Abstract
Celastrol is well known for its anti-inflammatory and anti-cancer effects. In this study, the efficacy of celastrol against dextran sulfate sodium (DSS)-induced inflammatory bowel disease (IBD) in mice was established and the mechanism was investigated using lipidomics. Celastrol treatment significantly alleviated DSS-induced colitis in mice, as revealed by the body weight, colon length, scores of rectal bleeding and diarrhea, serum TNF-α level, and histological analysis results. Lipidomics analysis based on UPLC/MS revealed characteristic changes in the metabolic profiles of the colitis mice, with altered levels of lipid markers associated with IBD, including LPC18 : 0, LPC18 : 1, LPC18 : 2, sphingomyelin (SM), and increased LPC18 : 0/LPC18 : 1 and LPC18 : 0/LPC18 : 2 ratios. For the celastrol-treated colitis mice, however, levels of the above lipid markers were restored, together with recovered saturated LPC/unsaturated LPC ratios. Accordingly, using GC–MS analysis, increased stearic acid (C18 : 0)/oleic acid (C18 : 1) and stearic acid (C18 : 0)/linoleic acid (C18 : 2) ratios were observed in colitis mice, which were later recovered after celastrol treatment. Quantitative real-time PCR analysis revealed that the liver expression of stearoyl-coenzyme A desaturase 1 (SCD1), the key enzyme controlling the desaturation of saturated fatty acid, was dramatically inhibited in IBD mice, and was obviously recovered after celastrol treatment. These results suggest that the increased saturated LPC/unsaturated LPC (and saturated fatty acid/unsaturated fatty acid) ratios associated with SCD1 down-regulation could be regarded as biomarkers of colitis, and celastrol alleviates DSS-induced colitis partially via up-regulation of SCD1, restoring the altered balance between stearic acid- and oleic acid-derived lipid species, which plays an important role in alleviating colitis. In all, this study provided the scientific basis for further development of celastrol in treating IBD.
1. Introduction
Inflammatory bowel disease (IBD) is a gastrointestinal tract disorder that mainly includes two phenotypes, namely Crohn’s disease (CD) and ulcerative colitis (UC). With the improvement of living standard, the incidence rate and the prevalence rate of IBD are increasingly high, especially in European and American countries.1,2 China, however, has the highest incidence rate of IBD in Asia.3 Moreover, with similar symptoms to irritable bowel syndrome (IBS), IBD is apt to be misdiagnosed.4,5 Currently the pathogenesis of IBD is still unclear, and a multitude of factors may have effects on IBD, such as environmental factors, infectious microbes, genetic susceptibility, and a dysregulated immune system.6–8
Nonsteroidal and steroidal anti-inflammatory drugs, immunosuppressants and immunomodulators are commonly used for the treatment of IBD. However, adverse reactions of these drugs were obvious and notable.9,10 For example, nonsteroidal anti-inflammatory drugs (NSAIDs) may produce some complications including intestinal strictures, enteropathy with anemia and the loss of protein, macroscopically inflammatory changes such as erosions and ulceration, aggravated diverticulosis, and recurrences of ulcerative colitis.11 Therefore, the safety of NSAIDs for treating IBD has been questioned.12 Adverse reactions of anti-TNF agents are hypersensitive reactions, skin manifestations, arthralgia, cytopenia, heart failure and autoimmune liver injury.13 Therefore, the research and development of new effective drugs for treating IBD is an area of great interest.
Celastrol, an active ingredient extracted from Trypterygium wilfordii Hook F., is well known for its anti-inflammatory and anti-cancer effects.14,15 In recent years, celastrol has been reported to suppress IBD.16,17 For example, celastrol was reported to improve experimental colitis in IL-10 deficient mice by up-regulating autophagy via the PI3K/Akt/mTOR signaling pathway.16 Others reported that celastrol improved dextran sulfate sodium (DSS)-induced colitis via modulating oxidative stress, inflammatory cytokines and intestinal homeostasis.17 Celastrol was also reported to inhibit the secretion of proinflammatory cytokines including IL-1β, TNF-α, IL-6, and IL-8 in inflammatory human colonic biopsies.18 However, these reported molecular targets may be involved in many signaling pathways,19 hence further investigation is still required to explore the specific mechanisms and targets of celastrol in treating colitis.
Metabolomics, in a holistic way, provides an efficient tool for investigating the mechanism of action of new drugs and identifying their potential therapeutic targets using biofluids (serum/plasma, urine), tissues, or other biological extracts.20,21 In our previous study, the anti-cancer effects of celastrol on human cervical cancer cells were investigated using a gas chromatography–mass spectrometry (GC–MS) based metabolomic approach, and several significant metabolites involved in energy, amino acid and nucleic acid metabolism were revealed, which were believed to be closely related to the therapeutic efficacy of celastrol.22 This work provides valuable information on the systemic effects of celastrol to further illustrate its anti-cancer mechanism. However, there have been no metabolomics investigations into the effect of celastrol on IBD.
A number of disrupted pathways in IBD have been revealed by metabolomics investigations, including amino acid metabolism, the methionine–homocysteine cycle, the TCA cycle, uric acid metabolism, fatty acid metabolism, and purine metabolism.23–31 Recently, using a lipidomics approach, significant alterations in lipid metabolism, mainly sphingomyelin (SM) and ceramide (CER) metabolism, were revealed in IBD mice.25 Since lipids have emerged as important signaling molecules regulating many cell functions including inflammatory responses, in this study, lipidomics based on ultraperformance liquid chromatography/mass spectrometry (UPLC–MS) was employed to investigate the efficacy of celastrol on DSS-induced IBD mice, to help elucidate the mechanisms of celastrol against colitis.
2. Materials and methods
2.1. Chemicals and reagents
Lipid markers were obtained from Avanti Polar Lipids (Alabaster, AL). All the authentic standards were purchased from Sigma-Aldrich (St. Louis, MO, US). Methanol, isopropanol and acetonitrile were purchased from TEDIA (Fairfield, OH, USA). Dextran sulfate sodium (DSS) was purchased from MP Biomedicals (Solon, OH, USA) (MW = 36000–50000).
2.2. Mice and treatments
All animal treatments in this study were approved by the Administrative Committee of Experimental Animal Care and Use of the Second Military Medical University (SMMU), and were in accordance with National Institute of Health guidelines on the ethical use of animals. Six- to 8-week-old male C57BL/6J mice were purchased from Shanghai Slac Laboratory Animal Co. Ltd (Shanghai, China) and were housed under standard conditions maintained at 25 ± 1 °C and 55 ± 5% humidity with a light/dark cycle of 12 h. All mice were acclimatized for one week prior to the study. Colitis was induced by adding DSS to the drinking water (3%, wt/vol) for 7 days. All mice were divided into four groups (n = 5 for each group) and had free access to the standard diet: (1) control group: the mice had free access to drinking water; (2) colitis group: the mice were given 3% DSS in drinking water for 7 days; (3) mesalazine (positive control drug) group: the mice were given 3% DSS in drinking water and were simultaneously treated with mesalazine (100 mg kg−1) once a day (by gavage) for 7 days; and (4) celastrol group: the mice were given 3% DSS in drinking water and were simultaneously treated with celastrol (1 mg kg−1) once a day (by gavage) for 7 days. The mice were fasted for 12 h before operation. Serum samples were collected through retro-orbital bleeding. On day 7 liver and colon tissues were harvested immediately following CO2 euthanization. All serum, liver and colon samples were stored at −80 °C before analysis.
2.3. Colitis evaluation
Body weights of the mice were recorded at the same time every day. Diarrhea, rectal bleeding, and bloody stool daily were assessed and reported as a score from 0 to 4 according to the disease activity index.32 The histological examination of the colon tissue was performed by blinded analysis and the severity of colon damage was determined by a routine hematoxylin and eosin (H&E)-stained section according to the morphological criteria described previously.32,33
2.4. Metabolomics analysis
2.4.1. Sample preparation.
For serum lipidomics analysis 20 μl serum were mixed with 4-fold chloroform/methanol (2 : 1 v/v) solution containing 2 μM LPC17 : 0, PC17 : 0, SM17 : 0 and CER17 : 0 as internal standards. The mixed solution was vortexed for 30 s and then allowed to stand for 5 min at room temperature. The mixture was centrifuged at 10 000 rpm for 10 min, then the lower organic phase was collected and evaporated at room temperature under vacuum, and the residue was dissolved in chloroform/methanol (2 : 1 v/v), followed by diluting with isopropanol : acetonitrile :H2O (2 : 1 : 1) before UPLC–MS analysis. For tissue lipidomics analysis, approximately 30 mg weighted tissues were homogenized with 14-fold methanol : H2O (4 : 3) solution and then 350 μl of the supernatant was mixed with 400 μl chloroform containing 2 μM LPC17: 0, PC17 : 0, SM17: 0 and CER17 : 0 as internal standards. The mixed solution was vortexed for 30 min at room temperature, and then centrifuged at 13 000 rpm for 20 min, and the lower organic phase was collected and evaporated at room temperature under vacuum. The residue was suspended with 100 μl chloroform:methanol (1 : 1) solution and then diluted with isopropanol : acetonitrile : H2O (2 : 1 : 1) solution before injection. Quality control (QC) samples were prepared by pooling equal volumes of each analyzed sample.
2.4.2. UPLC–MS analysis.
Ultra-performance liquid chromatography-quadrupole/time-of-flight mass spectrometry (Agilent Technologies 6538 UHD Accurate-Mass Q-TOF LC-MS and 1290 Infinity) was used for lipidomics analysis. An Waters Acquity CSH C18 column (1.7 μm particle size, 2.1 × 100 mm, Waters, Milford, MA, USA) was used as the stationary phase at 50 °C. Analysis conditions of UPLC : mobile phase A is acetonitrile/water (6/4) with 10 mM ammonium acetate and 0.1% formic acid; mobile phase B is 90% isopropanol/acetonitrile (9/1) with 10 mM ammonium acetate and 0.1% formic acid and 10% water. Gradient elution conditions were initially 30% B to 40% B at 3 min, and 43% B at 4 min and then were increased to 50% B at 6 min, to 53% B at 10 min, to 70% B at 12 min and to 99% B at 18 min before returning to initial conditions at 18.5 min with equilibration for 2 additional minutes for next injection. The flow rate was 0.3 ml min−1.
2.4.3. Data processing and multivariate data analysis.
Chromatographic and spectral data were deconvoluted using XCMSOnline (https://xcmsonline.scripps.edu/), from which a multivariate data matrix of sample identity, ion identity (retention time and m/z), and ion abundance was generated through centroiding, deisotoping, filtering, peak recognition, and integration. The variables presenting in at least 80% of the groups were kept. The intensity of each ion was calculated by normalizing the single ion counts vs. the total ion counts in the whole chromatogram. The data matrix was further exported into SIMCA-P11 software (Umetrics, Kinnelon, NJ) and then transformed by mean-centering and pareto scaling. Statistical models including principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) were established to represent the major latent variables in the data matrix. The goodness of the fit and prediction ability of the models were validated from cumulative R2Y and Q2 values. Permutation tests were performed to further confirm the models.
2.4.4. Biomarker identification and quantitation.
The differentiating ions among various groups were obtained using a statistically significant threshold of variable influence on projection (VIP) values from the PLS-DA model and two-tailed Student’s t test on the normalized raw data at the univariate analysis level, where the ions with VIP values larger than 1.5 and P-values less than 0.05 were selected. The potential biomarkers were then identified by comparing with the authentic standards based on their MS/MS fragmentation patterns and retention times. Concentrations of the metabolites were determined using multiple reaction-monitoring mass spectrometry based on standard curves using authentic standards.
2.4.5. Data analysis.
Data are expressed as mean ± SD. Statistical analysis was performed using Prism 5.0 (GraphPad Software, San Diego, CA) via two-tailed Student’s t-test or Mann–Whitney test to evaluate the significance of differences between the groups where necessary. A P-value below 0.05 was considered statistically significant.
2.5. ELISA analysis
A commercially available mouse tumor necrosis factor α (TNF-α) ELISA Kit (JRDUN Biotechnology, Shanghai, China) was used to determine serum TNF-α levels in the mice. A Pierce BCA Protein Assay Kit (Thermo Scientific) was used to normalize the absorbance values from each ELISA.
2.6. RNA analysis
The TRIzol reagent (Invitrogen) was used to extract RNA. cDNA generated from 1 μg total RNA using the SuperScript II Reverse Transcriptase kit (Invitrogen) was used for quantitative real-time PCR (qPCR). Primers were designed for qPCR using Primer Express software (Applied Biosystems, Foster City, CA), and sequences are available upon request. qPCRs were carried out using SYBR green PCR master mix (Applied Biosystems) in an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Values were quantified using the comparative threshold cycle method, and samples were normalized to β-actin.
3. Results
3.1. Celastrol attenuates DSS-induced colitis
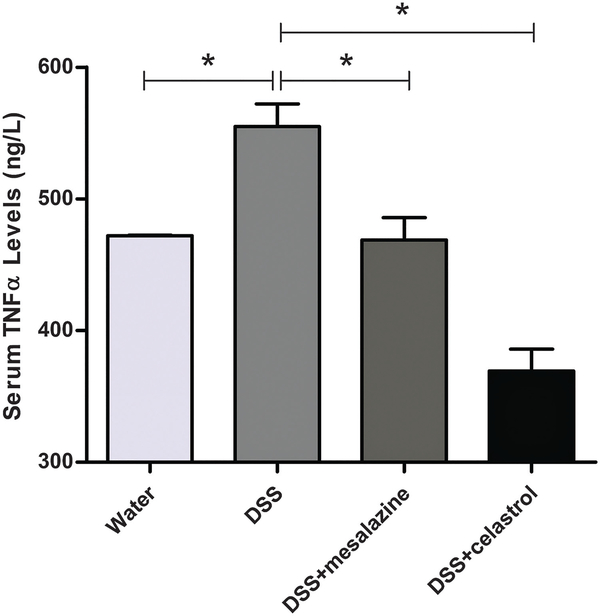
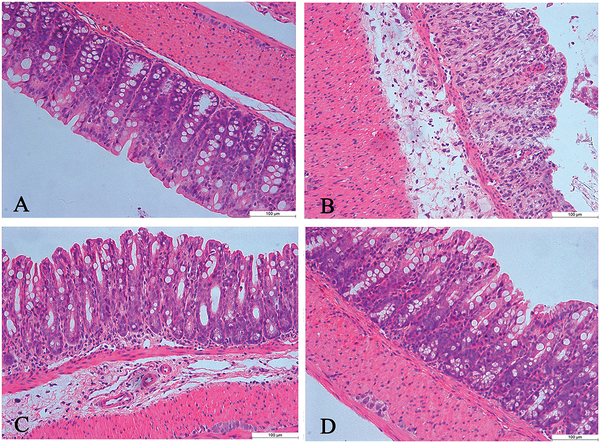
Administration of 3% DSS in drinking water to rodents has been widely used as an experimental animal model of IBD.32,33 The symptoms and histopathological characteristics of colitis induced by DSS are similar to human UC. In the present study, compared with the normal control group, the colitis mice had significantly lower body weight (Fig. 1A), and other obvious inflammatory signs of colitis, such as shorter colon length, rectal bleeding, and diarrhea (Fig. 1B–D). After celastrol treatment, however, the colitis mice had increased body weight, longer colon lengths, and decreased rectal bleeding and diarrhea scores compared to the untreated colitis group (Fig. 1). According to ELISA results, the serum TNF-α level was up-regulated in DSS-treated mice, and then significantly decreased in celastrol-treated colitis mice (Fig. 2). Histological analysis further confirmed severe inflammation with loss of crypts and surface epithelia in the colitis group, which was later attenuated after celastrol treatment (Fig. 3). These indicate that celastrol significantly reduced DSS-induced colitis, at a considerably low dosage (1 mg kg−1).
Fig. 1.
Evaluation of colitis. (A) Celastrol-treated colitis group (DSS + celastrol) had less body weight loss compared with the colitis group (DSS). (B) Comparison of the colon lengths of control (water), colitis (DSS), mesalazine-treated colitis (DSS + mesalazine), and celastrol-treated colitis (DSS + celastrol) groups. (C and D) Diarrhea scores and rectal bleeding scores of the four groups. (n = 5 for each group, ++P < 0.01 and +P < 0.05, comparing colitis and control groups; **P < 0.01, comparing celastrol-treated colitis group and colitis group; ##P < 0.01 and #P < 0.05, comparing mesalazine-treated colitis group and colitis group).
Fig. 2.
Serum TNF-α levels in the control, colitis, mesalazine-treated colitis group and celastrol-treated colitis group using ELISA analysis. Data were expressed as mean ± SD. *P < 0.05.
Fig. 3.
(A–D) Representative images of hematoxylin and eosin-stained colon tissues from control, colitis, mesalazine-treated colitis and celastrol-treated colitis groups, respectively.
3.2. Lipidomics of celastrol treated-colitis mice
Lipidomics analysis was performed on serum and colon samples of the mice. PLS-DA, a supervised classification method based on PLS (a regression technique), was employed for data analysis. In the scores plot, the celastrol-treated colitis group was separated from the colitis group and the mesalazine-treated colitis group (Fig. 4), in accordance with the phenotypes as shown in Fig. 1–3. Based on PLS-DA models discriminating the control and colitis mice, significantly altered ions with the VIP values above 1.5 and P values less than 0.05 were revealed, which were later identified as lipid metabolite markers including LPC18 : 1, LPC18 : 2, SM(d18 : 1/16 : 0), and SM(d18 : 1/18 : 0) using MS/MS and commercial standards (Table 1).
Fig. 4.
(A) Partial least squares-discriminant analysis (PLS-DA) of serum samples (with the cumulative R2X (0.82), R2Y (0.906), and Q2 (0.522)) for all the four examined groups. (B) Partial least squares-discriminant analysis (PLS-DA) of colon samples (with the cumulative R2X (0.907), R2Y (0.985), and Q2 (0.476)) for all the four examined groups.
Table 1.
Identified metabolite markers in the serum and colon samples of colitis mice
| No. | Retention time (min) | m/z | Identification results | B vs. A | C vs. B | D vs. B |
|---|---|---|---|---|---|---|
| 1 | 12.66 | 675.5446 | SM(d18 : 1/14 : 0) | ↑* | ↓ | ↓ |
| 2 | 14.06 | 703.5765 | SM(d18 : 1/16 : 0) | ↑* | ↓ | ↓* |
| 3 | 15.21 | 731.6073 | SM(d18 : 1/18 : 0) | ↑* | ↓ | ↓* |
| 4 | 14.28 | 729.5912 | SM(d18 : 1/18 : 1) | ↑ | ↓ | ↓ |
| 5 | 15.52 | 787.6061 | SM(d18 : 1/22 : 0) | (↑) | (↑) | (↑) |
| 6 | 3.37 | 522.3568 | LPC18 : 1 | ↓**(↓*) | ↓(↓) | ↑*(↑) |
| 7 | 2.50 | 520.3411 | LPC18 : 2 | ↓*(↓*) | ↓(↓) | ↑**(↑**) |
A, B, C and D represent control mice, colitis mice, mesalazine-treated colitis mice and celastrol-treated colitis mice, respectively.↑ or ↓ represents the up- or down-regulation of the metabolites in serum; ↑ or ↓ in brackets represent the up- or down-regulation of the metabolite levels in colon
P < 0.05,
P < 0.01.
3.3. Quantitation of the significant metabolites
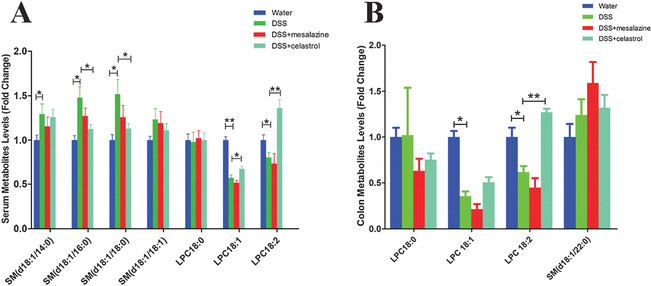
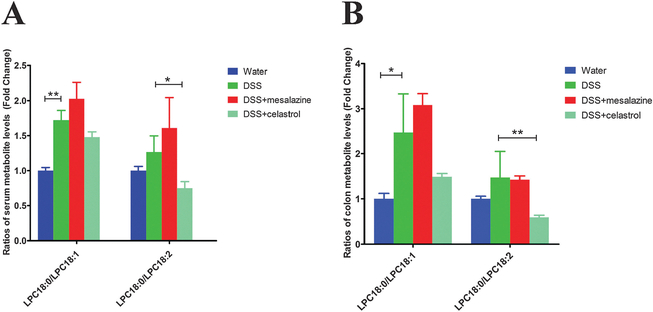
Next, levels of the above metabolite markers together with LPC18 : 0 (stearoyl-LPC), a saturated LPC (m/z = 524.3724 at 4.74 min), were quantitated based on standard curves using authentic standards. Compared to the control group, two unsaturated LPCs, namely LPC18 : 1 (oleoyl-LPC) and LPC18 : 2 (linoleoyl-LPC), were significantly decreased in serum and colon samples of colitis mice; whereas LPC18 : 0 was slightly increased in the colon samples of colitis mice (Fig. 5). As saturated LPCs can promote the release of proinflammatory cytokines in vivo and exacerbate the pathological signs of inflammation,34–38 while unsaturated LPCs are potentially protective with anti-inflammatory benefits for diseases associated with inflammation,39 the perturbance of IBD on these lipids coincided the inflammatory phenotypes in colitis mice. Accordingly, compared to the control mice, increased LPC18 : 0/LPC18 : 1 and LPC18 : 0/LPC18 : 2 ratios were observed in the colitis mice (Fig. 6), which were consistent with previous findings,40 and could be regarded as symbols of colitis.
Fig. 5.
Serum (A) and colon (B) SM and LPC levels in the four groups, i.e. control, colitis, mesalazine-treated and celastrol-treated colitis groups. Data were expressed as mean ± SD. *P < 0.05 and **P < 0.01.
Fig. 6.
Ratios of LPC18 : 0/LPC18 : 1 and LPC18 : 0/LPC18 : 2 in serum (A) and colon (B) samples of the mice. Data were expressed as mean ± SD. *P < 0.05 and **P < 0.01.
The increased ratio between stearoyl-LPC and oleoyl-LPC in colitis mice may be related to down-regulated liver stearoyl-coenzyme A desaturase 1 (SCD1), which resembles the phenotype of SCD1 null mice with deficient SCD1.41 SCD1 is the key enzyme controlling the desaturation of saturated fatty acid (SFA) to generate monounsaturated fatty acid (MUFA), after which saturated fatty acid (e.g. stearic acid C18 : 0) and monounsaturated fatty acid (e.g. oleic acid C18 : 1) participate in LPC metabolic balance through a series of reactions.40,42,43 To explore whether the above involved fatty acids had altered levels in colitis mice, we then determined their serum levels using GC/MS as described by Patterson et al. with slight modifications44 (for a typical chromatogram see Fig. S1, ESI†). As expected, serum unsaturated fatty acids (oleic acid C18 : 1 and linoleic acid C18 : 2) were decreased in the colitis mice compared to the control group, and C18 : 0/C18 : 1 and C18 : 0/C18 : 2 ratios were obviously increased (Fig. S2, ESI†), supporting our lipidomics findings and in accordance with previous reports.45 Taken together, as the rate-limiting enzyme in the synthesis of monounsaturated fatty acids from saturated fatty acid, the down regulation of SCD1 induced by DSS could therefore exacerbate colitis. Hence, SCD1 may serve as a potential target for the intervention or the treatment of inflammatory diseases including IBD.40
After celastrol treatment, compared with the colitis group, LPC18 : 1 and LPC18 : 2 levels were increased in celastrol-treated colitis mice (Fig. 5); and the ratios of LPC18 : 0/LPC18 : 1 and LPC18 : 0/LPC18 : 2 were decreased, which, however, were not observed in the mesalazine-treated colitis group (Fig. 6). Accordingly, increased levels of unsaturated fatty acids (C18 : 1 and C18 : 2) were observed after celastrol treatment, together with restored C18 : 0/C18 : 1 and C18 : 0/C18 : 2 ratios (Fig. S2, ESI†). These results implied a possible correlation between celastrol treatment and SCD1 up-regulation in alleviating colitis. Besides, the increased levels of SM, e.g. serum levels of SM(d18 : 1/16 : 0) and SM(d18 : 1/18 : 0) in the DSS-treated group, were consistent with a previous study,25 which were also restored after celastrol treatment (Fig. 5).
3.4. Analysis of SCD1 mRNA expression
Compared to the control group, the expression of Scd1 mRNA in liver was markedly decreased in colitis mice, consistent with a previous report.40 Decreased expression of Scd1 mRNA in colon was also observed, in accordance with the dramatically declined SCD1 expression in liver where SCD1 is highly expressed43 (Fig. 7). After celastrol treatment, however, up-regulation of Scd1 mRNA expression was observed in both colon and liver of colitis mice, in accordance with the alleviated pathological symptoms of colitis for this group, as shown in Fig. 1–3. Though in liver, celastrol treatment did not recover the SCD1 expression to as high as the normal level, it did significantly increase the SCD1 levels of the colitis mice compared to the DSS-only mice. Our lipidomics findings that celastrol treatment decreased the ratios of saturated/unsaturated LPCs and those of saturated/unsaturated fatty acids were quite supportive of these qPCR observations.
Fig. 7.
Real-time PCR measurement of Scd1 mRNA expression in the colon (A) and liver (B) of control, colitis, and celastrol-treated colitis groups. Data are represented as mean ± SD.
4. Discussion
Celastrol has long been known for its anti-inflammatory and anti-cancer effects.14,15 In recent years, celastrol was reported to ameliorate murine colitis via modulating oxidative stress, inflammatory cytokines and intestinal homeostasis,17 and improve experimental colitis in IL-10 deficient mice via the inhibition of the Akt/mTOR pathway.16 This study confirmed that celastrol can significantly reduce DSS-induced colitis, as revealed by body weight, colon length, scores of rectal bleeding and diarrhea, serum TNF-α level, and histological analysis results.
Currently several molecular targets of celastrol have been reported, such as TNF-α, NF-κB, COX-2, VEGF, Hsp90, and Akt/mTOR signaling.46–52 For example, celastrol was shown to inhibit the expression of the inflammatory cytokine IL-6 through the NF-kB-dependent pathway in prostate carcinoma cells.53 Celastrol was also found to induce rapid degradation of CIP2A (cancerous inhibitor of protein phosphatase 2A) in non-small-cell lung cancer cells by directly binding CIP2A protein and promoting CIP2A-CHIP (Hsp70-interacting protein) interaction.54 Another report indicated that celastrol showed anti-cancer effects on human triple negative breast cancer through the PI3k/Akt/mTOR axis.55 Celastrol was also reported to exert inhibitory effects on platelets, which contribute to inflammatory events.56 In all, though there are quite a few studies aiming to reveal the molecular targets of celastrol, its specific mechanism and targets in ameliorating colitis are largely unclear.
Metabolomics seeks to characterize the metabolic profile of a biological system. The identities and concentrations of the downstream metabolites are the products of biomolecular processes and can hence provide biochemical signatures for exploring drug efficacy and toxicity.20,57,58 Lipidomics is a metabolomic platform that specifically focuses on lipid species and plays an essential role in defining the biochemical mechanisms of lipid-related disease processes through identifying alterations in cellular lipid metabolism, trafficking, and homeostasis.59–61 In some reports, lipidomics has revealed a potential link between lipid profiling and colitis.23,25,40,62,63 In the present study, the levels of two unsaturated LPCs were significantly decreased whereas one saturated LPC was slightly increased in colon of colitis mice, and LPC18 : 0/LPC18 : 1 and LPC18 : 0/LPC18 : 2 ratios were increased for this group. Some SMs, such as SM(d18 : 1/16 : 0) and SM(d18 : 1/18 : 0), were found to have increased levels in colitis mice, consistent with previous findings.25 The perturbation of colitis on LPC and SM metabolism were later recovered after celastrol treatment. This illustrates the efficacy of celastrol in treating colitis via the intervention of LPC and SM metabolism.
In this study, Scd1 mRNA expression in liver of colitis mice was dramatically inhibited compared to the control mice, consistent with a previous report.40 As the sole enzyme responsible for the biogenesis of MUFA from SFA, SCD1 plays an important role in the homeostasis of MUFA and SFA.42 Our GC–MS quantitative analysis revealed decreased unsaturated fatty acids as well as increased SFA/MUFA ratios in colitis mice; accordingly, lipidomics analysis revealed increased saturated LPC/unsaturated LPC ratios in colitis mice. These were all recovered after celastrol treatment. As alterations in the balance of SFA and MUFA in lipids can influence a wide array of cellular functions, and celastrol treatment partially recovered Scd1 mRNA expression in colitis mice, our results thus reflect a possible correlation between celastrol and SCD1 regulation in treating IBD. Moreover, the intervention of celastrol upon alteration of SFA and MUFA levels in colitis mice may also be related to its antioxidative effect, which still awaits further study.64
SCD1 is regarded as an important enzyme in inflammatory progress40 and has been implicated in the regulation of adipocyte inflammation, macrophage inflammation, myocyte, and endothelial cell function in distinct cell types.65 As far as colitis is concerned, a previous metabolomics study revealed that down-regulation of liver SCD1 could accelerate colitis in mice;40 another study on colonic mucosa from patients with active UC revealed a markedly declined SCD1 gene expression compared to remission UC and healthy control groups.66 According to our qPCR analysis and consistent with the previous reports, the expression of SCD1 mRNA in the liver was markedly decreased in colitis mice. After celastrol treatment, however, SCD1 expression was obviously recovered in colitis mice compared to the DSS-only group. The present results not only confirmed the previous findings, but also indicated that celastrol alleviated IBD possibly via the up-regulation of SCD1, thus supporting the view that SCD1 and its related lipid species may serve as potential targets for the intervention or treatment of inflammatory diseases.40 However, in which way celastrol correlates with SCD1, whether directly or indirectly, is still unknown, which is an interesting question beyond this lipidomics investigation. Therefore, further work is still needed to uncover the pathways by which SCD1 expression is recovered after celastrol treatment.
5. Conclusions
In this study, anti-inflammatory effects of celastrol on IBD were confirmed and its mechanism was demonstrated with the assistance of lipidomics. Celastrol likely intervenes the LPC and SM metabolism of colitis mice, partially by up-regulating SCD1 expression and restoring the altered balance between stearic acid- and oleic acid-derived lipid species against proinflammatory signaling. This is the first lipidomics report exploring the mechanism of celastrol in treating IBD, which provided the scientific basis for further development of celastrol in treating IBD.
Supplementary Material
Acknowledgements
This study was supported by Project Funded by China Postdoctoral Science Foundation (2015M581534), and Li-Shi-Zhen Young Scientist Project of School of Pharmacy at the Second Military Medical University, Shanghai, China.
Abbreviations
- DSS
Dextran sulfate sodium
- IBD
Inflammatory bowel disease
- PLS-DA
Partial least squares-discriminant analysis
- PCA
Principal components analysis
- SCD1
Stearoyl-coenzyme A desaturase 1
- SM
Sphingomyelin
- CER
Ceramide
- LPC
Lysophosphatidylcholine
- PC
Phosphatidylcholine
- SFA
Saturated fatty acid
- MUFA
Monounsaturated fatty acid
- UPLC/MS
Ultra performance liquid chromatography/mass spectrometry
- GC/MS
Gas chromatography/mass spectrometry
- qPCR
Quantitative real-time PCR
- TNF-α
Tumor necrosis factor α
Footnotes
Disclosures
The authors who have taken part in this study declared that they have nothing to disclose regarding funding or conflict of interest.
Electronic supplementary information (ESI) available. See DOI : 10.1039/c5mb00864f
References
- 1.Abramson O, Durant M, Mow W, Finley A, Kodali P, Wong A, Tavares V, McCroskey E, Liu L, Lewis JD, Allison JE, Flowers N, Hutfless S, Velayos FS, Perry GS, Cannon R and Herrinton LJ, J. Pediatr, 2010, 157, 233–239.e1. [DOI] [PubMed] [Google Scholar]
- 2.Tursi A, Elisei W and Picchio M, Eur. J. Intern. Med, 2013, 24, 852–856. [DOI] [PubMed] [Google Scholar]
- 3.Ng SC, Tang W, Ching JY, Wong M, Chow CM, Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, Li MF, Ng KK, Kamm MA, Studd C, Bell S, Leong R, de Silva HJ, Kasturiratne A, Mufeena MNF, Ling KL, Ooi CJ, Tan PS, Ong D, Goh KL, Hilmi I, Pisespongsa P, Manatsathit S, Rerknimitr R, Aniwan S, Wang YF, Ouyang Q, Zeng Z, Zhu Z, Chen MH, Hu PJ, Wu K, Wang X, Simadibrata M, Abdullah M, Wu JC, Sung JJY and Chan FKL, Gastroenterology, 2013, 145, 158–165.e2. [DOI] [PubMed] [Google Scholar]
- 4.Vavricka SR, Spigaglia SM, Rogler G, Pittet V, Michetti P, Felley C, Mottet C, Braegger CP, Rogler D, Straumann A, Bauerfeind P, Fried M, Schoepfer AM and Swiss IBDCSG, Inflammatory Bowel Dis., 2012, 18, 496–505. [DOI] [PubMed] [Google Scholar]
- 5.Talley NJ, Phillips SF, Melton LJ, Mulvihill C, Wiltgen C and Zinsmeister AR, Gut, 1990, 31, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumgart DC and Carding SR, Lancet, 2007, 369, 1627–1640. [DOI] [PubMed] [Google Scholar]
- 7.Baumgart DC and Sandborn WJ, Lancet, 2012, 380, 1590–1605. [DOI] [PubMed] [Google Scholar]
- 8.Ordás I, Eckmann L, Talamini M, Baumgart DC and Sandborn WJ, Lancet, 2012, 380, 1606–1619. [DOI] [PubMed] [Google Scholar]
- 9.Kvasnovsky CL, Aujla U and Bjarnason I, Scand. J. Gastroenterol, 2015, 50, 255–263. [DOI] [PubMed] [Google Scholar]
- 10.Bjarnason I, Hayllar J, MacPherson AJ and Russell AS, Gastroenterology, 1993, 104, 1832–1847. [DOI] [PubMed] [Google Scholar]
- 11.Stadnicki A and Frysz-Naglak D, Wiad. Lek, 2007, 60, 286–290. [PubMed] [Google Scholar]
- 12.Kefalakes H, Stylianides TJ, Amanakis G and Kolios G, Eur. J. Clin. Pharmacol, 2009, 65, 963–970. [DOI] [PubMed] [Google Scholar]
- 13.Sousa P and Allez M, Curr. Opin. Gastroenterol, 2015, 31, 296–302. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Chen D, Cui QC, Yuan X and Dou QP, Cancer Res., 2006, 66, 4758–4765. [DOI] [PubMed] [Google Scholar]
- 15.Allison AC, Cacabelos R, Lombardi VRM, Álvarez XA and Vigo C, Prog. Neuropsychopharmacol. Biol. Psychiatry, 2001, 25, 1341–1357. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Sun Y, Shi P, Dong JN, Zuo LG, Wang HG, Gong JF, Li Y, Gu LL, Li N, Li JS and Zhu WM, Int. Immunopharmacol, 2015, 26, 221–228. [DOI] [PubMed] [Google Scholar]
- 17.Shaker ME, Ashamallah SA and Houssen ME, Chem. Biol. Interact, 2014, 210, 26–33. [DOI] [PubMed] [Google Scholar]
- 18.Pinna GF, Fiorucci M, Reimund J-M, Taquet N, Arondel Y and Muller CD, Biochem. Biophys. Res. Commun, 2004, 322, 778–786. [DOI] [PubMed] [Google Scholar]
- 19.Perkins ND, Nat. Rev. Mol. Cell Biol, 2007, 8, 49–62. [DOI] [PubMed] [Google Scholar]
- 20.Shah NJ, Sureshkumar S and Shewade DG, Indian J. Clin. Biochem, 2015, 30, 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Zhu SF, Zhao XL, Liu YX, Wan MH, Guo H, Liu YL, Gong HL, Chen GY and Tang WF, Pancreatology, 2015, 15, 337–343. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Qi Y, Liu H, Fan G and Chai Y, Biochim. Biophys. Acta, Gen. Subj, 2013, 1830, 2779–2789. [DOI] [PubMed] [Google Scholar]
- 23.Gu X, Song Y, Chai Y, Lu F, Gonzalez FJ, Fan G and Qi Y, Mol. BioSyst, 2015, 11, 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Gunther U and Nielsen OH, Metabolomics, 2015, 11, 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Y, Jiang C, Tanaka N, Krausz KW, Brocker CN, Fang Z-Z, Bredell BX, Shah YM and Gonzalez FJ, Am. J. Physiol.: Gastrointest. Liver Physiol, 2014, 307, G564–G573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawiskiba T, Deja S, Mulak A, Zabek A, Jawien E, Pawelka D, Banasik, Mastalerz-Migas A, Balcerzak W, Kaliszewski K, Skora J, Barc P, Korta K, Pormanczuk K, Szyber P, Litarski A and Mlynarz P, World J. Gastroenterol, 2014, 20, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams HRT, Willsmore JD, Cox IJ, Walker DG, Cobbold JFL, Taylor-Robinson SD and Orchard TR, Dig. Dis. Sci, 2012, 57, 2157–2165. [DOI] [PubMed] [Google Scholar]
- 28.Schicho R, Shaykhutdinov R, Ngo J, Nazyrova A, Schneider C, Panaccione R, Kaplan GG, Vogel HJ and Storr M, J. Proteome Res, 2012, 11, 3344–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer J, Liebisch G, Hofmann C, Huy C, Schmitz G, Obermeier F and Bock J, PLoS One, 2009, 4, e7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggiero C, Cherubini A, Ble A, Bos AJG, Maggio M, Dixit VD, Lauretani F, Bandinelli S, Senin U and Ferrucci L, Eur. Heart J, 2006, 27, 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heimerl S, Moehle C, Zahn A, Boettcher A, Stremmel W, Langmann T and Schmitz G, Biochim. Biophys. Acta, Mol. Basis Dis, 2006, 1762, 341–350. [DOI] [PubMed] [Google Scholar]
- 32.Cooper HS, Murthy SN, Shah RS and Sedergran DJ, Lab. Invest, 1993, 69, 238–249. [PubMed] [Google Scholar]
- 33.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y and Nakaya R, Gastroenterology, 1990, 98, 694–702. [DOI] [PubMed] [Google Scholar]
- 34.Stock C, Schilling T, Schwab A and Eder C, J. Immunol, 2006, 177, 8560–8568. [DOI] [PubMed] [Google Scholar]
- 35.Huang F, Subbaiah PV, Holian O, Zhang J, Johnson A, Gertzberg N and Lum H, Am. J. Physiol.: Lung Cell. Mol. Physiol, 2005, 289, L176–L185. [DOI] [PubMed] [Google Scholar]
- 36.Takabe W, Kanai Y, Chairoungdua A, Shibata N, Toi S, Kobayashi M, Kodama T and Noguchi N, Arterioscler., Thromb., Vasc. Biol, 2004, 24, 1640–1645. [DOI] [PubMed] [Google Scholar]
- 37.Liu-Wu Y, Hurt-Camejo E and Wiklund O, Atherosclerosis, 1998, 137, 351–357. [DOI] [PubMed] [Google Scholar]
- 38.Asaoka Y, Oka M, Yoshida K, Sasaki Y and Nishizuka Y, Proc. Natl. Acad. Sci. U. S. A, 1992, 89, 6447–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcón de la Lastra C, Barranco MD, Motilva V and Herrerías JM, Curr. Pharm. Des, 2001, 7, 933–950. [DOI] [PubMed] [Google Scholar]
- 40.Chen C, Shah YM, Morimura K, Krausz KW, Miyazaki M, Richardson TA, Morgan ET, Ntambi JM, Idle JR and Gonzalez FJ, Cell Metab, 2008, 7, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyazaki M, Kim YC, Gray-Keller MP, Attie AD and Ntambi JM, J. Biol. Chem, 2000, 275, 30132–30138. [DOI] [PubMed] [Google Scholar]
- 42.Flowers MT and Ntambi JM, Curr. Opin. Lipidol, 2008, 19, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodson L and Fielding BA, Prog. Lipid Res, 2013, 52, 15–42. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Hatzakis E, Nichols RG, Hao R, Correll J, Smith PB, Chiaro CR, Perdew GH and Patterson AD, Environ. Sci. Technol, 2015, 49, 8067–8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodson L, Skeaff CM and Fielding BA, Prog. Lipid Res, 2008, 47, 348–380. [DOI] [PubMed] [Google Scholar]
- 46.Kannaiyan R, Shanmugam MK and Sethi G, Cancer Lett, 2011, 303, 9–20. [DOI] [PubMed] [Google Scholar]
- 47.Zhu H, Liu X-W, Ding W-J, Xu D-Q, Zhao Y-C, Lu W, He Q-J and Yang B, Cancer Lett, 2010, 297, 155–164. [DOI] [PubMed] [Google Scholar]
- 48.Zhu H, Ding W-J, Wu R, Weng Q-J, Lou J-S, Jin R-J, Lu W, Yang B and He Q-J, Cancer Invest, 2010, 28, 23–32. [DOI] [PubMed] [Google Scholar]
- 49.Zhang D, Xu L, Cao F, Wei T, Yang C, Uzan G and Peng B, Cell Stress Chaperones, 2010, 15, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salminen A, Lehtonen M, Paimela T and Kaarniranta K, Biochem. Biophys. Res. Commun, 2010, 394, 439–442. [DOI] [PubMed] [Google Scholar]
- 51.Pang X, Yi Z, Zhang J, Lu B, Sung B, Qu W, Aggarwal BB and Liu M, Cancer Res, 2010, 70, 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu F, Han M and Wilson JX, Br. J. Pharmacol, 2009, 157, 1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang KC, Tsui KH, Chung LC, Yeh CN, Chen WT, Chang PL and Juang HH, PLoS One, 2014, 9, e93151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Ma L, Wen Z-S, Hu Z, Wu F-Q, Li W, Liu J and Zhou G-B, Carcinogenesis, 2014, 35, 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shrivastava S, Jeengar MK, Reddy VS, Reddy GB and Naidu VG, Exp. Mol. Pathol, 2015, 98, 313–327. [DOI] [PubMed] [Google Scholar]
- 56.Hu H, Straub A, Tian Z, Bassler N, Cheng J and Peter K, J. Cardiovasc. Pharmacol, 2009, 54, 240–245. [DOI] [PubMed] [Google Scholar]
- 57.Fiehn O, Comp. Funct. Genomics, 2001, 2, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji P, Wei Y, Sun H, Xue W, Hua Y, Li P, Zhang W, Zhang L, Zhao H and Li J, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci, 2014, 973, 45–54. [DOI] [PubMed] [Google Scholar]
- 59.Murphy SA and Nicolaou A, Mol. Nutr. Food Res, 2013, 57, 1336–1346. [DOI] [PubMed] [Google Scholar]
- 60.Hartler J, Tharakan R, Köfeler HC, Graham DR and Thallinger GG, Briefings Bioinf, 2013, 14, 375–390. [DOI] [PubMed] [Google Scholar]
- 61.Hu C, van der Heijden R, Wang M, van der Greef J, Hankemeier T and Xu G, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci, 2009, 877, 2836–2846. [DOI] [PubMed] [Google Scholar]
- 62.Andersson D, Kotarsky K, Wu J, Agace W and Duan R-D, Dig. Dis. Sci, 2009, 54, 1440–1448. [DOI] [PubMed] [Google Scholar]
- 63.El Alwani M, Wu BX, Obeid LM and Hannun YA, Pharmacol. Ther, 2006, 112, 171–183. [DOI] [PubMed] [Google Scholar]
- 64.Wang C, Shi C, Yang X, Yang M, Sun H and Wang C, Eur. J. Pharmacol, 2014, 744, 52–58. [DOI] [PubMed] [Google Scholar]
- 65.Liu X, Strable MS and Ntambi JM, Adv. Nutr, 2011, 2, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bueno-Hernandez N, Dominguez-Lopez A, Barreto-Zuniga R, Sanchez Munoz F and Yamamoto-Furusho JK, Inflammatory Bowel Dis, 2011, 17, E155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.