS residue T178, forming a 3-Å hydrogen bond (Figure 1D) (Walklate et al., 2016). FSS residue T178 is located near the N-terminus of the P-loop of myosin (red in Figure 1A), a highly conserved loop that binds the gamma-phosphate of ATP (black in Figure 1A). The P-loop forms part of the transducer region that controls the release of products from the nucleotide pocket during force generation (Coureux et al., 2004).
Molecular modeling predicts that important interactions of myosin residues identified above are eliminated by FSS mutations. The Y583S mutation abolishes the interaction of residue 583 with G699 (Figure 1C). The R672C mutation disrupts the hydrogen bond with T178 (Figure 1E) and also eliminates interaction with the SH1–SH2 helix in the post rigor state (Walklate et al., 2016). The T178I mutation eliminates interaction with R672 (Figure 1G). Overall, our modeling shows that the close proximity of the FSS mutations to the SH1–SH2 helix and/or the P-loop is likely to disrupt nucleotide hydrolysis and/or communication of the nucleotide state through the SH1–SH2 helix that results in lever arm rotation and force generation.
Production and verification of T178I, Y583S, and R672C transgenic lines
To create disease models with FSS myosin mutations T178I, Y583S, and R672C, we altered the wild-type Mhc gene of Drosophila using site-directed mutagenesis and produced transgenic lines by P element–mediated transformation. Viable transgenic lines were crossed into an Mhc10 IFM myosin-null background and lines for each transgene were assayed for myosin accumulation (Table 1). Lines expressing approximately wild-type myosin protein levels were verified to contain the expected myosin mutations through sequencing of reverse transcribed and PCR-amplified RNA (three independent lines each for Y583S and R672C and two independent lines for T178I). We also confirmed that the alternative exons flanking the region of each mutation were correctly spliced (for Y583S and R672C, exons 9a and 11e; for T178I, exons 3b and 7d). Notably, all R672C and T178I homozygous lines displayed poor viability and were sterile, thereby necessitating their propogation as heterozygotes over a balancer chromosome. The similar recessive phenotypes seen in R672C and T178I homozygotes correlate with the observation that each mutation disrupts interaction with the other residue.
TABLE 1:
Transgenic lines and relative FSS myosin expression levels.
| Line name | Chromosome location | Protein accumulation ± SEM |
|---|---|---|
| PwMhc2 | X | 1.00 ± 0.04 |
| PwMhcY583S-5M | 3 | 0.98 ± 0.03 |
| PwMhcY583S-9M | 3 | 1.07 ± 0.08 |
| PwMhcY583S-11F | X | 0.95 ± 0.03 |
| PwMhcR672C-17 | 3 | 0.92 ± 0.06 |
| PwMhcR672C-19 | 3 | 0.94 ± 0.04 |
| PwMhcR672C-20 | 3 | 0.93 ± 0.06 |
| PwMhcT178I-3 | 3 | 0.93 ± 0.01 |
| PwMhcT178I-27 | 3 | 0.94 ± 0.01 |
Insertion sites and myosin expression levels for homozygous transgenic lines of T178I, Y583S, and R672C constructs in the Mhc10 (IFM myosin null) background. Protein was isolated from 2-d-old females. Myosin protein accumulation was determined relative to actin for each transgene by SDS–PAGE and densitometry. Two-three independent lines from for each FSS transgene are shown. Values are compared with PwMhc2 (wild type) control. Protein amounts are means ± SEM.
FSS mutations eliminate flight in homozygotes but differentially affect flight ability in heterozygotes
Flight ability was determined for each homozygous FSS transgenic line in the Mhc10 null background by scoring the ability of individual adults to fly upward toward a light source (flight index = 6), horizontally (4), downward (2), or not at all (0). PwMhc2 control flies that express the wild-type Mhc transgene had an average flight index of 4.60 ± 0.02 at 2 d posteclosion (Table 2). In contrast, homozygous T178I, Y583S, and R672C flies were completely flightless (flight index = 0) and typically displayed a wings-up phenotype. As mentioned previously, T178I and R672C homozygotes showed poor viability, which accounts for the low number of individuals tested (Table 2).
TABLE 2:
Flight ability of homozygous and heterozygous FSS mutants.
| Line name | Age (d) | Number tested | Up (%) | Horizontal (%) | Down (%) | Not at all (%) | Flight index ± SEM |
|---|---|---|---|---|---|---|---|
| Homozygotes | |||||||
| PwMhc2 | 2 | 148 | 50.1 | 24.3 | 17.6 | 4.0 | 4.6 ± 0.02 |
| PwMhcY583S-9M | 2 | 117 | 0 | 0 | 0 | 100 | 0 |
| PwMhcY583S-11F | 2 | 120 | 0 | 0 | 0 | 100 | 0 |
| PwMhcR672C-17 | 2 | 13 | 0 | 0 | 0 | 100 | 0 |
| PwMhcR672C-20 | 2 | 15 | 0 | 0 | 0 | 100 | 0 |
| PwMhcT178I-3 | 2 | 11 | 0 | 0 | 0 | 100 | 0 |
| PwMhcT178I-27 | 2 | 23 | 0 | 0 | 0 | 100 | 0 |
| Heterozygotes | |||||||
| PwMhc2/+ | 2 | 136 | 79.4 | 14.0 | 6.6 | 0 | 5.4 ± 0.02 |
| PwMhcY583S-9M/+ | 2 | 134 | 27.6 | 29.1 | 23.1 | 20.2 | 3.28 ± 0.01 |
| PwMhcY583S-11F/+ | 2 | 129 | 25.6 | 31.8 | 26.3 | 16.3 | 3.34 ± 0.01 |
| PwMhcR672C-17/+ | 2 | 113 | 0 | 0 | 0 | 100 | 0 |
| PwMhcR672C-20/+ | 2 | 125 | 0 | 0 | 0 | 100 | 0 |
| PwMhcT178I-3/+ | 2 | 129 | 0 | 15.5 | 55.8 | 28.7 | 1.73 ± 0.01 |
| PwMhcT178I-27/+ | 2 | 131 | 0 | 12.9 | 58.9 | 28.2 | 1.69 ± 0.01 |
| PwMhc2/+ | 7 | 107 | 72 | 23 | 5 | 0 | 5.3 ± 0.01 |
| PwMhcY583S-9M/+ | 7 | 144 | 19.4 | 26.4 | 29.2 | 25 | 2.81 ± 0.01 |
| PwMhcY583S-11F/+ | 7 | 134 | 19.4 | 29.9 | 27.6 | 23.1 | 2.90 ± 0.02 |
| PwMhcT178I-3/+ | 7 | 136 | 0 | 0 | 16.2 | 83.8 | 0.32 ± 0.01 |
| PwMhcT178I-27/+ | 7 | 137 | 0 | 0 | 13.9 | 86.1 | 0.27 ± 0.01 |
Control (PwMhc2, PwMhc2/+) and FSS homozygotes and heterozygotes were assayed for the ability to fly up (U), horizontal (H), down (D), or not at all (N). Flight abilities are expressed as percentages of total number of flies tested. Two lines for each transgene are shown. Flight index was determined as 6U/T + 4H/T + 2D/T + 0N/T; T is the total number of flies tested in each cohort of 8–14 flies. Flight index mean and SEM were determined for the cohorts from each genotype. All mutant flight abilities are significantly different from the appropriate control and between 2 and 7 d of age, as determined by one-way ANOVA with Tukey's post-hoc test (p < 0.001).
T178I, Y583S, and R672C FSS myosin mutations are autosomal dominant in patients, so determining the effects of the mutations in the heterozygous state is critical to testing the validity of the Drosophila models. We thus generated heterozygotes by crossing mutant lines to wild-type yw flies and then tested offspring flight ability (Table 2). All lines of R672C mutants were completely flightless as heterozygotes at 2 d of age. Flight ability of T178I heterozygotes was significantly reduced at 2 d of age (1.71 ± 0.01, average of two lines) compared with 2-d-old PwMhc2/+ wild-type heterozygote controls (5.6 ± 0.02; p < 0.001). We examined whether an age-dependent worsening of flight muscle function occurred in T178I heterozygotes by testing flight ability at 7 d of age. This yielded a further significant reduction in flight index (0.30 ± 0.01, average of two lines; p < 0.001), with controls retaining their flight ability at 5.3 ± 0.01. Flight ability of Y583S heterozygotes was significantly reduced at 2 d of age (3.31 ± 0.01, average of two lines) compared with 2-d-old controls (5.6 ± 0.02; p < 0.001). A small, but significant, age-dependent worsening of flight muscle function was observed in Y583S heterozygotes at 7 d of age (2.86 ± 0.02, average of two lines; p < 0.001). We conclude that each FSS allele yields a dominant defective flight phenotype, with R672C most severe and Y583S least severe. The greater severity of T178I and R672C heterozygote flight phenotypes correlates with the lack of vigor and sterility found in homozygotes. It is also notable that T178I and Y583S heterozygotes, which demonstrate some flight ability as young adults, show further reductions in flight ability on aging.
FSS mutations cause severe ultrastructural defects in IFMs of transgenic flies
To determine whether FSS mutations disrupt the ultrastructure of IFMs, we examined a transgenic line for each mutant using transmission electron microscopy near the end point of myofibril assembly (late pupal stage) and during adulthood. We analyzed transverse and longitudinal sections of homozygous and heterozygous transgenic flies along with appropriate age-matched controls (Figures 2 and 3). One myofibril is shown for each panel, and it is representative of the myofibril population at that given stage of development.
FIGURE 2:
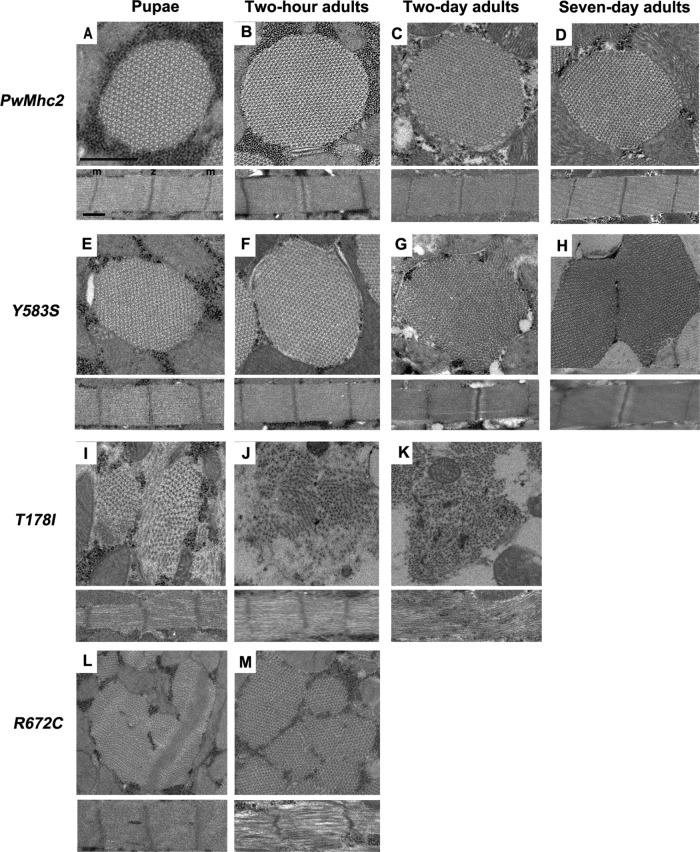
Homozygous FSS mutations Y583S, T1781, and R672C disrupt myofibril assembly and/or stability. Each panel contains transverse (top) and longitudinal (bottom) sections of a myofibril from homozygous transgenic lines. (A) Wild-type control (PwMhc2) late-stage pupa. Normal double-hexagonal packing of thick and thin filaments with oval shaped myofibril (top) as well as regular sarcomere structure (bottom) are observed. (B) Wild-type control (PwMhc2) 2-h-old adults. Normal structure is retained. (C) Wild-type control (PwMhc2) 2-d-old adult. Normal structure is retained. (D) Wild-type control (PwMhc2) 7-d-old adult. Normal structure is retained. (E) Y583S late-stage pupa resembles wild-type pupa, with normal double-hexagonal packing of thick and thin filaments and oval shaped myofibril (top) as well as regular sarcomere structure (bottom). (F) Y583S 2-h-old adult resembles wild-type 2-h-old adult, with normal double-hexagonal packing of thick and thin filaments and oval shaped myofibril (top) as well as regular sarcomere structure (bottom). (G) Y583S 2-d-old adult shows disrupted myofibril morphology with thick and thin filaments dispersing into neighboring myofibrils and gaps in the myofibrillar lattice. (H) Y583S 7-d-old adult shows continued disruption, with merging myofibrils. (I) T178I late-stage pupa shows poor myofibril morphology and disrupted hexagonal packing of myofilaments. Sarcomeres display gaps and fraying. (J) T178I 2-h-old adult shows further disruption of myofibril morphology, with myofilaments dispersed throughout the cell. (K) T178I 2-d-old adult shows disrupted morphology with Z-band material scattered within the myofibril. Sarcomeres lack defined M-lines and Z-bands. (L) R672C late-stage pupa shows irregularly shaped myofibrils, with some irregularities in sarcomere structure. (M) R672C 2-h-old adult shows variable sized myofibrils. Sarcomeres are poorly organized, with gaps and irregular borders. m, M-line; z, Z-band. Scale bars, 0.5 µm.
FIGURE 3:
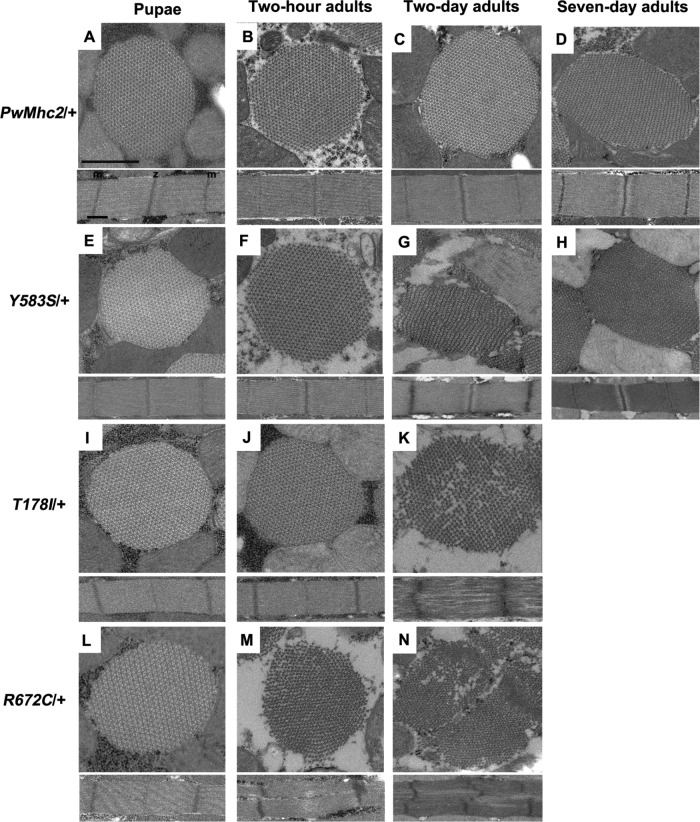
FSS mutations Y583S, T1781, and R672C show dominant disruption of myofibril stability. Each panel contains transverse (top) and longitudinal (bottom) sections of a myofibril from heterozygous transgenic lines. (A) Wild-type control (PwMhc2/+) late-stage pupa. Normal double-hexagonal packing of thick and thin filaments with oval-shaped myofibril (top) as well as regular sarcomere structure (bottom) are observed. (B) Wild-type control (PwMhc2/+) 2-h-old adult. Normal structure is retained. (C) Wild-type control (PwMhc2/+) 2-d-old adult. Normal structure is retained. (D) Wild-type control (PwMhc2/+) 7-d-old adult. Normal structure is retained. (E) Y583S/+ late-stage pupa resembles control pupa, with normal double-hexagonal packing of thick and thin filaments and oval-shaped myofibril (top) as well as regular sarcomere structure (bottom). (F) Y583S/+ 2-h-old adult resembles control 2-h-old adult, with normal double-hexagonal packing of thick and thin filaments and oval-shaped myofibril (top) as well as regular sarcomere structure (bottom). (G) Y583S/+ 2-d-old adult shows fragmented myofibril morphology. (H) Y583S/+ 7-d-old adult shows continued disruption, with merging myofibrils as seen in homozygous Y583S 7-d-old adults (Figure 2H). (I) T178I/+ late-stage pupa resembles control pupa, with normal double-hexagonal packing of thick and thin filaments and oval-shaped myofibril (top) as well as regular sarcomere structure (bottom). (J) T178I/+ 2-h-old adult resembles control 2-h-old adult, with normal double-hexagonal packing of thick and thin filaments and oval-shaped myofibril (top) as well as regular sarcomere structure (bottom). (K) T178I/+ 2-d-old adult shows disrupted morphology with abnormal hexagonal packing of myofilaments within the myofibril. Sarcomeres are poorly organized. (L) R672C/+ late-stage pupa resembles control pupa, with normal double-hexagonal packing of thick and thin filaments and oval-shaped myofibril (top) as well as regular sarcomere structure (bottom). (M) R672C/+ 2-h-old adult shows myofibrillar degeneration with some loss of myofilament packing regularity. Sarcomeres are poorly organized. (N) R672C/+ 2-d-old adult. Disruption in myofibril morphology is more intense, with scattered Z-band material present. Sarcomeres show branching and fusing phenotypes with irregular M-lines/Z-bands. m, M-line; z, Z-band. Scale bars, 0.5 µm.
Sections of the PwMhc2 transgenic control homozygotes at the late pupal stage revealed regularly packed myofibrils, with six thin filaments around each thick filament and well-formed sarcomeres (Figure 2A). These structures remained intact in 2-h-old, 2-d-old, and 7-d-old adults (Figure 2, B–D). The ultrastructure of homozygous Y583S late-stage pupae and 2-h-old adults (Figure 2, E and F) resembles that of PwMhc2 controls during the same developmental stages (Figure 2, A and B). Homozygous Y583S 2-d-old adults show some disruption in myofibril morphology with thick and thin filaments dispersing into neighboring myofibrils (Figure 2G). Y583S 7-d-old homozygotes show continued filament dispersion, with some fusion of neighboring myofibrils (Figure 2H). For T178I, homozygous pupae display assembly defects with disrupted myofibril morphology. There are myofilament subdomains within myofibrils that are poorly aligned with each other and fraying of filaments occurs in peripheral regions (Figure 2I). Two-hour-old T178I homozygous adults show extreme disruption in morphology with thick filaments dispersed in myofibril remnants and reduction in regular sarcomere patterns (Figure 2J). Homozygous T178I 2-d-old adults show continued disruption in myofibril morphology with essentially no regular sarcomeric structures and Z-band material scattered throughout the myofibril remnants (Figure 2K). For R672C, homozygous pupae display severe assembly defects with abnormally shaped and sized myofibrils. Z-band material is aberrantly localized, with poor myofilament organization (Figure 2L). Homozygous R672C 2-h-old adults show even more heterogeneity in myofibril size, with poor sarcomere morphology (Figure 2M). Note that IFMs in 2- and 7-d-old R672C homozygotes, as well as 7-d-old T178I homozygotes, were so severely disrupted and atrophied that preparation of electron micrographs of adequate resolution was not feasible. Overall, the structural abnormalities of myofibrils observed in 2-d-old IFMs (Figure 2) should account for the absence of flight ability (Table 2).
We next examined IFMs from control and FSS mutant heterozygotes to determine whether the functional defects observed in heterozygotes correlate with ultrastructural abnormalities. Control heterozygotes (PwMhc2/+) display wild-type filament packing and sarcomere structures at the late pupal stage and in 2-h-old, 2-d-old, and 7-d-old adults (Figure 3, A–D). Heterozygous Y583S late-stage pupae and 2-h-old adults show normal hexagonal packing of thick and thin filaments and sarcomere structures (Figure 3, E and F) that resemble controls (Figure 3, A and B). Heterozygous Y583S 2-d-old adults have disrupted myofibril morphology with thick and thin filaments beginning to disperse from myofibrils and possibly fuse with neighboring myofibrils (Figure 3G). Heterozygous Y583S 7-d-old adults display thick and thin filaments merging into neighboring myofibrils (Figure 3H). Heterozygous T178I late-stage pupae and 2-h-old adults display normal hexagonal packing of thick and thin filaments (Figure 3, I and J) that resembles control ultrastructure (Figure 3, A and B). Heterozygous T178I 2-d-old adults show disruption in hexagonal packing of thick and thin filaments, with filaments missing from the lattice and sarcomere structural elements that are disrupted (Figure 3K). Heterozygous T178I 7-d-old adults were not examined due to severe muscle degeneration. Heterozygous R672C late-stage pupae display normal hexagonal packing of thick and thin filaments (Figure 3L) that resemble control pupae (Figure 3A). Heterozygous R672C 2-h-old adults show myofibrils with disrupted myofilament arrays and branching sarcomeres with wavy Z- and M-line material (Figure 3M). Heterozygous R672C 2-d-old adults show disrupted myofibril morphology with scattered Z-band material; sarcomeres are further disordered as well (Figure 3N). Heterozygous R672C 7-d-old adults were not examined due to severe muscle degeneration. The ultrastructural disruption in FSS heterozygotes (Figure 3) correlates well with the flight defects observed in young and older adults (Table 2). Y583S heterozygotes retain the most normal structure and preserve the ability to fly to some extent, with some flies showing upward flight at 7 d of age. In contrast, R672C mutants show no flight ability as heterozygotes and display substantial ultrastructural defects as early as two h into adulthood. T178I shows both intermediate flight ability and ultrastructural integrity. In sum, the FSS models show dominant defects in both muscle function and myofibril morphology.
FSS mutations affect the biochemical and biophysical properties of myosin
We next assessed the steady-state ATPase parameters of the FSS myosins obtained from dissected IFMs of homozygotes (Table 3). All three mutant myosins showed statistically significant decreases in Ca-ATPase activity, with Y583S and R672C decreased by 30% and T178I decreased by 78%. In contrast, the more physiologically relevant basal Mg-ATPase showed no significant difference for Y583S. Interestingly, this rate was halved for R672C and doubled for T178I. However, while the latter rate increased, there was virtually no actin stimulation of Mg-ATPase activity for T178I, resulting in a Vmax that was only ∼1/3 of the control. Y583S actin-stimulated Vmax was also reduced, to approximately two-thirds of the control, whereas R672C Vmax was unaffected. The affinity constant (Km) for actin relative to actin-stimulated Mg-ATPase activity was significantly reduced for Y583S myosin (lower actin concentration needed for half-maximal ATPase activity) and significantly enhanced for R672C, with no change for T178I. As an overall assessment tool, the catalytic efficiency quantifies the ability of each myosin to hydrolyze ATP relative to its affinity for actin (Vmax/Km). The catalytic efficiency for Y583S myosin (3.44 ± 0.33 s-1/µM) is unchanged relative to control (3.48 ± 0.19 s-1/µM), whereas both R672C (2.16 ± 0.25 s-1/µM) and T178I (1.46 ± 0.32 s-1/µM) catalytic efficiencies are significantly reduced relative to control. This parameter agrees with the molecular modeling outcomes, which predict that both R672C and T178I disrupt the R672-T178 interaction and might therefore lead to similar phenotypes. It also correlates well with the flight-testing data and ultrastructural results showing that R672C and T178I mutations are more severe than Y583S.
TABLE 3:
ATPase and in vitro motility values for wild-type control (PwMhc2) and FSS mutant myosins.
| Myosin isoform (n for ATPase/motility) | Ca-ATPase (s-1) | Basal Mg-ATPase (s-1) | Actin-stimulated Vmax (s-1) | Actin-stimulated Km (µM) | Catalytic efficiency (s-1/µM) | Motility (µm/s) |
|---|---|---|---|---|---|---|
| PwMhc2 (n = 14/14) | 14.57 ± 2.46 | 0.24 ± 0.09 | 1.74 ± 0.37 | 0.54 ± 0.16 | 3.48 ± 0.19 | 5.08 ± 0.73 |
| Y583S (n = 6/5) | 10.16 ± 2.09** | 0.32 ± 0.10 | 1.14 ± 0.26** | 0.35 ± 0.14* | 3.44 ± 0.33 | 3.25 ± 0.41*** |
| T178I (n = 7/4) | 3.15 ± 0.99*** | 0.54 ± 0.14*** | 0.63 ± 0.17*** | 0.53 ± 0.24 | 1.46 ± 0.32*** | 2.53 ± 0.22*** |
| R672C (n = 5/5) | 9.94 ± 3.18* | 0.13 ± 0.04** | 2.22 ± 0.61 | 1.07 ± 0.33* | 2.16 ± 0.25** | 2.67 ± 0.16*** |
Biological replicates (n values) are composed of two technical replicates for ATPase or at least 30 motile filaments for in vitro motility. Data are shown as mean ± SD. Statistical significance was determined using an unpaired t test with Welch's correction. Significant differences were assumed for p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).
Finally, we used the in vitro motility assay to assess the ability of myosin molecules bound to a coverslip to move fluorescently labeled actin filaments in the presence of ATP. All three FSS mutations reduced actin velocity significantly, with Y583S decreasing it by 36% and both T178I and R672C reducing it by ∼50%. While the in vitro motility analysis suggests that Y583S is the least severe of the three alleles, in agreement with our assessments of flight ability, ultrastructural integrity and ATPase catalytic efficiency, the differences among the three alleles for actin filament velocity were not statistically significant. However, the significant reduction in unloaded velocity of actin filaments for all three FSS mutant myosins indicates that the percentage of time myosin remains bound to actin during the mechanochemical cycle (duty ratio) could be increased, as the drag induced in this ensemble system would reduce filament velocity (Harris and Warshaw, 1993). This excessive cross-bridge binding could be an important contributor to the contractures observed in patients.
DISCUSSION
We succeeded in creating three disease models for myosin-based FSS in an effort to probe the mechanistic basis of the disease. The Drosophila system has the advantages of producing adequate quantities of pure protein, which allows assessment of the biochemical and biophysical properties of the FSS myosins. Further, it permits evaluation of both recessive and dominant phenotypes in vivo. Such studies can define the root causes of FSS and their immediate consequences but not necessarily all the sequelae that arise in mammals, such as compensation through expression of other MYH isoforms, excessive development of other fiber types, or skeletal abnormalities arising from the primary muscle defects.
Our locomotory assays revealed the effect of the mutant myosin on the function of IFMs and thereby defined the severity and dominance of the disease alleles (Table 2). All three mutations result in the absence of flight in the homozygous state and two alleles (T178I and R672C) induce sterility and dramatically reduce viability. The latter is noteworthy in that the two mutant alleles are expressed in the presence of two wild-type myosin alleles in all muscles (except IFM and jump muscle) in the Mhc10 background (Collier et al., 1990). Hence the mutations are extremely deleterious and would be unlikely to allow survival in the absence of wild-type myosin in flies or humans. The flight tests of FSS heterozygotes indicate that the flight abilities of the transgenic lines can be partially rescued by the presence of a wild-type copy of the gene, depending on the severity of the FSS allele. Two-d old Y583S heterozygotes showed some flight ability, which was retained to some extent at day 7. In contrast, T178I showed severe flight defects at the younger age and essentially no flight at 7 d, whereas R672C heterozygotes lacked flight ability at 2 d (Table 2). These results illustrate the dominant nature of the mutations and the fact that different mutations show different degrees of severity, as is the case in patients. In contrast to our observations, studies in human heterozygotes suggest that the R672C phenotype is markedly less severe than that of T178I (Beck et al., 2014), although a great degree of phenotypic variability due to environmental or genetic factors has also been reported (Toydemir et al., 2006). In this regard the T178I mutation was originally classified as a mutation shared by FSS and SHS (Toydemir et al., 2006) but was subsequently reclassified as FSS (Beck et al., 2014). The standardization of growth conditions and genetic background with the Drosophila model obviates concerns with environmental and genetic influences that can confound human studies.
We tested the hypotheses that FSS mutant myosin affects myofibril assembly and stability. While myofibril assembly proved defective in homozygotes for the two severe alleles (T178I and R672C), this process proceeded normally for Y583S homozygotes (Figure 2) and for heterozygotes carrying any of the three alleles (Figure 3). This corresponds well with the normal myosin localization and myofibrillar architecture reported for myotubes derived from cultured myoblasts of a T178I/+ patient (Pokrzywa et al., 2015). Delays in cultured T178I/+ human myoblast development were noted, but these may have arisen from reduced expression levels of MYH3 (Pokrzywa et al., 2015). For Drosophila heterozygotes, all alleles eventually led to myofibrillar breakdown, with the degree of disarray correlating with level of flight ability (Figure 3 and Table 2). Analyses of biopsied embryonic tissue from FSS patients have not been reported to our knowledge, so it unclear whether myofibrillar degeneration occurs in utero. Racca et al. found relatively normal muscle architecture at the light microscope level for adult heterozygotes (Racca et al., 2015). While adults express MYH3 (Racca et al., 2015), it is not to the degree found in fetal skeletal muscle, where it represents 81% of all MYH transcripts (Racca et al., 2013). Hence effects of FSS mutations on myofibrillar structure may not be evident in human adults due to the presence of additional MYH isoforms.
The observation of normal myofibril assembly followed by degeneration in Drosophila FSS heterozygotes is reminiscent of the hypercontraction phenotype found previously in several contractile protein mutants, where muscle destruction occurs as a result of excessive cross-bridge activity (Beall and Fyrberg, 1991; Kronert et al., 1995; Naimi et al., 2001; Montana and Littleton, 2004; Barton et al., 2007; Cammarato et al., 2008; Viswanathan et al., 2015). This physiological phenotype correlates well with the mechanical studies of Racca et al. on fibers from two unrelated R672C/+ adults (Racca et al., 2015). These investigators observed reduced muscle relaxation and elevated tension that they attributed to a subpopulation of myosin cross-bridges displaying reduced kinetics of detachment. Studies of IFM and/or jump muscle mechanics (Swank, 2012) in Drosophila FSS heterozygotes should prove fruitful in determining whether similar phenotypes are found in our model system.
Our in vitro motility studies are the first reported for FSS myosins and these showed severely reduced actin gliding velocity. This has typically been interpreted to arise from an increased interaction time for the myosin cross-bridge (possibly limited by detachment kinetics, i.e., the rate of ADP release) (Uyeda et al., 1990; Harris and Warshaw, 1993). Given our in vitro motility observations, in conjunction with mechanical studies of fibers from adult patients (Racca et al., 2015), it seems plausible to postulate that excessive cross-bridge interactions are the basis of the disease phenotype. In contrast, however, transient kinetic studies on in vitro expressed FSS MYH3 human myosins with mouse light chains showed slowing of the ATP hydrolysis step and a longer detached state of cross-bridges as a common feature (Walklate et al., 2016). Enhanced ADP binding was found only for one of the three FSS myosins studied. However, increased affinity for actin in the presence of ADP was demonstrated in all cases, which could enhance cross-bridge stability (Walklate et al., 2016). In agreement with our ATPase results, Walklate et al. (2016) observed reductions in Vmax for FSS myosins. This was particularly consistent for T178I, which showed the poorest actin stimulation in both studies (Table 3). In contrast, we did not see a reduction in Vmax for R672C. However, it is important to note that in addition to differences in myosin species between the two studies, Walklate et al. (2016) analyzed S1 myosin head fragments, whereas we studied full-length dimeric myosins that are capable of forming head–head and head–rod interactions. We found that a significant biochemical predictor of severe phenotypic dysfunction is catalytic efficiency, which encapsulates both Vmax and Km values. The two severe mutant myosins (T178I and R672C) showed significantly reduced catalytic efficiencies, while Y583S myosin from organisms with the mildest phenotype showed no significant difference compared with wild type.
Homology modeling revealed significant interactions of the FSS residues with surrounding amino acids that are likely to impact myosin function on mutation. The Y583 residue is present in the loop connecting two β strands and interacts with G699 at the bend between the SH1 and SH2 helices (Figure 1B). Disruption of this interaction in the Y583S mutant (Figure 1C) is likely to adversely affect myosin function, as G699 plays an important role in motor activity and serves as the pivot point for the lever arm during the contractile cycle (Kinose et al., 1996). The G699A mutation in skeletal muscle myosin reduced actin-activated ATPase and in vitro motility activities by 5- and 100-fold, respectively (Kinose et al., 1996). This suggests a longer time spent in the actomyosin complex (Kinose et al., 1996). Likewise, a valine substitution in Dictyostelium myosin II yielded a 10-fold reduction in in vitro motility and a higher affinity of myosin for actin during the ADP-bound state (Patterson et al., 1997). Another study on Dictyostelium myosin containing an alanine substitution at the pivot point residue showed a 30-fold reduction in ATP binding to myosin, 11-fold higher affinity of ADP toward myosin, and a 180-fold higher affinity of ADP to the actomyosin complex (Batra et al., 1999). Since G699 mutant defects mirror those we observed in Y583S (reduced in vitro motility and actin-activated ATPase), it is reasonable to postulate that disrupted communication between the nucleotide-binding site and the lever arm pivot point within the SH1–SH2 helix domain is the major effect of the Y583S mutation. As the SH1–SH2 helix is hypothesized to act as a pathway that connects the nucleotide-binding pocket to the relay helix and the converter domain (Zheng and Brooks, 2005), it seems reasonable that the Y583S mutation yields defects in both ATPase activity and in vitro motility.
The R672 myosin residue is on a β-strand of myosin near to the SH1–SH2 helix, where it forms a 3-Å hydrogen bond with T178 located just before the N-terminus of the P-loop (Figure 1D). Mutation of either residue (R672C or T178I) disrupts this bond and thereby weakens the communication between the ATP terminal phosphate binding domain and the adjacent β-strand (Figure 1, E and G). Hence it might be expected that R672C and T178I would yield similar phenotypes, which is generally the case in our analysis (dominant flight dysfunction and myofibril disarray, similar reductions in catalytic efficiency and in vitro motility). However, we did find opposite effects on basal Mg-ATPase activity, and only T178I showed a decrease in Vmax (Table 3). The location of T178I adjacent to the P-loop might independently enhance nucleotide affinity, accounting for the stimulatory effect on basal Mg-ATPase. Point mutations in the P-loop region are known to disrupt ATPase activity and movement of actin filaments in vitro, possibly as a result of enhanced ADP retention (reviewed by Ruppel and Spudich [1996]). In addition to affecting the T178-R672 interaction, the R672C mutation is predicted to disrupt a π-cation interaction with relay helix residue F490 as well as disrupt or enhance interaction with the SH1–SH2 helix at residue N697 (Walklate et al., 2016). Overall, as was the case for the Y583S, the R672C and T178I mutations appear to disrupt a pathway that connects the nucleotide-binding pocket through the SH1–SH2 helix to the relay helix and converter domain.
In conclusion, our development of Drosophila models for FSS provided insights into understanding the basis of the human condition. As in humans, the models display dominant defects in muscle function, with varying severity depending on the allele expressed. Further, we found that myofibrillar stability is perturbed in the heterozygotes, which provides insights into how the phenotype might progress during human embryogenesis. The ability to isolate pure populations of mutant FSS myosins allowed us to detect abnormal ATPase activity. Further, we found that all the mutations significantly reduced in vitro motility, supporting the idea that increased cross-bridge binding is the basis of the disease. Finally, molecular modeling yielded insights into each of the mutations, with the general theme that they affect a pathway between the nucleotide-binding site and the relay helix and converter region, which is ultimately responsible for movement of the lever arm to generate force during the mechanochemical cycle. Testing the effects of the mutations on fiber mechanical parameters should yield insights into the basis of the observed functional defects. This can lead to rationally designed attempts to improve defective muscle function using pharmaceutical and/or genetic approaches.
MATERIALS AND METHODS
Protein structure analysis
The Drosophila IFM myosin isoform was modeled onto a Dictyostelium discoideum myosin II structure in the presence of the Mg.ATP complex (PDB #1FMW) using the SWISS-MODEL program tool (https://swissmodel.expasy.org) in the automated mode. We examined interactions of FSS myosin residues T178, Y582, and R672 (homologues of human T178, Y583, and R672, respectively) using the PyMOL program (PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC; www.pymol.org). IFM myosin sequences containing each mutant version of the FSS residues were also modeled and analyzed using this approach.
Molecular cloning
Construction of the mutant myosin genes was initiated by digesting the full-length myosin transgene (Swank et al., 2000) with EagI, which cuts at base 11,344 of the 5′ end, where base 1 is the start point of the cloned 5′ end fragment (XbaI site). This yields an 11.3 kb Mhc 5’ end fragment in the pCasper vector (Thummel and Pirrotta, 1992). For Y583S and R672C this 5’ Mhc fragment was cut with PstI and EagI at 8944 and 11,344, respectively, to subclone a 2.5-kb fragment in the pKS vector. This subclone was digested with NcoI and SacI at 10,535 and 11,344, respectively, to obtain an 809-bp fragment in the pLitmus vector. For T178I this 5′Mhc was cut with PstI and AvrII at 2570 and 6888, respectively, to obtain a 4.3-kb fragment in the pLitmus 28i vector. This subclone, pPA4.3, was digested with PstI and BamHI at 2570 and 4695, respectively, to obtain a 2.1-kb fragment in the pLitmus 28i vector. Site-directed mutagenesis using the QuickChange II kit (Agilent, Santa Clara, CA) was performed using sequence specific forward and reverse primers. For T178I, the exon-specific primer 5′-TCTATGTTGATCATCGGTGAGTCTGGT-3′ (nucleotide change shown as bold) was used in site-directed mutagenesis. For Y583S, the exon-specific primer 5′-GCCATTGCCCATTCGGCTGGTTGTGTG-3′ (nucleotide change shown as bold) was used in site-directed mutagenesis. For R672C, the exon specific primer used in site-directed mutagenesis was 5′-CCTCACTTCGTCTGTTGCATCATTCCC-3′ (nucleotide change shown as bold).
Site-directed mutagenesis products were sequenced (Eton Bioscience, San Diego, CA) to confirm each mutation, and each subfragment was cloned back into the larger fragments to obtain the full 5′Mhc fragment. Subsequently, the 19.1-kb 5′Mhc fragment with each mutation was digested with EagI. A 12.6-kb 3′Mhc fragment digested with EagI was gel isolated and then ligated to the 19.1 kb 5′Mhc fragment to generate the final genomic mutant construct in P element vector pCaSpeR 4 (Thummel and Pirrotta, 1992). This vector contains a β-lactamase gene allowing for selection of the final clones on ampicillin media and a Drosophila miniwhite gene (w+) as a selectable eye-color marker. The mutant constructs were purified using the QIAfilter Plasmid Maxi Kit (Qiagen Sciences, Germantown, MD). All ligation sites, splice junctions, and exons were verified by DNA sequencing to ensure that no cloning artifacts were introduced.
P element–mediated transformation of Mhc genes
Drosophila germline transformation was performed by BestGene Inc. (Chino Hills, CA) by injecting the transgene and a helper plasmid expressing transposase into Drosophila embryos of the yw strain (Rubin and Spradling, 1982). Around 1000 embryos were injected for each construct. Surviving adults (G0 generation) were individually crossed with the yw strain, and progeny were screened for orange eye color, indicating the presence of the miniwhite (w+) marker and the Mhc transgene. Orange-eyed flies from individual crosses were back crossed with the yw strain, and the offspring were screened for the presence of eye color and crossed to establish independent lines.
Transgenic lines were crossed with w1118; CyO/Bl1; TM2/TM6B balancer flies to map the chromosomal location of each transgene. For the 11 Y583S lines, three inserts mapped to the X chromosome, two to the second chromosome, and six to the third chromosome. Of the 26 R672C lines, two inserts mapped to the X chromosome, 12 to the second chromosome, and 12 to the third chromosome. Of the 27 T178I lines, four inserts mapped to the X chromosome, 20 to the second chromosome, and three to the third chromosome. Each of the X-linked and third chromosome lines was crossed into the Mhc10 background, which contains a second chromosome null allele for the indirect flight and jump muscle myosin isoforms (Collier et al., 1990). For the R672C and T178I constructs, all of the transgenic lines proved to be recessive sterile, and the mutant alleles were carried over balancer chromosomes.
Myosin expression levels
Protein expression levels from transgenes were determined by SDS–PAGE (Laemmli, 1970). Six upper thoraces from 2-d-old female flies from each transgenic line were homogenized in 180 μl of Laemmli sample buffer. yw flies were used as a control. Samples were boiled for 5 min and centrifuged for 2 min. Samples were loaded on precast 10% polyacrylamide Tris–glycine gels (Bio-Rad, Hercules, CA) at increasing concentration of the experimental samples and controls. Each transgenic line was examined at least five times, each time with a newly prepared sample. After running the gels at 110 V for 100 min in 1X SDS buffer, they were stained with Coomassie blue for 20–30 min and destained for 45 min. Gels were scanned and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). The myosin-to-actin ratio for each sample was determined in its linear range and relative expression was compared with control yw samples.
Reverse transcription and PCR
Reverse transcription and PCR (RT-PCR) were employed to confirm that each transgenic line expressed the appropriate site-directed nucleotide changes and to ensure that alternative splicing of Mhc RNA was not disrupted by the mutations. For RNA isolation (Becker et al., 1992), upper thoraces from 2-d-old adult female flies from each transgenic line were dissected and homogenized in 300 μl of 6 M urea and 3 M LiCl. After storage in a cold room overnight, samples were centrifuged for 10 min at 4°C. Each pellet was resuspended in 150 μl of 10 mM Tris–HCl, pH 7.5, 10 mM EDTA, and 1% SDS. This was followed by phenol:chloroform (1:1) extraction and ethanol precipitation of RNA. The RNA pellet was dried in a vacuum evaporator and resuspended in sterile RNase free water.
To generate cDNAs for each transgenic line, we employed a Protoscript cDNA synthesis RT-PCR kit (New England Biolabs, Ipswich, MA). Briefly, 0.5 μg of total RNA from each transgenic line was mixed with 3 µmol of reverse specific primer, and cDNA was synthesized according to the manufacturer's instructions. PCR was performed using 1 μl of cDNA and 3 µmol of each specific primer pair (as noted below) using the following conditions: 60 s at 94°C, then 30 cycles of 30 s at 94°C, 30 s at 55°C and 2 min at 68°C.
In addition to confirming the FSS mutations, appropriate splicing of alternative exons flanking the mutational sites was confirmed. For T178I, alternative exons 3 and 7 were examined. A reverse primer specific to exon 8 (5′-GTTCGTCACCCAGGGCCGTA-3′) and a forward specific primer to exon 2 (5′-TGGATCCCCGACGAGAAGGA-3′) were used to generate cDNA. For Y583S and R672C, alternative exons 9 and 11 were examined. A reverse primer specific to exon 12 (5′-GAGCTTCTTGAAGCCCTTACGG-3′) and a forward specific primer to exon 8 (5′-TCTGGATACCCAGCAGAAGCGT-3′) were used to generate cDNA. RT-PCR products were sequenced by Eton Bioscience.
Flight ability
Flight testing was performed on transgenic homozygous and heterozygous lines in a Mhc10 background along with age-matched transgenic controls (PwMhc2 or PwMhc2/+). Flight tests were executed at room temperature in a Plexiglas box (43 cm high × 27.5 cm wide × 43 cm long) with a light source at the top (Drummond et al., 1991). Ten cohorts of 8–14 female flies were collected on the day of eclosion and aged at room temperature for 2 d for homozygotes or 2 and 7 d for heterozygotes. Each fly was released individually in the center of the chamber and its ability to fly upward (U), horizontal (H), downward (D), or not at all (N) was scored. Each fly was assigned a flight index of 6 for upward flight, 4 for horizontal, 2 for downward, or 0 for no flight. Flight index was calculated using the formula 6*U/T+4*H/T+2*D/T+0*N/T, with T as the total number of flies tested for each cohort (Tohtong et al., 1995). Data are reported as mean ± SEM. Statistical comparisons were made by one-way analysis of variance (ANOVA) with Tukey's post-hoc test, with significance designated as p < 0.05.
Electron microscopy
Late-stage pupae as well as 2-h-old, 2-d-old, and 7-d-old adults were fixed and stained for ultrastructural analysis by electron microscopy according to an established protocol (O’Donnell and Bernstein, 1988). In brief, the head and abdomen were removed, and then thoracic regions not containing IFM were dissected away. Remaining tissue was placed in fixative containing 3% paraformaldehyde, 2% glutaraldehyde, 100 mM sucrose, 100 mM sodium phosphate, pH 7.2, and 2 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA). Samples were treated overnight at 4°C and then washed twice for 5 min each in 100 mM sodium phosphate, pH 7.2. This was followed by postfixative treatment (1.0% osmium, 100 mM sodium phosphate, pH 7.2) for 2 h at 4°C. Samples were then washed twice in distilled water for 5 min each at 4°C and dehydrated in acetone at room temperature. Samples were embedded in Spurr's medium. Sections were cut with a diamond knife and stained for 20 min in 2% aqueous uranyl acetate, pH 4.5, followed by lead citrate for 4 min. Sections were analyzed under a Tecnai 12 transmission electron microscope (FEI, Hillsboro, OR). At least three different organisms at each development stage were examined from the highest-expressing transgenic line of each mutant.
Myosin and actin isolation
Myosin was isolated as previously described (Swank et al., 2001; Kronert et al., 2008) with minor changes. Dorsolongitudinal IFMs were dissected from 120 to 180 split thoraces of young transgenic flies in York modified glycerol (20 mM potassium phosphate, pH 7.0, 2 mM MgCl2, 1 mM EGTA, 20 mM dithiothreitol (DTT), and a protease inhibitor mixture). Dissected muscles were placed in a microcentrifuge tube containing 1 ml of York-modified glycerol and centrifuged at 8500 × g in a Sorvall Microspin 24S centrifuge for 5 min at 4°C. Pellets were gently homogenized in 1 ml of York-modified glycerol containing 2% (vol/vol) reduced Triton X-100 and incubated for 30 min on ice. The permeabilized fibers were then centrifuged for 5 min as previously stated, and the pellet was washed and resuspended in 1 ml of York-modified glycerol lacking glycerol, followed by centrifugation. The fiber pellets were resuspended in 82 μl of high-salt myosin extraction buffer (1 M KCl, 50 mM potassium phosphate, pH 6.8, 5 mM MgCl2, 0.5 mM EGTA, 10 mM sodium pyrophosphate, 20 mM DTT, and a protease inhibitor mixture) and incubated on ice for 15 min. Insoluble material was removed by centrifugation, and the high-salt extract was then diluted 25-fold with nano-H2O containing 10 mM DTT. The sample was left on ice for myosin to precipitate overnight at 4°C. The precipitant was centrifuged at 100,000 × g for 20 min using a Beckman TLA 100.3 rotor. The pellet was resuspended in 13.5 μl of wash buffer (2.4 m KCl, 100 mM histidine, pH 6.8, 0.5 mM EGTA, and 20 mM DTT) and placed on ice for 30 min. The solution was slowly diluted eightfold with nano-H2O containing 10 mM DTT to a final salt concentration of 0.3 M KCl to precipitate residual actomyosin. The diluted solution was centrifuged at 60,000 × g for 25 min, and the supernatant was diluted 10-fold with nano-H2O containing 10 mm DTT. The solution was kept on ice for 1 h to precipitate myosin. The precipitant was centrifuged at 100,000 × g for 25 min, and the purified myosin pellet was dissolved in myosin storage buffer (0.5 M KCl, 20 mM MOPS, pH 7.0, 2 mM MgCl2, and 20 mM DTT). The myosin concentration was calculated by measuring the absorption at 280 and 310 nm and applying the following formula: 1 OD280-310/0.53 = 1 mg/ml (Margossian and Lowey, 1982). Purified myosin was used immediately for ATPase and in vitro motility assays.
G-actin was obtained by extraction of acetone powder of chicken breast muscle as described previously (Pardee and Spudich, 1982; Kronert et al., 2008). Soluble G-actin was recovered after dialysis against 2 mM Tris–HCl, pH 8.0, 0.2 mM ATP, 2 mM CaCl2, and 1 mM DTT. G-actin concentration was calculated by measuring the absorption at 290 and 310 nm and applying the following formula: 1 OD310-290/0.62 = 1 mg/ml. To prepare F-actin, one volume of 10 × polymerization buffer (50 mM Tris–HCl, pH 8.0, 0.5 M KCl, 20 mM MgCl2 and 10 mM ATP) was added to nine volumes of G-actin. The working F-actin solution was at a concentration of 30 μM in 1 × polymerization buffer. G-actin was polymerized and labeled with rhodamine-phalloidin for in vitro motility assays.
Steady-state ATPase activity and in vitro motility assays
ATPase activities for full-length myosin were determined using 2 µg of myosin and [γ-32P]-ATP as described previously (Swank et al., 2001; Kronert et al., 2008). Ca-ATPase activities was assessed in 10 mM imidazole, pH 6.0, 100 mM KCl, 10 mM CaCl2, 1 mM [γ-32P]-ATP at room temperature with 15 min incubation. The reaction was quenched using 1.8 N HClO4. For Mg2+ basal and actin-stimulated Mg-ATPase activities, myosin was added to Mg-ATPase solution (10 mM imidazole, pH 6.0, 20 mM KCl, 0.01 mM CaCl2, 1 mM MgCl2, 1 mM [γ-32P]-ATP) with increasing concentrations of F-actin (0.1–2 µM). The reaction was quenched using 1.8 N HClO4 after 25 min at room temperature prior to extraction and scintillation counting. Basal Mg2+ ATPase activities were subtracted from all data points in the actin-activated assay. Vmax and Km values were obtained by fitting data with the Michaelis–Menten equation using Prism (Graph Pad) software. Two technical samples were run for each biological replicate. ATPase values are presented as mean ± SD. Catalytic efficiency was determined by separately defining Vmax and Km values for each biological replicate and calculating the mean ± SD for their ratio (Vmax/Km). Values were assessed using unpaired t tests and Welch's correction, with significant differences assumed at p < 0.05.
In vitro motility assays were performed by adding ATP and fluorescently labeled F-actin to coverslips coated with purified myosin as previously described (Swank et al., 2001; Kronert et al., 2008). Video sequences were captured under fluorescence optics and were analyzed computationally to determine actin sliding velocity. At least 30 smoothly moving filaments were analyzed per biological replicate. In vitro motility values are presented as mean ± SD and were assessed using an unpaired t test and Welch's correction, with significant differences assumed at p < 0.05.
Acknowledgments
We appreciate the technical support provided by Jennifer A. Suggs (San Diego State University [SDSU]), aid with statistical analysis from Sahaana Chandran (SDSU), and helpful comments on the manuscript from Douglas Swank and Kaylyn Bell (Rensselaer Polytechnic Institute). Electron microscopy was carried out in the SDSU Electron Microscope Facility. This work was supported by National Institutes of Health grant R37GM032443 to S.I.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- DA
distal arthrogryposis
- FSS
Freeman–Sheldon syndrome
- IFM
indirect flight muscle
- MYH
myosin heavy chain
- SHS
Sheldon–Hall syndrome.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0526) on October 31, 2018.
REFERENCES
- Bamshad M, Van Heest AE, Pleasure D. (2009). Arthrogryposis: a review and update. J Bone Joint Surg Am (Suppl 4), 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton B, Ayer G, Maughan DW, Vigoreaux JO. (2007). Site directed mutagenesis of Drosophila flightin disrupts phosphorylation and impairs flight muscle structure and mechanics. J Muscle Res Cell Motil , 219–230. [DOI] [PubMed] [Google Scholar]
- Batra R, Geeves MA, Manstein DJ. (1999). Kinetic analysis of Dictyostelium discoideum myosin motor domains with glycine-to-alanine mutations in the reactive thiol region. Biochemistry , 6126–6134. [DOI] [PubMed] [Google Scholar]
- Beall CJ, Fyrberg E. (1991). Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J Cell Biol , 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AE, McMillin MJ, Gildersleeve HI, Shively KM, Tang A, Bamshad MJ. (2014). Genotype-phenotype relationships in Freeman-Sheldon syndrome. Am J Med Genet A , 2808–2813. [DOI] [PubMed] [Google Scholar]
- Becker KD, O’Donnell PT, Heitz JM, Vito M, Bernstein SI. (1992). Analysis of Drosophila paramyosin: identification of a novel isoform which is restricted to a subset of adult muscles. J Cell Biol , 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarato A, Dambacher CM, Knowles AF, Kronert WA, Bodmer R, Ocorr K, Bernstein SI. (2008). Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol Biol Cell , 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier VL, Kronert WA, O’Donnell PT, Edwards KA, Bernstein SI. (1990). Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev , 885–895. [DOI] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, Houdusse A. (2004). Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J , 4527–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DR, Hennessey ES, Sparrow JC. (1991). Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet , 70–80. [DOI] [PubMed] [Google Scholar]
- Fischer S, Windshugel B, Horak D, Holmes KC, Smith JC. (2005). Structural mechanism of the recovery stroke in the myosin molecular motor. Proc Natl Acad Sci USA , 6873–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman EA, Sheldon JH. (1938). Cranio-carpo-tarsal dystrophy. Arch Dis Child , 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EL, Ober MB, Emerson CP., Jr (1989). Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol , 2957–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DE, Warshaw DM. (1993). Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J Biol Chem , 14764–14768. [PubMed] [Google Scholar]
- Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. (1999). Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell , 459–470. [DOI] [PubMed] [Google Scholar]
- Kinose F, Wang SX, Kidambi US, Moncman CL, Winkelmann DA. (1996). Glycine 699 is pivotal for the motor activity of skeletal muscle myosin. J Cell Biol , 895–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk B, Felus J. (2016). Arthrogryposis: an update on clinical aspects, etiology, and treatment strategies. Arch Med Sci , 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronert WA, Dambacher CM, Knowles AF, Swank DM, Bernstein SI. (2008). Alternative relay domains of Drosophila melanogaster myosin differentially affect ATPase activity, in vitro motility, myofibril structure and muscle function. J Mol Biol , 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronert WA, O’Donnell PT, Fieck A, Lawn A, Vigoreaux JO, Sparrow JC, Bernstein SI. (1995). Defects in the Drosophila myosin rod permit sarcomere assembly but cause flight muscle degeneration. J Mol Biol , 111–125. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature , 680–685. [DOI] [PubMed] [Google Scholar]
- Margossian SS, Lowey S. (1982). Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol (Pt B), 55–71. [DOI] [PubMed] [Google Scholar]
- Montana ES, Littleton JT. (2004). Characterization of a hypercontraction-induced myopathy in Drosophila caused by mutations in Mhc. J Cell Biol , 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naimi B, Harrison A, Cummins M, Nongthomba U, Clark S, Canal I, Ferrus A, Sparrow JC. (2001). A tropomyosin-2 mutation suppresses a troponin I myopathy in Drosophila. Mol Biol Cell , 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell PT, Bernstein SI. (1988). Molecular and ultrastructural defects in a Drosophila myosin heavy chain mutant: differential effects on muscle function produced by similar thick filament abnormalities. J Cell Biol , 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee J, Spudich J. (1982). Purification of muscle actin. Methods Enzymol (Pt B), 164–181. [DOI] [PubMed] [Google Scholar]
- Patterson B, Ruppel KM, Wu Y, Spudich JA. (1997). Cold-sensitive mutants G680V and G691C of Dictyostelium myosin II confer dramatically different biochemical defects. J Biol Chem , 27612–27617. [DOI] [PubMed] [Google Scholar]
- Pokrzywa M, Norum M, Lengqvist J, Ghobadpour M, Abdul-Hussein S, Moslemi AR, Tajsharghi H. (2015). Developmental MYH3 myopathy associated with expression of mutant protein and reduced expression levels of embryonic MyHC. PLoS One , e0142094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racca AW, Beck AE, McMillin MJ, Korte FS, Bamshad MJ, Regnier M. (2015). The embryonic myosin R672C mutation that underlies Freeman-Sheldon syndrome impairs cross-bridge detachment and cycling in adult skeletal muscle. Hum Mol Genet , 3348–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racca AW, Beck AE, Rao VS, Flint GV, Lundy SD, Born DE, Bamshad MJ, Regnier M. (2013). Contractility and kinetics of human fetal and human adult skeletal muscle. J Physiol , 3049–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. (1982). Genetic transformation of Drosophila with transposable element vectors. Science , 348–353. [DOI] [PubMed] [Google Scholar]
- Ruppel KM, Spudich JA. (1996). Structure-function analysis of the motor domain of myosin. Annu Rev Cell Dev Biol , 543–573. [DOI] [PubMed] [Google Scholar]
- Swank DM. (2012). Mechanical analysis of Drosophila indirect flight and jump muscles. Methods , 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank DM, Bartoo ML, Knowles AF, Iliffe C, Bernstein SI, Molloy JE, Sparrow JC. (2001). Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J Biol Chem , 15117–15124. [DOI] [PubMed] [Google Scholar]
- Swank DM, Wells L, Kronert WA, Morrill GE, Bernstein SI. (2000). Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc Res Tech , 430–442. [DOI] [PubMed] [Google Scholar]
- Tajsharghi H, Kimber E, Kroksmark AK, Jerre R, Tulinius M, Oldfors A. (2008). Embryonic myosin heavy-chain mutations cause distal arthrogryposis and developmental myosin myopathy that persists postnatally. Arch Neurol , 1083–1090. [DOI] [PubMed] [Google Scholar]
- Tajsharghi H, Oldfors A. (2013). Myosinopathies: pathology and mechanisms. Acta Neuropathol , 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C, Pirrotta V. (1992). New pCaSpeR P element vectors. Dros Inf Serv , 150. [Google Scholar]
- Tohtong R, Yamashita H, Graham M, Haeberle J, Simcox A, Maughan D. (1995). Impairment of muscle function caused by mutations of phosphorylation sites in myosin regulatory light chain. Nature , 650–653. [DOI] [PubMed] [Google Scholar]
- Toydemir RM, Rutherford A, Whitby FG, Jorde LB, Carey JC, Bamshad MJ. (2006). Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nat Genet , 561–565. [DOI] [PubMed] [Google Scholar]
- Uyeda TQ, Kron SJ, Spudich JA. (1990). Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J Mol Biol , 699–710. [DOI] [PubMed] [Google Scholar]
- Viswanathan MC, Blice-Baum AC, Schmidt W, Foster DB, Cammarato A. (2015). Pseudo-acetylation of K326 and K328 of actin disrupts Drosophila melanogaster indirect flight muscle structure and performance. Front Physiol , 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walklate J, Vera C, Bloemink MJ, Geeves MA, Leinwand L. (2016). The most prevalent Freeman-Sheldon Syndrome mutations in the embryonic myosin motor share functional defects. J Biol Chem , 10318–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein A, Buchinger G, Braun A, von Bazan UB. (1980). A family with whistling-face-syndrome. Hum Genet , 177–189. [DOI] [PubMed] [Google Scholar]
- Zheng W, Brooks B. (2005). Identification of dynamical correlations within the myosin motor domain by the normal mode analysis of an elastic network model. J Mol Biol , 745–759. [DOI] [PubMed] [Google Scholar]