Abstract
Mouse embryonic stem cells (ESCs) proliferate with rapid cell cycle kinetics but without loss of pluripotency. The histone methyltransferase Dot1L is responsible for methylation of histone H3 at lysine 79 (H3K79me). We investigated whether ESCs require Dot1L for proper stem cell behavior. ESCs deficient for Dot1L tolerate a nearly complete loss of H3K79 methylation without a substantial impact on proliferation or morphology. However, shortly after differentiation is induced, Dot1L-deficient cells cease proliferating and arrest in G2/M phase of the cell cycle, with increased levels of aneuploidy. In addition, many aberrant mitotic spindles occur in Dot1L-deficient cells. Surprisingly, these mitotic and cell cycle defects fail to trigger apoptosis, indicating that mouse ESCs lack stringent cell cycle checkpoint control during initial stages of differentiation. Transcriptome analysis indicates that Dot1L deficiency causes the mis-regulation of a select set of genes, including many with known roles in cell cycle control and cellular proliferation as well as markers of endoderm differentiation. The data indicate a requirement for Dot1L function for early stages of ESC differentiation where Dot1L is necessary for faithful execution of mitosis and proper transcription of many genes throughout the genome.
Introduction
The chromatin of ESCs has been studied intensively since large sets of genes must be coordinately-regulated as ESCs differentiate or contribute to embryos [1]. For instance, bivalent domains comprised of overlapping regions of H3K27me and H3K4me mark key developmentally-regulated genes that are poised for later expression [2]. Another example is provided by the H3K9-specific histone methyltransferase G9a, which participates in the developmentally-regulated repression of Oct4 during early embryogenesis [3]. However, chromatin also is crucial for other cellular functions such as chromosome dynamics, silencing of heterochromatic regions, and DNA damage responses. These other features of chromatin have been less extensively studied in ESCs.
The highly conserved histone methyltransferase Dot1L methylates lysine 79 within the globular domain of histone H3 (H3K79me) despite lack of a SET domain [4]. Dot1L, and its homologs, have been implicated in a surprisingly diverse spectrum of chromatin functions. In both yeast and mice, Dot1L has been associated with RNA polymerase II during transcriptional elongation [5, 6]. In mixed lineage leukemia, an AF10/MLL fusion protein recruits DOT1L to the Hoxa9 gene, whose up-regulation may influence cancer progression [7]. Similarly, DOT1L is recruited to Hoxa5 by a translocation-induced CALM/AF10 fusion protein in acute T-cell lymphoblastic leukemia [8]. Dot1L also functions in the maintenance of telomeric heterochromatin in yeast and mouse ESCs [9, 10]. In yeast, H3K79me2 marks double-stranded DNA breaks and recruits the checkpoint protein 53BP1 to sites of DNA damage [11, 12]. Recombinationally-active chromatin of immunoglobulin genes are enriched in H3K79 methylation [13]. Other studies implicate Dot1L in chromosome stability and faithful progression through the cell cycle. For example, in Trypanosoma brucei, depletion of Dot1 is lethal, and leads to formation of haploid cells and cells devoid of DNA via a mechanism that involves cytokinesis without prior DNA replication [14]. H3K79me2 levels transiently increase at M-phase of the cell cycle, again suggesting a role for Dot1L in the cell cycle [4, 15]. In preimplantation mouse embryos, H3K79me3 localizes to pericentric heterochromatin, suggesting a role for H3K79me3 in centromere function [15].
Mouse embryos homozygous for a gene-trap allele of Dot1L exhibit prenatal lethality [16]. In addition, embryos homozygous for a targeted mutation in Dot1L exhibit a mid-gestation developmental arrest at E10.5 with several developmental phenotypes including abnormal angiogenesis, yolk sac abnormalities, and cardiac malformation [9]. The importance of chromatin for chromosome stability and cell cycle progression has been relatively understudied in ESCs. Since Dot1L is suspected to be involved in aspects of transcription as well as chromosomal and cell cycle function across several species and cell-types, we performed a comprehensive investigation of the importance of Dot1L for murine ESCs and their differentiated progenitors.
Materials and Methods
Chromatin Immunoprecipitation and QPCR
All chromatin immunoprecipitation (ChIP) experiments were performed in triplicate for J1 ESCs and MEFs essentially as described by Upstate (Cat # 17–295). For detailed protocol see supp. file S1. Primer pairs available upon request.
RNA Interference
shRNAs against mouse Dot1L mRNA, TRC-Mm1.0 shRNA plasmids were purchased from Open Biosystems. Plasmids encoding shRNAs against mouse Dot1L mRNA were as follows: TRCN0000125102, TRCN0000125103, TRCN0000125099, TRCN0000125100, TRCN0000125101, and pLKO.1 (empty vector). Plasmids were linearized and electroporated into J1 ESCs, selected in puromycin, and expanded as previously described [17].
ES Cell Culture, Differentiation and Proliferation Assays
J1 ESCs harboring shRNAs were grown in standard ESC media on DR4 feeders in the presence of puromycin. RA differentiation of shDot1L ESCs was performed as previously described [18]. Standardized EBs were prepared as described [19].
Cell Cycle Analysis
Cell cycle analysis was performed by flow cytometry on PI-stained cells on a Becton-Dickinson FACS Calibur flow cytometer. Briefly, cells were fixed in absolute ethanol and stained with 40μg/ml PI and 200ug/mL of DNase-free RNase A for 1 hour. Cells were then vortexed and filtered through a 30μM nylon mesh, and subjected to flow cytometry.
Apoptosis Assays
Assays used APC-conjugated Annexin-V (Invitrogen) and PI using flow cytometry according to the manufacturers instructions. Empty vector ESCs were treated with 12 μM camptothecin (Sigma-Aldrich) for 4 hours as a positive apoptosis control. Flow cytometry was performed using a Becton-Dickinson FACS Calibur Flow Cytometer and Cell Quest software.
Karyotype Analysis
Chromosome spreads were prepared as previously described [17].
Immunofluorescence, Microarray Analysis, and Real Time PCR
See Supp. File S1.
Results
A Chromatin Signature that Marks the Pluripotency Genes Oct4 and Sox2 in ES Cells is Reversed in Differentiated Somatic Cells
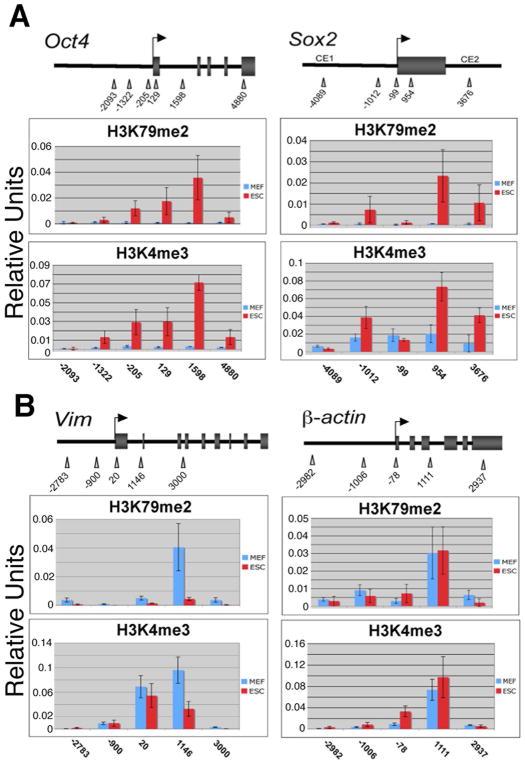
Histone H3 that is dimethylated at lysine 79 (H3K79me2) associates with active genes in several cell-types [20, 21], but has not been well studied in mouse ESCs. H3K4me3 also marks many actively transcribed genes [22–24]. To investigate the relative distribution of H3K79me2 and H3K4me3 upon the key pluripotency genes Pou5f1 (Oct4) and Sox2 in ESCs, we performed chromatin immunoprecipitation (ChIP) and assayed the resulting ChIP DNA with quantitative real-time PCR (ChIP-QPCR). To determine the distribution of both histone modifications across the Oct4 gene, QPCR primer pairs were tiled across a 6 kb region centered upon the Oct4 transcriptional start site (TSS). An interesting and unique pattern of H3K79me2 was uncovered in ESCs and differentiated cells. H3K79me2 levels were negligible 2 kb upstream and 4.8 kb downstream from the Oct4 TSS. However, high levels of H3K79me2 were found near the TSS, and peaked 1.5 kb downstream (Fig. 1A). We found a similar pattern for H3K4me3 upon Oct4 in ESCs. We also examined the distribution of H3K79me2 and H3K4me3 across Oct4 in mouse embryonic fibroblasts (MEFs), a differentiated somatic cell-type in which Oct4 is transcriptionally silent. We found essentially undetectable levels of H3K79me2 and H3K4me3 across Oct4 in MEFs. This pattern of H3K79me2 and H3K4me3 was also highly similar across the Sox2 gene, where peak enrichment occurred roughly 1kb downstream of the transcriptional start site in ESCs, but not MEFs (Fig. 1A). This location of peak enrichment downstream of the TSS is consistent with previous studies on active chromatin [6, 25].
Figure 1.
Distribution of H3K79me2 and H3K4me3 across transcribed genes in ESCs and MEFs. ChIP samples were collected from ESCs and MEFs using antibodies specific for H3K79me2 and H3K4me3 then assayed by quantitative real-time PCR using primer pairs tiled across the Oct4 and Sox2 promoters and flanking regions. (A) Oct4 chromatin is enriched for H3K79me2 and H3K4me3 ~1.5kb downstream of the TSS in ESCs (red bars), with virtually no enrichment in MEFs (blue bars). Sox2 chromatin was assayed by ChIP-QPCR for the same histone modifications. (B) H3K79me2 and H3K4me3 distribution across the promoter and flanking regions of the Vimentin gene (which is highly expressed in MEFs as compared to ESCs) in MEFs and ESCs. H3K79me2 and H3K4me3 distribution across the promoter and flanking regions of the β-actin housekeeping gene, which is constitutively expressed in all cell-types. Data points represent the mean of 9 PCR reactions, +/− the standard deviation.
In addition, we analyzed the chromatin of Vimentin, a gene expressed in differentiated cells. Peak levels of H3K79me2 and H3K4me3 enrichment were found downstream of the TSS in MEFs, but not ESCs, a pattern that closely resembled that of Oct4 and Sox2 (Fig. 1B). As a control for the specificity of our ChIP assay in detecting methylated histones across active genes, β-actin chromatin was analyzed since this gene is ubiquitously expressed in both ESCs and differentiated cells. Enrichment of both H3K79me2 and H3K4me3 was found downstream of the β-actin TSS in both MEFs and ESCs (Fig. 1B). These data demonstrate that H3K79me2 enrichment is highly similar to H3K4me3, a hallmark of transcriptionally active chromatin.
Dot1L-Deficient ESCs Proliferate Readily in an Undifferentiated State
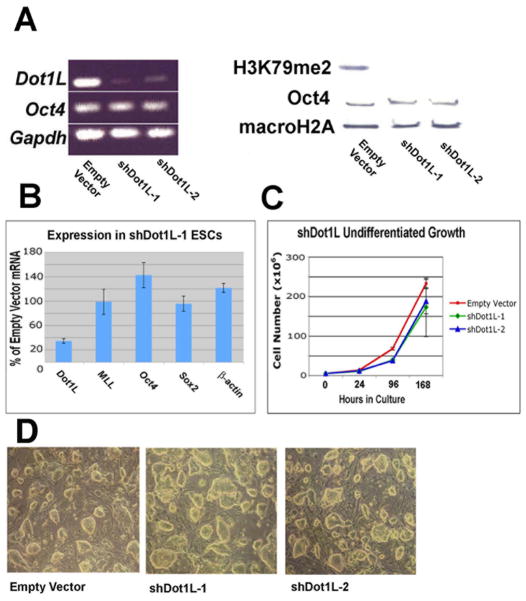
Based on the ChIP results for H3K79me2 distribution across Oct4 and Sox2, we hypothesized that loss of H3K79me2 may alter the transcriptional activity of these genes since histone modifications have been shown to affect the expression of Oct4 [3]. To better understand the consequences of Dot1L-deficiency in ESCs and its role in regulating pluripotency gene expression, we employed a short hairpin RNA (shRNA) interference strategy to disrupt Dot1L expression in ESCs. Four independent puromycin-selectable plasmids encoding U6 promoter-driven shRNAs directed against Dot1L were stably integrated into the genome of mouse ESCs. After 10 days of selection, colonies were picked and expanded in standard ESC medium containing leukemia inhibitory factor (LIF). Two independent cell lines (shDot1L-1 and shDot1L-2) exhibited substantial knockdown as compared to a cell line with an integrated empty vector (Fig. 2A). Global levels of H3K79me2 were drastically lower in Dot1L knockdown ESCs (Figure 2A). Surprisingly, levels of Oct4 mRNA and protein were essentially unchanged. MacroH2A was used as a loading control since it is a histone that is abundantly expressed in ESCs. The results indicate that Dot1L is solely responsible for dimethylation of H3K79. However, reduction of H3K79me2 to undetectable levels failed to greatly perturb Oct4 mRNA and protein as well as Sox2 mRNA levels (Fig. 2A and B). Oct4 and β-actin mRNA increased with decreased Dot1L knockdown, albeit not significantly. The expression of MLL, which is also involved in methylating histones, was unaffected (Fig. 2B). We also tested whether knockdown of Dot1L affected the proliferation of ESCs. To do this, triplicate cultures of shDot1L-1 and shDot1L-2 were grown under standard ESC culture conditions (in the presence of LIF and puromycin). After 3 passages and 168 total hours in culture, shDot1L-1 and shDot1L-2 ESCs displayed only a modest reduction in proliferation as compared to empty vector control ESCs (Fig. 2C). Also, the colony morphology of Dot1L-deficient ESCs was essentially normal (Fig. 2D).
Figure 2.
Analysis of Dot1L deficiency in undifferentiated ESCs harboring stably-integrated shRNA vectors. (A) RT-PCR of RNA from ESC lines shDot1L-1 and shDot1L-2 exhibited robust knockdown of Dot1L, but not Oct4 or Gapdh mRNAs. Also, Western blot analysis shows greatly diminished H3K79 methylation in ESC lines shDot1L-1 and shDot1L-2 without substantial alteration of Oct4 protein levels. The histone macroH2A was used as a loading control. (B) Quantitative real-time PCR was used to assay the expression of several genes in Dot1L-deficient ESCs. Dot1L mRNA is decreased in Dot1L knockdown ESCs compared to the empty vector control. MLL, a gene which encodes a histone methyltransferase specific for H3K4, is unaffected, as well as mRNAs expressed from Oct4, Sox2, or β-actin, which were not downregulated. Data is shown as the mean of 6 PCR reactions +/− the standard deviation. (C) Growth curve analysis of two Dot1L-deficient ESC lines as compared to a wild-type ESC control line harboring an integrated empty vector. The plotted data is the mean of 3 cell culture counts; error bars indicate standard deviation. (D) Cellular morphology of shDot1L ESCs compared to empty vector ESCs.
Dot1L-Deficient ESCs Exhibit Severe Proliferation Defects After Induced Differentiation
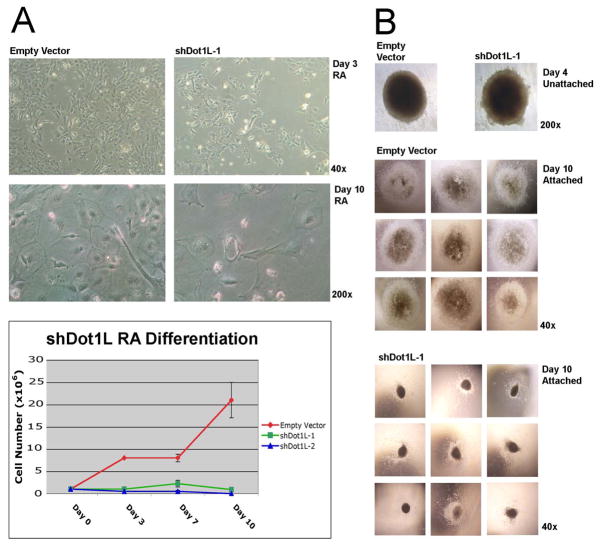
Two reports indicate that Dot1L is essential for murine embryogenesis [9, 16], yet Dot1L-deficient ESCs are viable and key pluripotency genes are not downregulated with loss of Dot1L. We therefore investigated the ability of shDot1L ESCs to undergo induced differentiation in vitro. To do this we subjected triplicate cultures of shDot1L and empty vector only ESCs (plated at low density without feeder cells or LIF) to all-trans retinoic acid (RA) [26, 27]. The empty vector control ESCs proliferated during the 10-day course of differentiation, but shDot1L cells failed to proliferate (Fig. 3A). We also subjected Dot1L-deficient cells and controls to an embryoid body (EB) differentiation procedure that utilizes a standardized number of cells to initiate EB formation [19]. EBs were formed by aggregating 2×104 ESCs in a conical 1.5 ml polypropylene tube under non-adherent conditions. After 4 days of non-adherent growth, the shDot1L-1 EBs were the same diameter as empty vector EBs, but the borders of shDot1L EBs were ragged (Fig. 3B). After adhering the EBs to gelatinized tissue culture wells, the shDot1L EBs failed to grow substantially as compared to empty vector control EBs, which became several times larger than Dot1L knockdown EBs after 10 days (Fig. 3B). The empty vector controls also developed areas that contained beating/contractile tissue, whereas the shDot1L EBs did not (data not shown).
Figure 3.
Differentiation of ESCs deficient for Dot1L. (A) Empty vector control and shDot1L (Dot1L-deficient) ESCs were subjected to RA-induced differentiation in the absence of LIF and feeder cells. Equal numbers of both cell lines were plated prior to RA-induced differentiation. Representative images after 3 and 10 days of RA-induced differentiation. RA differentiation experiments were performed in triplicate for two stable knockdown ESC lines (shDot1L-1 and shDot1L-2) and empty vector ESCs, and cells were counted over the course of 10 days to yield growth curves. (B) shDot1L-1 and empty vector ESCs subjected to a protocol designed to yield EBs of standardized size. Each EB was formed by aggregating 2×104 ESCs and imaged after 4 days in suspension culture, and again after attachment and 10 additional days in culture.
shDot1L ESCs exhibited severe differentiation defects in vitro, however we also assessed their ability to differentiate into all 3 germ layers in vivo using teratoma assays. The flanks of NOD/SCID mice were injected subcutaneously with approximately 2 million empty vector and shDot1L ESCs and teratomas were allowed to grow for 3–4 weeks. Both empty vector and shDot1L ESCs formed teratomas after this time period and differentiated into cell types of all 3 germ layers (Fig. S1A–D), however shDot1L teratomas appeared to have more structures with a glandular/epithelial appearance than empty vector control teratomas. We made sure that the shDot1L ESCs used to make teratomas exhibited robust Dot1L knockdown immediately before injection into NOD-SCID mice (Fig. S1E). However, we detected low-level expression of Dot1L in teratomas of both shDot1L and empty vector origin (Fig S1F), possibly due to silencing of the Dot1L-directed shRNA during the course of teratoma development. We also found that teratomas formed from shDot1L ESCs exhibited higher levels of residual Oct4 expression (Fig. S1F).
Dot1L is Critical for Normal Progression Through the Cell Cycle and Genome Stability
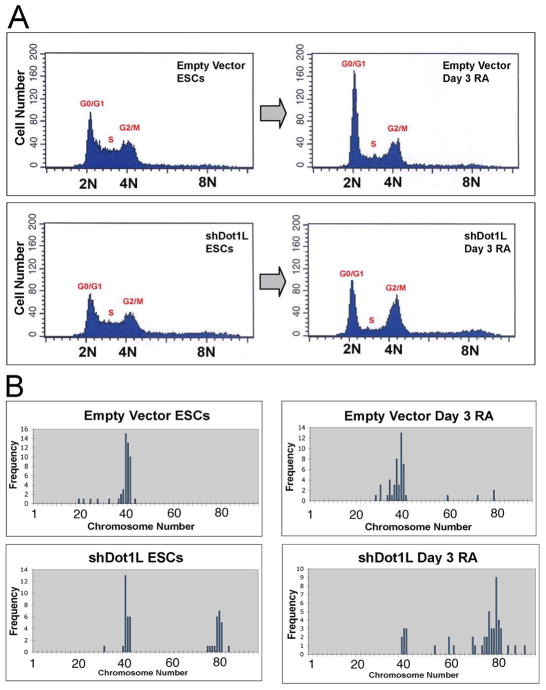
Previous studies suggest that Dot1L and H3K79me2 are regulated during progression of the cell cycle in a variety of eukaryotic cells including yeast, trypanosomes, and mammalian somatic cells [4, 11, 14, 15]. Therefore, we assessed Dot1L-deficient ESCs and their differentiated derivatives for alterations in cell cycle. In undifferentiated empty vector and shDot1L ESCs, the percentages of cells in G1, S, and G2/M-phases of the cell cycle were relatively similar, with a high percentage of cells in S-phase, as is normal for rapidly cycling mouse ESCs (Fig. 4A). However, careful inspection of the cytometry results indicated that Dot1L-deficient ESCs have 14% of cells in a state defined by higher than G2 (>4N) content of DNA, compared to just 4% in the empty vector cells (Fig. S2). When shDot1L and empty vector ESCs were differentiated for 3 days in RA, striking differences in the cell cycle profiles of these cells became apparent. The differentiated empty vector cells contained a high percentage of cells in G0/G1, fewer cells in S-phase, and a similar number of cells in G2/M (Fig. 4A). In contrast, when shDot1L cells were differentiated, the percentage of cells in G0/G1 was 16% lower than in the control, the percentage of cells in S-phase was 5% lower, and the percentage in G2/M increased 10% as compared to the control (Fig. 4A, Fig. S2). These numbers were similar on day 6 of differentiation (data not shown). In addition, we observed a substantial increase in the number of hyperploid cells (>4N) in the differentiated shDot1L cells, which made up 17.3% of the population (Fig. S2). These results show that upon differentiation, many Dot1L knockdown cells arrest in G2/M-phase, but an increased proportion of cells acquire a >4N content of DNA. To further investigate the apparent differentiation-induced hyperploidy of Dot1L knockdown cells, we prepared chromosome spreads from all samples and collected karyotype data (Fig. 4B). We found a modal number of 40 chromosomes in empty vector ESCs regardless of their differentiation state, as expected for mouse cells. We observed some spreads that had fewer than 40 chromosomes, even in wildtype ESCs, and these are possibly spreads in which one or more chromosomes were lost during the karyotyping procedure. However, undifferentiated Dot1L knockdown ESCs yielded many spreads with roughly tetraploid karyotypes, and this effect became more pronounced in shDot1L cells differentiated for 3 days in RA (Fig. 4B).
Figure 4.
Cell cycle and karyotype analysis of empty vector and shDot1L cell lines. (A) Undifferentiated empty vector and shDot1L ESCs had a similar proportion of cells in each stage of the cell cycle (left panels). shDot1L ESCs exhibited a relatively normal cell cycle, except for a small but detectable population of hyperploid (>4N) cells. After 3 days of RA differentiation, shDot1L cultures contained increased cells in G2/M-phase, and the number of hyperploid (>4N) cells also increased. (B) Karyotype Analysis. Undifferentiated empty vector ESCs had a modal number of 40 chromosomes, as did empty vector cells after 3 days of RA differentiation. Undifferentiated shDot1L ESCs yielded bimodally-distributed karyotypes (40, diploid; 80, tetraploid) when grown in standard ESC conditions. After 3 days of RA differentiation of shDot1L ESCs, the modal count changed to 80, with many spreads containing 60–90 chromosomes.
We surmised that Dot1L deficient ESCs may contain aberrant mitotic spindles, leading to mitotic arrest and tetraploidy. Therefore we analyzed spindle morphology using immunofluorescence. shDot1L and empty vector ESCs were stained for α-tubulin, histone H3 phosphorylated on serine 10 (H3S10ph), a marker of mitotic cells as well as with CREST antiserum which recognizes kinetochores (Fig. S3). Empty vector ESCs predominantly contained normal mitotic spindles (Fig. S3A). However, Dot1L deficient cells contained many mitotic spindles with aberrant morphology (Fig. S3B, C and D), which may be one cause of these cells becoming tetraploid. In addition, shDot1L ESCs have higher levels of H3S10ph protein than empty vector ESCs, albeit a minor difference, further suggesting that some cells stall or arrest in mitosis with loss of Dot1L (Fig. S3E).
Differentiating Dot1L-Deficient ESCs Fail to Initiate Apoptosis
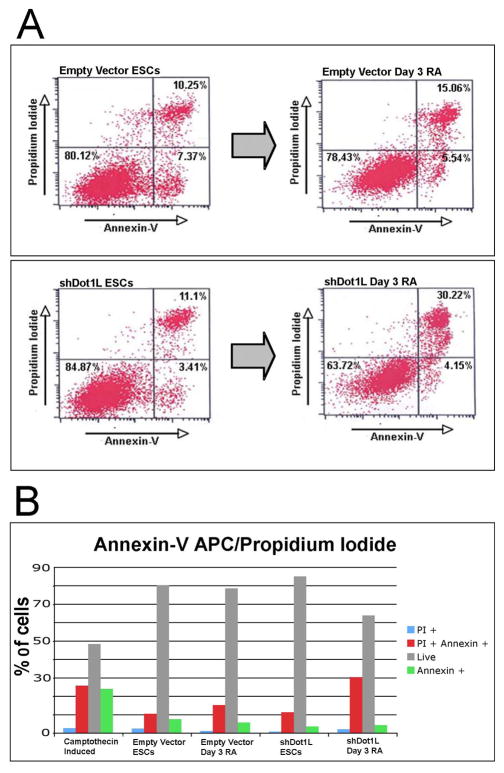
We hypothesized that Dot1L deficiency induces apoptosis in ESCs, leading to a failure of such cells to proliferate. A precedent for such arrest is provided by the example of MLL2-deficient ESCs, which readily undergo apoptosis when induced to differentiate [28]. To investigate if shDot1L cells undergo differentiation-induced apoptosis, we performed apoptosis assays on Dot1L-deficient ESCs subjected to RA-induced differentiation. We used a flow cytometry-based assay to detect early apoptotic cells using fluorescently-labeled Annexin-V, which binds phosphatidylserine on the plasma membrane exterior, which is an early marker of apoptosis [29]. To distinguish dead from early apoptotic cells, propidium iodide (PI) was used as a live counterstain to detect disrupted cells. We validated the assay by exposing empty vector ESCs to camptothecin, a drug known to induce apoptosis. We readily detected apoptosis in camptothecin-treated ESCs (Fig. S4). We found that undifferentiated empty vector and shDot1L ESCs contained 7.37% and 3.41% early apoptotic cells, respectively, and 10.25% and 11.1% disrupted cells (doubly-positive for Annexin-V and PI) (Fig. 5). This assay was repeated with highly similar results, demonstrating no increase in apoptosis with reduced Dot1L, contrary to what others have shown in ESCs after loss of Dot1L [9]. After 3 days of RA differentiation, the percentage of apoptotic cells did not increase significantly, even in Dot1L-deficient ESCs. However, the shDot1L cells (differentiated in RA for 3 days) accumulated a significantly increased number (30%) of Annexin-V/PI doubly-positive cells as compared to the empty vector control (Fig. 5B). We hypothesized that apoptosis might already have occurred in this 30% population, so we assayed apoptosis after only 2 days of differentiation, but found no increase in apoptotic cells (data not shown). We conclude that some differentiating Dot1L-deficient ESCs succumb by non-apoptotic mechanisms such as necrosis, resulting in disrupted cells with DNA accessible to PI. The expression of double-stranded RNA can induce the interferon response pathway, which can cause apoptosis [30, 31]. To this end we assayed empty vector and shDot1L cells for differentiation-induced up-regulation of markers of the interferon response pathway. We saw no induction of interferon-response genes (Fig. S5), a finding that is consistent with the failure to detect increased apoptosis in shRNA-induced Dot1L deficient cells. We conclude that the arrested proliferation of Dot1L-deficient cells after exposure to RA occurs by mechanisms other than apoptosis.
Figure 5.
Annexin-V flow cytometry assays for early apoptotic cells. (A) Undifferentiated and day 3 RA differentiated empty vector and shDot1L cells were stained with Annexin-V (AV) and propidium iodide (PI) and analyzed by flow cytometry. Three classes of cells were observed in each sample: low AV, low PI (live non-apoptotic cells), high AV, low PI (early apoptotic cells), and high AV, high PI (disrupted, presumably dead cells). Undifferentiated empty vector and shDot1L ESCs had similar profiles. (B) Flow cytometry results in graph form for each cytometry quadrant shown.
Transcriptome Responses to Dot1L Deficiency
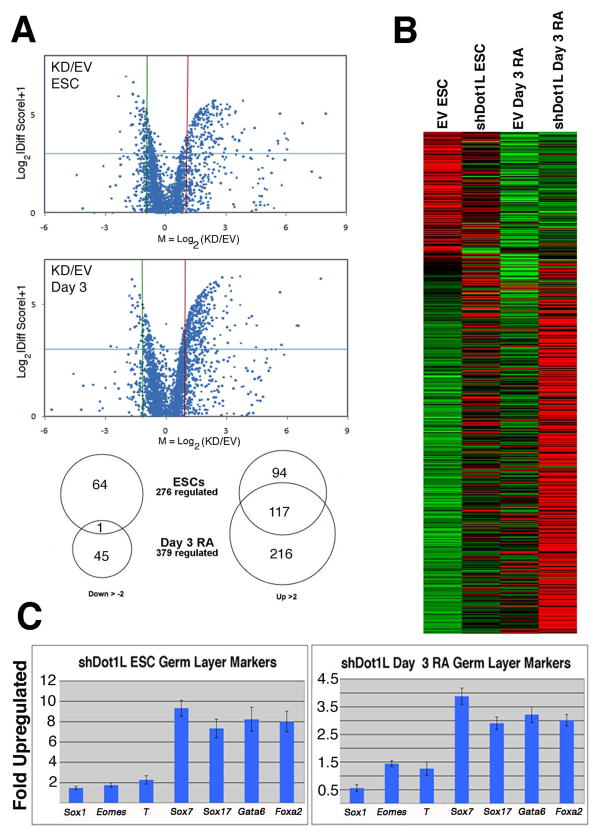
To determine the consequences of Dot1L deficiency for the transcriptome as opposed to limiting our analysis to several select genes, we prepared RNA from triplicate cultures of empty vector and shDot1L knockdown ESCs grown under standard ESC conditions and after 3 days of RA-induced differentiation. In all cases, feeder cells were removed prior to RNA preparation (see materials and methods). We subjected the triplicate RNA samples to microarray analysis using Illumina mouse Whole Genome™ Version 2 bead arrays that contain 45,281 probesets for detection of 31,492 genes. The transcriptomes of Dot1L knockdown and empty vector ESCs were highly similar, but we detected a small set of genes whose expression was perturbed by Dot1L deficiency (Fig. 6, Table S1). We prepared “volcano” plots of the data, which show expression values of genes with greater than two-fold up- or down-regulation in Dot1L knockdown cells as compared to empty vector controls (Fig. 6A). Surprisingly, substantially more genes were up-regulated by loss of Dot1L than down-regulated in both undifferentiated and differentiated cultures. Using this analysis, we found 211 up-regulated and 65 down-regulated genes in undifferentiated samples, and 333 up-regulated and 46 down-regulated genes in cells differentiated in RA for 3 days (Fig. 6A). A substantial proportion of genes were up-regulated by loss of Dot1L in both undifferentiated and differentiated cells, but little overlap occurred between the sets of genes down-regulated in the two cell-types. In total, 535 genes exhibited greater than two-fold differential expression in this analysis. We also prepared a heat map to indicate the degree of mis-regulation of the 535 genes (Fig. 6B).
Figure 6.
Transcriptome analysis of normal and Dot1L-deficient ESCs and cells after 3 days of RA-induced differentiation. (A) Volcano plots of Illumina transcriptome array results. The upper plot shows genes whose expression is affected by disruption of Dot1L in undifferentiated ESCs as compared to wild-type empty vector ESCs. The lower plot shows genes de-regulated by disruption of Dot1L in cells subjected to 3 days of RA-induced differentiation. Each dot indicates the log of the knockdown/wild-type expression ratio. Genes greater than 2-fold up-regulated lie to the right of the vertical red line, and genes greater than 2-fold down-regulated lie to the left of the vertical green line. Genes with statistically-significant responses to Dot1L disruption lie above the horizontal blue line. Also, a Venn diagram of Dot1L-regulated genes was derived from the volcano plot analysis. (B) A heat map that shows the expression (indicated by graded color hues) of the 534 genes identified as regulated by Dot1L. Red is highly expressed and green is lowly expressed. (C) Taqman QPCR validation of upregulated endoderm markers in shDot1L ESCs and RA differentiated cells.
We confirmed the validity of the microarray analysis by subjecting several up- and down-regulated genes to quantitative real time PCR validation analyses (Fig. 6C and S6). Interestingly, many genes thought to be involved in endoderm development were upregulated in shDot1L ESCs and day 3 RA samples compared to the empty vector cells whereas a set of genes from ectoderm (Sox1), trophectoderm (Eomes) and mesoderm (T) were not upregulated in either cell type (Fig. 6C). The results of Q-PCR agreed with the Illumina array results to a high degree. Finally, we subjected the 535 affected genes to gene ontology (GO) analysis using the GOEAST [32] software toolkit for GO enrichment analysis (Table 1, Table S2). This analysis yielded GO gene function classes that were over-represented in response to Dot1L knockdown. Of particular note, we found that a surprisingly high preponderance of Dot1L-regulated genes have known functions in the cell cycle, cellular proliferation, and differentiation.
Table 1.
Over-represented GO classes of Dot1L-regulated genes
| GOID | Term | p-value1 | regulated genes/GOID2 | genes/GOID3 | |
|---|---|---|---|---|---|
| Up-regulated in Dot1L-deficient cells | GO:0006898 | receptor-mediated endocytosis transmembrane receptor tyrosine kinase signaling | 0.000 | 4 | 24 |
| GO:0007169 | pathway | 0.000 | 9 | 217 | |
| GO:0006940 | regulation of smooth muscle contraction | 0.000 | 3 | 16 | |
| GO:0045596 | negative regulation of cell differentiation | 0.001 | 5 | 109 | |
| GO:0019319 | hexose biosynthetic process | 0.002 | 3 | 26 | |
| GO:0042744 | hydrogen peroxide catabolic process | 0.002 | 2 | 8 | |
| GO:0030279 | negative regulation of ossification | 0.002 | 2 | 8 | |
| GO:0008285 | negative regulation of cell proliferation | 0.002 | 6 | 146 | |
| GO:0019229 | regulation of vasoconstriction | 0.004 | 2 | 10 | |
| GO:0040015 | negative regulation of multicellular organism growth | 0.004 | 2 | 10 | |
|
| |||||
| Down-regulated in Dot1L-deficient cells | GO:0007050 | cell cycle arrest | 0.001 | 3 | 67 |
| GO:0030308 | negative regulation of cell growth | 0.006 | 2 | 36 | |
| GO:0007549 | dosage compensation | 0.015 | 1 | 5 | |
| GO:0030307 | positive regulation of cell growth | 0.015 | 1 | 5 | |
| GO:0048103 | somatic stem cell division | 0.015 | 1 | 5 | |
| GO:0045749 | negative regulation of S phase of mitotic cell cycle | 0.015 | 1 | 5 | |
| GO:0006703 | estrogen biosynthetic process | 0.015 | 1 | 5 | |
| GO:0007507 | heart development | 0.016 | 4 | 181 | |
| GO:0042416 | dopamine biosynthetic process | 0.018 | 1 | 6 | |
| GO:0006590 | thyroid hormone generation | 0.018 | 1 | 6 | |
p-values less than 0.050 indicate gene ontology classes that are more than 95% likely to be over-represented.
Number of genes in the indicated GOID class regulated by Dot1L.
Total number of genes in the indicated GOID class present on the microarray.
Discussion
We present in this report that Dot1L depletion leads to pleiotropic consequences in both ESCs and to a greater degree, differentiating cells through shRNA mediated gene knockdown. Even though H3K79me2 is enriched on 2 genes critical in maintaining pluripotency (Oct4 and Sox2) in undifferentiated ESCs, their expression is not decreased by a nearly complete loss of H3K79me2. Alternatively, Oct4 mRNA levels increased approximately 1.5 fold. Dot1L-deficient ESCs exhibit only modest proliferation defects when grown using standard ESC culture conditions designed to maintain pluripotency. This observation is surprising based on a recent finding that Dot1L 1lox/1lox as well as Dot1L 1lox/+ ESCs fail to proliferate as rapidly as wild type ESCs during a shorter time period than presented here (5 days vs. 7 days respectively) [9]. This difference may be due to the vastly different methods for disrupting Dot1L levels. In undifferentiated cell culture, Dot1L-deficient ESCs displayed similar overall proportions of cells in G1, S, and G2/M phases of the cell cycle as compared to wild-type controls. Subtle phenotypes included a small population of aneuploid cells with DNA content >4N, and a tendency of shDot1L cells to produce tetraploid chromosome spreads after treatment with colcimid. These findings are similar to those observed in ESCs derived from blastocysts bearing a homozygous deletion of Dot1L [9]. However, Dot1L-deficient ESCs underwent a profound proliferative failure soon after differentiation was induced. After exposure to RA for only 3 days, Dot1L-deficient cells accumulated in G2/M phase of the cell cycle. The apparent differentiation-induced G2/M arrest was accompanied by increased levels of hyperploidy, with a detectable peak centered on an 8N DNA content. In addition, differentiated Dot1L-deficient cells produced mostly tetraploid karyotypes after treatment with colcimid. These phenotypes are in stark contrast to the differentiation behavior of control wild-type ESCs. To determine the effects of reduced Dot1L expression in vivo, teratomas were formed using knockdown and control ESCs. We found that both cell types differentiated into teratomas containing cells of all 3 germ layers, and retained expression of Oct4 and Sox2. Though teratomas readily formed from Dot1L-deficient ESCs, these had higher levels of residual Oct4 expression as compared to wildtype teratomas, and also seemed to contain more clusters of differentiated cells with a glandular/epithelial histology. Though we confirmed knockdown of Dot1L in the cells used to make teratomas, we found low level Dot1L expression in the harvested teratomas. Therefore, it is possible that the Dot1L shRNA transgene was silenced during the course of teratoma formation. However, the apparently delayed kinetics of Oct4 repression as well possible histological differences, suggest that an early deficit of Dot1L expression altered the course of subsequent teratoma formation. Also of note, other evidence exists to suggest that the requirement for Dot1L in vivo may be less stringent, since Dot1L null embryos develop up to embryonic day 10.5 [9].
In addition to these effects, we find evidence that Dot1L participates in the transcriptional regulation of a select set of genes. The results indicate that many more genes are de-repressed by loss of Dot1L than are repressed in ESCs, supporting a recent finding that Dot1L is involved in maintaining silent heterochromatin [9]. Interestingly, an unusually high proportion of genes that are mis-regulated by loss of Dot1L have known functions related to the cell cycle and cellular proliferation. Also of note is our observation that many endoderm specific markers are upregulated in undifferentiated ESCs. The mechanism by which this occurs is unknown, however it may be directly linked to losing H3K79me2 if this histone modification acts to repress these genes in some manner, possibly in conjunction with other histone modifications. Further studies will be aimed at determining the precise role that H3K79me2 plays in their expression, as well as other highly misregulated genes found in our array analysis.
Together, the results indicate that Dot1L is dispensable for ESC viability, but Dot1L becomes crucial for proliferation soon after differentiation is initiated. Furthermore, the results indicate that Dot1L plays a critical role for proper progression of the cell cycle in differentiating ESCs. Based on the findings, we propose the following mechanism for Dot1L action in differentiating ESCs: Undifferentiated Dot1L cells exhibit some degree of chromosomal instability. Furthermore, after exposure of these cells to colcimid, a microtuble destabilizing agent and disruptor of mitotic spindles, tetraploid karyotypes are recovered. We observed many karyotypes with slightly less than a diploid number of chromosomes in undifferentiatied ESCs. This could easily be caused by loss of chromosomes during the course of chromosome spread preparation. However, it may also be that ESCs may inherently have a less stringent control of their chromosomal content as compared to other cell types [33–36]. Our results show that Dot1L is necessary for normal fidelity of chromosome transmission through mitosis. Consistent with this notion is our finding that abnormal spindles can be detected in proliferating Dot1L-deficient ESCs. Once differentiation is induced, Dot1L-deficient cells arrest with a G2 content of DNA, but fail to undergo significant levels of apoptosis. These findings suggest that a mitotic checkpoint exists in differentiating ESCs that causes arrest in G2, but fails to induce apoptosis. This checkpoint is present after only 3 days of differentiation in RA. Most proliferating mammalian cells have a spindle assembly checkpoint (SAC) that triggers mitotic arrest and eventual apoptosis in cells with improperly or incompletely formed spindles in M-phase [35]. Interestingly, a recent report shows that mouse and human ESCs are tolerant of increased levels of polyploidy after spindle disruption caused by extended exposure to microtubule-destabilizing agents, and exhibit an uncoupling of the SAC from apoptosis [35]. Yet ESCs do not simply lack the capacity to undergo apoptosis since ESCs lacking the histone methyltransferase Mll2 [28], or exposed to the drug camptothecin (this study) readily undergo apoptosis. Mitotic defects in cells lacking histone modification enzymes have been observed in other cell-types. For example, aberrant chromosome segregation is seen in cells deficient in H3K9me3, and HDAC3 RNAi leads to malformed spindles [37, 38]. Finally, the Trypanosoma brucei homolog of Dot1L is required for progression through mitosis with the proper number of chromosomes [14]. Therefore, the results presented here strongly suggest that Dot1L is required for mitotic spindle integrity and faithful execution of mitosis in ESCs. Furthermore, our results show that the SAC is active after only 3 days of RA-induced differentiation, and loss of Dot1L can cause SAC-induced proliferative arrest (very likely due to an inability to complete mitosis). However, the proliferation arrest in early differentiated cells fails to induce apoptosis, allowing some cells to reenter the cell cycle after failed mitosis with a roughly tetraploid complement of chromosomes. The transcription profiles of Dot1L-deficient ESCs and their differentiated derivatives contain a high proportion of mis-regulated genes with known functions in the cell cycle and cellular proliferation. These may represent direct targets of Dot1L regulation. Alternatively, numerous cell cycle and proliferation genes may be mis-regulated in response to Dot1L-induced mitotic defects.
Future stem cell therapies hope to alleviate human degenerative disease by transplantation of differentiated ESCs or other pluripotent cells. However, safety concerns exist that arise from the inherent teratogenicity of ESCs. This study shows that ESCs have a tendency to tolerate aneuploidy, and Dot1L is a histone methyltransferase whose activity is critical for chromosomal stability during the initial steps of ESC differentiation. In addition, it is clear that ESCs have unusually lax cell cycle checkpoint control. For these reasons, a better understanding of the ESC cell cycle, checkpoint control, and chromosomal stability is necessary if safe ESC-based therapeutics are to be developed for human treatments.
Supplementary Material
Acknowledgments
We thank Anupinder Kaur and Brenton Graveley for microarray facility services and Therese Doherty for proofreading. This work was supported in part by National Institutes of Health Grant RO1AG23687 and support from the Connecticut Stem Cell Research Program, and a USDA Hatch grant for animal stem cell research.
Footnotes
Author contributions:
Evan Barry: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Winfried Krueger: Assembly of data, data analysis
Caroline Jakuba: Collection and/or assembly of data
Eric Veilleux: Collection and/or assembly of data
Dominic Ambrosi: Collection and/or assembly of data
Craig Nelson: data analysis and interpretation
Theodore Rasmussen: Conception and design, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript
References
- 1.Rasmussen T. Developmentally-poised chromatin of embryonic stem cells. Front Biosci. 2008;13:1568–1577. doi: 10.2741/2781. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 3.Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8(2):188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 4.Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 5.Krogan NJ, Dover J, Wood A, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 6.Steger DJLM, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 8.Okada Y, Jiang Q, Lemieux M, et al. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006;8(9):1017–1024. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones B, Su H, Bhat A, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4(9):e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng HH, Feng Q, Wang H, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16(12):1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wysocki R, Javaheri A, Allard S, et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25(19):8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huyen Y, Zgheib O, Ditullio RA, Jr, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432(7015):406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 13.Ng HH, Ciccone DN, Morshead KB, et al. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci U S A. 2003;100(4):1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janzen CJ, Hake SB, Lowell JE, et al. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell. 2006;23(4):497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 15.Ooga M, Inoue A, Kageyama S, et al. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod. 2008;78(3):413–424. doi: 10.1095/biolreprod.107.063453. [DOI] [PubMed] [Google Scholar]
- 16.Botuyan MV, Lee J, Ward IM, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ambrosi DJ, Tanasijevic B, Kaur A, et al. Genome-wide reprogramming in hybrids of somatic cells and embryonic stem cells. Stem Cells. 2007;25(5):1104–1113. doi: 10.1634/stemcells.2006-0532. [DOI] [PubMed] [Google Scholar]
- 18.Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25(10):2567–2574. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- 19.Kurosawa H, Imamura T, Koike M, et al. A simple method for forming embryoid body from mouse embryonic stem cells. J Biosci Bioeng. 2003;96(4):409–411. doi: 10.1016/S1389-1723(03)90148-4. [DOI] [PubMed] [Google Scholar]
- 20.Im H, Park C, Feng Q, et al. Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. J Biol Chem. 2003;278(20):18346–18352. doi: 10.1074/jbc.M300890200. [DOI] [PubMed] [Google Scholar]
- 21.Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. Embo J. 2005;24(2):347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider R, Bannister AJ, Myers FA, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 23.Santos-Rosa H, Schneider R, Bannister AJ, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 24.Liang G, Lin JC, Wei V, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101(19):7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakoc CR, Sachdeva MM, Wang H, et al. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen TP, Mastrangelo MA, Eden A, et al. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J Cell Biol. 2000;150(5):1189–1198. doi: 10.1083/jcb.150.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5(4):695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 28.Lubitz S, Glaser S, Schaft J, et al. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol Biol Cell. 2007;18(6):2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G, Gurtu V, Kain SR, et al. Early detection of apoptosis using a fluorescent conjugate of annexin V. Biotechniques. 1997;23(3):525–531. doi: 10.2144/97233pf01. [DOI] [PubMed] [Google Scholar]
- 30.Bridge AJ, Pebernard S, Ducraux A, et al. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34(3):263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds A, Anderson EM, Vermeulen A, et al. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. Rna. 2006;12(6):988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36(Web Server issue):W358–363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Wu H, Loring J, et al. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev Dyn. 1997;209(1):85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Longo L, Bygrave A, Grosveld FG, et al. The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res. 1997;6(5):321–328. doi: 10.1023/a:1018418914106. [DOI] [PubMed] [Google Scholar]
- 35.Mantel C, Guo Y, Lee MR, et al. Checkpoint-apoptosis uncoupling in human and mouse embryonic stem cells: a source of karyotpic instability. Blood. 2007;109(10):4518–4527. doi: 10.1182/blood-2006-10-054247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebuzzini P, Neri T, Zuccotti M, et al. Chromosome number variation in three mouse embryonic stem cell lines during culture. Cytotechnology. 2008;58(1):17–23. doi: 10.1007/s10616-008-9164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McManus KJ, V, Biron L, Heit R, et al. Dynamic changes in histone H3 lysine 9 methylations: identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J Biol Chem. 2006;281(13):8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- 38.Ishii S, Kurasawa Y, Wong J, et al. Histone deacetylase 3 localizes to the mitotic spindle and is required for kinetochore-microtubule attachment. Proc Natl Acad Sci U S A. 2008;105(11):4179–4184. doi: 10.1073/pnas.0710140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.