Abstract
Prostate cancer is the most common non-skin related cancer affecting 1 in 7 men in the United States. Treatment of patients with prostate cancer still remains a difficult decision-making process that requires physicians to balance clinical benefits, life expectancy, comorbidities, and treatment-related side effects. Gleason score (a sum of the primary and secondary Gleason patterns) solely based on morphological prostate glandular architecture has shown as one of the best predictors of prostate cancer outcome. Significant progress has been made on molecular subtyping prostate cancer delineated through the increasing use of gene sequencing. Prostate cancer patients with Gleason score of 7 show heterogeneity in recurrence and survival outcomes. Therefore, we propose to assess the correlation between histopathology images and genomic data with disease recurrence in prostate tumors with a Gleason 7 score to identify prognostic markers. In the study, we identify image biomarkers within tissue WSIs by modeling the spatial relationship from automatically created patches as a sequence within WSI by adopting a recurrence network model, namely long short-term memory (LSTM). Our preliminary results demonstrate that integrating image biomarkers from CNN with LSTM and genomic pathway scores, is more strongly correlated with patients recurrence of disease compared to standard clinical markers and engineered image texture features. The study further demonstrates that prostate cancer patients with Gleason score of 4+3 have a higher risk of disease progression and recurrence compared to prostate cancer patients with Gleason score of 3+4.
Keywords: Prostate Cancer, Gleason Grading, Histopathology Image Analysis, Pathway Analysis
1. INTRODUCTION
Prostate cancer is the most common non-skin related cancer affecting 1 in 7 men in the United States.1 In 2016, the estimated numbers of death from prostate cancer was 26,120 and new cases account for 21% of all newly cancer diagnoses in men each year.1 Gleason grading is a strong predictor of survival among men with prostate cancer. Gleason score is a sum of the primary and secondary patterns with a range of 2 to 10. Studies show that patients with Gleason 4+3 tumors compared to Gleason 3+4 tumors had an increased risk of prostate cancer specific mortality and increased risk of recurrence and progression.2–4 As such, these studies indicate that delineating prostate cancer patients with Gleason score 7 into 3+4 and 4+3 has a prognostic significance for patients disease management and survival.3–5
There have been many studies on computer-aided Gleason grading.6–12 In general there are four approaches that have been used to assess prostate Gleason pattern grading including color-statistical based,6, 9 texture- based,8, 10, 13 structure-based8, 14, 15 and tissue-component-based.16–18 Gleason pattern 3 has a relative clear glandular shape while the Gleason pattern 4 consists of a range of various glandular patterns including glomeru-loid glands, cribriform glands, poorly formed and fused glands and irregular cribriform glands. Convolutional Neural Networks (CNN) based histopathology image analyses have demonstrated significant advances in tissue and cellular segmentation19–21 and classifications.21–24 Due to a large size of a WSI (typically a few GB/per WSI) that leads to not practicable for Gleason grading directly, we model the computational image biomarkers extracted from automatically cropped patches within the WSI to differentiate the images with different Gleason patterns.
To further explore, the Microarray-based gene expression signatures have been used in various studies to identify cancer subtypes, determine the recurrence of disease and response to specific therapies.25 Multiple studies have also shown that gene expression signatures can be used to analyze oncogenic pathways and these signatures have been used to identify differences between specific cancer types and tumor subtypes. Moreover, patterns of oncogenic pathway activity have been used to identify differences in underlying molecular mechanisms and shown to correlate with clinical outcome of specific cancers.26–28
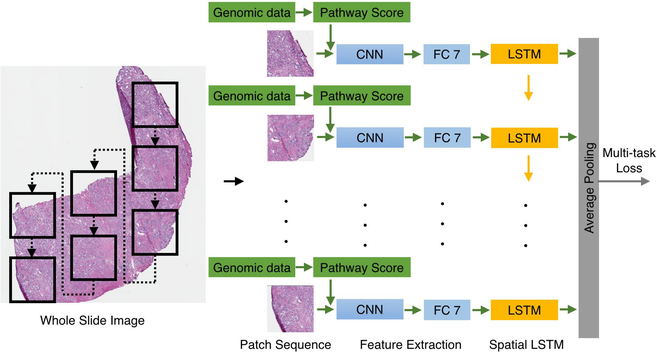
In the study, we apply the prostate cancer patients disease free time (months) since their initial treatment as the time-to-recurrence as the time variable for Cox survival model. Using the survival model, we assess the image biomarkers and genetic biomarkers related to recurrence hazard risk scores for prostate cancer patients with Gleason score of 7 in the context of other clinical prognostic factors, including the primary and the secondary Gleason patterns, patients’ prostate-specific antigen (PSA), age and clinical tumor stage by using our approach of neural networks analysis. The architecture of the neural networks is illustrated in Figure 1
Figure 1.
The architecture of convolutional neural networks with LSTM using WSI and genomic data for quantifying image and genetic biomarkers.
2. METHODS
2.1. Patch Sequence Modeling
In the study, we identify image biomarkers within tissue whole-slide images (WSIs) by modeling the spatial relationship of patches as a temporal sequence by adopting a recurrence network (RNN) model ,29 namely the long short-term memory (LSTM)30 to learn the fine-grained discriminative information among patches (here Gleason patterns 3 and 4) and the global representation of the WSIs.
2.2. Pathway Score
We examine genomic-based patterns of oncogenic pathway activity, the tumor microenvironment and other important features in human prostate tumors using a panel of 270 previously published gene expression signatures. We apply each signature to the prostate cancer gene expression microarray data from the TCGA/GDC data portal, for which the Gleason subtype have been determined. Consistent patterns of pathway activity emerge for each subtype, and we quantitatively assesses these patterns using a Student’s two tailed t-test, where a p-value of 0.01 or less is considered significant. To apply a given signature to a new data set, the expression data are filtered to contain only those genes that met the previous criteria, and the mean expression values are calculated using all genes within a given signature. The methods detailed in the original studies are followed as closely as possible.27, 28
In conjunction with clinical prognostic features including the primary and secondary Gleason patterns, patients’ prostate specific antigen (PSA), age and tumor stage, image biomarkers and pathway scores are fed into a Cox survival model for studying patients recurrence to advanced disease.
3. EXPERIMENTS AND RESULTS
The tissue WSIs used in the study are from Cancer Genome Atlas (TC-GA) and genetic data are from Genomic Data Commons (GDC). Patients with Gleason score 7 (3+4 and 4+4) are included in the study for the primary Gleason pattern 4 is a significant cut-off point for patients with prostate cancer, who wont be recommended active surveillance, but surgery instead. The image patches within each WSI are automatically cropped under 40X objective magnification with a patch size 4096×4096. No tissue, less tissue and low signal-to-noise ratio (SNR) patches are automatically eliminated from the experiments due to the heterogeneous quality of the GDC prostate WSIs. The patches with the tissue accounting for at least 20% of the whole area with high signal-to-noise ratio (SNR) are selected as the input of our experiments. This data preparation process keeps the integrity of tissue region meanwhile and has no discontinuity impact on LSTM. In total, 146 Gleason 3+4 and 101 Gleason 4+3 WSIs include 4753 and 2997 patches respectively. The patches inter-spatial relationship as an image sequence within a given WSI are modeled by CNN-based LSTM. The patches are warped to 256×256 and fed into the convolutional neural networks (CNN) with multi-task loss. Each patch is assigned two labels; one is the primary pattern and the other is the sum of the Gleason score. The momentum and weight decay of the CNN are 0.9 and 5x10−5 respectively. The initial learning rate is 10−3 and annealed by 0.1 after 100 iterations. The train the LSTM, we apply the same momentum, weight decay and initial learning rate. The CNN-LSTM implementation is based on Caffe toolbox.31
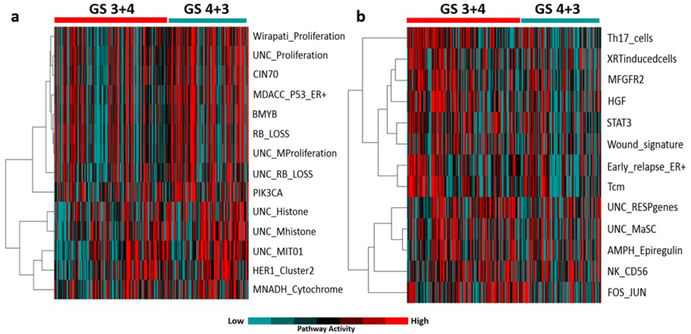
Illumina-RNAseq gene expression data including 20,437 genes for 247 human prostate cancer samples are acquired from GDC data portal. Our results determined that prostate cancer patients with a Gleason score of 4+3 are characterized by high expression of proliferation, BMYB, RB-loss and histone modification pathway.
The pathway scores are forward to LSTM layers with the neurons from fully-connected layer 7 together. The LSTM is built on top of the CNN for modeling long-term spatial modeling of the activation sequence. An average pooling layer maps the activations into one feature vector. For comparisons, three image texture feature methods including SURF,32 HOG33 and LBP34 are applied for biomarker quantification in conjunction with clinical prognostic features for patients recurrence analysis activity compared to prostate cancer patients with Gleason score of 3+4. Moreover, prostate cancer patients with Gleason score of 3+4 represent higher expression of immune system related pathways including Th17 cells, Tcm and STAT3 pathway activity compared to prostate cancer patients with Gleason score of 4+3 Figure 2.
Figure 2.
Patterns of pathway activity (a) prostate cancer patients with Gleason score of 4+3 show higher expression of proliferation, BMYB, RB and histone modification signatures (b) prostate cancer patients with Gleason score of 3+4 patients show lower expression of immune system related pathway signatures including Th17 cells, Tcm and STAT3.
Table 1 shows the hazard ratios of recurrence of the disease using different image features (LBP, HOG, SURF, neurons) with pathway scores. From the result, image biomarkers using CNN-LSTM outperforms other image feature quantification methods. With pathway score, image and genetic biomarkers show the highest recurrence hazard ratio compared to other clinical prognostic features.
Table 1.
Recurrent hazard ratios of clinical prognostic features and biomarkers using different image feature quantification methods with pathway scores. Here LBP, HOG and SURF are texture features within the WSIs. Here LBP+PS, HOG+PS, SURF+PS, CNN+PS and CNN-LSTM+PS are image biomarkers quantified from LBP, HOG, SURF, CNN and CNN with LSTM methods with pathway scores; CNN-LSTM is only use image biomarkers from convolutional neural network with LSTM alone.
| Methods | Primary Pattern | Secondary Pattern | PSA | Age | Tumor Stage | Biomarkers |
|---|---|---|---|---|---|---|
| LBP+PS | 1.0433 | 0.9986 | 0.8673 | 0.9990 | 1.0230 | 1.0759 |
| HOG+PS | 1.0433 | 0.9986 | 0.8673 | 0.9990 | 1.0230 | 1.0759 |
| SURF+PS | 1.0666 | 0.9768 | 0.8603 | 1.0017 | 1.0329 | 1.0703 |
| CNN+PS | 1.1091 | 1.1289 | 0.802 | 0.9998 | 1.1671 | 2.5796 |
| CNN-LSTM | 1.6953 | 1.0568 | 0.8049 | 0.9857 | 1.2612 | 5.0554 |
| CNN-LSTM+PS | 2.5598 | 0.6328 | 0.6643 | 1.0095 | 1.0474 | 5.7325 |
4. CONCLUSION
Our preliminary results demonstrate that integrating image features with genomic pathway scores, are more strongly correlated with patients recurrence of disease compared to standard clinical markers and engineered image texture features. The study further demonstrates that prostate cancer patients with Gleason score of 4+3 have higher risk of disease progression and recurrence compared to prostate cancer patients with Gleason score of 3+4. As such, we propose that patients with a Gleason score of 7 should not be considered as a homogeneous group for disease management and prognosis.
REFERENCES
- [1].Siegel RL, Miller KD, and Jemal A, “Cancer statistics, 2016,” CA: a cancer journal for clinicians 66(1), 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- [2].Wright JL, Salinas CA, Lin DW, Kolb S, Koopmeiners J, Feng Z, and Stanford JL, “Differences in prostate cancer outcomes between cases with gleason 4+ 3 and gleason 3+ 4 tumors in a population-based cohort,” The Journal of urology 182(6), 2702 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Amin A, Partin A, and Epstein JI, “Gleason score 7 prostate cancer on needle biopsy: relation of primary pattern 3 or 4 to pathological stage and progression after radical prostatectomy,” The Journal of urology 186(4), 1286–1290 (2011). [DOI] [PubMed] [Google Scholar]
- [4].Burdick MJ, Reddy CA, Ulchaker J, Angermeier K, Altman A, Chehade N, Mahadevan A, Kupelian PA, Klein EA, and Ciezki JP, “Comparison of biochemical relapse-free survival between primary gleason score 3 and primary gleason score 4 for biopsy gleason score 7 prostate cancer,” International Journal of Radiation Oncology* Biology* Physics 73(5), 1439–1445 (2009). [DOI] [PubMed] [Google Scholar]
- [5].Stark JR, Perner S, Stampfer MJ, Sinnott JA, Finn S, Eisenstein AS, Ma J, Fiorentino M, Kurth T, Loda M, et al. , “Gleason score and lethal prostate cancer: does 3+ 4= 4+ 3?,” Journal of Clinical Oncology 27(21), 3459–3464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huang P-W and Lee C-H, “Automatic classification for pathological prostate images based on fractal analysis,” IEEE transactions on medical imaging 28(7), 1037–1050 (2009). [DOI] [PubMed] [Google Scholar]
- [7].Doyle S, Feldman M, Tomaszewski J, and Madabhushi A, “A boosted bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies,” IEEE transactions on biomedical engineering 59(5), 1205–1218 (2012). [DOI] [PubMed] [Google Scholar]
- [8].Doyle S, Hwang M, Shah K, Madabhushi A, Feldman M, and Tomaszeweski J, “Automated grading of prostate cancer using architectural and textural image features,” in [Biomedical imaging: from nano to macro, 2007. ISBI 2007. 4th IEEE international symposium on], 1284–1287, IEEE; (2007). [Google Scholar]
- [9].Tabesh A, Teverovskiy M, Pang H-Y, Kumar VP, Verbel D, Kotsianti A, and Saidi O, “Multifeature prostate cancer diagnosis and gleason grading of histological images,” IEEE transactions on medical imaging 26(10), 1366–1378 (2007). [DOI] [PubMed] [Google Scholar]
- [10].Khurd P, Bahlmann C, Maday P, Kamen A, Gibbs-Strauss S, Genega EM, and Frangioni JV, “Computer-aided gleason grading of prostate cancer histopathological images using texton forests,” in [Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on], 636–639, IEEE; (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ren J, Sadimin ET, Wang D, Epstein JI, Foran DJ, and Qi X, “Computer aided analysis of prostate histopathology images gleason grading especially for gleason score 7,” in [Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE], 3013–3016, IEEE; (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ren J, Sadimin E, Foran DJ, and Qi X, “Computer aided analysis of prostate histopathology images to support a refined gleason grading system,” in [SPIE Medical Imaging], 101331V–101331V, International Society for Optics and Photonics (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jafari-Khouzani K and Soltanian-Zadeh H, “Multiwavelet grading of pathological images of prostate,” IEEE Transactions on Biomedical Engineering 50(6), 697–704 (2003). [DOI] [PubMed] [Google Scholar]
- [14].Khurd P, Grady L, Kamen A, Gibbs-Strauss S, Genega EM, and Frangioni JV, “Network cycle features: Application to computer-aided gleason grading of prostate cancer histopathological images,” in [Biomedical Imaging: From Nano to Macro, 2011 IEEE International Symposium on], 1632–1636, IEEE; (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nguyen K, Sabata B, and Jain AK, “Prostate cancer grading: Gland segmentation and structural features,” Pattern Recognition Letters 33(7), 951–961 (2012). [Google Scholar]
- [16].Nguyen K, Jain AK, and Allen RL, “Automated gland segmentation and classification for gleason grading of prostate tissue images,” in [Pattern Recognition (ICPR), 2010 20th International Conference on], 1497–1500, IEEE; (2010). [Google Scholar]
- [17].Riddick A, Shukla C, Pennington C, Bass R, Nuttall R, Hogan A, Sethia K, Ellis V, Collins A, Maitland N, et al. , “Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues,” British journal of cancer 92(12), 2171–2180 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gorelick L, Veksler O, Gaed M, Gomez JA, Moussa M, Bauman G, Fenster A, and Ward AD, “Prostate histopathology: Learning tissue component histograms for cancer detection and classification,” IEEE transactions on medical imaging 32(10), 1804–1818 (2013). [DOI] [PubMed] [Google Scholar]
- [19].Pan X, Li L, Yang H, Liu Z, Yang J, Zhao L, and Fan Y, “Accurate segmentation of nuclei in pathological images via sparse reconstruction and deep convolutional networks,” Neurocomputing 229, 88–99 (2017). [Google Scholar]
- [20].Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, and Tomaszewski J, “Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology,” in [Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008. 5th IEEE International Symposium on], 284–287, IEEE; (2008). [Google Scholar]
- [21].Xu J, Luo X, Wang G, Gilmore H, and Madabhushi A, “A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images,” Neurocomputing 191, 214–223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hou L, Singh K, Samaras D, Kurc TM, Gao Y, Seidman RJ, and Saltz JH, “Automatic histopathology image analysis with cnns,” in [Scientific Data Summit (NYSDS), 2016. New York: ], 1–6, IEEE; (2016). [Google Scholar]
- [23].Cruz-Roa A, Basavanhally A, González F, Gilmore H, Feldman M, Ganesan S, Shih N, Tomaszewski J, and Madabhushi A, “Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks,” in [SPIE medical imaging], 9041, 904103–904103, International Society for Optics and Photonics (2014). [Google Scholar]
- [24].Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, and Saltz JH, “Patch-based convolutional neural network for whole slide tissue image classification,” in [Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition], 2424–2433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sotiriou C and Piccart MJ, “Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care?,” Nature Reviews Cancer 7(7), 545–553 (2007). [DOI] [PubMed] [Google Scholar]
- [26].Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, Datto ΜB, Kelley M, Mathey-Prevot B, Potti A, et ah, “A pathway-based classification of human breast cancer,” Proceedings of the National Academy of Sciences 107(15), 6994–6999 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi M-B, Harpole D, Lancaster JM, Berchuck A, et ah, “Oncogenic pathway signatures in human cancers as a guide to targeted therapies,” Nature 439(7074), 353–357 (2006). [DOI] [PubMed] [Google Scholar]
- [28].Gatza ML, Silva GO, Parker JS, Fan C, and Perou CM, “An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer,” Nature genetics 46(10), 1051–1059 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bengio Y, Simard P, and Frasconi P, “Learning long-term dependencies with gradient descent is difficult,” IEEE transactions on neural networks 5(2), 157–166 (1994). [DOI] [PubMed] [Google Scholar]
- [30].Hochreiter S and Schmidhuber J, “Long short-term memory,” Neural computation 9(8), 1735–1780 (1997). [DOI] [PubMed] [Google Scholar]
- [31].Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, and Darrell T, “Caffe: Convolutional architecture for fast feature embedding,” in [Proceedings of the 22nd ACM international conference on Multimedia,], 675–678, ACM; (2014). [Google Scholar]
- [32].Bay H, “Surf: Speeded up robust features,” Computer vision and image understanding 110(3), 346–359 (2008). [Google Scholar]
- [33].Dalal N and Triggs B, “Histograms of oriented gradients for human detection,” in [Computer Vision and Pattern Recognition, 2005. CVPR 2005. IEEE Computer Society Conference on], 1, 886–893, IEEE; (2005). [Google Scholar]
- [34].Ojala T, Pietikainen M, and Maenpaa T, “Multiresolution gray-scale and rotation invariant texture classification with local binary patterns,” IEEE Transactions on pattern analysis and machine intelligence 24(7), 971–987 (2002). [Google Scholar]