Abstract
The widespread use of monoclonal antibodies for therapeutic applications has led to intense interest in optimizing several of their natural properties (affinity, specificity, stability, solubility and effector functions) as well as introducing non-natural activities (bispecificity and cytotoxicity mediated by conjugated drugs). A common challenge during antibody optimization is that improvements in one property (e.g., affinity) can lead to deficits in other properties (e.g., stability). Here we review recent advances in understanding trade-offs between different antibody properties, including affinity, specificity, stability and solubility. We also review new approaches for co-optimizing multiple antibody properties and discuss how these methods can be used to rapidly and systematically generate antibodies for a wide range of applications.
Keywords: mAb, Fab, Fc, CDR, aggregation, developability
1. Introduction
Monoclonal antibodies (mAbs) are being used in diverse therapeutic and diagnostic applications due to several of their attractive properties (Fig. 1) [1–3]. The most important antibody properties relate to their natural functions, such as their high binding affinity and specificity mediated by their complementarity-determining regions (CDRs) within the variable regions (variable heavy, VH, and variable light, VL). Other key natural antibody properties include their effector functions – such as antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) – which are mediated by their constant regions (fragment crystallizable, Fc).
Figure 1.
Overview of the key properties of monoclonal antibodies. The lines connecting different antibody properties highlight their interdependence and that optimization of any one property can lead to defects in other properties.
However, many other attractive properties of mAbs have contributed to their success as therapeutics (Fig. 1). For example, the favorable biophysical properties of antibodies – such as their high conformational (folding) stability and colloidal stability (solubility) – enable the formulation and extended storage of concentrated antibody solutions with minimal aggregation [4, 5]. Moreover, the relative ease of manufacturing different mAbs using similar platform processes has enabled the generation of large numbers of lead antibodies for rapid mAb optimization and analysis. Advances in antibody discovery methods (e.g., immunization, phage and yeast surface display) have enabled the generation of mAbs against almost any target [6, 7]. These and other attractive antibody properties featured in Figure 1 are key to the success of antibody therapeutics.
Nevertheless, most antibodies identified during the initial discovery process are not suitable for therapeutic use and require additional optimization [8]. For example, the binding affinities of some lead antibodies are not high enough for therapeutic applications and must be enhanced through in vitro antibody display methods. However, these methods have an increased risk of producing antibodies with poor biophysical properties. An outstanding challenge in the field is that optimizing properties such as antibody affinity can lead to defects in other properties such as antibody stability, specificity and solubility. The resulting trade-offs between improvements in some antibody properties and reductions in others highlight that they are often interdependent and cannot be easily separated. Therefore, it is important to understand the molecular determinants that mediate trade-offs between various antibody properties in order to improve the generation of optimized antibody therapeutics. Here we review recent findings related to trade-offs between antibody affinity and three other key antibody properties (stability, specificity and solubility), and discuss important areas of future work aimed at overcoming these trade-offs.
2. Antibody affinity/stability trade-offs
One challenge associated with optimizing antibody affinity is that increases in affinity can lead to decreases in antibody stability. Natural antibody affinity maturation relies on the introduction of somatic mutations followed by clonal selection of antibody variants with improved affinity. However, not all somatic mutations contribute to antibody affinity. Previous reports have suggested that antibodies accumulate some somatic mutations to increase affinity and others to compensate for the destabilizing effects of affinity-enhancing mutations [9–12]. These studies raise the intriguing possibility that the natural process of reshaping the antigen-binding site during antibody affinity maturation involves chemical and structural changes that are destabilizing to the antibody framework, and compensatory mutations are required to maintain thermodynamic stability.
The importance of affinity/stability trade-offs during antibody affinity maturation was recently highlighted for single-domain (VH) antibodies [13, 14]. The investigators introduced mutations throughout the VH frameworks and CDRs using error-prone PCR and displayed the antibody libraries on the surface of yeast. The libraries were sorted to identify antibody variants with high antigen binding (Alzheimer’s Aβ42 peptide) and expression (anti-myc tag). After a single round of mutagenesis and selection, an antibody variant was identified with three mutations that displayed increased affinity but significantly reduced stability (reduction in apparent melting temperature of 18 °C). Moreover, additional rounds of mutagenesis and selection for antigen binding and antibody expression led to selection of VH antibody variants that displayed large reductions in antibody stability and significant increases in affinity. In fact, highly mutated variants with six to twelve mutations were partially unfolded when evaluated as soluble proteins, revealing the strongly destabilizing effects of the affinity-enhancing mutations.
These results both highlight that trade-offs can occur during affinity maturation and raise questions about how to overcome them. It is unexpected that antibody affinity maturation using yeast surface display would lead to the isolation of destabilized antibodies [13, 14] because the sophisticated quality control mechanisms of yeast are expected to reduce the likelihood of displaying destabilized antibodies [15, 16]. However, it appears that the yeast secretory pathway fails to efficiently recognize and degrade destabilized forms of small and highly soluble VH antibodies. Therefore, the investigators sought to develop a method for co-selecting sets of mutations that increase antibody affinity while maintaining stability in order to overcome the previously observed affinity/stability trade-offs [14]. The key advance was to replace the conventional antibody (anti-myc tag) used to select for high antibody expression with a conformational probe (Protein A) that is selective for folded antibodies (VH3 antibodies have a Protein A binding site on their framework [17]). The investigators found that the relative binding of Protein A to VH antibodies on the surface of yeast displayed a large dynamic range (~12-fold change in Protein A binding for a ~18 °C change in apparent melting temperature of the VH antibodies) that was strongly correlated with antibody stability (R2 of 0.92) [14]. When the investigators repeated the directed evolution process by co-selecting for antigen and Protein A binding, they identified VH antibodies with significantly increased affinities and high stabilities.
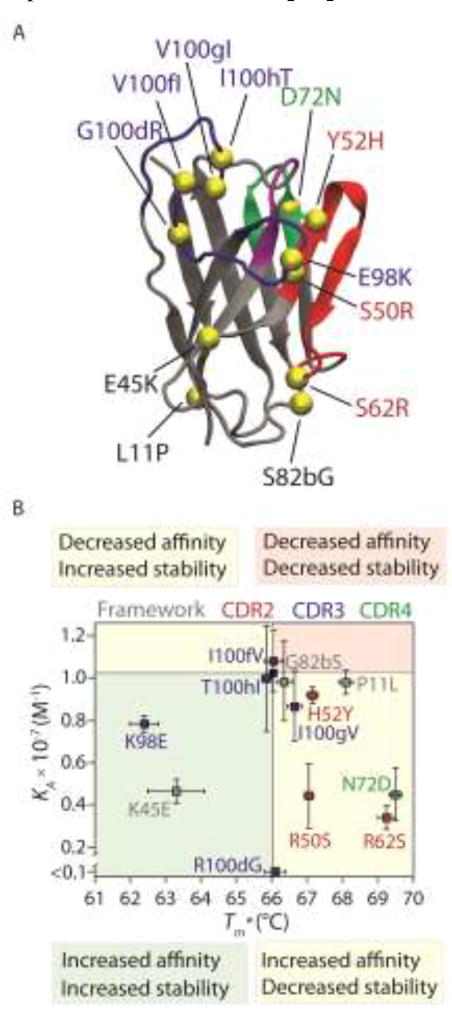
An example of one of the evolved VH antibodies with twelve acquired mutations is shown in Fig. 2A [13]. To understand how this evolved antibody maintained thermodynamic stability despite mutating almost 10% of its sequence during affinity maturation, the investigators generated twelve single reversion mutants to investigate the contribution of each mutation to stability (apparent melting temperature, Tm*) and affinity (association constant, KA; Fig. 2B). The majority of the mutations that increased affinity (decrease in KA when reverted to the wild-type residue) resulted in a decrease in stability (increase in Tm* when reverted to the wildtype residue), suggesting that affinity-enhancing mutations may generally have a higher risk of destabilization. For example, reversion of the N72 mutation to the wild-type residue (D72) led to a decrease in affinity (KA of evolved VH antibody is greater than KA of N72D reversion mutant) and an increase in stability (Tm* of N72D reversion mutant is greater than Tm* of evolved VH antibody). Two stabilizing mutations (K45 and K98) compensated for the loss in VH stability due to the affinity-enhancing mutations. Interestingly, these stabilizing mutations were also beneficial for antibody affinity. Moreover, mutations in the third CDR (CDR3) of the VH antibody generally displayed the lowest affinity/stability trade-offs. For example, the most important affinity mutation (R100d in CDR3) did not impact stability, which is consistent with the high sequence variability of heavy chain CDR3 for antibodies generated by the immune system. This suggests that antibody libraries designed with sequence variation only in heavy chain CDR3 have a reduced risk for displaying affinity/stability trade-offs. More generally, these and other findings [18–20] suggest that affinity-enhancing mutations have an increased risk of destabilization and that compensatory mutations are frequently required to maintain antibody thermodynamic stability.
Figure 2.
Efficient affinity maturation of antibody variable (VH) domains requires co-selection of stabilizing mutations that compensate for the destabilizing effects of affinity-enhancing mutations. (A) Structural model of a human variable domain specific for the Alzheimer’s Aβ42 peptide that was co-evolved for enhanced affinity and stability using directed evolution methods. The evolved antibody domain acquired 12 mutations in the framework and complementarity-determining regions (CDRs). (B) Analysis of the contribution of each acquired mutation to affinity and stability. Single reversion mutations that reduce the equilibrium association constant (KA) indicate mutations that are beneficial for affinity, while those that increase KA correspond to mutations that are detrimental to affinity. Likewise, single reversion mutations that reduce the apparent melting temperature (Tm*) indicate mutations that are beneficial for stability, while those that increase Tm* correspond to mutations that are detrimental to stability. Several of the key affinity mutations (R50, R62 and N72) are strongly destabilizing and are compensated for by two key stabilizing mutations that also enhance antibody affinity (K45 and K98). The solid lines are the values of KA (~1 × 10−7 M−1) and Tm* (66 °C) for the evolved VH domain with 12 mutations. The CDRs are defined using Kabat numbering, and CDR4 is defined as residues 71–78. The figure is adapted from Reference [13].
Although relatively few studies have investigated trade-offs between antibody affinity and stability, analogous trade-offs have been observed for non-antibody affinity proteins. For example, directed evolution of designed ankyrin repeat proteins (DARPins) against HER2 led to the isolation of mutants with significantly improved affinity but reduced stability [21]. The evolved DARPin variant with the greatest gain in affinity (>700-fold) displayed large losses in stability (30 °C reduction in apparent melting temperature). Single reversion mutants revealed the mutations that resulted in the greatest increases in affinity were most destabilizing. Similar results were found for an affinity-maturated fibronectin domain specific for lysozyme [22], and protein engineering approaches have been developed to overcome these trade-offs [23]. These and related findings [24, 25] demonstrate the generality of affinity/stability trade-offs during affinity maturation.
3. Antibody affinity/specificity trade-offs
Another particularly important and challenging antibody property to optimize is antibody specificity. In this review, improved antibody specificity refers to reduced binding to non-target molecules. It is expected that most mutations that increase affinity – such as those that simply increase hydrophobicity or charge – also reduce specificity. The isolation of rare mutations that increase affinity while maintaining or even increasing specificity – as observed for antibodies generated by the immune system – is particularly important and typically possible only through selection for high specificity in addition to high affinity.
Given that the maximal chemical diversity of antibody CDRs is unimaginably large (>1078 antibody variants based on 20 different amino acids at ~60 sites in the CDRs), it is extremely challenging to define the sequence determinants of antibody specificity. Nevertheless, Sidhu and co-workers have elegantly dissected this complex problem by generating antibody libraries with highly restricted amino acid diversity in the CDRs [26, 27]. The investigators evaluated the capacity of different types of amino acids in the CDRs to mediate high affinity while maintaining high specificity. One of their most striking findings is that antibodies with high levels of tyrosine in their CDRs – even for antibody variants with >10 tyrosines in heavy chain CDR3 – have unusually low levels of non-specific interactions. This finding – along with the fact that tyrosine is one of the most common amino acids in antibody CDRs [28] – strongly suggests that tyrosine is a critical amino acid for mediating specific antigen recognition [29].
These studies also revealed several other key CDR sequence determinants of antibody specificity [26, 27]. First, the presence of arginine in the CDRs was the greatest risk factor for non-specific interactions. Antibody variants without arginine residues in heavy chain CDR3 displayed the lowest levels of non-specific binding. Moreover, serine was identified as an important hydrophilic amino acid that fails to induce non-specific interactions even at the highest levels evaluated (e.g., up to nine serines in heavy chain CDR3). Importantly, the same investigators demonstrated that combinations of only serine and tyrosine mutations in antibody CDRs are sufficient to mediate high-affinity binding. These unusual antibodies also have extremely low levels of nonspecific interactions [30]. It will be important in the future to expand these invaluable studies to further elucidate the sequence and structural determinants of antibody specificity.
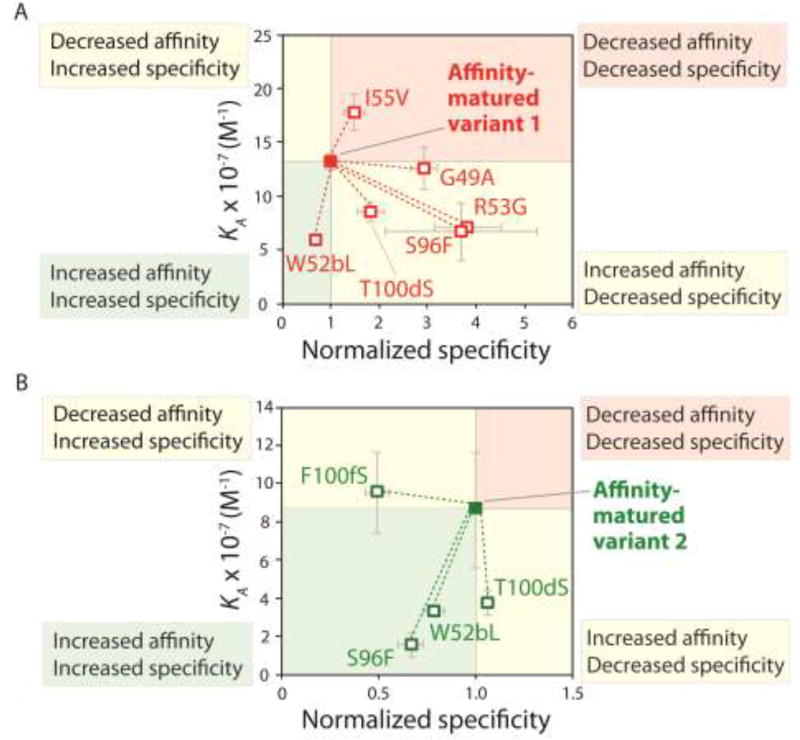
Another example of affinity/specificity trade-offs during antibody affinity maturation was highlighted in a recent directed evolution study [31] that involved the optimization of a camelid single-domain (VHH) antibody specific for α-synuclein [32]. The investigators first used alanine scanning mutagenesis to identify sites in the CDRs of the VHH antibody that were permissive to mutation without large detrimental impacts on affinity [31]. Next, they diversified 14 permissive sites in CDR2 and CDR3 in a manner in which the wild-type residue at each site was sampled as well as 1–5 of the most commonly occurring residues at each site in natural antibodies [33]. Finally, they displayed the antibody library on the surface of yeast and selected antibody variants with increased affinity using magnetic-activated and fluorescence-activated cell sorting [31]. Two of the affinity-matured antibodies isolated from this library are highlighted in Figure 3 [31]. The investigators generated single reversion mutations to evaluate the impact of each mutation on antibody affinity and specificity. Most of the affinity-enhancing mutations (three out of four) acquired by the first antibody variant reduced specificity (Fig. 3A). For example, the arginine mutation at position 53 (R53) in CDR2 increased affinity but reduced specificity. A notable exception is the tryptophan mutation at position 52b (W52b) in CDR2, which increased both affinity and specificity. Conversely, the second antibody variant displayed much more favorable properties, as two of the three affinity-enhancing mutations (W52b and S96) also increased specificity, and the third affinity-enhancing mutation (T100d) weakly impacted specificity (Fig. 3B). It is also notable that the affinity/specificity trade-offs of individual mutations were highly context dependent, as the same affinity-enhancing mutation (S96) led to opposite impacts on antibody specificity in the two closely related variants (Fig. 3).
Figure 3.
Mutational analysis of the contributions of antibody (VHH) mutations accumulated during affinity maturation to affinity and specificity. Single reversion mutations were generated for two affinity-matured variable domains specific for α-synuclein to determine the contribution of each acquired mutation to affinity and specificity. Values of the equilibrium association constant (KA) were measured using fluorescence polarization. Normalized specificity was measured as the binding of the parent antibody to milk proteins divided by that for the reversion mutant (parent/reversion mutant). Reductions in affinity or specificity for the reversion mutants indicate that the original mutations acquired during affinity maturation contribute positively to either property. The figure is adapted from Reference [31].
The identification of mutations that increase both affinity and specificity – especially for antibodies that are affinity matured in vitro – is rare and methods for improving the identification of these types of mutations are particularly important. There are two main approaches for improving the selection of antibodies with increased specificity. The first and most common one is to perform positive selections for antigen binding and negative selections for non-specific binding to eliminate variants with low specificity [34–37]. Several types of polyspecificity reagents have been developed for eliminating non-specific variants and enabling the isolation of antibodies with specificities that rival those of natural antibodies [37, 38]. For example, one particularly effective polyspecificity reagent is composed of soluble membrane proteins generated from mammalian cell lysates [34, 37, 39, 40]. Negative selections performed with this polyspecificity reagent and positive selections performed with the target antigen led to the isolation of antibodies with greatly reduced levels of non-specific binding relative to antibodies obtained without negative selections [37]. Moreover, chaperone proteins (e.g., Hsp90) have also been shown to be effective and well-defined polyspecificity reagents [38]. These examples demonstrate that various types of polyspecificity reagents are invaluable tools for performing negative selections to remove non-specific antibody variants during library sorting.
A second key approach for identifying antibodies with increased specificity during in vitro selections is to perform positive selections for antigen binding in the presence of high concentrations of non-target molecules. Complex mixtures such as serum or milk are particularly effective at blocking non-specific interactions. The use of complex mixtures of non-target molecules during positive selections was recently evaluated during the affinity maturation of single-chain (scFv) antibodies against the Alzheimer’s Aβ42 peptide [41]. The investigators generated antibody libraries with sequence diversity restricted to heavy chain CDR3, displayed the libraries on the surface of yeast, and selected for antibody variants with increased affinity. The investigators first performed antibody selections in the absence of high concentrations of non-target molecules. Interestingly, the selected antibodies had CDRs enriched in arginine residues (~50% of the mutations were arginine), which is likely due to the fact that the Aβ peptide is negatively charged at neutral pH (isoelectric point of ~pH 5.5) and hydrophobic. The highest affinity antibody variant selected using this method (A10) contained three arginine mutations in heavy chain CDR3 and lost all antigen binding activity in the presence of high concentrations of non-target molecules (1% milk). Alanine scanning mutagenesis of the evolved antibody revealed that the majority of the binding affinity was mediated by the arginine mutations. These findings are consistent with previous results that arginine CDR mutations are a key risk factor for poor antibody specificity [26, 27, 42].
A strikingly different result was obtained when the antibody selections were repeated against the Aβ42 peptide in the presence of high concentrations of non-target molecules (1% milk) [41]. In this case, the selected antibodies had increased specificity despite that some of the antibody variants had several arginine CDR mutations. For example, the highest affinity antibody variant obtained using the more stringent approach (B2) had multiple arginine CDR mutations but retained antigen binding in the presence of 1% milk. Surprisingly, alanine scanning mutagenesis revealed that the B2 antibody was not dependent on the arginine mutations for affinity, suggesting that these positively charged mutations were selected for other purposes (e.g., antibody expression and solubility). These results suggest that overreliance on arginine CDR residues for affinity is linked to increased affinity/specificity trade-offs, and that arginine CDR mutations display context-dependent impacts on antibody specificity.
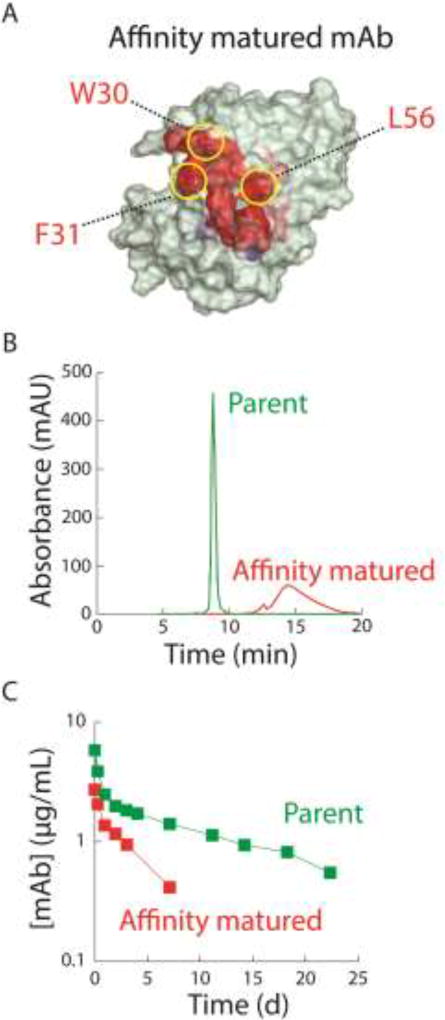
One notable consequence of poor antibody specificity is that it can lead to poor pharmacokinetics and fast antibody clearance [40, 43–45]. The challenges of antibody affinity/specificity trade-offs and their connection to fast antibody clearance were elegantly highlighted in a recent study involving the affinity maturation of an antibody specific for nerve growth factor [44]. The investigators increased the affinity of a lead antibody by an order of magnitude via several CDR mutations. However, the affinity-matured variant – which had several hydrophobic CDR mutations (Fig. 4A) – displayed increased levels of non-specific interactions with various non-target molecules and chromatography matrices (Fig. 4B). Moreover, the affinity-matured variant was cleared much faster in vivo than the parent antibody. The investigators were able to overcome these affinity/specificity trade-offs by replacing three of the hydrophobic CDR mutations that were not involved in antigen binding with polar residues. Impressively, the optimized antibody displayed high specificity and excellent pharmacokinetic properties without reductions in affinity. These and related findings [46–49] provide valuable insights for overcoming affinity/specificity trade-offs by engineering antibody CDRs and frameworks to achieve high specificity while maintaining high antibody affinity.
Figure 4.
Affinity maturation of an anti-nerve growth factor antibody leads to the accumulation of mutations that result in non-specific interactions and fast antibody clearance. (A) Structural model of the variable regions of the affinity-matured antibody. The residues responsible for low specificity (W30 and F31 in heavy chain CDR1 and L56 in heavy chain CDR2) are highlighted. (B) Size-exclusion chromatography reveals that the affinity-matured antibody interacts with the column matrix and displays abnormally long elution times relative to the parental antibody. (C) Antibody clearance rates from rats (dose of 0.3 mg/kg) reveals that the affinity-matured antibody is cleared much faster than the parental antibody. The figure is redrawn and adapted from Reference [44].
4. Antibody affinity/solubility trade-offs
Another important challenge in developing antibody therapeutics is maintaining high antibody solubility during the accumulation of affinity-enhancing mutations in the CDRs and framework regions [4, 5]. Some affinity-enhancing mutations – such as hydrophobic mutations that are solvent-exposed in the CDRs – are expected to be beneficial for affinity but detrimental for solubility. Nevertheless, it is critical to identify and engineer highly soluble antibodies because antibody aggregation is linked to immunogenicity [50].
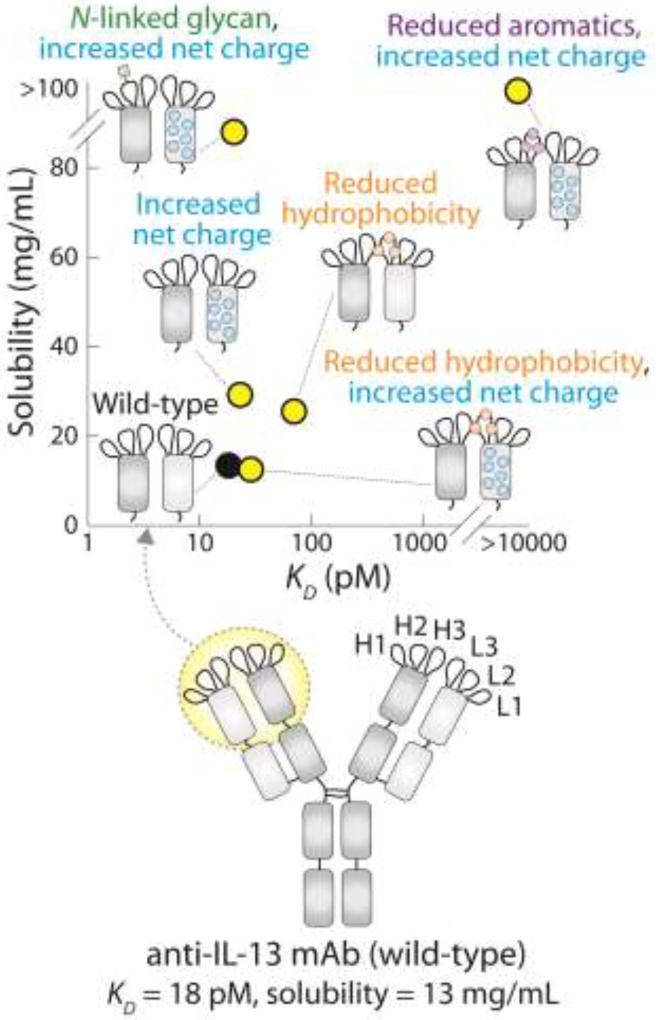
One particularly insightful study related to antibody affinity/solubility trade-offs evaluated several protein engineering strategies to overcome solubility problems for a monoclonal antibody (CNTO607) specific for IL-13 (Fig. 5) [51]. The poor solubility (~13 mg/mL) of the wild-type, high-affinity antibody (KD of 18 pM) was due to a hydrophobic hotspot in heavy chain CDR3 that involved a Phe-His-Trp motif. Mutation of these residues to alanine greatly increased solubility (>164 mg/mL) but also greatly reduced affinity (KD > 45000 pM). Given that these hydrophobic CDR residues were critical for affinity, the investigators evaluated alternative strategies for increasing antibody solubility without mutating heavy chain CDR3. First, they evaluated the impact of increasing antibody net charge by introducing positively charged residues into the frameworks of the variable domains. This strategy resulted in a modest increase in solubility (~29 mg/mL) without significantly altering antibody affinity. Second, they identified multiple CDRs (heavy chain CDR1 and light chain CDR3) that were not directly involved in antigen binding and evaluated mutations in these CDRs that increased their hydrophilicity (especially light chain CDR3). However, these mutations resulted in only moderate improvements of antibody solubility (~25–29 mg/mL).
Figure 5.
Mutations in an anti-IL-13 mAb display trade-offs between antibody affinity and solubility. Key residues that mediate high affinity while compromising solubility are three consecutive aromatic and histidine residues (Phe-Trp-His) in heavy chain CDR3. The most effective combination of mutations that increase antibody solubility while maintaining affinity are a single mutation that introduces an N-linked glycosylation site in heavy chain CDR2 and multiple mutations in the variable light (VL) domain that increase the overall net (positive) charge. The data are from Reference [51].
The third and most impactful approach that increased the solubility of CNTO607 was the introduction of an N-linked glycosylation site into heavy chain CDR2 (Fig. 5) [51]. While the conventional N-linked glycosylation site in antibodies is located in the Fc fragment, it is possible for antibodies to acquire non-conventional glycosylation sites in the CDRs due to asparagine mutations with the appropriate neighboring residues (Asn-X-Ser/Thr). CNTO607 was originally identified with an N-linked glycan in heavy chain CDR2, but this was removed due to the analytical and manufacturing challenges associated with developing antibodies with nonconventional glycosylation sites. However, the investigators found that re-introducing this glycosylation site dramatically increased solubility without impacting antibody affinity (Fig. 5). It appears that the proximity of this glycan to the hydrophobic hotspot in heavy chain CDR3 enables it to increase antibody hydrophilicity without reducing affinity. This and a follow-up study [52] powerfully demonstrate both the challenges of affinity/solubility trade-offs and methods for overcoming them.
Another excellent example of affinity/solubility trade-offs and how to overcome them was demonstrated in the study of an antibody (Li33) specific for LINGO-1 (a glycoprotein involved in demyelination disorders) [53]. This potent antibody (EC50 of 0.12 nM) has unusually low solubility (<1 mg/mL). Crystal structures of the Fab and F(ab')2 forms of the antibody revealed hydrophobic residues in the CDRs that form CDR-CDR and CDR-framework contacts, and appear to mediate antibody self-association and aggregation. Therefore, the investigators tested several protein engineering strategies to increase antibody solubility without reducing affinity. One surprisingly effective strategy was reformatting the poorly soluble IgG1 antibody (<1 mg/mL) to IgG2 (>50 mg/mL) and IgG4 (>30 mg/mL) antibodies. The precise mechanism for this enhanced solubility is unclear, as simple explanations based on antibody charge are not consistent with the increased solubility. Instead, it appears that subtle differences in molecular geometry linked to the hinge sequences and patterns of disulfide bonding may be involved in the large solubility enhancements.
Two other protein engineering strategies also proved to be useful for increasing the solubility of the anti-LINGO-1 antibody [53]. First, hydrophobic CDR residues that were not involved in antigen binding were replaced with smaller hydrophobic or hydrophilic residues, which greatly increased solubility (from <1 mg/mL to >10 mg/mL) without reducing affinity. Second, N-linked glycosylation sites were introduced (one at a time) at four sites in the CH1 domain with the goal of disrupting hydrophobic interactions involving the CDRs. Interestingly, glycosylation at two sites was highly effective at increasing solubility (from <1 mg/mL to >50 mg/mL) without impacting affinity. The two most effective glycosylation sites were those closest to the crystal contacts involving the hydrophobic CDRs, which is consistent with the glycans disrupting hydrophobic interactions. These and related studies [44, 54–66] demonstrate powerful protein engineering methods for overcoming trade-offs between antibody affinity and solubility.
5. Future directions
There are several important areas of future research that are key to minimizing trade-offs between different antibody properties during the discovery and development of antibody therapeutics. First, it will be important to develop methods for predicting antibody specificity based on antibody sequence and structure. The fact that measurements of antibody specificity (non-specific binding and self-association) are the best biophysical descriptors of the likelihood of antibody success in the clinic [67] highlights the importance of developing computational and bioinformatics methods for predicting antibody specificity. Some of the key factors that determine antibody specificity are becoming clearer, including the numbers of charged, hydrophobic and hydrophilic residues in CDRs [26, 27, 39, 41, 42, 44, 68–70] as well as the net charge of the variable regions [46, 47, 49]. However, methods are needed to collectively describe these disparate findings and provide guidelines for identifying antibody variants with high specificity based only on their amino acid sequences or their combined sequences and structures. The development of such predictive methods would have an immediate impact on the field by enabling the identification of high-affinity antibody candidates that also have high specificity.
It will also be important in the future to improve methods for designing antibody libraries for in vitro antibody discovery and affinity maturation to minimize trade-offs between antibody affinity and other key biophysical properties. The fact that only a small fraction of maximal chemical (amino acid) diversity can be sampled in any antibody library means that it is critical to sample mutations that will most likely maintain or enhance antibody biophysical properties while improving affinity [39]. Future work in antibody library design will need to address many outstanding challenges, especially those related to developing constraints for focusing amino acid diversity on mutations that are most likely to lead to high antibody specificity, solubility and stability in addition to high affinity. This is non-trivial because some mutations (e.g., positively charged mutations in the CDRs) that are helpful for one biophysical property (e.g., solubility) can be detrimental for other properties (e.g., specificity) [5, 26, 41]. Therefore, it will be especially important to generate guidelines for predicting antibody CDR sequences that maximize several biophysical properties (e.g., specificity and solubility) without eliminating key interactive residues that contribute to antibody affinity. The library design methods should also consider amino acid diversity in antibody frameworks (in addition to the CDRs) to enable the identification of both affinity-enhancing mutations (that can be destabilizing) and compensatory stabilizing mutations [13, 14]. Previous work suggests that library design methods that follow patterns of natural antibody diversity for both framework regions [71] and CDRs [31, 72–74] may be most effective.
A third key area of future research is to develop more efficient experimental screening methods for identifying antibodies with minimal trade-offs between affinity and other biophysical properties. For antibodies discovered or matured using in vitro methods (e.g., phage and yeast surface display), it is key to develop better methods for eliminating suboptimal variants during the library sorting process prior to evaluating them as soluble (non-fusion) proteins. It will be important to further develop effective and well-defined polyspecificity reagents for negative selections to eliminate antibodies with low specificity and solubility. It is not clear to what extent this is possible using a single polyspecificity reagent [37, 38] or whether multiple reagents are needed to eliminate antibodies with defects in different properties. It will also be important to identify new conformational ligands that recognize diverse classes of human variable regions to achieve robust selection of antibody variants with high stability.
Another key area of future work is to use deep sequencing methods to identify antibody mutations that improve a given property (e.g., affinity) without compromising other properties (e.g., stability and specificity). Previous studies have demonstrated the power of this methodology for identifying antibody sites that are highly tolerant to mutation as well as antibody mutations that lead to higher affinity without stability trade-offs [75, 76]. Similarly, deep sequencing methods have been used to finely tune antibody specificity – without compromising affinity – to remove undesirable binding to non-target molecules that have high homology to the target antigens [77]. It will be important in the future to use these powerful methods along with advanced data analytics to better understand trade-offs between various key antibody properties.
A final key area of future work is the development of improved computational methods for predicting mutations in antibody CDRs and frameworks that co-optimize multiple antibody properties. Predictions of mutations that improve one property typically do not consider their potential negative effects on other properties. Nevertheless, encouraging results are emerging for computational design methods to optimize antibody affinity, stability and/or solubility [56, 78–80]. Future efforts will also need to improve structural predictions of antibody CDRs [81, 82] – especially the long and highly variable heavy chain CDR3 – to accurately predict CDR mutations that are beneficial to different antibody properties. The continued development of such computational methods is expected to make experimental screening methods more efficient and enable the rapid generation of antibodies with optimized properties for diverse therapeutic applications.
Highlights.
Antibodies display trade-offs between key properties during affinity maturation
Antibody mutations that increase affinity are commonly destabilizing
Affinity-enhancing mutations can reduce antibody specificity and solubility
New methods are reported for minimizing trade-offs between key antibody properties
Acknowledgments
We thank members of the Tessier lab for their helpful suggestions. This work was supported by the National Institutes of Health (R01GM104130 to P.M.T.), National Science Foundation (CBET 1159943 and 1605266 to P.M.T., Graduate Research Fellowship to L.A.R.), and the Albert M. Mattocks Chair (to P.M.T).
Footnotes
Conflicts of interest
P.M.T. has received honorariums and/or consulting fees for presentations of this and/or related research findings at MedImmune, Eli Lilly, Bristol-Myers Squibb, Janssen, Merck, Genentech, Amgen, Pfizer, Adimab, Abbvie, Abbott, DuPont, Schrödinger and Novo Nordisk.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tiller KE, Tessier PM. Advances in antibody design. Annu Rev Biomed Eng. 2015;17:191–216. doi: 10.1146/annurev-bioeng-071114-040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maynard J, Georgiou G. Antibody engineering. Annu Rev Biomed Eng. 2000;2:339–376. doi: 10.1146/annurev.bioeng.2.1.339. [DOI] [PubMed] [Google Scholar]
- 3.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 4.Perchiacca JM, Tessier PM. Engineering aggregation-resistant antibodies. Annu Rev Chem Biomol Eng. 2012;3:263–286. doi: 10.1146/annurev-chembioeng-062011-081052. [DOI] [PubMed] [Google Scholar]
- 5.Lowe D, Dudgeon K, Rouet R, Schofield P, Jermutus L, Christ D. Aggregation, stability, and formulation of human antibody therapeutics. Adv Protein Chem Struct Biol. 2011;84:41–61. doi: 10.1016/B978-0-12-386483-3.00004-5. [DOI] [PubMed] [Google Scholar]
- 6.Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 7.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 8.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10:345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]
- 9.Sun SB, Sen S, Kim NJ, Magliery TJ, Schultz PG, Wang F. Retraction of "Mutational Analysis of 48G7 Reveals that Somatic Hypermutation Affects Both Antibody Stability and Binding Affinity". J Am Chem Soc. 2018;140:1976. doi: 10.1021/jacs.7b08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Sen S, Zhang Y, Ahmad I, Zhu X, Wilson IA, Smider VV, Magliery TJ, Schultz PG. Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation. Proc Natl Acad Sci U S A. 2013;110:4261–4266. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Wang F, Sen S, Zhang Y, Ahmad I, Zhu X, Wilson IA, Smider VV, Magliery TJ, Schultz PG. Retraction for Wang et al., Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation. Proc Natl Acad Sci U S A. 2017;114:E7855. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Sun SB, Sen S, Kim NJ, Magliery TJ, Schultz PG, Wang F. Mutational analysis of 48G7 reveals that somatic hypermutation affects both antibody stability and binding affinity. J Am Chem Soc. 2013;135:9980–9983. doi: 10.1021/ja402927u. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Julian MC, Li L, Garde S, Wilen R, Tessier PM. Efficient affinity maturation of antibody variable domains requires co-selection of compensatory mutations to maintain thermodynamic stability. Sci Rep. 2017;7:45259. doi: 10.1038/srep45259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julian MC, Lee CC, Tiller KE, Rabia LA, Day EK, Schick AJ, 3rd, Tessier PM. Co-evolution of affinity and stability of grafted amyloid-motif domain antibodies. Protein Eng Des Sel. 2015;28:339–350. doi: 10.1093/protein/gzv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J Mol Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 16.Kowalski JM, Parekh RN, Mao J, Wittrup KD. Protein folding stability can determine the efficiency of escape from endoplasmic reticulum quality control. J Biol Chem. 1998;273:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- 17.Potter KN, Li Y, Capra JD. Staphylococcal protein A simultaneously interacts with framework region 1, complementarity-determining region 2, and framework region 3 on human VH3-encoded Igs. J Immunol. 1996;157:2982–2988. [PubMed] [Google Scholar]
- 18.Zabetakis D, Anderson GP, Bayya N, Goldman ER. Contributions of the complementarity determining regions to the thermal stability of a single-domain antibody. PLoS One. 2013;8:e77678. doi: 10.1371/journal.pone.0077678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson GP, Teichler DD, Zabetakis D, Shriver-Lake LC, Liu JL, Lonsdale SG, Goodchild SA, Goldman ER. Importance of Hypervariable Region 2 for Stability and Affinity of a Shark Single-Domain Antibody Specific for Ebola Virus Nucleoprotein. PLoS One. 2016;11:e0160534. doi: 10.1371/journal.pone.0160534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldman ER, Liu JL, Zabetakis D, Anderson GP. Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview. Front Immunol. 2017;8:865. doi: 10.3389/fimmu.2017.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houlihan G, Gatti-Lafranconi P, Lowe D, Hollfelder F. Directed evolution of anti-HER2 DARPins by SNAP display reveals stability/function trade-offs in the selection process. Protein Eng Des Sel. 2015;28:269–279. doi: 10.1093/protein/gzv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackel BJ, Kapila A, Wittrup KD. Picomolar affinity fibronectin domains engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling. J Mol Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porebski BT, Conroy PJ, Drinkwater N, Schofield P, Vazquez-Lombardi R, Hunter MR, Hoke DE, Christ D, McGowan S, Buckle AM. Circumventing the stability-function trade-off in an engineered FN3 domain. Protein Eng Des Sel. 2016 doi: 10.1093/protein/gzw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Aha P, Gu K, Kuimelis RG, Kurz M, Lam T, Lim AC, Liu H, Lohse PA, Sun L, Weng S, Wagner RW, Lipovsek D. Directed evolution of high-affinity antibody mimics using mRNA display. Chem Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]
- 25.Karanicolas J, Corn JE, Chen I, Joachimiak LA, Dym O, Peck SH, Albeck S, Unger T, Hu W, Liu G, Delbecq S, Montelione GT, Spiegel CP, Liu DR, Baker D. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42:250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birtalan S, Fisher RD, Sidhu SS. The functional capacity of the natural amino acids for molecular recognition. Mol Biosyst. 2010;6:1186–1194. doi: 10.1039/b927393j. [DOI] [PubMed] [Google Scholar]
- 27.Birtalan S, Zhang Y, Fellouse FA, Shao L, Schaefer G, Sidhu SS. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 28.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217:133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 29.Fellouse FA, Barthelemy PA, Kelley RF, Sidhu SS. Tyrosine plays a dominant functional role in the paratope of a synthetic antibody derived from a four amino acid code. J Mol Biol. 2006;357:100–114. doi: 10.1016/j.jmb.2005.11.092. [DOI] [PubMed] [Google Scholar]
- 30.Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J Mol Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 31.Tiller KE, Chowdhury R, Li T, Ludwig SD, Sen S, Maranas CD, Tessier PM. Facile affinity maturation of antibody variable domains using natural diversity mutagenesis. Front Immunol. 2017;8:986. doi: 10.3389/fimmu.2017.00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Genst EJ, Guilliams T, Wellens J, O'Day EM, Waudby CA, Meehan S, Dumoulin M, Hsu ST, Cremades N, Verschueren KH, Pardon E, Wyns L, Steyaert J, Christodoulou J, Dobson CM. Structure and properties of a complex of alpha-synuclein and a single-domain camelid antibody. J Mol Biol. 2010;402:326–343. doi: 10.1016/j.jmb.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Swindells MB, Porter CT, Couch M, Hurst J, Abhinandan KR, Nielsen JH, Macindoe G, Hetherington J, Martin AC. abYsis: Integrated Antibody Sequence and Structure-Management, Analysis, and Prediction. J Mol Biol. 2017;429:356–364. doi: 10.1016/j.jmb.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Kelly RL, Zhao J, Le D, Wittrup KD. Nonspecificity in a nonimmune human scFv repertoire. MAbs. 2017;9:1029–1035. doi: 10.1080/19420862.2017.1356528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 36.Ackerman M, Levary D, Tobon G, Hackel B, Orcutt KD, Wittrup KD. Highly avid magnetic bead capture: an efficient selection method for de novo protein engineering utilizing yeast surface display. Biotechnol Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vasquez M, Wittrup KD, Krauland E. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel. 2013;26:663–670. doi: 10.1093/protein/gzt047. [DOI] [PubMed] [Google Scholar]
- 38.Kelly RL, Geoghegan JC, Feldman J, Jain T, Kauke M, Le D, Zhao J, Wittrup KD. Chaperone proteins as single component reagents to assess antibody nonspecificity. MAbs. 2017;9:1036–1040. doi: 10.1080/19420862.2017.1356529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly RL, Le D, Zhao J, Wittrup KD. Reduction of Nonspecificity Motifs in Synthetic Antibody Libraries. J Mol Biol. 2018;430:119–130. doi: 10.1016/j.jmb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly RL, Sun T, Jain T, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Vasquez M, Wittrup KD, Xu Y. High throughput cross-interaction measures for human IgG1 antibodies correlate with clearance rates in mice. mAbs. 2015;7:770–777. doi: 10.1080/19420862.2015.1043503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiller KE, Li L, Kumar S, Julian MC, Garde S, Tessier PM. Arginine mutations in antibody complementarity-determining regions display context-dependent affinity/specificity trade-offs. J Biol Chem. 2017;292:16638–16652. doi: 10.1074/jbc.M117.783837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 43.Hotzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, Fielder P, Carter PJ, Kelley RF. A strategy for risk mitigation of antibodies with fast clearance. mAbs. 2012;4:753–760. doi: 10.4161/mabs.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobson CL, Devine PW, Phillips JJ, Higazi DR, Lloyd C, Popovic B, Arnold J, Buchanan A, Lewis A, Goodman J, van der Walle CF, Thornton P, Vinall L, Lowne D, Aagaard A, Olsson LL, Ridderstad Wollberg A, Welsh F, Karamanos TK, Pashley CL, Iadanza MG, Ranson NA, Ashcroft AE, Kippen AD, Vaughan TJ, Radford SE, Lowe DC. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci Rep. 2016;6:38644. doi: 10.1038/srep38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dostalek M, Prueksaritanont T, Kelley RF. Pharmacokinetic de-risking tools for selection of monoclonal antibody lead candidates. MAbs. 2017;9:756–766. doi: 10.1080/19420862.2017.1323160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bumbaca Yadav D, Sharma VK, Boswell CA, Hotzel I, Tesar D, Shang Y, Ying Y, Fischer SK, Grogan JL, Chiang EY, Urban K, Ulufatu S, Khawli LA, Prabhu S, Joseph S, Kelley RF. Evaluating the Use of Antibody Variable Region (Fv) Charge as a Risk Assessment Tool for Predicting Typical Cynomolgus Monkey Pharmacokinetics. J Biol Chem. 2015;290:29732–29741. doi: 10.1074/jbc.M115.692434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, Nanami M, Sekimori Y, Nabuchi Y, Aso Y, Hattori K. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng Des Sel. 2010;23:385–392. doi: 10.1093/protein/gzq009. [DOI] [PubMed] [Google Scholar]
- 48.Boswell CA, Tesar DB, Mukhyala K, Theil FP, Fielder PJ, Khawli LA. Effects of charge on antibody tissue distribution and pharmacokinetics. Bioconjug Chem. 2010;21:2153–2163. doi: 10.1021/bc100261d. [DOI] [PubMed] [Google Scholar]
- 49.Sharma VK, Patapoff TW, Kabakoff B, Pai S, Hilario E, Zhang B, Li C, Borisov O, Kelley RF, Chorny I, Zhou JZ, Dill KA, Swartz TE. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci U S A. 2014;111:18601–18606. doi: 10.1073/pnas.1421779112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberg AS. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8:E501–507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu SJ, Luo J, O'Neil KT, Kang J, Lacy ER, Canziani G, Baker A, Huang M, Tang QM, Raju TS, Jacobs SA, Teplyakov A, Gilliland GL, Feng Y. Structure-based engineering of a monoclonal antibody for improved solubility. Protein Eng Des Sel. 2010;23:643–651. doi: 10.1093/protein/gzq037. [DOI] [PubMed] [Google Scholar]
- 52.Bethea D, Wu SJ, Luo J, Hyun L, Lacy ER, Teplyakov A, Jacobs SA, O'Neil KT, Gilliland GL, Feng Y. Mechanisms of self-association of a human monoclonal antibody CNTO607. Protein Eng Des Sel. 2012;25:531–537. doi: 10.1093/protein/gzs047. [DOI] [PubMed] [Google Scholar]
- 53.Pepinsky RB, Silvian L, Berkowitz SA, Farrington G, Lugovskoy A, Walus L, Eldredge J, Capili A, Mi S, Graff C, Garber E. Improving the solubility of anti-LINGO-1 monoclonal antibody Li33 by isotype switching and targeted mutagenesis. Protein Sci. 2010;19:954–966. doi: 10.1002/pro.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kehoe JW, Whitaker B, Bethea D, Lacy ER, Boakye K, Santulli-Marotto S, Ryan MH, Feng Y, Wheeler JC. Isolation and optimization for affinity and biophysical characteristics of anti-CCL17 antibodies from the VH1-69 germline gene. Protein Eng Des Sel. 2014;27:199–206. doi: 10.1093/protein/gzu012. [DOI] [PubMed] [Google Scholar]
- 55.Lee CC, Julian MC, Tiller KE, Meng F, DuConge SE, Akter R, Raleigh DP, Tessier PM. Design and optimization of anti-amyloid domain antibodies specific for beta-amyloid and islet amyloid polypeptide. J Biol Chem. 2016;291:2858–2873. doi: 10.1074/jbc.M115.682336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sormanni P, Amery L, Ekizoglou S, Vendruscolo M, Popovic B. Rapid and accurate in silico solubility screening of a monoclonal antibody library. Sci Rep. 2017;7:8200. doi: 10.1038/s41598-017-07800-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sormanni P, Aprile FA, Vendruscolo M. The CamSol Method of Rational Design of Protein Mutants with Enhanced Solubility. J Mol Biol. 2015;427:478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 58.Lauer TM, Agrawal NJ, Chennamsetty N, Egodage K, Helk B, Trout BL. Developability index: a rapid in silico tool for the screening of antibody aggregation propensity. J Pharm Sci. 2012;101:102–115. doi: 10.1002/jps.22758. [DOI] [PubMed] [Google Scholar]
- 59.Buck PM, Kumar S, Wang X, Agrawal NJ, Trout BL, Singh SK. Computational methods to predict therapeutic protein aggregation. Methods Mol Biol. 2012;899:425–451. doi: 10.1007/978-1-61779-921-1_26. [DOI] [PubMed] [Google Scholar]
- 60.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perchiacca JM, Lee CC, Tessier PM. Optimal charged mutations in the complementarity-determining regions that prevent domain antibody aggregation are dependent on the antibody scaffold. Protein Eng Des Sel. 2014;27:29–39. doi: 10.1093/protein/gzt058. [DOI] [PubMed] [Google Scholar]
- 62.Perchiacca JM, Ladiwala AR, Bhattacharya M, Tessier PM. Aggregation-resistant domain antibodies engineered with charged mutations near the edges of the complementarity-determining regions. Protein Eng Des Sel. 2012;25:591–601. doi: 10.1093/protein/gzs042. [DOI] [PubMed] [Google Scholar]
- 63.Dudgeon K, Rouet R, Kokmeijer I, Schofield P, Stolp J, Langley D, Stock D, Christ D. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc Natl Acad Sci U S A. 2012;109:10879–10884. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perchiacca JM, Bhattacharya M, Tessier PM. Mutational analysis of domain antibodies reveals aggregation hotspots within and near the complementarity determining regions. Proteins. 2011;79:2637–2647. doi: 10.1002/prot.23085. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Singh SK, Kumar S. Potential aggregation-prone regions in complementarity-determining regions of antibodies and their contribution towards antigen recognition: a computational analysis. Pharmaceutical research. 2010;27:1512–1529. doi: 10.1007/s11095-010-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, Das TK, Singh SK, Kumar S. Potential aggregation prone regions in biotherapeutics: A survey of commercial monoclonal antibodies. mAbs. 2009;1:254–267. doi: 10.4161/mabs.1.3.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y, Cao Y, Lynaugh H, Brown M, Baruah H, Gray LT, Krauland EM, Xu Y, Vasquez M, Wittrup KD. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci U S A. 2017;114:944–949. doi: 10.1073/pnas.1616408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Datta-Mannan A, Thangaraju A, Leung D, Tang Y, Witcher DR, Lu J, Wroblewski VJ. Balancing charge in the complementarity-determining regions of humanized mAbs without affecting pI reduces nonspecific binding and improves the pharmacokinetics. MAbs. 2015;7:483–493. doi: 10.1080/19420862.2015.1016696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Datta-Mannan A, Lu J, Witcher DR, Leung D, Tang Y, Wroblewski VJ. The interplay of non-specific binding, target-mediated clearance and FcRn interactions on the pharmacokinetics of humanized antibodies. MAbs. 2015;7:1084–1093. doi: 10.1080/19420862.2015.1075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong G, Chappey O, Niel E, Scherrmann JM. Enhanced cellular uptake and transport of polyclonal immunoglobulin G and fab after their cationization. J Drug Target. 2000;8:67–77. doi: 10.3109/10611860008996853. [DOI] [PubMed] [Google Scholar]
- 71.Lombana TN, Dillon M, Bevers J, 3rd, Spiess C. Optimizing antibody expression by using the naturally occurring framework diversity in a live bacterial antibody display system. Sci Rep. 2015;5:17488. doi: 10.1038/srep17488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baek DS, Kim YS. Construction of a large synthetic human Fab antibody library on yeast cell surface by optimized yeast mating. Journal of microbiology and biotechnology. 2014;24:408–420. doi: 10.4014/jmb.1401.01002. [DOI] [PubMed] [Google Scholar]
- 73.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, Pluckthun A, Virnekas B. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 74.Zhai W, Glanville J, Fuhrmann M, Mei L, Ni I, Sundar PD, Van Blarcom T, Abdiche Y, Lindquist K, Strohner R, Telman D, Cappuccilli G, Finlay WJ, Van den Brulle J, Cox DR, Pons J, Rajpal A. Synthetic antibodies designed on natural sequence landscapes. J Mol Biol. 2011;412:55–71. doi: 10.1016/j.jmb.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 75.Koenig P, Lee CV, Walters BT, Janakiraman V, Stinson J, Patapoff TW, Fuh G. Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc Natl Acad Sci U S A. 2017;114:E486–E495. doi: 10.1073/pnas.1613231114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koenig P, Lee CV, Sanowar S, Wu P, Stinson J, Harris SF, Fuh G. Deep Sequencing-guided Design of a High Affinity Dual Specificity Antibody to Target Two Angiogenic Factors in Neovascular Age-related Macular Degeneration. J Biol Chem. 2015;290:21773–21786. doi: 10.1074/jbc.M115.662783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koenig P, Sanowar S, Lee CV, Fuh G. Tuning the specificity of a Two-in-One Fab against three angiogenic antigens by fully utilizing the information of deep mutational scanning. MAbs. 2017;9:959–967. doi: 10.1080/19420862.2017.1337618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baran D, Pszolla MG, Lapidoth GD, Norn C, Dym O, Unger T, Albeck S, Tyka MD, Fleishman SJ. Principles for computational design of binding antibodies. Proc Natl Acad Sci U S A. 2017;114:10900–10905. doi: 10.1073/pnas.1707171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van der Kant R, Karow-Zwick AR, Van Durme J, Blech M, Gallardo R, Seeliger D, Assfalg K, Baatsen P, Compernolle G, Gils A, Studts JM, Schulz P, Garidel P, Schymkowitz J, Rousseau F. Prediction and Reduction of the Aggregation of Monoclonal Antibodies. J Mol Biol. 2017;429:1244–1261. doi: 10.1016/j.jmb.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Durme J, De Baets G, Van Der Kant R, Ramakers M, Ganesan A, Wilkinson H, Gallardo R, Rousseau F, Schymkowitz J. Solubis: a webserver to reduce protein aggregation through mutation. Protein Eng Des Sel. 2016;29:285–289. doi: 10.1093/protein/gzw019. [DOI] [PubMed] [Google Scholar]
- 81.Weitzner BD, Jeliazkov JR, Lyskov S, Marze N, Kuroda D, Frick R, Adolf-Bryfogle J, Biswas N, Dunbrack RL, Jr, Gray JJ. Modeling and docking of antibody structures with Rosetta. Nat Protoc. 2017;12:401–416. doi: 10.1038/nprot.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Norn CH, Lapidoth G, Fleishman SJ. High-accuracy modeling of antibody structures by a search for minimum-energy recombination of backbone fragments. Proteins. 2017;85:30–38. doi: 10.1002/prot.25185. [DOI] [PMC free article] [PubMed] [Google Scholar]