Abstract
This paper is a concise review aiming to assemble the most relevant topics presented by the authors at ORS‐Philadelphia Spine Research Society Fourth International Spine Research Symposium. It centers on the latest advances in disc development, its main structural entities, and the populating cells, with emphasis on the advances in pivotal molecular pathways responsible for forming the intervertebral discs (IVD). The objective of finding and emphasizing pathways and mechanisms that function to control tissue formation is to identify and to explore modifications occurring during normal aging, disease, and tissue repair. Thus, to comprehend that the cellular and molecular basis of tissue degeneration are crucial in the study of the dynamic interplay that includes cell‐cell communication, gene regulation, and growth factors required to form a healthy and functional tissue during normal development.
Keywords: development, stem cell, tissue‐specific progenitor cells
1. INTRODUCTION
This paper is a concise review aiming to assemble the most relevant topics presented by the authors at ORS‐Philadelphia Spine Research Society Fourth International Spine Research Symposium. It centers on the latest advances in disc development, its main structural entities and the populating cells, with an emphasis on the advances in pivotal molecular pathways responsible for forming the intervertebral discs (IVD). The objective of finding and emphasizing pathways and mechanisms that function to control tissue formation is to identify and to explore modifications occurring during normal aging, disease, and tissue repair. Thus, a precise understanding of normal tissue development—including cell‐cell communication, gene regulation, and growth factor dynamics—is imperative in identifying the cellular and molecular processes contributing to tissue degeneration, and for designing therapeutic interventions to curb and/or reverse these processes.
The development of the axial skeleton is a multistep process initiated by the formation of the notochord during early embryonic development. The notochord is laid down along the rostral‐caudal axis, providing a primitive axial skeleton as well as secreted signals for the patterning of surrounding tissues. The vertebral column is formed by aggregation of the somitic mesenchyme around the notochord, which undergoes progressive patterning and differentiation to form the annulus fibrosus (AF), vertebral bodies, cartilage endplates, and ligaments. The notochord disappears where the vertebral bodies form but expands within the perichordal disc to form the nucleous pulposus (NP). In the following sections, we will provide a detailed overview of IVD development, transcription factors, growth factors and/or morphogens, and the cell types that regulate the formation of the IVD.
2. THE NOTOCHORD
2.1. Embryonic origins of the notochord
conserved tissue that is necessary and sufficient for axis induction during gastrulation, and contributes to cells of the notochord and prechordal mesendoderm.1 The dorsal organizer was originally identified in amphibians by Spemann and Mangold,2 leading to the identification of homologous structures with conserved function in other species, including the embryonic shield in teleosts,3 Hensen's node in the chick,4 and the node in the mouse embryo.5
During gastrulation of the mouse embryo, a population of progenitor cells emerge from the anterior aspect of the primitive streak, termed axial mesoderm (also referred to as chordamesoderm or mesendoderm) and ingress to form the node.6, 7 The node is a transient, late organizer population consisting a few hundreds of cells that form a teardrop‐shaped pit at the distal tip of the murine embryo at embryonic day (E) 7.5, responsible for establishing the left‐right asymmetry of the body plan.8, 9 Motile cilia localized on the apical surface of cells within the node beat in a clockwise rotation to drive a leftward flow of extraembryonic fluid containing morphogens, such as Nodal, secreted by the columnar epithelial cells of ventral node (termed “nodal flow”).10, 11, 12 Disruption of either ciliogenesis13, 14, 15 or cilia motility16 results in abnormal left‐right patterning of the mouse embryo. Although essential for proper embryo patterning, nodal flow is required only during a brief window of development, from 1 to 6 somite stage, which spans 6 to 7 hours of development in the mouse.17, 18 The TGFβ family proteins Nodal and Lefty‐2, as well as the homeobox protein Pitx2 are essential to the establishment of left‐right asymmetry in vertebrates.18 Studies using targeted gene deletion in the mouse have identified key transcription factors expressed in the node and required for node morphogenesis and/or function, including FoxH1,19 brachyury (T),20 Lhx1,21 FoxA2,22 Tead,23 Otx2,24 and Noto.25 Recent studies applied genome‐wide analyses to characterize the gene regulatory networks driving the formation and function of the node and notochord,26 demonstrating dramatic alterations in gene expression patterns as cells transition between developmental states and identifying key pathways and matrix components that may define these distinct stages.27
As the embryo elongates, the notochord development is initiated as trunk notochord precursors emerge from the node to form the notochordal plate at E8.0‐E8.5 in the mouse.28 The notochordal plate is continuous with the dorsal gut endoderm and positioned in the axial midline of the embryo; it is formed by three distinct cell types derived from the axial mesoderm—the prechordal plate, the anterior head process, and the node‐derived notochordal precursors.6, 29, 30 At E9.0, the notochord plate folds off the gut endoderm; cells of the prechordal plate contribute to the forebrain and rostral hindbrain, while cells of the anterior head process form the anterior notochord which rests in a central position in the mouse embryo flanked by the dorsal ridge of the neural tube (the floor plate) and ventrally by the gut endoderm (the endoderm plate).31, 32 Laterally, the notochord is flanked by the paraxial mesoderm, which will give rise to the somites and subsequently the AF and vertebrae. Live imaging of notochord formation in the mouse highlighted further differences in its cellular origins; the trunk notochord is derived from the node by mediolateral intercalation while the tail notochord is formed by node‐derived cells that actively migrate toward the posterior and are maintained at the caudal end of the trunk notochord until incorporation at a later stage.9
The notochord forms a continuous rod‐like structure in which cells display homogeneous morphology and gene expression patterns along the A/P axis. The distinction between the anterior head process and prechordal plate in the notochord is marked in the mouse by specific differences in genetic regulation, including dependence of the trunk notochord on expression of the transcription factors Noto9 and T.33 Conversely, loss of Nodal signaling34 or loss of expression of the transcription factor Lim121 leads to complete loss of notochord formation. Formation of all levels of the notochord (including the anterior head process and prechordal plate) are dependent on the activity of the transcription factors Foxh119, 35 and Foxa2.22, 36
Similar to the organizer from which it is derived, the notochord is a transient structure in the developing embryo that serves at least two essential functions.37 First, the notochord forms the primitive anterior/posterior axis of the embryo; a continuous rod‐like structure that runs along the midline of the embryo, surrounded by the peri‐notochordal basement membrane composed of extracellular matrix proteins.38 The correct deposition and organization of this extracellular matrix is essential for notochord morphogenesis and maintenance of the rod‐like structure of the notochord during early embryonic development.39 In the mouse, formation of the peri‐notochordal sheath is dependent on hedgehog signaling.40 In its structural role, the notochord resembles cartilage; cells express the Sry‐related HMG‐box transcription factors Sox5, Sox6, and Sox941, 42 and secrete an extracellular matrix rich in collagens, laminins, and aggrecan.43, 44, 45 However, while chondrocytes secrete a hydrophilic extracellular matrix enabling the tissue to remain hydrated and resist compressive load,46 studies in zebrafish and Xenopus demonstrate that notochord cells secrete a thick basement membrane sheath but retain hydrated materials in large intracellular vacuoles thereby allowing cells to resist compression.47 Notochord vacuoles are generated by post‐Golgi trafficking pathways and considered as the final step in notochord differentiation, relying on the preceding chordamesoderm specification, convergent extension, formation of the notochord sheath, and the spatiotemporal activation of vacuolating signals within the axial notochord.48 The notochord remains in place until the development of the permanent axial skeleton. The second role of the notochord is to secrete morphogens, such as Shh and noggin, through which it regulates the patterning of surrounding tissues, including the neural tube,49 the sclerotome of the somites,50 the pancreas,51 and the aorta.52 It is important to underscore that our understanding of the pathways that regulate the formation and function of the embryonic notochord are largely based on studies in model organisms (including mouse, chick, zebrafish, and Xenopus); although several characteristics appear to be conserved, functional differences may exist between species, particularly, in humans where less detailed investigation has been undertaken.
2.2. Fate of notochord‐derived cells
During development of the axial skeleton, the notochord disappears in areas where the vertebral bodies form but expands within the perichordal disc, to form the central nucleus pulposus (NP).53 Within the newly formed NP, notochord cells proliferate and produce a glycosaminoglycan‐rich extracellular matrix that separates the original cell mass into a network of cell clusters. Using whole transcriptome analysis, recent studies have highlighted remarkable differences in signaling and cell biosynthesis associated with the transition from notochord (E12.5) to NP (postnatal day 0) in mice, including decreased expression of the Shh pathway and increased expression of the TGFβ and IGF‐1 pathways.27 In mice, the transcription factors Foxa1 and Foxa2 are required for proper formation of the NP from the embryonic notochord.54 In most vertebrates, there is a progressive postnatal loss of large vacuolated notochord cells and the NP becomes instead populated by small chondrocyte‐like cells.55, 56 In humans, cells of the NP change markedly with age; by skeletal maturity, despite maintaining expression of notochord markers (including brachyury, galectin‐3, and CD24),57 cells of the NP assume distinct phenotypic and molecular characteristics.58, 59, 60, 61, 62 The loss of notochord cells from the NP is associated with the onset of degenerative changes in the IVD, suggesting that these cells are required for NP maintenance.55, 63, 64 A number of recent studies have begun to characterize the NP cell molecular phenotype and changes associated with age and degeneration in humans.57, 65, 66
The fate of the notochord cells within the NP has long been debated. It has been proposed that small chondrocyte‐like NP cells were mesenchyme‐derived, populating the NP following migration from the surrounding cartilage endplate (CEP)67 or originating from transient amplifying cells in the perichondrium at the periphery of the disc.68 In this context, notochord cells were postulated to direct mesenchyme cell migration and stimulate matrix synthesis prior to undergoing apoptosis or necrosis at the completion of disc formation.69, 70 Alternatively, notochord cells were proposed to serve as IVD‐specific progenitors undergoing terminal differentiation to give rise to the small cells of the NP.63, 71, 72 In the mouse, genetic strategies for lineage‐tracing have demonstrated that all cells of the adult NP are notochord‐derived.53, 73
3. THE ANNULUS FIBROSUS
Carlier74 evaluated the embryonic development of the sheep IVD, and described the tissue surrounding the notochord to be composed of undifferentiated embryonic cells that produce a matrix, which becomes irregular‐shaped and localized in fibrils, and subsequently converts into fibrous, arranging themselves in lamellae. This structure was called as the AF. Besides the NP, the AF is another crucial component of the IVD. It forms orthogonal layers of collagen‐rich fibrils surrounding the proteoglycan‐rich and gelatinous NP while connecting the two adjacent levels of the vertebral bodies forming a strong joint.
3.1. Embryonic origin of AF
AF originates from the somitocoele. The dorsal epithelium of the somite gives rise to the dermo‐myotome, while the ventral region gives rise to the sclerotome starting at stage III of somitogenesis.75 Before giving rise to the sclerotome, the somitocoele undergoes epithelium to mesenchymal transition (EMT). The dorsolateral domain of early sclerotome gives rise to another compartment of the somite called “syndetome”.76 Syndetome is characterized by the expression of Scleraxis (Scx).76, 77 Scx is a basic helix‐loop‐helix transcription factor. All tendon/ligament lineage of cells express Scx.78 Using Scx Cre ; R26 LacZ reporter line for fate‐mapping studies, it was demonstrated that entire AF is derived from Scx‐expression cells.79 Scx GFP expression was detected as early as E12.5 in the AF of the mouse embryo. The Scx Cre ‐R26 Ali4 fluorescent reporter also marked the Scx GFP cells at E12.5 suggesting their lineage from syndetome.80 Vertebral bodies did not have Scx+ cells showing the specificity of these reporters. Although previously it was thought that AF arises from the sclerotome region of the somite, Murchison et al,81 detected endogenous Scx GFP expression in the AF of the E18.5 mouse embryo, showing that it stays on as the AF develops This conclusion was mainly due to the markers used to fate‐map, which were not exclusive to sclerotome. In one such study using Tbx18 Cre ; R26 LacZ line for fate‐mapping studies, Tbx18‐derived cells were observed in AF at E16.5.82 However, Tbx18 Cre marks several other embryonic structures including myocardial cells in early embryo at E9.5.83 Also, a few NP cells were observed to be Tbx18+, although Choi et al53 and McCann et al.73 have shown that all NP cells derive from a homogenous population of notochordal cells. The Tbx18 Cre used in the Bruggeman et al82 study is not inducible, and hence, will also mark syndetome, which in turn originates from the sclerotome. Therefore, it is not clear from the Bruggeman study whether the Tbx18+ cells in the AF came from syndetome or sclerotome. Similarly, other studies also used markers that did not distinguish between the cells derived exclusively from sclerotome and/or syndetome. However, fate‐mapping studies using Scx driver lines, which markers only “syndetome” compartment of the somite, clearly shows that the Scx‐derived cells form the AF.
3.2. Regulation of AF development and its maintenance
3.2.1. Sonic hedgehog signaling
The induction of somitocoele to form sclerotome is driven by Sonic hedgehog (Shh) produced by the notochord and floorplate which upregulates Pax1 expression in the sclerotome.50, 84, 85, 86, 87 However, Choi et al showed that Shh expression from the notochord and not floor plate is sufficient for maintenance of Pax1 expression in the ventral sclerotome.88 Pax1 is a marker of ventral sclerotome, while Pax9 is a marker of dorsal sclerotome. Bone morphogenetic protein (BMP) signaling antagonizes Shh signaling in this process, while Noggin and Gremlin1, in turn, antagonizes BMP, allowing Shh to regulate sclerotome differentiation.89, 90 In addition to Pax1, Shh also regulates the expression of genes required for the development of axial skeleton like Pax9, NKx3.2, and Sox9.85, 91, 92, 93, 94 Pax1 and Pax9 are crucial for axial skeleton development, as mutations in these genes cause defects in the formation of the vertebral column.95, 96, 97 Bapx1 is known to be downstream of Sox9,98 and is activated by Pax1 and Pax9 in the sclerotome.99, 100 Deletion of Bapx1 resulted in vertebral defects. With the induction of sclerotome, Pax3 and Pax7 expression is downregulated. The notochord‐derived NP continues to express Shh in the postnatal stages, and its blockade by small molecule cyclopamine in vitro, or conditional targeting in vivo, decreased the expression of hedgehog signaling targets PTCH1 and GLI1 in the AF, suggesting that AF responds to Shh produced by the NP cells.101
3.2.2. HMG‐box transcription factors
Both embryonic notochord and ventral sclerotome express Sox9 by E8.5 and 10.5, respectively, during mouse development.102 Brent et al73 showed that the syndetome in early mouse embryo had mixed cell population, some expressed Scx, while others were Sox9, Sox5, and Sox6 positive.77 It is known that Sox9 is upstream of Sox5 and Sox6 in chondrogenesis.103, 104 While Sugimoto et al79 showed overlapping Scx and Sox9 expression in syndetome at E10.5, Brent et al showed that in Sox5/Sox6 mutants the Sox9 expression remains unchanged, and the Scx expression, as well as the tendon progenitors, increased in number, suggesting that cartilage differentiation mediated through Sox5/ Sox6 is stimulated by suppressing tendon development.77 In 2003, Brent et al found that Shh negatively regulates tendon lineage through Pax1.76 By targeting Sox9 by ScxCre, only in the syndetome‐derived AF, it was shown that Sox9 is crucial for the formation of proper AF.79 However, Ck19Cre‐mediated targeting of Sox9 in the notochord, showed the absence of AF from the IVD by E15.5.102 This effect could be due to loss of rostrocaudal notochord from the CK19 Cre; Sox9 flox/flox mutants E10.5 onwards, and failure to initiate AP‐segmentation of perinotochordal sclerotome by E11.5.102 Conversely, short fragments of functional notochord can start cartilage differentiation in Sox9+ perinotochordal sclerotome resulting in a metameric‐like pattern resembling regular vertebral column during development. These studies point out the importance of notochord, and notochordal signals in the maintenance of AF. Sox9 continues to play an essential role in the IVD after skeletal maturity. Conditional targeting of Sox9 using Agc1CreERT2 in 2‐month‐old mice severely affected the entire disc structure and extracellular matrix remodeling 1 month later.105 Yet, Agc1CreERT2 targets Sox9 in the NP, AF, cartilaginous endplate and adjacent growth plate chondrocytes. By Chip‐on‐Chip analysis, Ctgf was identified as a direct target of Sox9 in rat NP cells. The role of Sox9 in the regulation of Ctgf was validated by conditional targeting of Sox9, at 2 weeks of age in mice, using Col2CreER driver line and analyzing the CTGF expression by 2 months of age and severe structural defects were also observed. Loss of Sox9 and CTGF resulted in severe structural defects in these mice.106 Blockade of Shh in vitro and targeting its conditional allele in vivo, in the NP, resulted in loss of Sox9, and extracellular matrix markers like collagen 1, collagen 2, chondroitin sulfate, and keratan sulfate in the AF, indicating that Shh, from notochord‐derived NP, continues to regulate AF development and differentiation in the postnatal stages.
3.2.3. TGFβ signaling
Using Col2a Cre Baffi et al targeted Tgfβr2 using its conditional allele to block response to TGFβ signaling and showed that the development of IVD and AF was affected at E13.5, E15.5, and E17.5.107 Profiling studies from cultured sclerotome at E11.5 to identify the targets of TGFβ and BMP signaling in vitro showed that Scx, Sox5, Sox6, and Sox9 were few of TGFβ signaling targets. This study showed that TGFβ signaling is crucial for differentiation of AF from sclerotome. Jin et al108 used the tamoxifen‐inducible allele of Col2 CreER to targeted TGFβr2 expressing cells in the neonatal stages and showed that Col10a1, MMP13, ADAMTS4, and ADAMTS5 are negatively by TGFβ signaling. Based on the reporter data Cre‐mediated recombination in this allele was observed in the inner AF and growth plate chondrocytes only. In the postnatal stages, TGFβ signaling is downstream of Shh signaling in neonatal mouse IVDs.101 Hayes and Ralphs109 showed that TGFβ1 alone or in combination with IGF1 stimulated sulphated glycosaminoglycan, Col1, and Col2 secretion by AF cells.109 Using the mouse model of spondylocarpotarsal synostosis (SCT) Zieba et al106 showed that Filamin b (Flnb) null mice have early onset of degenerative disc defects, especially in the AF. SCT is an autosomal recessive disorder with loss of function mutation in Filamin B (FLNB) gene resulting in progressive vertebral fusions. FLNB was detected as early as E14.5 in mouse IVD. This study also showed an increase in TGFβ and BMP signaling in the AF of Flnb‐null mice. This raises the issue of the specific role of TGFβ in postnatal disc maintenance.106 Figure 1 is a schematic illustration depicting key stages of IVD development, highlighting the growth factors, morphogens, and transcription factors.
Figure 1.

Schematic illustration depicting key stages of intervertebral disc development, highlighting the growth factors, morphogens, and transcription factors. Depiction of key stages in axial skeletogenesis, including (A) node formation and elongation in the early embryo; (B) aggregation of the somatic mesenchyme around the notochord to form a continuous perichordal tube with metameric condensation of the axial mesenchyme (depicted by darker blue bands) leading to spine segmentation; and (C) formation of intervertebral discs. Notochord/nucleus pulposus derived structures are colored in red, and structures contributing to the annulus fibrosus are colored in blue. At each stage, selected growth factors, morphogens, and transcription factors (TFs) known to be required for IVD development are indicated, with notochord/nucleus pulposus associated factors indicated in red, and annulus fibrosus associated factors indicated in blue
4. PROGENITOR CELLS IN THE INTERVERTEBRAL DISC
Tissue maintenance and repair require the presence of local progenitor cells or stem cells, or the recruitment of appropriate cell types into the damaged site(s) for repair. In tissues that requires a constant turnover of cells in normal maintenance, replenishment is strictly controlled from specialized region(s) within the tissue, referred to as the stem cell niche. Example of localized niche is the intestinal crypt110, 111 with stem cells and supporting cells that continue to replenish the enterocytes as they are shed. Others include stem cells in the hair follicles112, 113 and the kidney nephrons.114 These are self‐renewing and proliferative stem cells with specialized requirements in tissues for high‐throughput cell replenishment. In tissues that do not require a high maintenance, stem cells are activated or recruited into the damaged sites as required. Bone fracture is a good example where progenitor cells are recruited from multiple regional sites to the damaged site for repair.115 Like articular cartilage, IVD do not repair well once damaged and the reason is unknown.116, 117 It could be related to the avascular nature of these tissues or the presence, location and ability to activate regional stem cells. There are many studies addressing the presence of stem/progenitor cells in the IVD in human and animal models, mostly using in vitro approaches, with limited in vivo findings. In addition, while the developmental and cellular processes in the formation of the IVD may be conserved in the different mammalian models, the cellular compositions of the adult discs are vastly different in the animal models analyzed to that of human; in particular the NP.118 Thus, in understanding of these studies, one needs to place the findings in context, from the perspective of the detection and cell isolation methods, source of the cells, and their differentiation potentials.
4.1. Presence of mesenchymal stem cells in the IVD
Potential stem cells with mesenchymal stem cells' (MSC) characteristics have been isolated from human IVDs. Given the difficulties of obtaining healthy tissues and the need to address the repair potentials of IVDs, many studies have used degenerated human IVD tissues in the search for progenitor cells. A seminal study used an explant culture approach in the isolation of cells with MSC characteristics from both the AF and NP.119 This approach relied on the migration and proliferation of cells selected as they detached from the explants and adhered to the plastic surface of the culture plate. From this, cells expressing typical MSC markers, such as CD105, CD166, CD63, CD49a, CD90, CD73, p75 low‐affinity nerve growth factor receptor, and CD133/1, were identified.119
It is interesting that MSCs with very similar characteristics can be isolated from both AF and NP tissues, even though their developmental origin is different,120 suggesting that these MSCs may not be of embryonic origin persisting from the development of the IVD. This would be consistent with the finding that pericytes, cells lining blood vessels shared similar cell surface markers with MSCs in situ,121 and the conceptual commentary that all MSCs may be of pericyte origin.122 Whether pericytes could find their way into the NP is questionable as the IVD is an avascular tissue. One possibility is external recruitment as exogenous human MSCs were shown to have a capacity to migrate through the CEP to the NP in ex vivo whole organ cultures using bovine IVDs, under culturing conditions that would induce degeneration such as excessive mechanical loading, limiting nutrition, and physical injury from a needle puncture.123, 124 However, CD146, a marker for MSCs with pericyte origin was not identified in progenitor cells isolated from mouse and human IVDs,125 but the existence of MSCs of pericyte origin cannot be excluded.
4.1.1. MSC markers in the healthy and degenerate IVD
The identification of CD133 positive (CD133+) cells is interesting as CD133 is a cell marker for many types of progenitor cells. However, it seems that CD133+ cells isolated from degenerated human IVD cells did not exhibit better adipogenic or chondrogenic differentiation capacity than CD133 negative (CD133−) cells,119 suggesting that in a degenerative state of an IVD, the environment could alter the functionality of the progenitor cells. The notion that a “local niche” could influence MSC potential is supported by a study comparing progenitor cells isolated from the NP and bone marrow (BM) MSCs from the same individual with degenerated NP that show a diminished adipogenic differentiation potential of the MSCs isolated from the degenerated NP.126 Furthermore, while using a common method for the isolation of MSCs from explant cultures of human IVDs, variation exists in the differentiation potential of the MSCs isolated.119, 126 While the reason is unclear, it is again likely to be related to changing niche of a degenerating IVD with age and severity of degeneration as confounding factors. Adding to the complexity is the identification of a small population of cells with MSC characteristics (CD105, CD90, and STRO‐1) from degenerated human IVDs that also co‐express OCT3/4 (a primitive marker for multipotency) and NOTCH1 (a signaling marker associated with cell fate determination).127 This represents only a minor population of NP cells which is as expected for progenitor cells in adult tissues.
4.2. Progenitor cells' isolation and functional assays
The explant approach to isolated MSCs presents a heterogeneous population of cells derived from degenerated IVDs that can differentiate along the mesenchyme lineage.119, 126, 127 Thus, it is also important to assess whether there are single‐cell progenitors in the IVD. This requires the approach of the colony‐forming assay (CFA), and requires the dissociation of the tissues to release cells as singletons and assess their potential to form colony‐forming units (CFUs) from a single cell.
A recent study assessed the CFUs of MSCs isolated from porcine NP of normal and degenerated (induced by annular puncture) conditions from the same animal.128 While both sources of MSCs (expressing MSC markers, CD29, CD90, and CD44) could differentiate into the classic mesenchymal lineages (osteoblasts, chondrocytes, and adipocytes), the CFUs were different, being higher from degenerated NP, and with increased proliferation rates in vitro. On the other hand, MSCs from the healthy NP showed a better chondrogenic differentiation potential and higher expression of NP extracellular matrix such as aggrecan and type II collagen.128 As the IVD degenerates, the changes in mechanical property can also influence the local environment/niche of progenitor cells. Indeed, adjusting extracellular matrix stiffness and elasticity can influence the differentiation lineage of progenitor cells in vitro,129, 130 and the fate of NP derived progenitor cells.131
Potential progenitor cells with MSCs characteristics have been isolated from the CEP from degenerated human IVDs, harvested through posterior discectomy procedure.132 Cells were cultured in 2% low‐melting‐point agarose in a 3D environment, allowing the formation of cell clusters. Large cell clusters were selected and expanded in monolayer culture. These cells express the typical markers for MSCs including CD105, CD73, CD90, CD44, CD166, and Stro‐1,132 and able to differentiated efficiently long the mesoderm lineage to osteoblasts, chondrocytes, and adipocytes. It is important to bear in mind that these are not clonal cells, but cells that form a cluster. Given the potential IVD progenitor cell niche is adjacent to the CEP and the connection between CEP and AF, it is possible that the progenitor cells in CEP132 and AF119, 133, 134 identified in the in vitro studies are originated from the IVD niche. More recently, cells expressing CD146 was found to be localized along the surface of the outer AF, a potential migratory route of the progenitor cells in the IVD niche and as part of the progression for differentiation.135 Interestingly, CD146 appears to define a commitment of AF cells for a contractile phenotype in vitro, in their ability to contract a collagen gel. This may be related to the CD146 expression in response to TGFβ1 stimulation and higher expression of SM22α and elastin, both associated with contractile property of tissues.135
4.2.1. Disialoganglioside 2 and tyrosine kinase receptor
Two novel NP progenitor cell markers: A detailed analysis of cells isolated from the NP of mouse and human IVDs showed the presence of potential “stem cells” with self‐renewal potentials in vivo, and progenitor cells with specific cell surface markers that could inform a hierarchical differentiation progression for progenitor cells to a mature NP cell.125 These were initially identified in the mouse NP assessed using the colony‐forming assay with isolated NP cells cultured in a methylcellulose semi‐solid medium136 that identified adhesive fibroblast colonies and nonadhesive sphere forming colonies. Focusing on cells from the sphere‐forming colonies (positive for type II collagen and aggrecan expression), two novel NP progenitor cell markers, disialoganglioside 2 (GD2) and tyrosine kinase receptor (Tie2) were identified. Importantly, it was shown that Tie2+/GD2− cells behave as dormant stem cells; Tie2+/GD2+ double positive cells have stem cell properties with self‐renewal potentials, with Tie2−/GD2+ as potential NP cell progenitors; the same Tie2−/GD2+ population showed expression of the “NP marker” CD24, suggesting possible committed NP progenitors; and finally, loosing expression for both Tie2 and GD2, but maintaining CD24 expression, define mature cells in the NP.125 Interestingly, in human IVDs, the combined number of Tie2+/GD2− and Tie2+/GD2+ cells, as well as the CFUs of NP cells, decline rapidly with age at around 40 years old, coincide with the age of onset and severity of IVD degeneration.125 It was further shown that angiopoietin‐1 as a ligand of Tie2 may have an important in maintaining these progenitor cells and protecting cells in the NP from apoptosis.125 These findings by Sakai et al125 are supported by two additional studies. From young bovine coccygeal discs, Tekari et al137 sorted NP cells for Tie2 and showed that Tie2+ cells characteristics of progenitors able to differentiate into the osteogenic, chondrogenic, and adipogenic lineages in vitro, forming spheroid colonies although with a decline during expansion.137 In another study, NP‐derived cells harvested from patients undergoing discectomy were subjected to cell sorting based on the Tie2 and GD2 co‐expression. The analyzed Tie2+/GD2+ population showed similar properties in colony‐forming ability, cell proliferation, and stem cell gene expression compared to BM‐derived MSCs from the same subjects. Interestingly, Tie2+/GD2+ cells differentiated into osteoblasts similar to BM‐MSCs, were found to be superior in chondrogenic differentiation but inferior in adipogenesis, compared to BM‐MSCs.138 Of interest is the work reported by Rodrigues‐Pinto et al.66 Human embryo and fetal spines (notochord and somites/sclerotome) were isolated by microdissection to follow the spatiotemporal expression of the believed human notochordal markers. Expression of Tie2, as well as KRT8, KRT18, KRT19, T, GAL3, CD24, CD55, CD90, BASP1, CTGF, and E‐Cad was assessed by immunohistochemistry. Their findings showed that Tie2, but also CD90 and E‐Cad, were not expressed in the early developing spine between the studied period of 3.5 to 18 weeks post‐conception, suggesting Tie2 is expressed later in IVD development and may be considered as an NP progenitor cell marker.
To summarize, these “progenitor cells” do show heterogeneity in their differentiation potential, and their presence in “healthy” human IVD has yet to be thoroughly studied, and their relationship to the adhesive MSCs in this and other studies also need to be addressed. It is possible that they are from notochord or sclerotome source and differ in embryonic or postnatal origins. In Figure 2, we summarize the to date identified cells in the healthy IVD, and their evolution in function of time.
Figure 2.
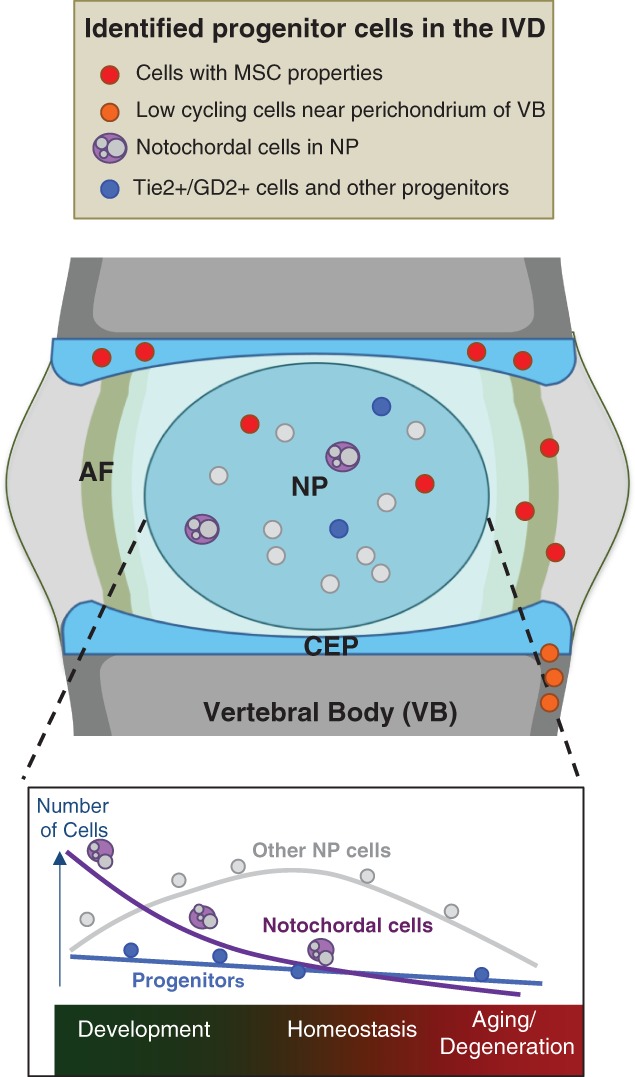
Identified progenitor cells populating the IVD and the changes in their numbers as a function of time, from development, through homeostasis and to aging. Depiction of cells with MSC properties, low‐cycling cells near the perichondrium of the vertebral body, notochordal and Tie2+/GD2+ and other progenitor cells
4.3. In vivo cell tracing
Using pulse‐chase labeling with BrDU has been reported for the IVD in New Zealand white rabbits.139 Few proliferating cells can be identified in the NP and AF during the labeling period, but following a prolong chase period, BrDU label‐retaining cells (potential stem/progenitor cells) are detected, concentrating in a region close to the perichondrium at the junction of the outer AF and the vertebral body,139 suggesting the presence of a stem cell niche. This niche is analogous to a region known as the “Groove of Ranvier” in long bones, where progenitor cells have been identified to reside in this region that serves to replenish chondrocytes in the cartilage growth plate.140, 141 These cells, identified in the IVD express MSCs markers, including Notch1, Stro‐1, and c‐KIT.139 Similar cells were identified in Sprague‐Dawley rats, Gottinger minipigs and degenerated human IVDs.139 A model was proposed for the presence of progenitor cells in this niche that undergo a transition of amplifying cells, and finally, differentiated cells of the IVD. Furthermore, it may be possible that these cells migrate into the IVD during growth and repair. Indeed, possible “migration routes” of these progenitor cells in the outer AF were studied, from an analysis of a cell adhesion and migration marker (β1 integrin) and EMT markers (Snail‐1 and ‐2) in young and aged rabbits.142 Activation of EMT would be consistent with a change in the migratory property of a cell. In EMT, the cytoskeleton of the cells is rearranged to a flattened phenotype to facilitate the migration of cells to a different location whereby members of the SNAIL superfamily of transcription factors are activated. As such, this pool of cells is suggested to be a source of progenitor cells for the maintenance of the AF during adult life.142 However, how this relates to the MSCs identified in degenerative human IVDs from the in vitro analyses is unclear, that will require careful cell tracing analysis using specific genetic tools in mice.
4.4. Summary
Together, the in vitro and in vivo studies support the presence of resident progenitor cell populations in the IVD. While in vitro studies can provide useful information, it is important to understand the limitation in the isolation and expansion approaches, as the possibility of cellular dedifferentiation cannot be excluded. Bona fide dedifferentiation of human articular chondrocytes was showed with a gradual unregulated expression of MSCs markers in during monolayer cultures; including CD90, CD166, CD49c, CD44, CD10, CD26, CD49e, CD151, CD51/61, and CD81,143 that can be accelerated by FGF2 supplement.144 Whether of IVD cells dedifferentiate in monolayer culture has not been specifically addressed. Furthermore, the relevance of the progenitor cells in human needs substantial validation as many studies uses degenerated IVDs, and why the IVD continues to degenerate even in the presence of these progenitor cells is not clear at all. The opportunities are available to decipher the presence, location, and fate of these progenitor cells in animal models prior to validation in human tissues. Given the vast mouse tools available to study cell fate, it is highly feasible if appropriate gene markers can be identified to track and localize potential progenitor cells or cells providing homeostasis support of the IVD tissues. A golden opportunity is the application of single‐cell transcriptomic analyses to interrogate the cell types present in the different components of the IVD, and to identified presence of potential progenitor cells without in vitro culture and cell expansion. The technologies are available and the cost is becoming affordable.145, 146 Once identified, the in situ identification and localization in the IVD tissue can be validated. This is clearly important in the formulation of therapeutic treatments of symptomatic IVD degeneration, for exogenous cell therapy, or activation of endogenous progenitor cells for repair.
ACKNOWLEDGEMENTS
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Awards Number RO1AR065530 (CLD) and R01AR066517 (ZG). The content is solely under the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The Natural Sciences and Engineering Research Council of Canada and Canadian Institutes of Health Research funded C.A.S., who is also supported by a CIHR New Investigator Award and Early Researcher Award from the Ontario Ministry of Research and Innovation. The Research Council of Hong Kong under the Theme‐based Research Scheme (T12‐708/12‐N) provided funding support to DC. The authors thank Dr Wilson Chan for his help in the preparation of Figure 2.
Author contributions
C.A.S., D.C., C.L.D., and Z.G. contributed to the concept of the paper and wrote the manuscript. All authors have read and approved the final submitted manuscript.
Conflict of interest
The authors declare not to have any conflict of interest.
Séguin CA, Chan D, Dahia CL, Gazit Z. Latest advances in intervertebral disc development and progenitor cells. JOR Spine. 2018;1:e1030. 10.1002/jsp2.1030
Funding information National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: R01AR065530R01AR066517; The Natural Sciences and Engineering Research Council of Canada and Canadian Institutes of Health Research, Grant/Award Number: CIHR New Investigator Award and Early Researcher A; The Research Council of Hong Kong, Grant/Award Number: T12‐708/12‐N
REFERENCES
- 1. Niehrs C. Regionally specific induction by the Spemann‐Mangold organizer. Nat Rev Genet. 2004;5(6):425‐434. [DOI] [PubMed] [Google Scholar]
- 2. Mangold H. Über die Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Roux Arch Entw Mech Org. 1924;100:599‐638. [Google Scholar]
- 3. Oppenheimer JM. Transplantation experiments on developing teleosts (Fundulus and Perca). J Exp Zool. 1936;72:409‐437. [Google Scholar]
- 4. Waddington CH. Developmental mechanics of chick and duck embryos. Nature. 1930;125:924‐925. [Google Scholar]
- 5. Beddington RS. Induction of a second neural axis by the mouse node. Development. 1994;120(3):613‐620. [DOI] [PubMed] [Google Scholar]
- 6. Tam PP, Beddington RS. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development. 1987;99(1):109‐126. [DOI] [PubMed] [Google Scholar]
- 7. Lee JD, Anderson KV. Morphogenesis of the node and notochord: the cellular basis for the establishment and maintenance of left‐right asymmetry in the mouse. Dev Dyn. 2008;237(12):3464‐3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sulik K, Dehart DB, Inagaki T, et al. Morphogenesis of the murine node and notochordal plate. Dev Dyn. 1994;201(3):260‐278. [DOI] [PubMed] [Google Scholar]
- 9. Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13(6):884‐896. [DOI] [PubMed] [Google Scholar]
- 10. Davidson BP, Tam PP. The node of the mouse embryo. Curr Biol. 2000;10(17):R617‐R619. [DOI] [PubMed] [Google Scholar]
- 11. Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left‐right asymmetry. Cell. 2006;125(1):33‐45. [DOI] [PubMed] [Google Scholar]
- 12. Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left‐right asymmetry. Genes Dev. 2002;16(18):2339‐2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Babu D, Roy S. Left‐right asymmetry: cilia stir up new surprises in the node. Open Biol. 2013;3(5):130052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanaka Y, Okada Y, Hirokawa N. FGF‐induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left‐right determination. Nature. 2005;435(7039):172‐177. [DOI] [PubMed] [Google Scholar]
- 15. Nonaka S, Tanaka Y, Okada Y, et al. Randomization of left‐right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95(6):829‐837. [DOI] [PubMed] [Google Scholar]
- 16. McGrath J, Brueckner M. Cilia are at the heart of vertebrate left‐right asymmetry. Curr Opin Genet Dev. 2003;13(4):385‐392. [DOI] [PubMed] [Google Scholar]
- 17. Shiratori H, Hamada H. The left‐right axis in the mouse: from origin to morphology. Development. 2006;133(11):2095‐2104. [DOI] [PubMed] [Google Scholar]
- 18. Schlueter J, Brand T. Left‐right axis development: examples of similar and divergent strategies to generate asymmetric morphogenesis in chick and mouse embryos. Cytogenet Genome Res. 2007;117(1–4):256‐267. [DOI] [PubMed] [Google Scholar]
- 19. Yamamoto M, Meno C, Sakai Y, et al. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior‐posterior patterning and node formation in the mouse. Genes Dev. 2001;15(10):1242‐1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rashbass P, Wilson V, Rosen B, Beddington RS. Alterations in gene expression during mesoderm formation and axial patterning in Brachyury (T) embryos. Int J Dev Biol. 1994;38(1):35‐44. [PubMed] [Google Scholar]
- 21. Shawlot W, Behringer RR. Requirement for Lim1 in head‐organizer function. Nature. 1995;374(6521):425‐430. [DOI] [PubMed] [Google Scholar]
- 22. Ang SL, Rossant J. HNF‐3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78(4):561‐574. [DOI] [PubMed] [Google Scholar]
- 23. Sawada A, Nishizaki Y, Sato H, et al. Tead proteins activate the Foxa2 enhancer in the node in cooperation with a second factor. Development. 2005;132(21):4719‐4729. [DOI] [PubMed] [Google Scholar]
- 24. Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996;122(1):243‐252. [DOI] [PubMed] [Google Scholar]
- 25. Beckers A, Alten L, Viebahn C, Andre P, Gossler A. The mouse homeobox gene Noto regulates node morphogenesis, notochordal ciliogenesis, and left right patterning. Proc Natl Acad Sci USA. 2007;104(40):15765‐15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamplin OJ, Cox BJ, Rossant J. Integrated microarray and ChIP analysis identifies multiple Foxa2 dependent target genes in the notochord. Dev Biol. 2011;360(2):415‐425. [DOI] [PubMed] [Google Scholar]
- 27. Peck SH, McKee KK, Tobias JW, Malhotra NR, Harfe BD, Smith LJ. Whole Transcriptome analysis of notochord‐derived cells during embryonic formation of the nucleus Pulposus. Sci Rep. 2017;7(1):10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balmer S, Nowotschin S, Hadjantonakis AK. Notochord morphogenesis in mice: current understanding & open questions. Dev Dyn. 2016;245(5):547‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimelman D, Griffin KJ. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev. 2000;10(4):350‐356. [DOI] [PubMed] [Google Scholar]
- 30. Kinder SJ, Tsang TE, Wakamiya M, et al. The organizer of the mouse gastrula is composed of a dynamic population of progenitor cells for the axial mesoderm. Development. 2001;128(18):3623‐3634. [DOI] [PubMed] [Google Scholar]
- 31. Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. 1997;68(1–2):3‐25. [DOI] [PubMed] [Google Scholar]
- 32. Camus A, Davidson BP, Billiards S, et al. The morphogenetic role of midline mesendoderm and ectoderm in the development of the forebrain and the midbrain of the mouse embryo. Development. 2000;127(9):1799‐1813. [DOI] [PubMed] [Google Scholar]
- 33. Herrmann BG. Expression pattern of the Brachyury gene in whole‐mount TWis/TWis mutant embryos. Development. 1991;113(3):913‐917. [DOI] [PubMed] [Google Scholar]
- 34. Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17(13):1646‐1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoodless PA, Pye M, Chazaud C, et al. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 2001;15(10):1257‐1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weinstein DC, Ruiz i Altaba A, Chen WS, et al. The winged‐helix transcription factor HNF‐3 beta is required for notochord development in the mouse embryo. Cell. 1994;78(4):575‐588. [DOI] [PubMed] [Google Scholar]
- 37. Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132(11):2503‐2512. [DOI] [PubMed] [Google Scholar]
- 38. Gotz W, Osmers R, Herken R. Localisation of extracellular matrix components in the embryonic human notochord and axial mesenchyme. J Anat. 1995;186(Pt 1):111‐121. [PMC free article] [PubMed] [Google Scholar]
- 39. Trapani V, Bonaldo P, Corallo D. Role of the ECM in notochord formation, function and disease. J Cell Sci. 2017;130(19):3203‐3211. [DOI] [PubMed] [Google Scholar]
- 40. Choi KS, Harfe BD. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proc Natl Acad Sci USA. 2011;108(23):9484‐9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smits P, Lefebvre V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development. 2003;130(6):1135‐1148. [DOI] [PubMed] [Google Scholar]
- 42. Ng LJ, Wheatley S, Muscat GEO, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183(1):108‐121. [DOI] [PubMed] [Google Scholar]
- 43. Domowicz M, Li H, Hennig A, Henry J, Vertel BM, Schwartz NB. The biochemically and immunologically distinct CSPG of notochord is a product of the aggrecan gene. Dev Biol. 1995;171(2):655‐664. [DOI] [PubMed] [Google Scholar]
- 44. Zhao Q, Eberspaecher H, Lefebvre V, de Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209(4):377‐386. [DOI] [PubMed] [Google Scholar]
- 45. Parsons MJ, Pollard SM, Saúde L, et al. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129(13):3137‐3146. [DOI] [PubMed] [Google Scholar]
- 46. Knudson CB, Knudson W. Cartilage proteoglycans. Semin Cell Dev Biol. 2001;12(2):69‐78. [DOI] [PubMed] [Google Scholar]
- 47. Adams DS, Keller R, Koehl MA. The mechanics of notochord elongation, straightening and stiffening in the embryo of Xenopus laevis. Development. 1990;110(1):115‐130. [DOI] [PubMed] [Google Scholar]
- 48. Wang F, Gao ZX, Cai F, et al. Formation, function, and exhaustion of notochordal cytoplasmic vacuoles within intervertebral disc: current understanding and speculation. Oncotarget. 2017;8(34):57800‐57812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamada T, Placzek M, Tanaka H, Dodd J, Jessell TM. Control of cell pattern in the developing nervous system: polarizing activity of the floor plate and notochord. Cell. 1991;64(3):635‐647. [DOI] [PubMed] [Google Scholar]
- 50. Fan CM, Tessier‐Lavigne M. Patterning of mammalian somites by surface ectoderm and notochord: evidence for sclerotome induction by a hedgehog homolog. Cell. 1994;79(7):1175‐1186. [DOI] [PubMed] [Google Scholar]
- 51. Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124(21):4243‐4252. [DOI] [PubMed] [Google Scholar]
- 52. Meadows SM, Fletcher PJ, Moran C, et al. Integration of repulsive guidance cues generates avascular zones that shape mammalian blood vessels. Circ Res. 2012;110(1):34‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237(12):3953‐3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maier JA, Lo Y, Harfe BD. Foxa1 and Foxa2 are required for formation of the intervertebral discs. PLoS One. 2013;8(1):e55528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hunter CJ, Matyas JR, Duncan NA. The notochordal cell in the nucleus pulposus: a review in the context of tissue engineering. Tissue Eng. 2003;9(4):667‐677. [DOI] [PubMed] [Google Scholar]
- 56. Roberts S. Disc morphology in health and disease. Biochem Soc Trans. 2002;30(Pt 6):864‐869. [DOI] [PubMed] [Google Scholar]
- 57. Richardson SM, Ludwinski FE, Gnanalingham KK, Atkinson RA, Freemont AJ, Hoyland JA. Notochordal and nucleus pulposus marker expression is maintained by sub‐populations of adult human nucleus pulposus cells through aging and degeneration. Sci Rep. 2017;7(1):1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sive JI, Baird P, Jeziorsk M, Watkins A, Hoyland JA, Freemont AJ. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol Pathol. 2002;55(2):91‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Minogue BM, Richardson SM, Zeef LAH, Freemont AJ, Hoyland JA. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 2010;62(12):3695‐3705. [DOI] [PubMed] [Google Scholar]
- 60. Minogue BM, Richardson SM, Zeef LAH, Freemont AJ, Hoyland JA. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther. 2010;12(1):R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Power KA, Grad S, Rutges JPHJ, et al. Identification of cell surface‐specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum. 2011;63(12):3876‐3886. [DOI] [PubMed] [Google Scholar]
- 62. Risbud MV, Schoepflin ZR, Mwale F, et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the spine research interest group at the 2014 annual ORS meeting. J Orthop Res. 2015;33(3):283‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age‐related changes in lumbar intervertebral discs: 2002 Volvo award in basic science. Spine (Phila Pa 1976). 2002;27(23):2631‐2644. [DOI] [PubMed] [Google Scholar]
- 64. Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246(1):129‐137. [DOI] [PubMed] [Google Scholar]
- 65. Tang X, Jing L, Richardson WJ, et al. Identifying molecular phenotype of nucleus pulposus cells in human intervertebral disc with aging and degeneration. J Orthop Res. 2016;34(8):1316‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rodrigues‐Pinto R, Berry A, Piper‐Hanley K, Hanley N, Richardson SM, Hoyland JA. Spatiotemporal analysis of putative notochordal cell markers reveals CD24 and keratins 8, 18, and 19 as notochord‐specific markers during early human intervertebral disc development. J Orthop Res. 2016;34(8):1327‐1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vujovic S, Henderson S, Presneau N, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209(2):157‐165. [DOI] [PubMed] [Google Scholar]
- 68. Henriksson H et al. Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: a study in four species. Spine (Phila Pa 1976). 2009;34(21):2278‐2287. [DOI] [PubMed] [Google Scholar]
- 69. Kim KW, Lim TH, Kim JG, Jeong ST, Masuda K, An HS. The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine (Phila Pa 1976). 2003;28(10):982‐990. [DOI] [PubMed] [Google Scholar]
- 70. Trout JJ, Buckwalter JA, Moore KC. Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat Rec. 1982;204(4):307‐314. [DOI] [PubMed] [Google Scholar]
- 71. Pazzaglia UE, Salisbury JR, Byers PD. Development and involution of the notochord in the human spine. J R Soc Med. 1989;82(7):413‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liebscher T, Haefeli M, Wuertz K, Nerlich AG, Boos N. Age‐related variation in cell density of human lumbar intervertebral disc. Spine (Phila Pa 1976). 2011;36(2):153‐159. [DOI] [PubMed] [Google Scholar]
- 73. McCann MR, Tamplin OJ, Rossant J, Séguin CA. Tracing notochord‐derived cells using a Noto‐cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5(1):73‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Carlier EW. Fate of the notochord and development of the intervertebral disc in the sheep, with observations on the structure of the adult disc in these animals. J Anat Physiol. 1890;24(Pt 4):573‐584.1. [PMC free article] [PubMed] [Google Scholar]
- 75. Christ B, Ordahl CP. Early stages of chick somite development. Anat Embryol (Berl). 1995;191(5):381‐396. [DOI] [PubMed] [Google Scholar]
- 76. Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113(2):235‐248. [DOI] [PubMed] [Google Scholar]
- 77. Brent AE, Braun T, Tabin CJ. Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development. 2005;132(3):515‐528. [DOI] [PubMed] [Google Scholar]
- 78. Cserjesi P, Brown D, Ligon KL, et al. Scleraxis: a basic helix‐loop‐helix protein that prefigures skeletal formation during mouse embryogenesis. Development. 1995;121(4):1099‐1110. [DOI] [PubMed] [Google Scholar]
- 79. Sugimoto Y, Takimoto A, Akiyama H, et al. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013;140(11):2280‐2288. [DOI] [PubMed] [Google Scholar]
- 80. Sugimoto Y, Takimoto A, Hiraki Y, Shukunami C. Generation and characterization of ScxCre transgenic mice. Genesis. 2013;51(4):275‐283. [DOI] [PubMed] [Google Scholar]
- 81. Murchison ND, Price BA, Conner DA, et al. Regulation of tendon differentiation by scleraxis distinguishes force‐transmitting tendons from muscle‐anchoring tendons. Development. 2007;134(14):2697‐2708. [DOI] [PubMed] [Google Scholar]
- 82. Bruggeman BJ, Maier JA, Mohiuddin YS, et al. Avian intervertebral disc arises from rostral sclerotome and lacks a nucleus pulposus: implications for evolution of the vertebrate disc. Dev Dyn. 2012;241(4):675‐683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cai CL, Martin JC, Sun Y, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454(7200):104‐108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Borycki AG, Mendham L, Emerson CP Jr. Control of somite patterning by Sonic hedgehog and its downstream signal response genes. Development. 1998;125(4):777‐790. [DOI] [PubMed] [Google Scholar]
- 85. Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13(2):225‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dockter J, Ordahl CP. Dorsoventral axis determination in the somite: a re‐examination. Development. 2000;127(10):2201‐2206. [DOI] [PubMed] [Google Scholar]
- 87. Marcelle C, Ahlgren S, Bronner‐Fraser M. In vivo regulation of somite differentiation and proliferation by Sonic Hedgehog. Dev Biol. 1999;214(2):277‐287. [DOI] [PubMed] [Google Scholar]
- 88. Choi KS, Lee C, Harfe BD. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech Dev. 2012;129(9–12):255‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stafford DA, Brunet LJ, Khokha MK, Economides AN, Harland RM. Cooperative activity of noggin and gremlin 1 in axial skeleton development. Development. 2011;138(5):1005‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rider CC, Mulloy B. Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem J. 2010;429(1):1‐12. [DOI] [PubMed] [Google Scholar]
- 91. Buttitta L, Mo R, Hui CC, Fan CM. Interplays of Gli2 and Gli3 and their requirement in mediating Shh‐dependent sclerotome induction. Development. 2003;130(25):6233‐6243. [DOI] [PubMed] [Google Scholar]
- 92. Murtaugh LC, Zeng L, Chyung JH, Lassar AB. The chick transcriptional repressor Nkx3.2 acts downstream of Shh to promote BMP‐dependent axial chondrogenesis. Dev Cell. 2001;1(3):411‐422. [DOI] [PubMed] [Google Scholar]
- 93. Zeng L, Kempf H, Murtaugh LC, Sato ME, Lassar AB. Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes Dev. 2002;16(15):1990‐2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang XM, Yang XJ. Temporal and spatial effects of sonic hedgehog signaling in chick eye morphogenesis. Dev Biol. 2001;233(2):271‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wallin J, Wilting J, Koseki H, Fritsch R, Christ B, Balling R. The role of Pax‐1 in axial skeleton development. Development. 1994;120(5):1109‐1121. [DOI] [PubMed] [Google Scholar]
- 96. Koseki H, Wallin J, Wilting J, et al. A role for Pax‐1 as a mediator of notochordal signals during the dorsoventral specification of vertebrae. Development. 1993;119(3):649‐660. [DOI] [PubMed] [Google Scholar]
- 97. Wilm B, Dahl E, Peters H, Balling R, Imai K. Targeted disruption of Pax1 defines its null phenotype and proves haploinsufficiency. Proc Natl Acad Sci USA. 1998;95(15):8692‐8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yamashita S, Andoh M, Ueno‐Kudoh H, Sato T, Miyaki S, Asahara H. Sox9 directly promotes Bapx1 gene expression to repress Runx2 in chondrocytes. Exp Cell Res. 2009;315(13):2231‐2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rodrigo I, Hill RE, Balling R, Münsterberg A, Imai K. Pax1 and Pax9 activate Bapx1 to induce chondrogenic differentiation in the sclerotome. Development. 2003;130(3):473‐482. [DOI] [PubMed] [Google Scholar]
- 100. Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126(24):5699‐5711. [DOI] [PubMed] [Google Scholar]
- 101. Dahia CL, Mahoney E, Wylie C. Shh signaling from the nucleus pulposus is required for the postnatal growth and differentiation of the mouse intervertebral disc. PLoS One. 2012;7(4):e35944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Barrionuevo F, Taketo MM, Scherer G, Kispert A. Sox9 is required for notochord maintenance in mice. Dev Biol. 2006;295(1):128‐140. [DOI] [PubMed] [Google Scholar]
- 103. Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813‐2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Smits P, Li P, Mandel J, et al. The transcription factors L‐Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1(2):277‐290. [DOI] [PubMed] [Google Scholar]
- 105. Henry SP, Liang S, Akdemir KC, de Crombrugghe B. The postnatal role of Sox9 in cartilage. J Bone Miner Res. 2012;27(12):2511‐2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zieba J, Forlenza KN, Khatra JS, et al. TGFβ and BMP dependent cell fate changes due to loss of Filamin B produces disc degeneration and progressive vertebral fusions. PLoS Genet. 2016;12(3):e1005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Baffi MO, Slattery E, Sohn P, Moses HL, Chytil A, Serra R. Conditional deletion of the TGF‐beta type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Dev Biol. 2004;276(1):124‐142. [DOI] [PubMed] [Google Scholar]
- 108. Jin H, Shen J, Wang B, Wang M, Shu B, Chen D. TGF‐beta signaling plays an essential role in the growth and maintenance of intervertebral disc tissue. FEBS Lett. 2011;585(8):1209‐1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hayes AJ, Ralphs JR. The response of foetal annulus fibrosus cells to growth factors: modulation of matrix synthesis by TGF‐β1 and IGF‐1. Histochem Cell Biol. 2011;136(2):163‐175. [DOI] [PubMed] [Google Scholar]
- 110. Meran L, Baulies A, Li VSW. Intestinal stem cell niche: the extracellular matrix and cellular components. Stem Cells Int. 2017;2017:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kretzschmar K, Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev Cell. 2016;38(6):590‐600. [DOI] [PubMed] [Google Scholar]
- 112. Ji J, Ho BSY, Qian G, Xie XM, Bigliardi PL, Bigliardi‐Qi M. Aging in hair follicle stem cells and niche microenvironment. J Dermatol. 2017;44:1097‐1104. [DOI] [PubMed] [Google Scholar]
- 113. Rompolas P, Greco V. Stem cell dynamics in the hair follicle niche. Sem Cell Dev Biol. 2014;25:34‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Little MH. Growing kidney tissue from stem cells: how far from “party trick” to medical application? Cell Stem Cell. 2016;18(6):695‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Einhorn TA, Gerstenfeld LC. Fracture healing: mechanisms and interventions. Nat Rev Rheumatol. 2015;11(1):45‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Melrose J, Smith SM, Fuller ES, et al. Biglycan and fibromodulin fragmentation correlates with temporal and spatial annular remodelling in experimentally injured ovine intervertebral discs. Eur Spine J. 2007;16(12):2193‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Humzah MD, Soames RW. Human intervertebral disc: structure and function. Anat Rec. 1988;220(4):337‐356. [DOI] [PubMed] [Google Scholar]
- 118. Alini M, Eisenstein SM, Ito K, et al. Are animal models useful for studying human disc disorders/degeneration? Eur Spine J. 2008;17(1):2‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Risbud MV, Guttapalli A, Tsai TT, et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine. 2007;32(23):2537‐2544. [DOI] [PubMed] [Google Scholar]
- 120. Chan WC et al. Coming together is a beginning: the making of an intervertebral disc. Birth Defects Res C Embryo Today. 2014;102(1):83‐100. [DOI] [PubMed] [Google Scholar]
- 121. Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3(3):301‐313. [DOI] [PubMed] [Google Scholar]
- 122. Caplan AI. All MSCs are pericytes? Cell Stem Cell. 2008;3(3):229‐230. [DOI] [PubMed] [Google Scholar]
- 123. Illien‐Jünger S, Gantenbein‐Ritter B, Grad S, et al. The combined effects of limited nutrition and high‐frequency loading on intervertebral discs with endplates. Spine. 2010;35(19):1744‐1752. [DOI] [PubMed] [Google Scholar]
- 124. Illien‐Jünger S, Pattappa G, Peroglio M, et al. Homing of mesenchymal stem cells in induced degenerative intervertebral discs in a whole organ culture system. Spine. 2012;37(22):1865‐1873. [DOI] [PubMed] [Google Scholar]
- 125. Sakai D, Nakamura Y, Nakai T, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Blanco JF, Graciani IF, Sanchez‐Guijo FM, et al. Isolation and characterization of mesenchymal stromal cells from human degenerated nucleus pulposus: comparison with bone marrow mesenchymal stromal cells from the same subjects. Spine. 2010;35(26):2259‐2265. [DOI] [PubMed] [Google Scholar]
- 127. Brisby H, Papadimitriou N, Brantsing C, Bergh P, Lindahl A, Barreto Henriksson H. The presence of local mesenchymal progenitor cells in human degenerated intervertebral discs and possibilities to influence these in vitro: a descriptive study in humans. Stem Cells Dev. 2013;22(5):804‐814. [DOI] [PubMed] [Google Scholar]
- 128. Mizrahi O, Sheyn D, Tawackoli W, et al. Nucleus pulposus degeneration alters properties of resident progenitor cells. Spine J. 2013;13(7):803‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677‐689. [DOI] [PubMed] [Google Scholar]
- 130. Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Navaro Y, Bleich‐Kimelman N, Hazanov L, et al. Matrix stiffness determines the fate of nucleus pulposus–derived stem cells. Biomaterials. 2015;49:68‐76. [DOI] [PubMed] [Google Scholar]
- 132. Liu L‐T, Huang B, Li CQ, Zhuang Y, Wang J, Zhou Y. Characteristics of stem cells derived from the degenerated human intervertebral disc cartilage endplate. PloS One. 2011;6(10):e26285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Feng G, Yang X, Shang H, et al. Multipotential differentiation of human anulus fibrosus cells: an in vitro study. JBJS. 2010;92(3):675‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jin L, Liu Q, Scott P, et al. Annulus fibrosus cell characteristics are a potential source of intervertebral disc pathogenesis. PloS One. 2014;9(5):e96519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Nakai T, Sakai D, Nakamura Y, et al. CD146 defines commitment of cultured annulus fibrosus cells to express a contractile phenotype. J Orthop Res. 2016;34(8):1361‐1372. [DOI] [PubMed] [Google Scholar]
- 136. Kobayashi T, Nakano T, Nakahata T, et al. Formation of mast cell colonies in methylcellulose by mouse peritoneal cells and differentiation of these cloned cells in both the skin and the gastric mucosa of W/Wv mice: evidence that a common precursor can give rise to both" connective tissue‐type" and" mucosal" mast cells. J Immunol. 1986;136(4):1378‐1384. [PubMed] [Google Scholar]
- 137. Tekari A, Chan SCW, Sakai D, Grad S, Gantenbein B. Angiopoietin‐1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res Ther. 2016;7(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Li XC, Tang Y, Wu JH, Yang PS, Wang DL, Ruan DK. Characteristics and potentials of stem cells derived from human degenerated nucleus pulposus: potential for regeneration of the intervertebral disc. BMC Musculoskelet Disord. 2017;18(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Henriksson HB, Thornemo M, Karlsson C, et al. Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: a study in four species. Spine. 2009;34(21):2278‐2287. [DOI] [PubMed] [Google Scholar]
- 140. Karlsson C, Thornemo M, Henriksson HB, Lindahl A. Identification of a stem cell niche in the zone of Ranvier within the knee joint. J Anat. 2009;215(3):355‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Candela ME, Yasuhara R, Iwamoto M, Enomoto‐Iwamoto M. Resident mesenchymal progenitors of articular cartilage. Matrix Biol. 2014;39:44‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Henriksson HB, Svala E, Skioldebrand E, Lindahl A, Brisby H. Support of concept that migrating progenitor cells from stem cell niches contribute to normal regeneration of the adult mammal intervertebral disc: a descriptive study in the New Zealand white rabbit. Spine. 2012;37(9):722‐732. [DOI] [PubMed] [Google Scholar]
- 143. Diaz‐Romero J, Nesic D, Grogan SP, Heini P, Mainil‐Varlet P. Immunophenotypic changes of human articular chondrocytes during monolayer culture reflect bona fide dedifferentiation rather than amplification of progenitor cells. J Cell Physiol. 2008;214(1):75‐83. [DOI] [PubMed] [Google Scholar]
- 144. Lee J, Lee JY, Chae BC, Jang J, Lee EA, Son Y. Fully dedifferentiated chondrocytes expanded in specific Mesenchymal stem cell growth medium with FGF2 obtains Mesenchymal stem cell phenotype in vitro but retains chondrocyte phenotype in vivo. Cell Transplant. 2017;26(10):1673‐1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single‐cell RNA sequencing. Mol Cell. 2015;58(4):610‐620. [DOI] [PubMed] [Google Scholar]
- 146. Semrau S, Goldmann JE, Soumillon M, Mikkelsen TS, Jaenisch R, van Oudenaarden A. Dynamics of lineage commitment revealed by single‐cell transcriptomics of differentiating embryonic stem cells. Nat Commun. 2017;8(1):1096. [DOI] [PMC free article] [PubMed] [Google Scholar]


