Abstract
Drug-induced kidney injury, largely caused by proximal tubular intoxicants, limits development and clinical use of new and approved drugs. Assessing preclinical nephrotoxicity relies on animal models that are frequently insensitive; thus, potentially novel techniques — including human microphysiological systems, or “organs on chips” — are proposed to accelerate drug development and predict safety. Polymyxins are potent antibiotics against multidrug-resistant microorganisms; however, clinical use remains restricted because of high risk of nephrotoxicity and limited understanding of toxicological mechanisms. To mitigate risks, structural analogs of polymyxins (NAB739 and NAB741) are currently in clinical development. Using a microphysiological system to model human kidney proximal tubule, we exposed cells to polymyxin B (PMB) and observed significant increases of injury signals, including kidney injury molecule-1 KIM-1and a panel of injury-associated miRNAs (each P < 0.001). Surprisingly, transcriptional profiling identified cholesterol biosynthesis as the primary cellular pathway induced by PMB (P = 1.22 ×10–16), and effluent cholesterol concentrations were significantly increased after exposure (P < 0.01). Additionally, we observed no upregulation of the nuclear factor (erythroid derived-2)–like 2 pathway, despite this being a common pathway upregulated in response to proximal tubule toxicants. In contrast with PMB exposure, minimal changes in gene expression, injury biomarkers, and cholesterol concentrations were observed in response to NAB739 and NAB741. Our findings demonstrate the preclinical safety of NAB739 and NAB741 and reveal cholesterol biosynthesis as a potentially novel pathway for PMB-induced injury. To our knowledge, this is the first demonstration of a human-on-chip platform used for simultaneous safety testing of new chemical entities and defining unique toxicological pathway responses of an FDA-approved molecule.
Keywords: Nephrology
Keywords: Cell stress, Toxicology, Toxins/drugs/xenobiotics
A human kidney microphysiological system was used to define polymyxin antibiotic toxicity and for safety assessment of next generation polymyxin derivatives.
Introduction
The rise in multidrug resistant microorganisms is a global health challenge, and one strategy increasingly under consideration to combat the epidemic is the reconfiguration of older antibiotics (1). Discovered in the late 1940s from Paenibacillus polymyxa (Bacillus polymyxa), polymyxin B (PMB) has been prescribed alone or in combination with other antimicrobials to treat life-threatening gram-negative bacterial infections (2, 3). PMB belongs to the cyclic lipopeptide class of antibiotics and comprises a decapeptide sequence that contains a polycationic heptapeptide ring and a fatty acyl tail. The amphipathic chemistry of PMB is essential for antibacterial activity. Electrostatic interaction between the cationic residues of PMB with negatively charged lipid A phosphates at the outer membrane of gram-negative bacteria displaces divalent cations (e.g., Mg2+ and Ca2+) that are necessary for lipopolysaccharide leaflet stability and allows the hydrophobic tail of PMB to be inserted into the outer membrane (4). This stimulates “self-uptake” through the exchange of lipids between the inner and outer membranes (5). Together, these events disrupt lipid homeostasis and promote permeabilization of the membrane to produce bactericidal effects (4, 5).
The rise in multidrug resistant bacterial infections has renewed interest in the polymyxin family of antibiotics, yet PMB and colistin continue to be used only as a last-resort treatment because of dose-limiting nephrotoxicity (6). In a retrospective study of 73 patients receiving PMB, close to 60% developed acute kidney injury (AKI) (7). In mice and rats treated with PMB, the primary site of accumulation and injury within the kidney is the proximal tubule (8, 9). These data are in agreement with the pharmacokinetics of PMB in humans, where the renal clearance of PMB is only a small fraction of the expected filtration clearance, indicating that PMB undergoes extensive tubular reabsorption (10). Accumulation of PMB in proximal tubule epithelial cells (PTECs) can likely be attributed to the negative charge of the brush border, the high expression of transporters and receptors involved in reabsorption of solutes from the ultrafiltrate, or a combination of the two (11, 12). Consequently, the toxicity of PMB toward PTECs may be driven by its polycationic nature. In response, Northern Antibiotics Ltd (NAB739) and Spero Therapeutics (SPR741) are currently developing analogs of PMB with fewer cationic charges that exhibit improved renal safety profiles in both in vitro and in vivo models (13–20).
Despite their existence for 6 decades, the definitive mechanisms of PMB nephrotoxicity remain poorly understood. In vitro studies have implicated death receptor activation and perturbation of the mitochondrial membrane along with a loss of membrane potential and generation of ROS as potential pathways responsible for polymyxin-induced injury. However, these findings are based primarily on transformed cell lines that were exposed to supraphysiologic drug concentrations (21, 22). To better elucidate the mechanism of PMB nephrotoxicity and develop mitigating strategies, we cultured primary human PTECs in a 3-D microfluidic device (Figure 1A) to create a kidney tubule MPS and used it to (a) probe the mechanism of PMB nephrotoxicity and (b) compare the safety of PMB to its NAB analogs (Figure 1B). The kidney tubule MPS performs normal physiological functions of the proximal tubule and displays appropriate injury responses, such as induction of heme oxygenase-1 (HMOX1) and shedding of kidney injury molecule-1 (KIM-1) (23, 24). These properties, along with the ease of sampling, imaging, and harvesting of cells, make the kidney tubule MPS well suited for mechanistic studies and obviating the use of animal models or cells. To bridge clinical observations to in vitro tests, we coupled analysis of urinary protein and miRNA biomarkers of AKI with whole-transcriptome RNA-Seq MPS analysis. To our knowledge, this is the first demonstration of a human-on-chip MPS platform’s utility for both safety testing of new chemical entities while defining unique toxicological pathway responses of an FDA-approved parent molecule.
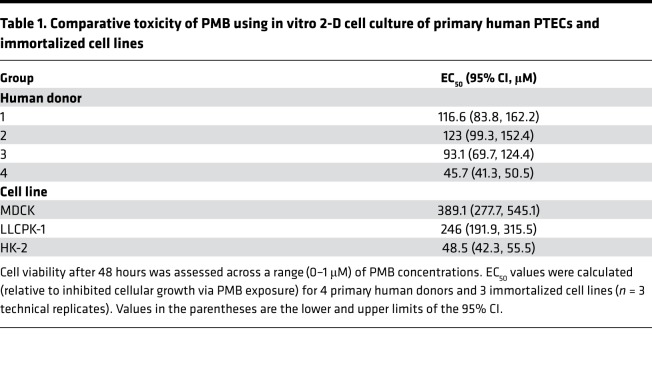
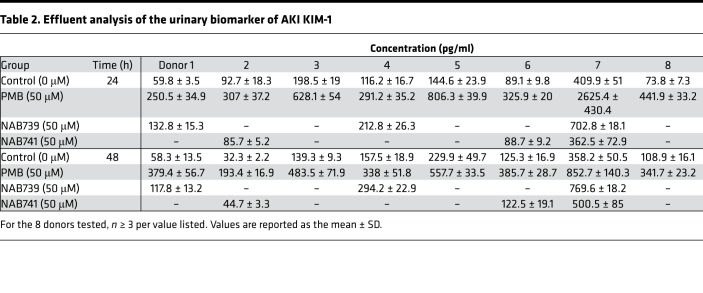
Figure 1. Kidney microphysiological system, PMB/analog structural variants, and PMB dosimetry in the microphysiological system.
(A) Single-channel Nortis microphysiological system (MPS). (B) Structural comparisons of PMB and polymyxin analogs (NAB739 and NAB741). (C) PMB dosimetry in the MPS after 48 hours of treatment with increasing PMB concentrations. Cell survival was assessed for differences in viability (green) and toxicity (red) with increasing concentrations of PMB. Devices exposed to 250 μM or higher resulted in severe cell death and loss of tubule integrity while 50 μM PMB caused detectable but modest injury (n = 1).
Results
PMB dose escalation in a 2-D culture and a 3-D kidney MPS.
Nephrotoxic properties of polymyxin antibiotics have traditionally been conducted using renal cell lines grown in 2-D cultures. To evaluate PMB toxicity in PTECs, we compared dose response profiles against transformed cell lines (LLCPK-1, MDCK, and HK-2) because the latter group have been traditionally used to measure PMB-induced cytotoxicity (25, 26). Following 48 hours of treatment, as indicated in Table 1, PTECs displayed increased sensitivity to PMB-induced cytotoxicity relative to LLCPK-1 and MDCK cells, while HK-2 cells were similar (PTEC EC50 range: 45.7 to 123 μM; LLCPK-1 EC50: 246 μM; MDCK EC50: 389.1 μM; HK-2 EC50: 48.5 μM). Comparing the average EC50 value of PTECs (94.6 μM), cytotoxicity sensitivity was 4-fold higher compared with MDCK, 2.6-fold higher compared with LLCPK-1, and approximately 50% lower compared with HK-2.
Table 1. Comparative toxicity of PMB using in vitro 2-D cell culture of primary human PTECs and immortalized cell lines.
For comparison, PTECs cultured in the MPS were exposed for 48 hours to increasing PMB concentrations (Figure 1C). After PMB treatment, concentrations at or above 250 μM resulted in severe cytotoxicity, as demonstrated by loss of tubular integrity and high percentage of dead/dying cells. Subsequent experiments focused on a dose of 50 μM of PMB because this dose resulted in a high degree of cell viability and approximates the maximum concentration observed in nonhuman primates treated with PMB and derivatives (Martti Vaara, unpublished observations).
Assessing polymyxin-induced nephrotoxicity using injury biomarkers.
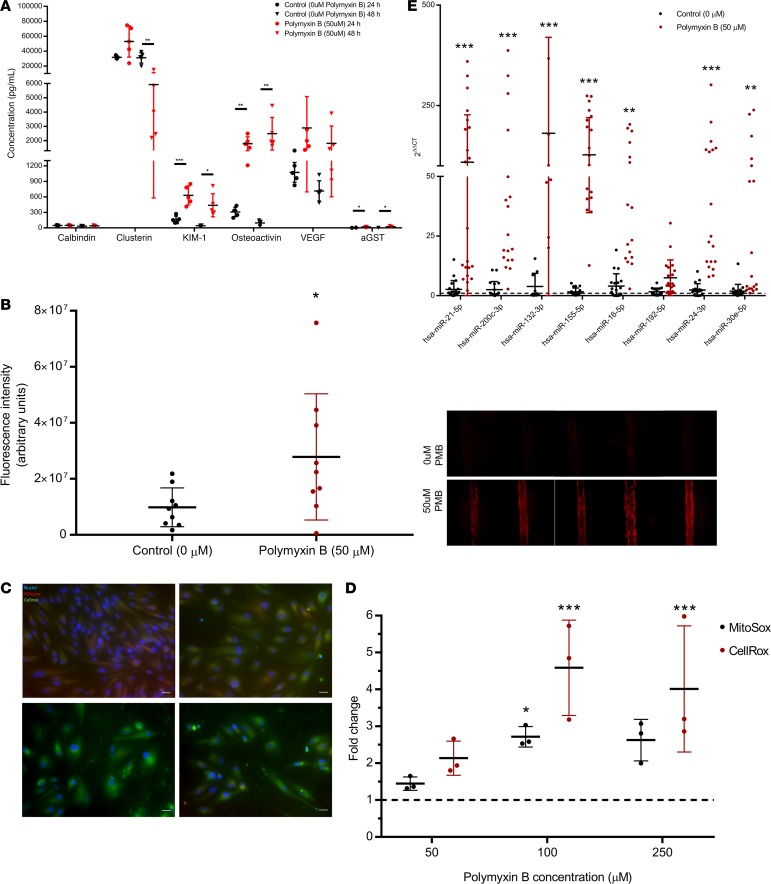
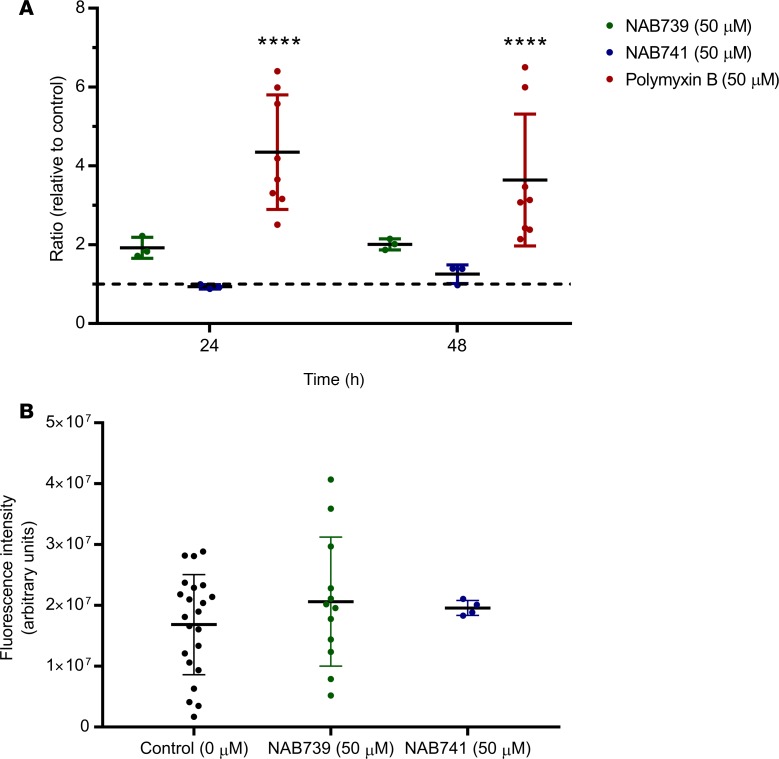
New biomarkers, particularly those of tubular origin, show excellent promise for detecting and reporting the severity of AKI. KIM-1 is a transmembrane glycoprotein that is absent from nonproliferative PTECs but becomes highly upregulated during AKI and is proteolytically cleaved at the plasma membrane to produce a soluble form that is detectable in urine (10, 27, 28). Surface expression of KIM-1 enables clearance of apoptotic bodies and necrotic debris, which is proposed to be a protective mechanism of tubular cells to resolve injury and prevent inflammation (29, 30). We quantified KIM-1 concentrations in MPS effluents after treatment with 50 μM of PMB, NAB739, or NAB741 after 24 and 48 hours of exposure. Results of the KIM-1 response with 8 human donor sources are summarized in Table 2. In response to PMB exposure at both 24 and 48 hours, the concentration of KIM-1 was approximately 4-fold higher relative to controls, which was statistically significant (P < 0.001). In addition to KIM-1, a panel of kidney injury biomarkers (calbindin, clusterin, osteoactivin, KIM-1, VEGF, and α-GST) was analyzed, revealing significant differences for osteoactivin, KIM-1, VEGF, and α-GST between PMB and controls (Figure 2A).
Table 2. Effluent analysis of the urinary biomarker of AKI KIM-1.
Figure 2. Effluent and cell-associated biomarkers of nephrotoxicity.
(A) Urinary kidney injury biomarkers (osteoactivin, KIM-1, VEGF, and α-GST) were assayed from MPS effluents. There was a significant difference between PMB-treated control MPS effluent for urinary biomarkers KIM-1, osteoactivin, and α-GST. (B) Cell-associated HMOX1. HMOX1 was detected by immunocytochemistry (left panel, representative donor) and quantified (right panel, n = 2 donors, 9 controls, 8 treated). A significant induction (P < 0.05) for devices treated with 50 μM PMB relative to controls was observed. Values are reported as MFI ± SD. (C) PTECs in 2-D culture for 72 hours were exposed to increasing concentrations (top left, 0 μM; top right, 50 μM; bottom left, 100 μM; bottom right, 250 μM) of PMB for 24 hours. Using cellular stains, MitoSOX (red) and CellROX (green), ROS was measured. (D) ROS quantitation. Fluorescence intensity was quantified and normalized to control intensity values. A significant (P < 0.001) increase of cellular ROS was observed at 100 μM and 250 μM PMB. MitoSOX, an indicator of mitochondrial-specific ROS, showed a significant increase at doses higher than 50 μM PMB (100 μM) (n = 3 fields of view/dose for image quantitation, P < 0.05). (E) Urinary miRNA biomarkers. The effluent at 48 hours showed significant induction of miRNA-21, -200c, -132, -155, -16, -24, and -30e across multiple human donor cells (n = 3) cultured within the 3-D kidney MPS. Values are reported as the mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001).
PTEC-associated HMOX1 was recently shown to be a relatively sensitive biomarker for predicting drug-induced nephrotoxicity in vitro compared with other biomarkers (23). To assess expression/induction of HMOX1, MPSs were processed by immunocytochemistry and the relative fluorescent signal intensity quantified. After 48 hours of PMB treatment, cell-associated HMOX1 was approximately 3-fold higher relative to controls across multiple donors (P < 0.05, Figure 2B).
Polymyxin antibiotics have previously been hypothesized to mediate adverse effects via overproduction of ROS (31). We measured ROS production in 2-D–cultured PTECs in response to increasing concentrations of PMB exposure using both CellROX (a free radical sensor that fluoresces when oxidized by ROS) and MitoSOX (a fluorogenic dye that detects intracellular mitochondrial superoxide). In response to PMB exposure, cell morphology and density significantly changed in a dose-dependent relationship. CellROX and MitoSOX levels were minimal in controls and increased in response to PMB (50–250 μM) (Figure 2C). Quantifying fluorescence intensity (relative to controls) showed significant (P < 0.001 or 0.05) production of both cellular ROS and higher mitochondrial ROS but only at higher concentrations (100 and 250 μM of PMB) than were demonstrated to upregulate injury biomarkers in the MPS, suggesting that increased ROS production is downstream of initiating toxic events (Figure 2D).
The presence of several miRNAs in the urine of patients are associated with AKI (32, 33). We tested the utility of these miRNAs in our MPS by quantifying effluent concentrations following 48 hours of PMB exposure. We identified miRNA-21, -200c, -132, -155, -16, -24, and -30e as being significantly (P < 0.001 or 0.01) higher in the MPS effluents (Figure 2E). This observation was consistent among multiple donors, with average miRNA induction being 250-fold higher than controls without PMB exposure.
The comparative safety of 2 structural analogs of PMB, NAB739 or NAB741, was supported by the lack of “urinary” biomarker response via KIM-1 induction (Figure 3A). Following treatment with NAB compounds, there was no significant change at 24 hours for NAB739 (P = 0.38) or NAB741 (P = 0.31). This was also the case at 48 hours of treatment for both NAB739 (P = 1.0) and NAB741 (P = 0.96). After 48 hours of treatment, MPSs were fixed and analyzed by immunocytochemistry. The lack of a KIM-1 safety signal for the NAB compounds was corroborated by a lack of an increase in the HMOX1 MPS-associated signal following 48 hours of treatment (Figure 3B) in contrast with what was observed following PMB treatment (Figure 2B).
Figure 3. PMB versus NAB739/NAB741 demonstrates mitigated toxicity with analogs.
(A) Effluents were collected at 24 and 48 hours of treatment and analyzed for KIM-1 levels (n = 8 donors). Significant accumulation of KIM-1 in the effluent was observed only in the PMB group. (B) Cell-associated HMOX1. HMOX1 expression was measured by immunostaining, with no significant differences observed for either NAB739 or NAB741 relative to controls across several donors treated with 50 μM NAB739 (n = 3) and 50 μM NAB741 (n = 3) relative to controls (n = 5). Values are reported as the mean ± SD (****P < 0.0001).
Transcriptional response to polymyxin exposure.
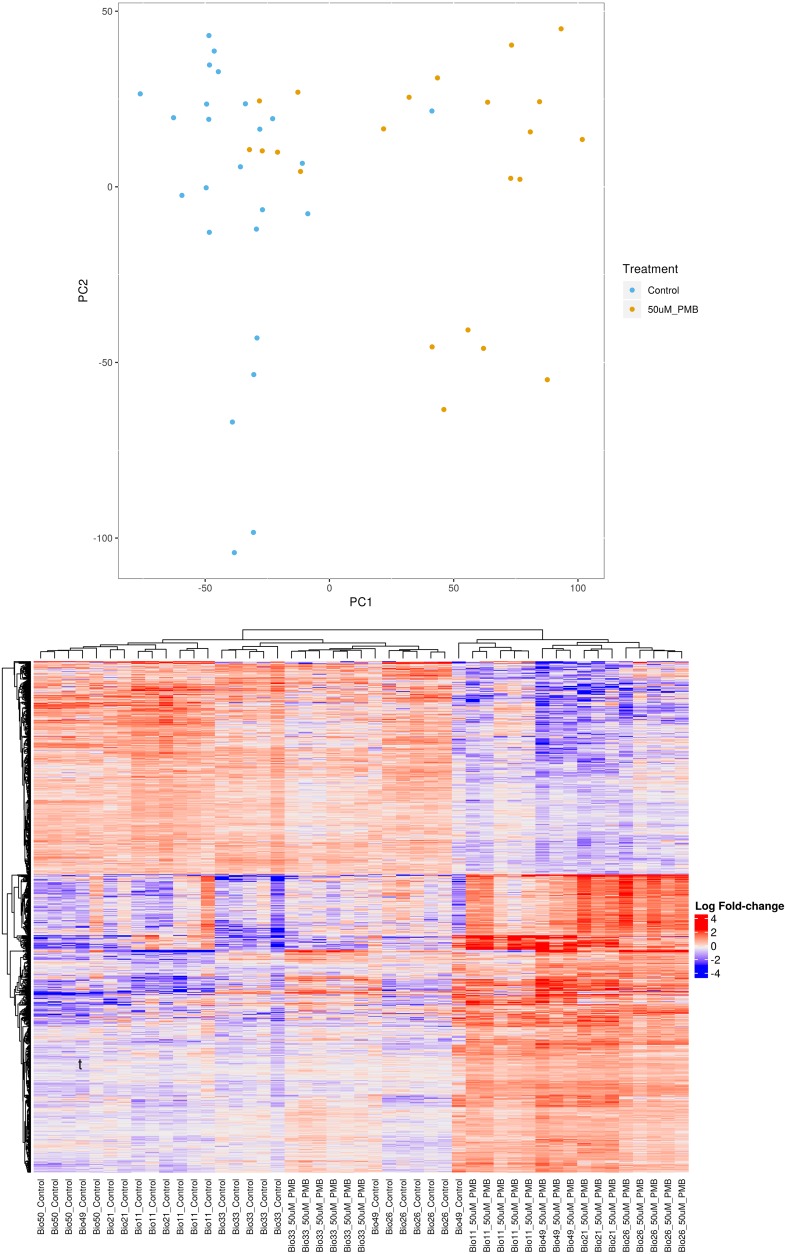
Global transcriptional changes were initially assessed via RNA-Seq, in MPSs exposed to either PMB or control media. In principal component analysis (PCA), principal component 1 separated groups based on PMB treatment into 2 clustered sections, indicating dominant transcriptional differences were in response to drug treatment rather than inter- and intra-individual donor response variation. With this approach, we identified 808 genes that were significantly different between control and PMB treatment groups that are visually delineated by heatmap analysis (Figure 4). It should be noted that 1 donor (BIO-33) did not fall within the PCA/heatmap groupings, with a transcriptional profile indicative of no response to PMB treatment (principal component 1), and 1 donor (BIO-26) did cluster differentially relative to other controls in principal component 2.
Figure 4. RNA-Seq and transcriptional response to PMB exposure.
PCA reveals clustering within respective groups of treatment (control versus 50 μM PMB) as well as a distinction of the treated and control (PC1) and between the individual donors (PC2). Heatmap analysis of individual MPSs grouped by donors and treatment groups.
Specifically, we observed the transcriptional upregulation of the metallothionein family (MT1G, MT1H, MT1M, MT1X, MT2A, MT1E, and MT1F); unfolded protein response (5/32 genes); nuclear factor (erythroid derived-2)–like 2–mediated (NRF2-mediated) antioxidant response (8/56); and cholesterol biosynthesis (16/25 genes), as described in Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123673DS1). Relative to controls, PMB exposure caused a 1.31 log2 fold change for HMOX1, a 2.62 log2 fold change for MT1G, a 2.58 log2 fold change for MT1H, a 2.23 log2 fold change for MT1M, a 2.21 log2 fold change for MT1X, a 1.82 log2 fold change for MT2A, a 1.51 log2 fold change for MT1E, and a 1.47 log2 fold change for MT1F. Given the proposed mechanism of PMB nephrotoxicity (ROS generation), canonical genes in the antioxidant NRF2-mediated antioxidant response pathway (NQO1, GCLM, FTH1, GPX6, KEAP1, and SOD1) were analyzed, but none was significantly changed in response to PMB. This observation is concordant with the lack of an increase in ROS production at 50 μM of PMB (Figure 2C).
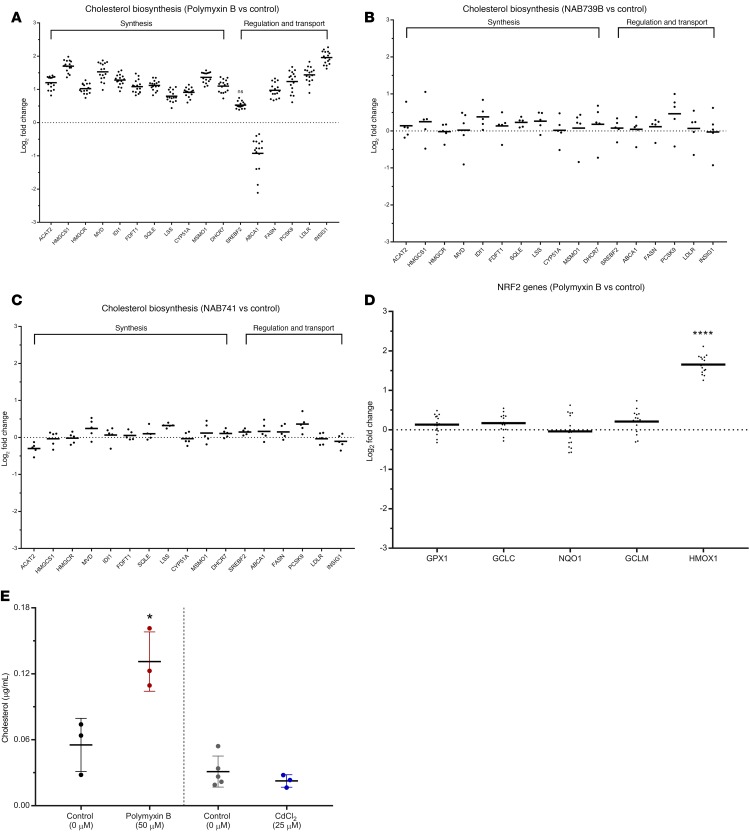
To identify the complex pathways significantly perturbed at the transcriptional level, we used Qiagen Ingenuity Pathway Analysis software, which summarizes the canonical pathways affected in response to PMB exposure (data not shown). The superpathway of cholesterol biosynthesis was the most significantly upregulated pathway (P = 1.22 × 10–16) followed by cholesterol biosynthesis I, II, and III (P = 8.26 × 10–13). The genes significantly affected by PMB treatment are grouped by cholesterol synthesis and regulation/transport in Figure 5A.
Figure 5. RNA-Seq identifies novel injury pathways that are PMB specific.
(A–C) Transcriptional response, via activation of cholesterol biosynthesis pathways, was visualized for the top 10 genes for PTECs in the MPS exposed to (A) PMB, (B) NAB739, and (C) NAB741. (D) PMB exposure shows little/no activation of the NRF2-mediated antioxidant response pathway. Control and PMB-treated MPS effluents were tested for the presence of cholesterol following 48 hours of treatment. (E) Effluent cholesterol analysis. In response to PMB exposure (left panel), cholesterol levels showed a significant increase at 48 hours (P < 0.05) while cadmium chloride exposure (right panel, 25 μM for 48 hours) exhibited similar levels of cholesterol to controls (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
We then compared the transcriptional response after exposure to PMB with the response after exposure to polymyxin structural analogs NAB739 or NAB741 (Figure 5, B and C) and the proximal tubule nephrotoxin cadmium chloride (Supplemental Figure 1). In PCA, the structural analogs NAB739 and NAB741 clustered tightly with the control group (data not shown), which was supported by the transcriptional response to the structural analogs NAB739 and NAB741 that identified only 18 genes and 1 gene, respectively, relative to the control group. Exposure to the proximal tubule nephrotoxin cadmium chloride revealed significant effects on the NRF2-mediated antioxidant response pathway (Supplemental Figure 1A) as well as the unfolded protein response pathway (data not shown) but no significant effects on cholesterol pathway genes (Supplemental Figure 1B), indicating differential modes of response to nephrotoxin exposures. Although Qiagen Ingenuity Pathway Analysis software implicated the NRF2 pathway for PMB, analysis of canonical genes associated with this pathway revealed that only HMOX1 was induced (Figure 5D).
Quantification of cholesterol and cholesterol precursors in effluents of the MPS.
To determine whether the transcriptional changes in cholesterol biosynthesis genes after PMB exposure have a functional consequence, we quantified the levels of cholesterol in the MPS effluents. PTECs exposed to PMB showed a significant increase (P < 0.01) in effluent cholesterol at 48 hours. In contrast, when effluents from cadmium chloride–treated MPSs were examined, no changes in cholesterol or cholesterol precursors were observed (Figure 5E).
Membrane binding of PMB and NAB741.
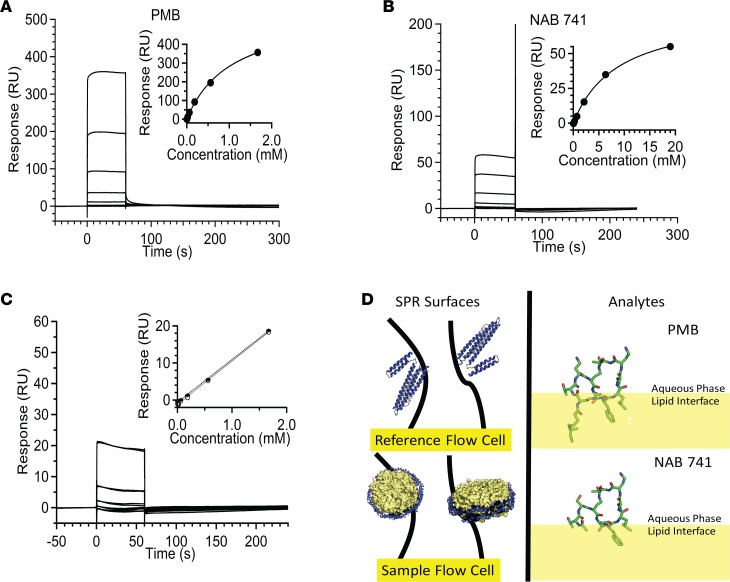
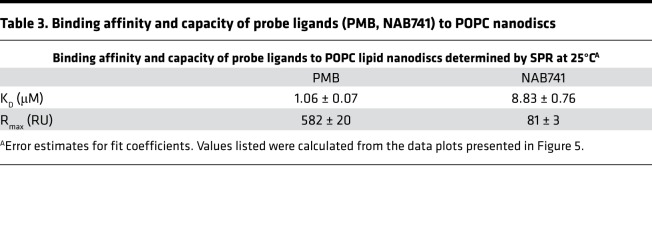
The improved safety profile of the PMB NAB analogs may be due to decreased interactions with the renal proximal tubule apical membrane because of their lower cationic charge (14). We hypothesized that the transcriptional activation of the cholesterol biosynthetic pathway after PMB exposure may be a direct response to cell membrane damage. Given that the first step in membrane damage is membrane binding, we elected to investigate this hypothesis in vitro. Nanodisc membrane assemblies were incubated with either PMB or NAB741, and binding parameters measured by surface plasmon resonance (SPR). As shown in Figure 6, A and B, and Table 3, NAB741 displayed an equilibrium dissociation constant (KD) that was approximately 8-fold higher than the parent PMB molecule. The linearity of the equilibrium response data at lower concentrations is presented in Figure 6C. Figure 6D includes a speculative model for this difference wherein the acyl “tail” extending from the cyclic core of PMB provides a membrane anchor that embeds partially into the lipid bilayer of the nanodisc membranes.
Figure 6. SPR experiments demonstrate high affinity and capacity of PMB–lipid nanodisc interactions.
(A) Binding data for PMB (2.3, 6.9, 20.6, 61.9, and 186.557.1670 μM) to nanodiscs. Equilibrium binding constant determined by fitting equilibrium responses to a steady-state binding model (inset). (B) Binding data for NAB741 (26, 78, 235.704.2111, 6,333, and 19,000 μM) to nanodiscs. Binding isotherm fit to a steady-state binding model for NAB741 (inset). (C) Representative sensorgrams showing overlapping duplicates. Linearity observed for equilibrium response data (duplicates), at lower concentrations (2.3, 6.9, 20.6, 61.9, 186.557.1670 μM) of NAB741 (dotted lines, inset). (D) Strategy for immobilization of 1-pal- mitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid nanodiscs on sample flow cell surface and MSP1D1 (Protein Data Bank 2A01, depicted as lipid-free ApoA1 with N-terminal region in yellow) on the reference surface (left panel). Partitioning of PMB and NAB741 into the lipid bilayer determined by the Orientation of Proteins and Membranes database (https://opm.phar.umich.edu; OPM database). Conformation of NAB741 is based on PMB (Protein Data Bank 5L3F) and is not intended to be an exhaustive representation of conformers. Values are reported as the mean ± SD.
Table 3. Binding affinity and capacity of probe ligands (PMB, NAB741) to POPC nanodiscs.
Discussion
The nephrotoxicity of polymyxin antibiotics limits their use in patients and is a significant concern to drug developers seeking to identify safer alternatives. To address these challenges, we used a human kidney MPS to investigate PMB-induced nephrotoxicity at drug concentrations within the physiological range (10). In this study, we recapitulated PMB-induced nephrotoxicity ex vivo and assessed the safety of structural PMB analogs through analysis of multiple biomarkers associated with AKI.
As compared with traditional cultures in 2-D, PTECs cultured in the 3-D kidney MPS exhibit both functional and structural aspects of the kidney proximal tubule (24). Transformed cell lines in toxicity screens are typically used as a standardized cell type because of their ease of use and robust growth characteristics. However, in this study we found that porcine and canine kidney cell lines are less sensitive than primary PTECs while the human HK2 cell line had a degree of sensitivity to PMB toxicity that fell within the range of primary PTECs. Furthermore, analysis of the same PTEC donors at the transcriptional level revealed that 2-D–cultured PTECs exhibit PCA plot clustering quite distinct from a 3-D MPS culture, indicating culture conditions play a significant role in gene expression (data not shown). There are caveats to using primary human cells to populate MPSs, including their finite propagation, limited sourcing for investigators, and inter-individual variability among donors. In support of this final point, 1 of the donors tested in this study did not exhibit the transcriptional responses shared by all other donors tested. Future directions to address these issues will rely on the development of robust protocols for differentiating pluripotent stem cells into mature cell types, including proximal tubule epithelial cells.
Using transformed cell lines, prior proposed mechanisms of PMB-induced toxicity were identified via a direct pathway to apoptosis from mitochondrial perturbation (with resultant ROS production) or an indirect pathway via death receptor activation in response to treatment with greater than or equal to 1 μM of PMB (21, 22). To address these findings, we exposed PTECs to increasing concentrations of PMB (up to 1 μM) and identified a distinct response compared with high-dose 2-D culture systems. We did not observe an increase in mitochondrial ROS generation until higher PMB concentrations were reached (100-μM and 250-μM concentrations), which are highly toxic in MPSs and greater than levels that induce MPS effluent biomarkers of AKI. Furthermore, interrogation of the global transcriptome (for PTECs grown in the 3-D kidney MPS) revealed a significant upregulation of HMOX1 and MT gene family members when cells were exposed to 50 μM of PMB. However, the primary pathways significantly affected involved upregulation of cholesterol biosynthesis. Interestingly, recognized NRF2-mediated antioxidant response genes (e.g., NQO1, GCLM, and GPX) were unaffected despite significant induction of HMOX1 and MT genes. This lack of induction of an antioxidant response is supported by the lack of increased ROS production in response to treatment with 50 μM of PMB. Thus, the induction of HMOX1 and MT genes represents a “generalized” stress response in the kidney MPS. In support of this, treatment of kidney MPS with the nephrotoxic heavy metal cadmium chloride induces HMOX1 and other NRF2 pathway–associated genes without affecting cholesterol biosynthetic transcriptome or cholesterol production (as measured in MPS effluent).
Upregulation of cholesterol biosynthesis is a unique pathway of PMB nephrotoxicity that has not previously been reported to our knowledge. We have shown that in response to PMB exposure, cholesterol accumulates to higher levels in the effluent of the MPS. A recent study suggests that PMB-induced membrane damage can be significantly reduced when kidney membrane analogs are loaded with cholesterol (34). These findings are in agreement with our observations showing PMB-exposed cells generating cholesterol (via upregulation of cholesterol biosynthesis) and attempting to load cellular membranes. Interestingly, clinical findings from Johnson et al show, in a cohort of patients with AKI, induction of HMG-CoA reductase (HMGCR) resulting in cholesterol loading within the proximal tubule, causing an increase in urinary cholesterol levels (35). In our study, we also observed a significant induction of HMGCR mRNA in response to toxicant, which likely explains the effluent cholesterol measurements. Furthermore, transcripts for ABCA1, an apical cholesterol efflux transport protein, were significantly downregulated in the PMB-treated MPS. Upregulation of HMGCR and downregulation of ABCA1 transcripts indicate that the cholesterol effluent response was likely not the result of transport-mediated sterol excretion, but rather the result of membrane-associated sterols being shed into the tubular lumen. Measuring urinary cholesterol levels (including precursors) in response to AKI has the potential to be a sensitive biomarker of injury that, when detected early on, could provide guidance on patient care using a commonly measured analyte.
The mechanisms of cellular uptake of PMB have been proposed to be primarily driven by megalin-mediated endocytosis (36, 37). However, it was recently shown that human oligopeptide transporter 2 (PEPT2) mediates cellular uptake of polymyxins in renal tubular cells (human embryonic kidney 293 cells) that overexpressed 15 essential solute carrier transporters (11). In our study, PEPT2 was significantly downregulated in response to PMB exposure (data not shown), which can indicate the transcriptional response to decrease cellular uptake of PMB via PEPT2. A hypothesis generated from our findings is that targeting inhibition of PEPT2-mediated transport may reduce the overall nephrotoxic effects of PMB accumulation.
The SPR experiments reveal a clear difference in the relative affinities of PMB versus NAB741 for model membranes. The SPR results also indicate that both compounds have very fast on and off rates, but PMB clearly accumulates in or on the membrane compared with NAB741. Because the association and dissociation are too fast to recover rate constants, the recovered KD values were obtained from the equilibrium response. The 8-fold lower apparent KD after PMB exposure is consistent with its ability to disrupt PTEC membranes. We have provided a speculative model for this difference wherein the acyl “tail” extending from the cyclic core of PMB provides a membrane anchor that embeds partially into the lipid. The difference in binding capacity of the 2 compounds, reflected in the maximum response (Rmax) values, suggests that many PMBs are able to pack together on the membrane surface, possibly due to peptide–peptide interactions orthogonal to the membrane surface. Speculatively, NAB741 would not participate in intermolecular interactions because it is not as tightly anchored in the membrane. As a result, the NAB741 may not associate on the membrane and afford a lower Rmax. This suggests that the difference in capacity for binding to the membrane is as important for the differential toxicity of PMB versus NAB741 as the difference in affinity. Regardless of the detailed mechanism, the results reveal a clear difference in the tendency of these peptides to bind to the membranes — the necessary first step before cellular internalization.
Compared with a recent study observing increases of KIM-1/urinary creatinine levels in patients after 72 hours of PMB exposure, we observed significant accumulation of KIM-1 at both 24 and 48 hours of exposure, demonstrating the sensitivity of the 3-D kidney MPS to detect early injury (38). Classical clinical biomarkers of AKI are continuously criticized for their lack of sensitivity (defined by receiver operating characteristic curves), highlighting the need to develop and identify new biomarkers of renal injury (40). Our results demonstrate the potential of KIM-1 as well as select miRNAs (miRNA-21, -200c, -132, -155, -16, -24, and -30e) to be significantly induced for detection of AKI in a sensitive and timely manner. With respect to the utility of these biomarkers when comparing 3-D and 2-D in vitro approaches, KIM-1 is expressed at significantly higher levels of our PTECs in 2-D compared with 3-D (C. Maass et al., Massachusetts Institute of Technology, personal communication). This mitigates the utility of KIM-1 as a sensitive biomarker of AKI in vitro.
In summary, we have taken an organ-on-a-chip technology (human kidney 3-D MPS), applied it to toxicological investigation of a known class of nephrotoxic antibiotics (polymyxins), and defined unique exposure responses. In addition, we have evaluated the safety of 2 structural variants of PMB (NAB739 and NAB741). NAB741, now being developed by Spero Therapeutics as SPR741, has recently completed a phase I clinical trial reporting, “Data from our Phase 1 SAD/MAD clinical trial of SPR741 demonstrate SPR741 was well tolerated at single doses up to and including 800 mg and multiple daily doses up to and including 600 mg every 8 hours for 14 consecutive days” (40). The clinical trial results are in concordance with our MPS assessment of NAB741, but it should be noted that we looked only at short-term exposures, warranting studies using treatment paradigms of lower doses and duration to most accurately emulate patient outcomes. Finally, sensitive detection of urinary biomarkers (KIM-1 and miRNAs) using the human kidney 3-D MPS highlights the implementation of new technologies for detection of AKI.
The human kidney 3-D MPS platform has demonstrated robust ability toward multiple preclinical development applications, including toxicological investigations (23, 41). New chemical entity advancement through the preclinical development pipeline is extremely costly when potential candidates are triaged based on inaccurate safety data. Preclinical test systems can often be the culprit behind false-positive results based on poor in vitro to in vivo predictions. The human kidney 3-D MPS bridges the in vitro to in vivo gap, providing a reliable system to detect human toxicity within 24 hours of exposure. Further implementation of organ-on-a-chip technologies, emphasizing the 3 Rs of toxicology (reduce, refine, replace), will make a positive impact by reducing the overall number of animals tested, reducing the usage of only relevant species for predictive toxicity testing, and advancing the ultimate goal of replacing animals in preclinical safety assessment (e.g., human on a chip) (42, 43).
Methods
Tissue acquisition and cell isolation.
Human kidney tissues were collected at the University of Washington Medical Center under a University of Washington Institutional Review Board–approved Human Subjects protocol. Tissue was processed within 12 hours as previously described (29). In brief, tissue was finely minced under sterile conditions, subjected to collagenase digestion with shaking at 37°C for 30 minutes, mixed 1:1 with horse serum, vortexed, and allowed to briefly settle, and supernatant was harvested. The cell suspension was centrifuged, washed, and plated in T25 flasks containing DMEM/F12 media supplemented with insulin, transferrin, selenium, and hydrocortisone.
Cell seeding in Nortis devices.
The Nortis devices were constructed with an extracellular matrix of type I collagen, and the cell chamber was coated with type IV collagen to facilitate cell adhesion. Cultures of primary renal epithelial cells, upon reaching greater than 80% confluency, were subjected to trypsin digestion and manual cell scraping to obtain single-cell suspensions. The cells were counted, washed, and resuspended at a concentration of 20 × 106 cells/ml, and 1–2 μl were injected into the Nortis device. Cells were incubated to allow matrix adhering for 12 hours before initiating flow at a rate of 0.5 μl/min. The integrity of the tubule cell structure was assessed grossly by light microscopy weekly.
PMB dosimetry in 2-D cell culture.
PTECs and transformed cell lines (LLCPK-1, MDCK, and HK-2) were seeded at a density of approximately 10,000 cells/well in a 96-well plate and allowed to culture overnight before dosing. PMB (National Drug Code 39822-0170-2, X-Gen Pharmaceuticals Inc) dosimetry was assessed across 10 concentrations, starting at concentration 1 μM and serial diluted down 1:1 with cell medium. Cells were exposed to PMB for 48 hours before cell cytotoxicity was measured using the Abcam ab112118 Cell Cytotoxicity Assay Kit. Each EC50 across multiple human donor PTECs and transformed cell lines was defined and plotted using Prism GraphPad.
Viability demonstrated using live/dead staining.
The LIVE/DEAD Viability/Cytotoxicity Kit and Hoechst 33342 stain (Life Technologies) were used to distinguish viable cells from dead cells according to the manufacturer’s specifications. Briefly, calcein AM (final concentration 2 μM), EthD-1 (final concentration 4 μM), and Hoechst 33342 (final concentration 0.1 μg/ml) were diluted in prewarmed D-PBS. MPS chips were perfused at 5 μl/min via a luminal port of a kidney MPS for 20 minutes and then incubated for 10 minutes at 37°C. After the staining procedure, chips were imaged using fluorescent microscopy to visualize live (green-stained cells) and dead cells (red-stained cells) with nuclei marker Hoechst 33342 (blue-stained cells).
Analysis of ROS generation.
Primary human PTECs were grown to approximately 75% confluency in a multichamber culture slide. Upon 75% confluency, PTECs were individually exposed to increasing concentrations of PMB (0, 50, 100, and 250 μM) for 48 hours then stained for ROS generation using both CellROX (C10444, Thermo Fisher Scientific) for 30 minutes and MitoSOX (M36008) for 10 minutes. After staining, PTECs were imaged using fluorescent microscopy with a Nikon Ti-S microscope, and both red and green intensity were quantified with Nikon Eclipse software. PMB-treated cells’ ROS intensity was normalized to the control chamber, and the results were expressed graphically (per nuclei).
Analysis of KIM-1.
Effluents of kidney MPS exposed to 50 μM of PMB, 50 μM of NAB739 or NAB741 (Northern Antibiotics Ltd), or control at both 24 and 48 hours were analyzed for human KIM-1 using the Meso Scale Discovery Human KIM-1 Kit (K141JHD-2).
Analysis of miRNA.
After 48 hours of exposure to PMB, 200 μl of effluents were collected (dose escalation study) and used for miRNA isolation using RNeasy Micro Kit (Qiagen catalog number 74004). miRNA-21, -200c, -432, -132, -155, -16, -192, -24, and -30e were quantified with custom miScript miRNA PCR Array. miRNA cycle threshold values were normalized using the geometric mean of technical controls (miRNA reverse transcription control and positive PCR control) for each sample. The 2–ΔΔCt was calculated compared with the control group.
Isolation of mRNA and RNA-Seq.
Cells cultured within the kidney MPS were isolated after 48 hours of treatment, and RNA was isolated using the Qiagen RNeasy Micro Kit. The RNA library was prepared using a SMARTer Stranded Total RNA Sample Prep Kit - Low Input Mammalian (Takara catalog number 634861), and RNA-Seq was performed at 300 cycles in 2 lanes using the Illumina NextSeq 500 High Output v2 Kit (GEO accession GSE123264).
Immunocytochemistry analysis of HMOX1.
Anti-HMOX1 antibody (rabbit polyclonal, Abcam) was used. Cells were fixed with 4% formaldehyde in PBS. Samples were permeabilized with PBS plus 0.1% Tween 20 and blocked with 1% BSA for 1 hour. The samples were then incubated with primary (1:100) and secondary (1:1,000) antibodies followed by washing, using a standard immunocytochemistry protocol. Secondary antibodies were goat anti–rabbit IgG (H+L) secondary antibody and Alexa Fluor 594 conjugate (both from Abcam). Diluted ProLong Gold Antifade Mountant with DAPI reagents (Thermo Fisher Scientific) was used in the final step of staining. Cells were imaged using fluorescent microscopy with a Nikon Eclipse Ti-S microscope, and red (Fluor 594) intensity was quantified with Nikon Eclipse software.
Quantification of cholesterol in the effluent.
Optima liquid chromatography/mass spectrometry grade solvents (water, methylene chloride, chloroform, formic acid, and methanol) and sodium chloride (American Chemical Society [ACS] certified) were purchased from Thermo Fisher Scientific. Materials and supplies used for lipid extraction and ultra–HPLC-mass spectrometry (UHPLC-MS) were purchased from Thermo Fisher Scientific.
Deuterated (d7 = [25, 26, 26, 26, 27, 27, 27-2H]) 7-dehydrocholesterol was prepared as reported previously (44). 13C3-desmosterol was a gift from Ned Porter and Keri Tallman and was prepared as reported (44). All other sterol standards and internal standards were purchased from Avanti Polar Lipids Inc. 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD) was purchased from MilliporeSigma.
Lipid extraction from kidney chip effluents.
Before lipid extraction, internal isotopically labeled sterol standards (d7-cholesterol [50 ng], d7-7-dehydrocholesterol [50 ng], and 13C3-desmosterol [10 ng]) were added to each sample. Folch solution (2 ml, chloroform: methanol = 2:1 v/v) was added directly to each kidney chip effluent sample (100 μl). NaCl aqueous solution (0.5 ml, 0.9% w/v) was then added, and the resulting mixture was briefly vortexed and then centrifuged for 5 minutes in a clinical tabletop centrifuge (at ambient temperature). The lower (organic) phase was recovered and transferred to a 15-ml polypropylene conical centrifuge tube, and the solvent was removed in vacuo using a Savant SpeedVac (Thermo Fisher Scientific). Finally, the resulting dried extracts were redissolved in methylene chloride (300 μl) before further processing.
UHPLC-MS/MS analyses of cholesterol in kidney chip effluents.
Analysis of cholesterol was performed by UHPLC-MS/MS using a triple quadrupole mass spectrometer (API 4000; AB SCIEX) equipped with atmospheric pressure chemical ionization. For analysis, 100 μl of reconstituted cholesterol was transferred to a liquid chromatography vial, dried under a stream of argon, and reconstituted in 20 μl of 90% methanol with 0.1% formic acid. Reverse-phase chromatography was performed with the following conditions: Kinetex C18 column (100 mm × 2.1 mm, 1.7-μm particle diameter; Phenomenex); flow rate, 0.4 ml/min; elution solvent, 90% methanol with 0.1% formic acid.
Mass spectrometry (MS) conditions were nebulizer current, 3 mA; curtain gas, 10 psi; ion source gas, 20 psi; collision gas, high; entrance potential, 10 V; collision energy, 25 V; declustering potential, 80 V; temperature, 300°C. For MS analysis, selective reaction monitoring (SRM) was used to monitor the dehydration process of the ion [M+H]+ or [M+H-H2O]+ as described previously (45–47). Cholesterol was quantified relative to the d7-cholesterol internal standard.
PTAD derivatization and UHPLC-MS/MS analyses of 7-dehydrocholesterol, lanosterol, and desmosterol in kidney chip effluents.
Analysis of 7-dehydrocholesterol (7-DHC), desmosterol, and lanosterol was performed by UHPLC-MS/MS using a triple quadrupole mass spectrometer (API 4000; AB SCIEX) equipped with electrospray ionization. The analytes were derivatized using the PTAD derivatization method (48). Then 90 μl of each sample was dried under a stream of argon and reconstituted in 25 μl methanol, and 5 μl of a 10 mg/ml PTAD stock solution in methylene chloride was added to the sample solution in methanol. The reaction was allowed to proceed for 10 minutes at ambient temperature before analysis. Reverse-phase chromatography was performed with the following conditions: C18 column (Kinetex, 100 mm × 2.1 mm, 1.7 μm particle diameter; Phenomenex); flow rate, 0.3 ml/min; elution solvent, methonal with 0.1% formic acid.
MS conditions: spray voltage, 4,500 V; curtain gas, 10 psi; ion source gas, 20 psi; collision gas, high; entrance potential, 10 V; collision energy, 23 V (for 7-DHC–PTAD adduct, 27 V); declustering potential, 94 V; temperature, 300°C. The PTAD adducts of 7-DHC, desmosterol, and lanosterol were analyzed by SRM. For 7-DHC and d7-7-DHC, the loss of the PTAD derivative from the sodium adduct produced the ion [M+Na-PTAD]+, which was monitored (7-DHC/d7-7-DHC, m/z 582.6/589.6 → 407.6/414.5). The predominant precursor ions for the PTAD derivatives of desmosterol and lanosterol were solvent addition products in which methanol was added covalently (5). In these cases, the loss of methonal from the PTAD derivative resulted in the ion [M+H-MeOH]+, which was monitored (12C/13C3-desmosterol, M/Z 592.6/595.6 → 560.5/563.6; 12C/13C3-lanosterol, M/Z 634.4/637.4 → 602.5/605.3).
Surface plasmon resonance.
SPR experiments were performed on a Biacore T200 optical biosensor instrument at 25°C, using a Series S Sensor Chip CM4, N-hydroxysuccinimide, N-ethyl-N′-(3-diethylaminopropyl) carbodiimide, and ethanolamine (GE Healthcare Life Sciences) for amine coupling and blocking reactions. POPC nanodiscs were immobilized onto the surface of a flow cell to reach density levels between 5,500 and 6,000 RU, using 200- to 300-nM nanodiscs in 10 μM sodium acetate pH 5.0, with a running buffer containing 10 μM HEPES and 150 μM NaCl (pH 7.4). Free MSP1D1 proteins were immobilized onto the reference surface, using 1 μM in 10 mM sodium acetate pH 5.5 to reach levels between 1,800 to 2,000 RU, taking into account the mass difference between a nanodisc and 2 MSP1D1s.
Direct binding measurements were performed by injecting analytes at a flow rate of 50 μl/min, in a PBS running buffer containing 50 μM sodium phosphate and 350 μM NaCl (pH 7.2). Association and dissociation phases of the analytes were monitored for 60 seconds and 180 to 600 seconds, respectively.
Raw sensorgrams were processed by Biacore T200 evaluation software to include double-referencing (subtracting data from reference cell and from blank injections) and fitting to a steady-state binding model. Fitting of the steady-state response during association was also performed using Igor Pro 6, which uses the Levenberg-Marquardt nonlinear least-squares fitting algorithm to obtain KD values.
Statistics.
Data are reported as mean ± SD. For comparison of means, statistical tests (Student’s t tests, 1-way and 2-way ANOVA with multiple comparisons test) were applied using GraphPad software with P value less than 0.05 as the statistical significance threshold.
Statistical analysis for global transcriptomics was carried out using R (version 3.4.0). Before fitting any models, we first excluded any genes that were expressed at consistently low levels across all samples. Because not all donors received all the treatments, we used linear mixed models, with the treatment as the fixed effect and the donor as the random effect, where the intradonor correlation was incorporated into the covariance (49). To use linear mixed models, we first converted the gene counts to log counts/million using the trimmed mean of M values normalization (50) and then estimated both observation-level and sample-specific weights using the voomWithQualityWeights function from the Bioconductor limma package (51–53). The observation-level weights allow us to use a linear mixed model (by accounting for the dependence between mean and variance), and the sample-specific weights enable us to weight individual samples up or down. We selected genes based on a 1.25-fold or greater difference in expression and a false discovery rate of 5%, meaning we estimate that at most 5% of the significant genes are false positives. Rather than using a post hoc fold change filtering criterion, we used the treat function from limma, which incorporates the fold change into the statistic, meaning that instead of testing for genes that have fold changes different from 0, we test whether the fold change is greater than 1.25-fold in absolute value (49).
Author contributions
EJW, EJK, and J. Himmelfarb conceived and supervised the overall project. MV and TV conceived the inclusion of novel polymyxin derivatives. EJW, KAL, LW, TKB, JWM, MJL, MR, WMA, CT, KMH, J. Herron, LX, MBM, SR, MV, TV, EJK, and J. Himmelfarb designed experiments and analyzed data. EJW, KAL, MJL, MR, CT, KMH, J. Herron, MBM, and SR performed experiments. EJW, KAL, LW, TKB, JWM, WMA, CT, KMH, J. Herron, LX, MBM, SR, J. Himmelfarb, VV, and EJK wrote and edited the manuscript.
Supplementary Material
Acknowledgments
The authors would like to acknowledge and thank Kiera Sullivan, John Ruzinski, Chris Frazer, and Sengkeo Srinouanprachanh for their assistance in sample analysis and handling. We would like to thank Northern Antibiotics and Spero Therapeutics for supplying the polymyxin structural analogs. We acknowledge help from the Biomarker Core Facility of the National Institute of Environmental Health Sciences Center for Exposures, Diseases, Genes and Environment (P30ES07033-EJK to LX and J. Himmelfarb and the University of Washington School of Pharmacy Mass Spectrometry Facility. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the NIH under award numbers 5UH3TR000504 and UG3TR002158 (to J. Himmelfarb). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional support was provided by R01HD092659 (to LX), Sao Paulo Research Foundation (FAPESP 2016/04935-2; to MBM), and Assistance Agreement 83573801 awarded by the US Environmental Protection Agency (EPA; to EJK). The content has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and do not necessarily reflect those of the agency. The EPA does not endorse any products or commercial services mentioned in this publication. This work was also supported by an unrestricted gift from the Northwest Kidney Centers to the Kidney Research Institute.
Version 1. 12/20/2018
Electronic publication
Footnotes
Conflict of interest: MV and TV are employees of Northern Antibiotics Ltd, Espoo, Finland. SR is an employee of AstraZeneca, and VV is an employee of Pfizer.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(24):e123673. https://doi.org/10.1172/jci.insight.123673.
Contributor Information
Lu Wang, Email: lwang3@uw.edu.
Michelle Redhair, Email: mredhair@uw.edu.
William M. Atkins, Email: winky@uw.edu.
Kelly M. Hines, Email: kmhines5@u.washington.edu.
Josi Herron, Email: jh0618@uw.edu.
Libin Xu, Email: libinxu@uw.edu.
Susanne Ramm, Email: Susanne_Ramm@hms.harvard.edu.
Vishal Vaidya, Email: vvaidya@partners.org.
Martti Vaara, Email: martti.vaara@northernantibiotics.com.
Timo Vaara, Email: timo.vaara@northernantibiotics.com.
Jonathan Himmelfarb, Email: himmej@u.washington.edu.
Edward J. Kelly, Email: edkelly@uw.edu.
References
- 1.Chakradhar S. What’s old is new: reconfiguring known antibiotics to fight drug resistance. Nat Med. 2016;22(11):1197–1199. doi: 10.1038/nm1116-1197. [DOI] [PubMed] [Google Scholar]
- 2.Bergen PJ, et al. Polymyxin combinations: pharmacokinetics and pharmacodynamics for rationale use. Pharmacotherapy. 2015;35(1):34–42. doi: 10.1002/phar.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storm DR, Rosenthal KS, Swanson PE. Polymyxin and related peptide antibiotics. Annu Rev Biochem. 1977;46:723–763. doi: 10.1146/annurev.bi.46.070177.003451. [DOI] [PubMed] [Google Scholar]
- 4.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol. 2013;8(6):711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausell A, Garcia-Subirats M, Pujol M, Busquets MA, Rabanal F, Cajal Y. Gram-negative outer and inner membrane models: insertion of cyclic cationic lipopeptides. J Phys Chem B. 2007;111(3):551–563. doi: 10.1021/jp064757+. [DOI] [PubMed] [Google Scholar]
- 6.Falagas ME, Kasiakou SK. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit Care. 2006;10(1):R27. doi: 10.1186/cc3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubin CJ, Ellman TM, Phadke V, Haynes LJ, Calfee DP, Yin MT. Incidence and predictors of acute kidney injury associated with intravenous polymyxin B therapy. J Infect. 2012;65(1):80–87. doi: 10.1016/j.jinf.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Abdelraouf K, Braggs KH, Yin T, Truong LD, Hu M, Tam VH. Characterization of polymyxin B-induced nephrotoxicity: implications for dosing regimen design. Antimicrob Agents Chemother. 2012;56(9):4625–4629. doi: 10.1128/AAC.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun B, et al. Imaging the distribution of polymyxins in the kidney. J Antimicrob Chemother. 2015;70(3):827–829. doi: 10.1093/jac/dku441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandri AM, et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis. 2013;57(4):524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, et al. Human oligopeptide transporter 2 (PEPT2) mediates cellular uptake of polymyxins. J Antimicrob Chemother. 2016;71(2):403–412. doi: 10.1093/jac/dkv340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manchandani P, Zhou J, Babic JT, Ledesma KR, Truong LD, Tam VH. Role of renal drug exposure in polymyxin B-Induced nephrotoxicity. Antimicrob Agents Chemother. 2017;61(4):e02391-16. doi: 10.1128/AAC.02391-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali FE, et al. Pharmacokinetics of novel antimicrobial cationic peptides NAB 7061 and NAB 739 in rats following intravenous administration. J Antimicrob Chemother. 2009;64(5):1067–1070. doi: 10.1093/jac/dkp331. [DOI] [PubMed] [Google Scholar]
- 14.Mingeot-Leclercq MP, Tulkens PM, Denamur S, Vaara T, Vaara M. Novel polymyxin derivatives are less cytotoxic than polymyxin B to renal proximal tubular cells. Peptides. 2012;35(2):248–252. doi: 10.1016/j.peptides.2012.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Vaara M, et al. Novel polymyxin derivatives carrying only three positive charges are effective antibacterial agents. Antimicrob Agents Chemother. 2008;52(9):3229–3236. doi: 10.1128/AAC.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaara M, Sader HS, Rhomberg PR, Jones RN, Vaara T. Antimicrobial activity of the novel polymyxin derivative NAB739 tested against Gram-negative pathogens. J Antimicrob Chemother. 2013;68(3):636–639. doi: 10.1093/jac/dks438. [DOI] [PubMed] [Google Scholar]
- 17.Vaara M, Vaara T. The novel polymyxin derivative NAB739 is remarkably less cytotoxic than polymyxin B and colistin to human kidney proximal tubular cells. Int J Antimicrob Agents. 2013;41(3):292–293. doi: 10.1016/j.ijantimicag.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Vaara M. New polymyxin derivatives that display improved efficacy in animal infection models as compared to polymyxin B and colistin. Med Res Rev. 2018;38(5):1661–1673. doi: 10.1002/med.21494. [DOI] [PubMed] [Google Scholar]
- 19.Vaara M, et al. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob Agents Chemother. 2010;54(8):3341–3346. doi: 10.1128/AAC.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaara M, Vaara T, Vingsbo Lundberg C. Polymyxin derivatives NAB739 and NAB815 are more effective than polymyxin B in murine Escherichia coli pyelonephritis. J Antimicrob Chemother. 2018;73(2):452–455. doi: 10.1093/jac/dkx394. [DOI] [PubMed] [Google Scholar]
- 21.Azad MA, et al. Significant accumulation of polymyxin in single renal tubular cells: a medicinal chemistry and triple correlative microscopy approach. Anal Chem. 2015;87(3):1590–1595. doi: 10.1021/ac504516k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azad MA, et al. Polymyxin B Induces Apoptosis in Kidney Proximal Tubular Cells. Antimicrob Agents Chemother. 2013;57(9):4329–4335. doi: 10.1128/AAC.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adler M, et al. A quantitative approach to screen for nephrotoxic compounds in vitro. J Am Soc Nephrol. 2016;27(4):1015–1028. doi: 10.1681/ASN.2015010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber EJ, et al. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int. 2016;90(3):627–637. doi: 10.1016/j.kint.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkinson SE, Chung GW, van Loon E, Bakar NS, Dalzell AM, Brown CD. The limitations of renal epithelial cell line HK-2 as a model of drug transporter expression and function in the proximal tubule. Pflugers Arch. 2012;464(6):601–611. doi: 10.1007/s00424-012-1163-2. [DOI] [PubMed] [Google Scholar]
- 26.Broadhead CL, Walker D, Skinner R, Simmons NL. Differential cytotoxicity of Ifosfamide and its metabolites in renal epithelial cell cultures. Toxicol In Vitro. 1998;12(3):209–217. doi: 10.1016/S0887-2333(97)00113-6. [DOI] [PubMed] [Google Scholar]
- 27.Bailly V, Zhang Z, Meier W, Cate R, Sanicola M, Bonventre JV. Shedding of kidney injury molecule-1, a putative adhesion protein involved in renal regeneration. J Biol Chem. 2002;277(42):39739–39748. doi: 10.1074/jbc.M200562200. [DOI] [PubMed] [Google Scholar]
- 28.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62(1):237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- 29.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118(5):1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, et al. KIM-1-mediated phagocytosis reduces acute injury to the kidney. J Clin Invest. 2015;125(4):1620–1636. doi: 10.1172/JCI75417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vattimo Mde F, Watanabe M, da Fonseca CD, Neiva LB, Pessoa EA, Borges FT. Polymyxin B nephrotoxicity: from organ to cell damage. PLoS One. 2016;11(8):e0161057. doi: 10.1371/journal.pone.0161057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavkovic M, et al. Detection of drug-induced acute kidney injury in humans using urinary KIM-1, miR-21, -200c, and -423. Toxicol Sci. 2016;152(1):205–213. doi: 10.1093/toxsci/kfw077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavkovic M, Vaidya VS. MicroRNAs and drug-induced kidney injury. Pharmacol Ther. 2016;163:48–57. doi: 10.1016/j.pharmthera.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khondker A, Alsop RJ, Dhaliwal A, Saem S, Moran-Mirabal JM, Rheinstädter MC. Membrane cholesterol reduces polymyxin B nephrotoxicity in renal membrane analogs. Biophys J. 2017;113(9):2016–2028. doi: 10.1016/j.bpj.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson AC, Ware LB, Himmelfarb J, Zager RA. HMG-CoA reductase activation and urinary pellet cholesterol elevations in acute kidney injury. Clin J Am Soc Nephrol. 2011;6(9):2108–2113. doi: 10.2215/CJN.02440311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelraouf K, Chang KT, Yin T, Hu M, Tam VH. Uptake of polymyxin B into renal cells. Antimicrob Agents Chemother. 2014;58(7):4200–4202. doi: 10.1128/AAC.02557-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moestrup SK, et al. Evidence that epithelial glycoprotein 330/megalin mediates uptake of polybasic drugs. J Clin Invest. 1995;96(3):1404–1413. doi: 10.1172/JCI118176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babic JT, Manchandani P, Ledesma KR, Tam VH. Evaluation of Urinary KIM-1 for Prediction of Polymyxin B-Induced Nephrotoxicity. Antimicrob Agents Chemother. 2017;61(11):e01735-17. doi: 10.1128/AAC.01735-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keirstead ND, et al. Early prediction of polymyxin-induced nephrotoxicity with next-generation urinary kidney injury biomarkers. Toxicol Sci. 2014;137(2):278–291. doi: 10.1093/toxsci/kft247. [DOI] [PubMed] [Google Scholar]
- 40. Spero Therapeutics. IV antibiotics potentiator platform for treating MDR gram-negative infections in the hospital. Spero website. https://sperotherapeutics.com/pipeline/spr741-spr206-iv-potentiator-platform/ Accessed November 11, 2018.
- 41.Chang SY, et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight. 2017;2(22):e95978. doi: 10.1172/jci.insight.95978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21(12):745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maschmeyer I, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15(12):2688–2699. doi: 10.1039/C5LC00392J. [DOI] [PubMed] [Google Scholar]
- 44.Korade Z, et al. The effect of small molecules on sterol homeostasis: measuring 7-dehydrocholesterol in Dhcr7-deficient neuro2a cells and human fibroblasts. J Med Chem. 2016;59(3):1102–1115. doi: 10.1021/acs.jmedchem.5b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L, Korade Z, Rosado DA, Liu W, Lamberson CR, Porter NA. An oxysterol biomarker for 7-dehydrocholesterol oxidation in cell/mouse models for Smith-Lemli-Opitz syndrome. J Lipid Res. 2011;52(6):1222–1233. doi: 10.1194/jlr.M014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L, Liu W, Sheflin LG, Fliesler SJ, Porter NA. Novel oxysterols observed in tissues and fluids of AY9944-treated rats: a model for Smith-Lemli-Opitz syndrome. J Lipid Res. 2011;52(10):1810–1820. doi: 10.1194/jlr.M018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu L, Sheflin LG, Porter NA, Fliesler SJ. 7-Dehydrocholesterol-derived oxysterols and retinal degeneration in a rat model of Smith-Lemli-Opitz syndrome. Biochim Biophys Acta. 2012;1821(6):877–883. doi: 10.1016/j.bbalip.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W, Xu L, Lamberson C, Haas D, Korade Z, Porter NA. A highly sensitive method for analysis of 7-dehydrocholesterol for the study of Smith-Lemli-Opitz syndrome. J Lipid Res. 2014;55(2):329–337. doi: 10.1194/jlr.D043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 50.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu R, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43(15):e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7:261. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.