Abstract
Impaired autophagy may be associated with normal and pathological aging. Here we explore a link between autophagy and domain function of Werner protein (WRNp). Werner (WRN) mutant cell lines AG11395, AG05229 and normal aged fibroblast AG13129 display a deficient response to tunicamycin mediated endoplasmic reticulum (ER) stress induced autophagy compared to clinically unaffected GM00637 and normal young fibroblast GM03440. Cellular endoplasmic reticulum (ER) stress mediated autophagy in WS and normal aged cells is restored after transfection with wild type full length WRN, but deletion of the acidic domain from wild type WRN fails to restore autophagy. The acidic domain of WRNp was shown to regulate its transcriptional activity, and here, we show that it affects the transcription of certain proteins involved in autophagy and aging. Furthermore, siRNA mediated silencing of WRN in normal fibroblast WI-38 resulted in decrease of age related proteins Lamin A/C and Mre11.
Keywords: Acidic domain, Aging, Autophagy, Beclin-1, Endoplasmic reticulum, RecQ helicase, Werner protein, Werner syndrome
1. Introduction
RecQ helicases are ubiquitous in life and are conserved throughout species to human cells [1]. There are five RecQ helicases in humans, and defects in three of these give rise to clinical disorders associated with cancer predisposition and/or variable symptoms of premature aging [2,3]. One of these disorders is Werner syndrome (WS), which is a recessive autosomal disease associated with premature aging, and the individuals develop age associated diseases including cancer [4,5]. WS shares many features of normal aging including the expression status of various genes within cells [6].
WRN protein (WRNp, 1432 aa) is well characterized for its participation in DNA metabolism, and it has ATP dependent 3′→5′ DNA helicase and single strand DNA annealing activities [2,7]. WRN has other conserved catalytic domains including 3′→5′ exonuclease, DNA binding RecQ C-terminal (RQC) [8–10], helicase-and-ribonuclease D/C-terminal (HRDC) and C-terminal NLS [11,12]. WRN also contains an acidic region (424–477 aa), which is composed of direct repeats of 27 amino acids. This domain is important for WRNp transcriptional acitivity [13,14]. Various WRN patient mutations have been shown to affect particular domain function. One of them is a mutation in the helicase domain at K577M which blocks its helicase activity [15,16]. Mutation in the N-terminal exonuclease domain at position E84A blocks the exonuclease activity [17]. Central region mutations in RQC domains R993A and F1037A result in very weak annealing, DNA binding and helicase activity [18]. Defects in transcription have also been observed in WS cells implicating that WRNp may have a role in transcriptional control [13,14,19]. WRNp participates in transcription of genes after stress induced by barbiturate Phenobarbital, an antiepileptic drug [20].
Autophagy, and specifically macroautophagy is an evolutionarily conserved catabolic process, which degrades cellular proteins and damaged or excess organelles from the cells [21]. Damaged macromolecules or organelles prevent proper cellular functions. After sensing the cellular status double membrane vacuoles known as autophagosomes are formed and they engulf the protein cargoes and transport them to the lysosomes for degradation. The acidic environment of the lysosomes digests this content [22–24]. With this machinery cells can maintain proper cellular homeostasis [25]. This process is evolutionary conserved from flies to mammals. It was observed that loss-of-functions or mutations in Atg1 (Unc-51), Atg7, Atg18, and Beclin-1 (Bec-1) cause of reduced life span of the nematode Caenorhabditis elegans [26]. Reduced expression of different atg genes resulted in decreased life span of the fruit fly Drosophila melanogaster [27]. Failure of autophagy leads to the accumulation of damaged cellular proteins, which may be responsible for the development of different neurodegenerative disorders [28] such early Alzheimer’s disease (AD), where expression of beclin-1 is reduced [29]. Fibroblasts’ from other neurodegenerative disease such as Machado Joseph Disease (MJD) showed autophagy impairment and accumulation of aggregated proteins [30]. Here the disease protein, ATXN3, is proposed to regulate many cellular pathways including aging [31,32]. Recent accumulating data suggest that autophagy plays a major role in cellular aging [33]. Autophagy generally decreases with age and this may result in the accumulation of damaged non-functional proteins, which cause oxidative stress [34]. This perturbs many cellular functions and contributes to the development of accelerated aging [25].
Alteration of transcriptional activities of WRNp through RNA pol I and RNA pol II have been reported [14,35]. Analysis by expression array showed that both normal old and WS cells have lower expression of Beclin-1. Additionally, ubiquitination and proteosomal degradation is also defective in WS cells [6], and this may lead to accumulation of damaged proteins within cells. Interestingly Beclin-1 plays a crucial role at the initial stage of autophagic vesicle formation [36–38]. In our previous study we showed poor response of starvation induced autophagy in WS cells. After transient overexpression of full length wild type WRNp in WS cells, upregulation of autophagic proteins were observed [39]. In an earlier study it was found that WRNp is elevated by tunicamycin mediated endoplasmic reticulum (ER) stress [40]. The ER stressor tunicamycin blocks N-linked glycosylation (N-glycans) of proteins which induces the unfolded protein response in the ER lumen, known as ER stress. Imbalance between misfolded proteins and their clearances resulted in more ER stress, which ultimately induces the self-degradative process or autophagy through ERAD pathways. Defective autophagy may lead to accumulation of non functional proteins inside cells and ultimately results in different diseases including aging. In our earlier studies we confirmed that WRNp is necessary for the ER stress induced autophagy.
Here, we investigate the autophagic response of WS cell after ER stress. We found that normal aged cells and WS cells have a very limited response to Endoplasmic reticulum (ER) stress induced autophagy. Restoration of autophagic proteins was complemented by transfection with full length WRN in these cell lines. We then mapped the involved region to the acidic domain of WRN, which was also involved in the transcription of genes necessary to combat cellular stress. Thus our results suggest that the acidic domain of WRNp plays a main role for the transcriptional activation of autophagic proteins, which are down regulated in WS cells and in normal aged cells.
2. Materials and methods
2.1. Materials
Minimal Essential Media (MEM) (ATO47), Dulbecco’s Modified Eagle Medium (DMEM) (AT007), L-glutamine (TCL012), MEM Non Essential Amino Acid (ACL006), MEM Amino Acids (ACL002). Agarose low EEO (MB002) were purchased from Himedia, India. Fetal bovine serum (FBS) (10270), penicillin-streptomycin (15140122), MEM Vitamin solution (11120–52) were obtained from Life Technologies, USA. Bovine serum albumin (BSA) (A9418), Tunicamycin (T7765), Monodancylcadaverin (MDC) (30432) were obtained from Sigma-Aldrich, USA. Protease inhibitor cocktail (M250–1 ml), Phosphatase inhibitor cocktail II (GX-0211AR) were from Genetix, India. Forward and reverse primers were synthesized from KRIC, India, Taq polymerase (BB-E0010), MgCl2 (BB-E0011), dNTPs (BB-C0021), from Bio Bharati, India. Q5 High-Fidility DNA Polymerase (M0491G) was obtained from New England Biolabs (NEB), England. Klenow enzyme (EP0054), T4 DNA ligase, 50% PEG (EL0011) solutions were purchased from fer-mentas, USA. Gel purification kit (28104) was from Qiagen,Germany. siRNA WRN (439240) and control siRNA (4390846) were purchased from Invitrogen, USA. Anti-LC3 B (ab48394) was purchased from Abcam, England), anti-Atg5 (9980S), anti-Beclin-1(3738S), anti-p62 (8025S), anti-NFKB (6956P), anti-Mre11(4847P), anti-CDC-2 (9112), were purchased from Cell Signaling Technologies, USA. Anti-Histone 3.3 B (GTX115549) was purchased from Gen Tex, USA. Anti-ß-Actin (SC-376421) Anti-Werner (SC-5626) antibodies, horse radish peroxidase-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology, USA. pBABE-puro mCherry-EGFP-LC3 B which encodes a fusion protein EGFP, mCherry and LC3B, was a generous gift from Dr. Jayanta Debnath (Department of Pathology, University of California, San Francisco).
2.2. Cell lines and culture conditions
WS cells AG11395 (SV40 transformed Fibroblast; Age: 60 Year, Biopsy source: Skin; Tissue source: Skin; Gender: Male; Ethnicity: Caucasia) and WS primary fibroblasts AG05229 (untransformed Fibroblast; Age: 25 Year, Biopsy source: Thigh; Tissue source: Skin; Gender: Male; Ethnicity: Caucasian) unaffected GM00637 (SV40- transformed fibroblast, Age: 18 Year, Biopsy source: Unspecified; Tissue source: Skin; Gender: Female; Ethnicity: Caucasia), untransformed primary fibroblast young (GM03440 - Age: 20 Year, Biopsy Source: Leg; Tissue source: Skin; Gender: Male; Ethnicity: Caucasia) and normal aged (AG13129) Age: 89 Year, Biopsy source: Arm; Tissue source: Skin; Gender: Male; Ethnicity: Caucasia) cells were cultured in Minimal Essential Medium (MEM) supplemented with 10% FBS, 1% peni-cillin_streptomycin, 1% L-glutamine, 1% MEM NEAA, 1% MEM Amino Acids, 1% MEM Vitamin solution. WI-38 (SV40 transformed fibroblast; Age: 3 months gestation fetus; Biopsy source: Lung; Tissue source: Lung; Gender: Female; Ethnicity: Caucasian) cells were cultured in Delbecco’s Modified Eagle Medium (DMEM) with 10% FBS. All the cells were maintained at 37 °C, 5% CO2 and 95% relative humidity (RH). ER stress was performed by incubating exponential growing cells with 2.5 μg/ml tunicamycin.
2.3. MDC staining
AG11395 and GM00637 cells (2 × 104) were seeded in 35 mm plate for overnight. After 16 h cells were treated with 2.5 μg/ml tunicamycin for different time points (2–24 h). The cells were then washed with 1× PBS thrice and incubated with 50 mmole/L MDC for 10 min at 37 °C. After washing with 1× PBS the cells were mounted on glass slides and viewed under a fluorescence microscope (Leica DM 2500).
2.4. Autophagic flux measurement
After 16 h of growth cells were transfected with pBABE-puro mCherry-EGFP-LC3 B using Xtrem gene as per manufacturer’s instructions (Roche). Next day cells were ER stressed with 2.5 μg/ml tunica-mycin for 8 h. Then cells were washed with 1×PBS thrice and mounted on glass slides followed by fluorescence microscopic observation (Leica DM 2500).
Autophagic flux of GM00637 and AG11395 and AG11395 cell transfected with full length wild type WRN and WRN Δ Acidic was also measured using anti-p62 antibody.
2.5. Western bloting
Whole cell lysates were prepared from the cells with lysis buffer containing 1% Triton X-100, 50 mM NaCl, 50 mM NaF, 20 mM Tris (pH 7.4), 1 mM EGTA, 1 mM sodium vanadate, 0.2 mM PMSF, 0.5% NP-40 and protease inhibitors (Bio vision). The supernatant was collected and protein concentration was estimated using Bradford’s reagent. Cell lysates containing equal amount of protein (80 μg) were solubilized in Lamellae buffer, boiled for 5 min, and electrophoresed on a 12% SDS-polyacrylamide gel in Tris-glycine buffer (pH 8.8). Proteins were then transferred to polyvinylidine difluoride (PVDF) membrane (Bio-Rad). Nonspecific binding was blocked with 5% non-fat dry milk and 0.05% Tween-20 in 20 mM Tris-Cl, pH 7.6 (TBS-T). After incubation with the appropriate primary antibody, membrane was washed with TBS-T and blot was reincubated with secondary antibodies conjugated with horse radish peroxidase (HRP). Bound antibodies were detected by the ECL detection reagent (Santa Crutz).
2.6. Cloning
The deletion of acidic domain from WRN (WRN A Acidic) was generated by inverse PCR using the pEGFP-C3WRN (1–1432) encoding plasmid as the template using final concentration 0.5 pM forward primer 5’-ATAGTGGCACGGTAGAACCAA-3′ and reverse primer 5’-GGGAGATAAATGCTCAGTAGA-3′, 0.02 U/pl Q5 High-Fidility DNA Polymerase (NEB) with 200 pM dNTPs. Total PCR protocol consisted of 35 cycle using reaction condition as initial denature at 98 °C for 30 secconds, denature at 98 °C for 10 s, annealing at 58.5 °C for 30 s, extention at 72 °C for 4.5 min, final extention 72 °C for 2 min, hold at 4 °C for 5 min on Biorad. Purification of PCR product and making blunt end using klenow enzyme as per manufacturer’s instructions. Again, purification of product and then ligation was done with T4 DNA ligase as per manufactuer’s protocol. 0.8 or 1.5% agarose gel monitoring was done after each step. Deletion of was confirmed after sequencing using chromas_lite software by Xcelris Lab Ltd., (India).
2.7. Statistical analysis
Two-tailed Student’s t-test was used to evaluate the statistical differences between the groups. Here P < 0.05 was considered as statistically significant. Error bars represent the means ± SD for all plots. Data analysis was performed using the Microsoft Office 2007 software.
3. Results
3.1. WS cells show lower endoplasmic reticulum stress induced autophagy
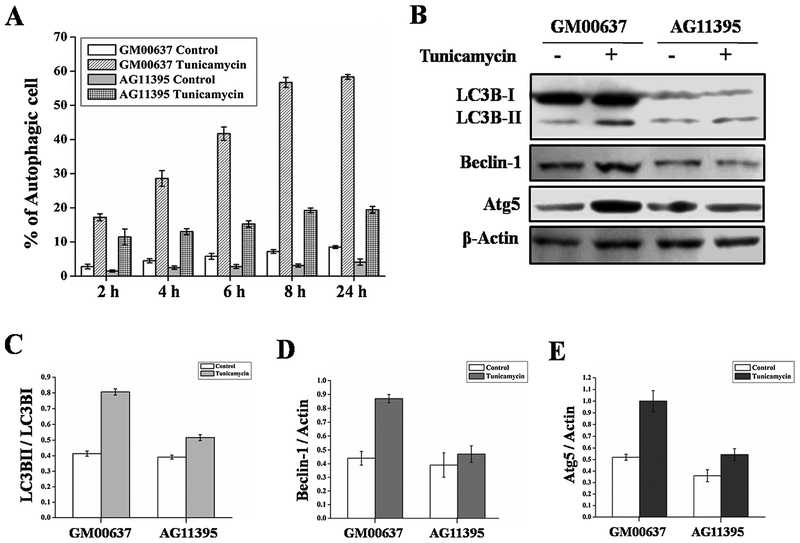
Endoplasmic reticulum (ER) stress induced autophagy was measured in WS cell (AG11395) and in normal skin fibroblast cells (GM00637) by MDC (Monodansylcadaverine) staining. ER stress was induced by incubating the cells with 2.5 μ g/ml tunicamycin. Cells were collected at different time points (Fig. 1A) and stained for autophagic vacuoles with MDC followed by fluorescence microscopic analysis. As seen in Fig. 1A, following ER stress there were fewer autophagic AG11395 cells than GM00637 cells at all time points tested. Further, we performed immunoblot from total cellular extract of WS and normal cell lines and analysed for autophagy marker proteins. As seen in Fig. 1B, the band corresponding to LC3BII is more intense in the normal cell line after ER stress compared to WS cells. The ratio of intensity LC3BII and LC3BI is shown in Fig. 1C. Similarly, autophagy marker proteins Beclin-1 and Atg5 increased more after ER stress in GM00637 than in WS cells (Fig. 1D and 1E respectively).
Fig. 1.
Normal fibroblast cell (GM00637) and Werner syndrome fibroblast cell (AG11395) were treated with ER stress inducer tunicamycin. Control cells were incubated with complete medium. (A) Graphical representation of MDC staining at different time points 2–24 h of all the cells. More than 600 cells were analysed for each condition. (B) Immunobloting of total cellular extract for autophagy marker proteins of normal fibroblast GM00637 and WS cell AG11395. (C) Graphical representation of ratio LC3BII and LC3BI of both cell lines. (D) Graphical representation of ratio Beclin-1 and Actin of both cell lines. (E) Graphical representation of ratio Atg5 and Actin of both cell lines. 3 western blots from 3 separate sets of experiments were used.
3.2. Transient expression of WRN rescues autophagy in WS cells
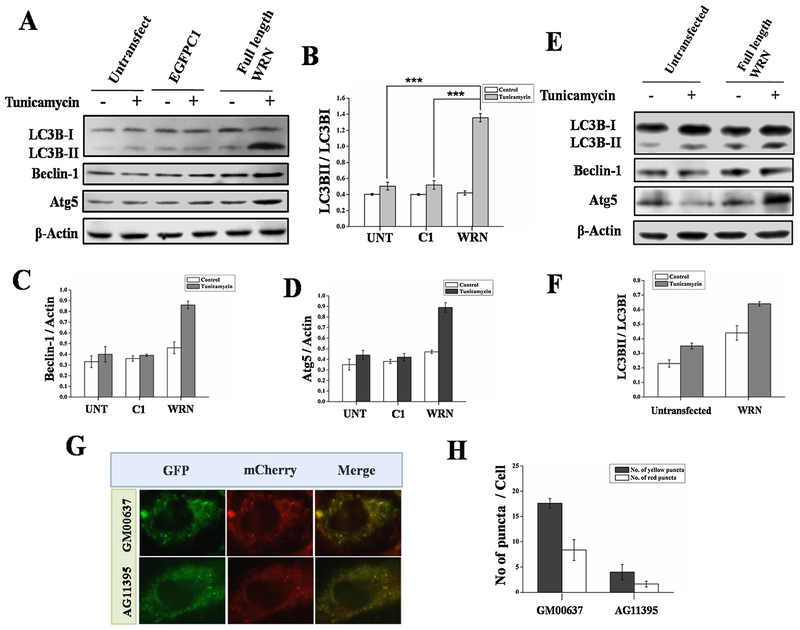
As seen in Fig. 2A and E, when we over-expressed WRN in WS cells (AG11395 and AG05229), respectively, ER stressed induced autophagy was restored as determined by the expression of autophagic marker proteins. The ratio of LC3BII and LC3BI is plotted for both the sets (Fig. 2B and F) and the level of Beclin-1, and Atg5 compared to ß-actin are plotted in (Fig. 2C and D respectively). Further, in WS cells the total amount of autophagic vacuole formation is much less than in normal fibroblasts, determined after transfecting the cells with mCherry-EGFP-LC3 plasmid followed by ER stress (Fig. 2G). As seen in (Fig. 2H), the ratio of yellow to red dots in GM00637 (0.47) and AG11395 (0.41) cells were almost similar indicating that the rate of subsequent fusion with acidic lysosomes was similar while the total amount of autophagic vacuoles were fewer in AG11395 cells compared to normal.
Fig. 2.
(A) AG11395 cells were transfected with empty vector (EGFPC1) or full length WRN plasmid and go for ER stress for 8 h. Total cell lysate was immunoblotted with anti-LC3B, anti-Beclin-1, anti-Atg5 antibodies. Here β-Actin was used as a loading control. (B–D) Bands intensity were measured and graphically represent the ratio of LC3B-II and LC3B-I, Beclin-1 and Atg5 at different time point respectively. (E) AG05229 cells were tranfected with full length WRN plasmid and go for ER stress for 8 h. Total cell lysate was immunoblotted with anti-LC3B, anti-Beclin-1, anti-Atg5 antibodies. Here β-Actin was used as a loading control. (F) Graphically represent the ratio of LC3B-II and LC3B-I. Normal (GM00637) and WS (AG11395) cells were transfected with pBABE-puro mCherry-EGFP-LC3 B and ER stressed for 8 h. (G) mCherry and EGFP signal were observed under fluorescence microscopy. Atleast 450 cells were examined for different conditions. (H) Graphical representation of autophagic flux of all cell lines. 3 western blots from 3 separate sets of samples were used. *** p < 0.0005, mean ± SD, n=3.
3.3. Functional domain of WRN responsible for the restoration of autophagic responses
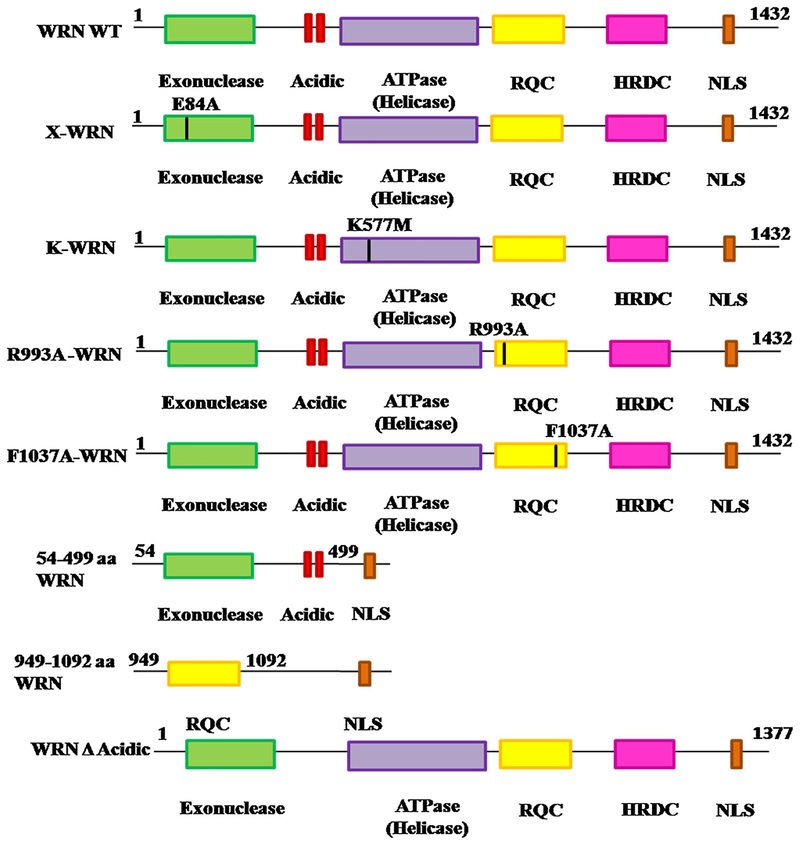
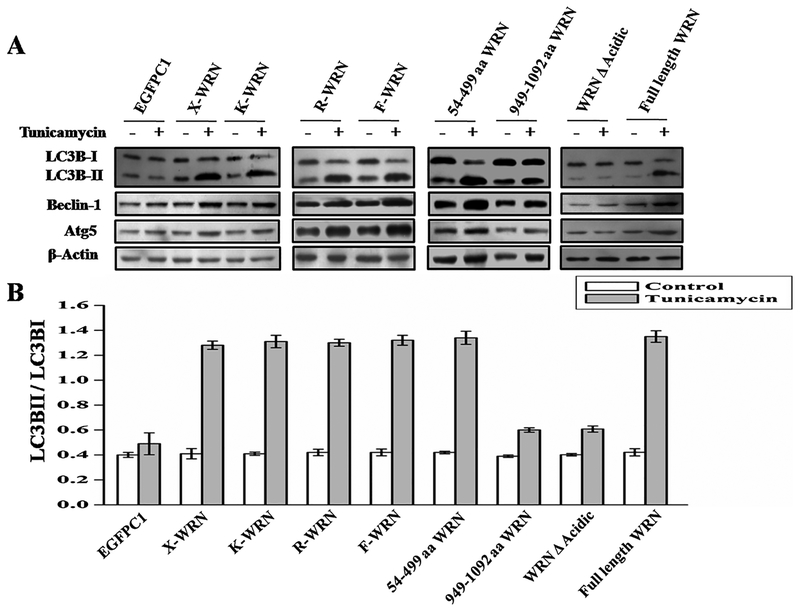
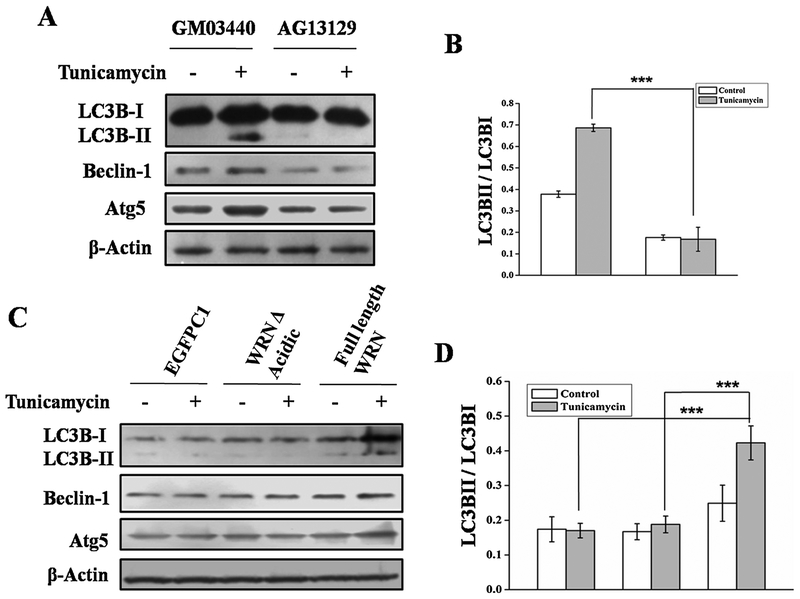
We then evaluated the role of various catalytic domains of WRNp in autophagy. Different clones of mutant WRN were used (Fig. 3). X-WRN (mutation at E84A) and K-WRN (mutation at K577M) is defective in exonuclease and helicase activity respectively [17]. Over expression of those mutants in WS cells resulted in ER stress induced autophagy similar to wild type WRN indicating that helicase and exonuclease functions of WRNp are not essential for autophagy (Fig. 4A). Similar results were observed for all the mutant WRN fragments including the one containing the RQC domain (949–1092 aa), which is known as interacting domain of WRNp. Interestingly, the fragment of WRN containing 54–499 aa, which encompasses the acidic and exonuclease domains were able to induce autophagy like wild type WRN (Fig. 4A). Finally, a deletion of the acidic domain (428–482 aa) was constructed from full length wild type WRN with the help of inverse PCR. Introduction of this WRN deleted acidic domain (WRN A Acidic) into cells was not able to induce autophagic proteins after ER stress in WS (Fig. 4A) and resulted in decreased expression of autophagy marker protein after ER stress (Fig. 4A and B). Expression levels of different WRN vectors in WS AG11395 cell line are shown in the supplementary S2.
Fig. 3.
Schematic diagram representing wild type WRN and different point and deletion mutant WRN used in this study.
Fig. 4.
AG11395 cells were transfected with different WRN mutant or fragmented WRN or full length WRN. Cells were then treated with tunicamycin for 8 h along with complete medium. (A) Immunobloting of total cellular extract for autophagy marker proteins. (B) Graphical representation of ratio LC3BII and LC3BI of all conditions. 3 western blots from 3 separate sets of samples were used.
3.4. Normal autophagic flux
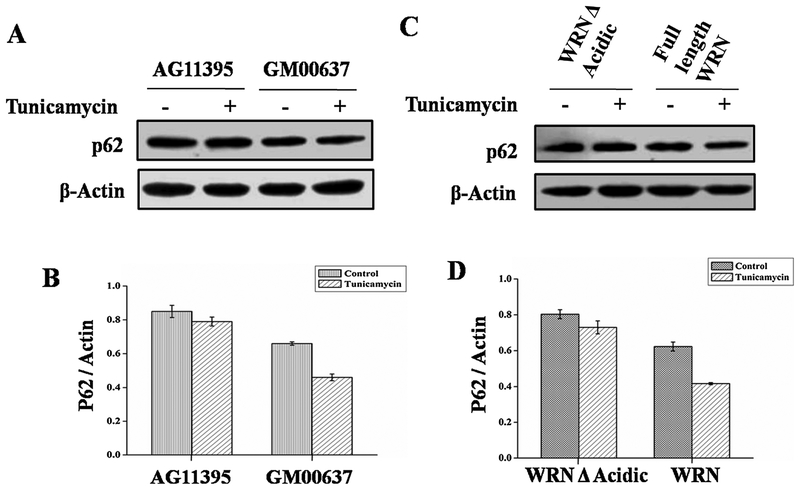
As p62 protein plays an important role in autophagy flux [41], we checked p62 protein status by immunobloting whole cell extract from WS and normal cell lines (Fig. 5A). The amount of p62 did not change in WS after ER stress but in normal fibroblast the amount of p62 decreased after ER stress (Fig. 5A and B). The higher amount of p62 in WS may be due to the impairment of early steps in autophagy. Similarly, when WS cells were transfected with full length WRN and WRN A acidic the amount of p62 increased after tunicamycin treatment in cells transfected with WRN A acidic only (Fig. 5C and D) indicating the role of the acidic domain in autophagy mediated clearance due to autophagy induction.
Fig. 5.
AG11395 and GM00637 cell lines were treated with ER stressor tunicamycin for 8 h along with complete medium. (A) Immunobloting of total cellular extract for autophagy flux marker protein p62. (B) Graphical representation of ratio of p62 and Actin. (C) AG11395 cell was transfected with wild type WRN and WRN Δ Acidic. Whole cell lysate was immunoblotted with anti-p62. (D) Graphical representation of p62 and Actin ratio. Here β-Actin was used as a loading control. 3 western blots from 3 separate sets of samples were used.
3.5. WRN restores ER stress induced autophagy in aged fibroblast
To determine the effect of full length WRNp in the aging process we used normal young (AG13129) and old (GM03440) fibroblast. As expected aged fibroblast showed reduced autophagy after ER stress compared to young fibroblast (Fig. 6A and B). When we expressed full length WRN in aged fibroblast, ER stress induced autophagy was restored (Fig. 6C and D). But when we transfected with WRN Δ Acidic, ER stress induced autophagy was not restored, indicating the significant role of the acidic domain of WRN.
Fig. 6.
Unaffected normal young (GM03440) and aged (AG13129) fibroblast were treated with ER stressor tunicamycin for 8 h along with complete medium. (A) Total cell lysate was immunoblotted with anti-LC3B, anti-Beclin-1, anti-Atg5 antibodies. Here β-Actin was used as a loading control. (B) Bands intensity were measured and graphically represent the ratio of LC3B-II and LC3B-I. (D) Graphical representation of represent the ratio of LC3B-II and LC3B-I. 3 western blots were examined for each condition. *** p < 0.0005, mean ± SD, n=3.
3.6. WRN acidic domain can influence the expression of other proteins
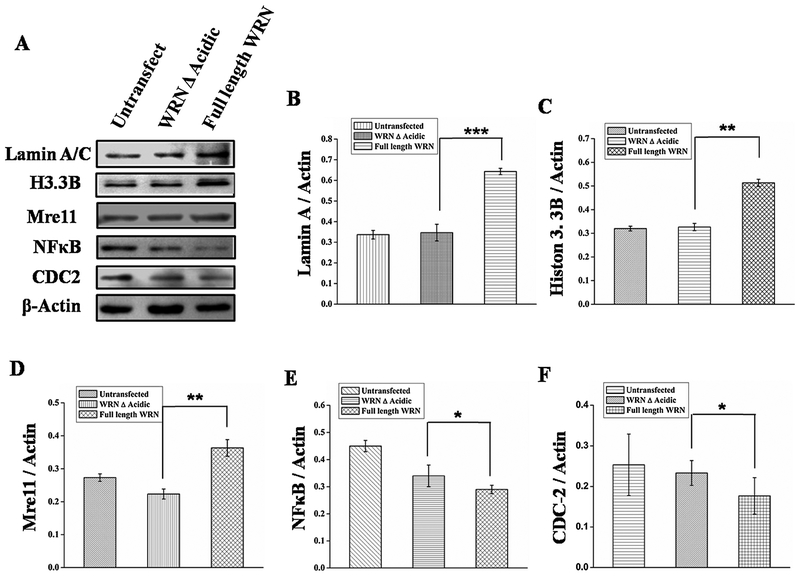
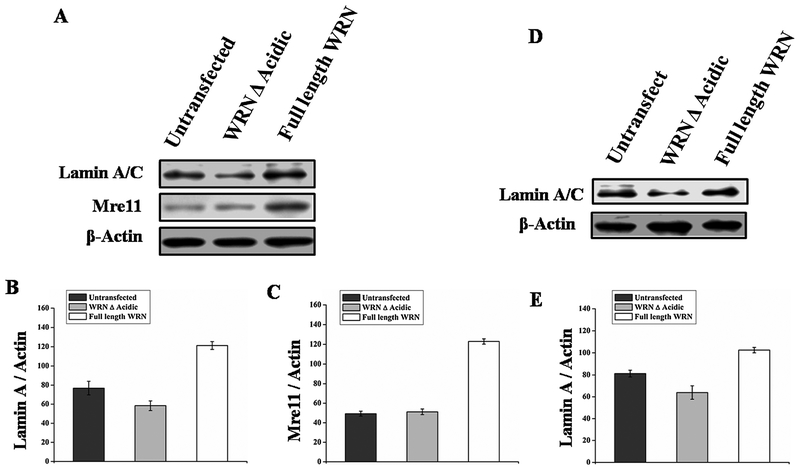
After finding a unique property of the acidic domain we assessed whether the increased expression of proteins due to the presence of the acidic domain is restricted to autophagy or whether it has involvement in other cellular processes. Some of the proteins including Lamin A/C, Mre11, Histone 3 B were found to have reduced expression in WS cells [19,6] and proteins like Lamin A/C, Mre11, Histone 3B are down regulated with aging [42–46] as seen in Fig. 7, transfection of full length WRN in WS cells resulted in enhanced expression of proteins Lamin, Mre11, Histone 3 B and decreased NFkB, CDC2. Simultaneously, it was observed that Lamin and Mre11 are upregulated after transient overexpression of WRN in normal fibroblast GM00637 (Fig. 8A). Moreover, upregulation of Lamin after WRN overexpression in WI38 cell (Fig. 8D) indicating overexpression of WRN can also enhance protein expression on WRN positive background.
Fig. 7.
AG11395 cell was transfected with wild type WRN and WRN A Acidic. Whole cell extract was immunoblotted with different proteins like anti-Lamin, anti-H3.3B, anti-Mre11, anti-NF-κB and anti-CDC2. Here ß-Actin was used as a loading control. (B-F) represents Graphical representation of the ratio of Lamin A, Histone 3.3B, Mre11, NFkB and CDC2 with actin respectively. 3 western blots were examined for each condition. * p < 0.05, ** p < 0.005, *** p < 0.0005 mean ± SD, n = 3.
Fig. 8.
Normal fibroblast GM00637 and WI-38 cell was transfected with wild type WRN and WRN Δ Acidic (A and D). Whole cell extract was immunoblotted with different proteins like anti-Lamin, anti-Mre11, Here β-Actin was used as a loading control. (B–C) represents graphical representation of the ratio of Lamin A, and Mre11. (E) Represents graphical representation of the ratio of Lamin A in WI-38 cell.
3.7. Inhibition of WRNp using siRNA decreases expression of age associated genes
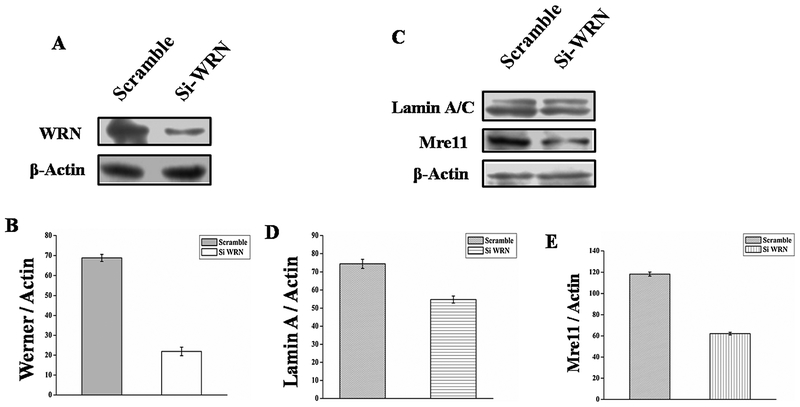
Finding that full length WRNp resulted in upregulation of some age associated genes in AG11395 cells, we wanted to determine the cellular fate after depleting WRN from WI-38 cells. Using siRNA we knocked down about 70% of WRNp in WI-38 cells (Fig. 9A and B). Protein levels of Lamin A/C and Mre11 were decreased in siRNA treated WI-38 cells compared to scrambled siRNA (Fig. 9C) in complete medium. The ratio of different proteins with actin in siRNA and scramble RNA treated WI-38 cells were compared and plotted (Fig. 9D Lamin A/C: Actin), (Fig. 9E Mre11: Actin).
Fig. 9.
(A) Silencing of WRN protein using WRN siRNA in Wi-38 cell. (A) Western blot analysis of expression of WRN protein in siWRN and scrambled siRNA transfected cell. (B) Graphical representation of inhibition of WRN protein. ** p < 0.005, mean ± SD, n=3. (C) After transfection with siWRN or scrambled siRNA WI-38 cells were allowed to grow in complete medium for 24 h. Western blot of total cell lysate using anti-Lamin A/C, anti-Mre11 antibodies. Here β-Actin was used as a loading control. (D–E) Quantitfication of band intensity was performed using Image J. ** p < 0.005, mean ± SD, n=3.
4. Discussion
In this study we find that WS cells display reduced ER stress induced autophagy. Transfection with full length WRN or the 54–499 aa fragment of WRN rescued the ER stress induced autophagy in WS cell by upregulating autophagic proteins. Moreover, this acidic domain can also influence the expression of other proteins not apparently related to autophagy.
In our previous work we found that WS cells are deficient in starvation induced autophagy and that transfection of full length WRN resulted in restoration of autophagy [39]. ER stress induced damage in different proteins and organelles invokes autophagy in order to maintain cellular homeostasis by clearing damaged proteins or organelles. Thus, the absence of autophagy results in accumulation of damaged proteins and the development of pathophysiological features including aging. Most of the work on WRNp has addressed its role in DNA repair, replication and how different domains of WRNp attributed to distinct functions, which are important for cellular metabolism. In this study we have identified that the acidic region of WRNp plays an important role in the regulation of autophagy. This acidic region was previously shown to stimulate RNA pol II and hence associated with transcription. Also, the role of WRNp in the transcription process has remained unclear, but overexpression of WRN in WS cells caused enhanced expression of different proteins including some autophagy related ones (LC3B, Atg5 and Beclin 1) [39]. Involvement of the acidic region of WRNp in transcription has been reported [13,14]. Here we also document that WRN transfection in WS cells resulted in increased expression of not only autophagy related proteins but also of others relevant to the aging process. Moreover, intense investigation showed lack of autophagy response in normal aged fibroblast [47]. From our study it seems that the presence of WRNp not only restores autophagy in WS cells but also in normal aged fibroblasts. This emphasizes the central role of WRN in aging. It is tempting to speculate that lack of autophagy may be associated with aging and that WRN may be involved in the process by upregulating some autophagy related proteins and thus accelerated aging becomes a hallmark of WS patients. Additionally, in the normal aging process, WRN may be inactivated as its catalytic functions are regulated by different modulations which are also affected in aged cells. These include post translational modifications including acetylation [48–50] phosphorylation [51,52] and oxidation [53].
IREα is a marker of ER stress mediated accumulation of unfolded proteins. A recent study showed a higher amount of IREa and of phosphorylated IREα in WRN null or defective WRN mice compared with wild type animals [54]. Hac1 is a transcription factor, which encodes proteins involved in the unfolded protein response (UPS) [55]. Inhibition of Hac1 blocks atg8 processing and prApe1 maturation thus inhibits autophagy [56]. WS cells are deficient in RNA pol I and II mediated transcription of various transcription factors. It has been known that there is a regulatory link between ER associated degradation (ERAD) and the UPR [57]. ERAD is leading mechanism which can handle misfolded protein [58] during ER stress. After ERAD, unfolded proteins that accumulate in the ER lumen are transported into the cytosol and then degraded by the proteasomes [59]. Ws cells cannot mitigate ER stress as the proteosomal protein level is low in these cells [6].
Recently, a yeast two-hybrid system study showed that WRN is covalently attached with SUMO-1 and that Ubc9 is an interactive partner of the N-terminal segment of WRNp [60]. Here, we found that the N-terminal fragment (54–499 aa) of WRN can rescue autophagy. Interestingly, Ubc9 mediated SUMOylation is sufficient to upregulate autophagy in cardiac myocytes [61]. We have demonstrated that the lower expression of Beclin-1 and Atg5 in WS cells after ER stress can be complemented by transfection with full length wild type WRN or its 54–499 aa fragment. Accordingly, deletion of this acidic domain (WRN Δ Acidic) from wild type WRN renders it incapable of upregulating autophagic proteins like LC3, BECN1, ATG-5 and thus autophagy.
Further, we observed that inhibition of WRN using siRNA resulted in diminished age related proteins LaminA/C and Mre11. Interestingly, we found that the acidic domain containing full length WRN can up-regulate the protein levels of Lamin A/C by 1.9 fold, Mre11 by 1.3 fold compared to WRN Δ acidic. Our results corroborate previous work, which showed that WRN knock down from cells decreased the amount of Lamin A/C and Mre11 in normal human fibroblast cells [19]. In Hutchinson-Gilford progeria syndrome (HGPS), mutation in Lamin A/C is well documented, though the involvement of Lamin A/C in the progression of the diseases is not yet known [42]. Involvement of Mre11 is important for DNA DSB repair and telomere length maintaining. Accumulation of damaged DNA may be another explanation for aging and age related diseases. Replicative senescence associated with telomere shortening is well documented and high expression of Mre11 along with other proteins may be essential for maintenance of telomeres, particularly during stress, to avoid faster aging [43]. Lack of Mre11 or some other DNA repair proteins may be cause of improper DNA repair. Additionally, we also found an increase of H3 histone family 3B protein by 1.5 fold after transfection with the acidic domain containing full length WRN. Interesting, experiments in budding yeast (Saccharomyces cerevisiae) reveal increased histone H3 supply by inactivation of the Hir (histone information regulator) complex or that overexpression of histones H3 dramatically extends lifespan [44]. Additionally, H3.3 is essential for the lifespan extension of Caenorhabditis elegans [45]. It has been shown that old human cells have reduced histone synthesis compared to young cells [46]. On the other hand, we found that WS contains a higher amount of CDC2 and NFkB protein. Introduction of full length WRNp results decreased amount of expression of these protein. Previous study also stated excess expression of CDC2 in WS [6]. Cdc2 is essential for cell cycle progression and remarkable reduction is connected with senescence. We found reduction of CDC2 after transfection of full length WRN. Lowering of CDC2 may cause of cell cycle arrest. Simultaneously, CDC2 inhibition is connected with autophagic cell death [62]. It was already noticed that the NF-κB pathway is upregulated in WS. Introduction of full length WRN reduces the amount of NFkB [19]. Transcriptional activity of NF-κB is increased in a variety of tissues with aging and is associated with many age-related degenerative diseases including Alzheimer’s, diabetes and osteoporosis [63].
Thus, taken together, it is possible that the acidic domain of WRNp helps to maintain the proper cellular function by upregulation of autophagy via the interaction with Ubc9 and or concomitant with trans activation of autophagic genes through its acidic domain.
5. Conclusion
Our report finds a relationship between WRNp and ER stress induced autophagy. WS cells are less responsive in autophagy. Full length wild type WRN can restore autophagy in WS cells. WRN knock down from normal fibroblast cells resulted downregulation age associated proteins Lamin A/C and Mre11. On the other hand, deletion of acidic domain of WRN shows reduced autophagy concomitant with down regulation of Beclin-1, Atg5 and LC3B. Further, the acidic domain has a global transcription property. Thus, we suggest that the acidic domain of WRNp is key for its transcriptional activity. Thus WRNp positively regulates autophagy related proteins and promotes autophagy to prevent cellular aging.
Supplementary Material
Acknowledgement
The authors would like to acknowledge for financial support for this research work the Council for Scientific and Industrial Research (CSIR), project no: [37(1673)/16/EMR-II]] and UGC, Government of India funded UPE-II programme of Jadavpur University. Also thankful to DST purse phase-II programme for the scholar fellowship. We are thankful to Mr. Subhadeep Das (Department of Life Science and Biotechnology, Jadavpur University, Kolkata, India) for his assistant. We sincerely acknowledge Dr. Jayanta Debnath (Department of Pathology, University California, San Francisco) for pBABE-puro mCherry-EGFP-LC3B plasmid.
Abbreviations:
- ATG
autophagy-related gene
- ERAD
endoplasmic reticulum associated degradation
- MDC
monodansylcadaverine
- mCherry-EGFP-LC3
mCherry enhanced green fluorescent protein-microtubule-associated protein-1 light chain 3
- WS
Werner syndrome
- WRNp
Werner protein
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.10167j.dnarep.2018.05.003.
References
- [1].Bachrati CZ, Hickson ID, RecQ helicases: suppressors of tumorigenesis and premature aging, J. Biol. Chem 374 (2003) 577–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Croteau DL, Popuri V, Opresko PL, Bohr VA, RecQ helicases in DNA repair, recombination, and replication, Annu. Rev. Biochem 83 (2014) 519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hickson ID, RecQ helicases: caretakers of the genome, Nat. Rev. Cancer 3 (2003) 169–178. [DOI] [PubMed] [Google Scholar]
- [4].Rezazadeh S, RecQ helicases; at the crossroad of genome replication, repair, and recombination, Mol. Biol. Rep 39 (2012) 4527–4543. [DOI] [PubMed] [Google Scholar]
- [5].Ozgenc A, Loeb LA, Current advances in unraveling the function of the Werner syndrome protein, Mutat. Res 577 (2005) 237–251. [DOI] [PubMed] [Google Scholar]
- [6].Kyng KJ, May A, Kolvraa S, Bohr VA, Gene expression profiling in Werner syndrome closely resembles that of normal aging, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 12259–12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Machwe A, Ganunis R, Bohr VA, Orren DK, Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA, Nucleic Acids Res. 28 (2000) 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brosh RM Jr., Karmakar P, Sommers JA, Yang Q, Wang XW, Spillare EA, Harris C, Bohr VA, P53 modulates the exonuclease activity of werner syndrome protein, J. Biol. Chem 276 (2001) 35093–351029. [DOI] [PubMed] [Google Scholar]
- [9].Laine JP, Opresko PL, Indig FE, Harrigan JA, Kobbe CV, Bohr VA, Werner protein stimulates topoisomerase I DNA relaxation activity, Cancer Res. 63 (2003) 7136–7146. [PubMed] [Google Scholar]
- [10].Opresko PL, Cheng WH, Bohr VA, Junction of RecQ helicase biochemistry and human disease, J. Biol. Chem 279 (2004) 18099–18102. [DOI] [PubMed] [Google Scholar]
- [11].Kamath-Loeb AS, Johansson E, Burgers PMJ, Loeb LA, Functional interaction between the Werner Syndrome protein and DNA polymerase delta, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kamath-Loeb AS, Shen JC, Loeb LA, Fry M, Werner syndrome protein II: characterization of the integral 3′→5′ DNA exonuclease, J. Bio. Chem 273 (1998) 34145–34150. [DOI] [PubMed] [Google Scholar]
- [13].Ye L, Nakura J, Morishima A, Miki T, Transcriptional activation by the Werner syndrome gene product in yeast, Exp. Gerontol 33 (1998) 805–812. [DOI] [PubMed] [Google Scholar]
- [14].Balajee AS, Machwe A, May A, Gray MD, Oshima J, Martin GM, Nehlin JO, Brosh RM Jr., Orren DK, Bohr VA, The Werner syndrome protein is involved in RNA polymerase II transcription, Mol. Biol. Cell 10 (1999) 2655–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J, The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease, Nat. Genet 20 (1998) 11411–11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Aggarwala M, Sommersa JA, Shoemakerb RH, Brosh RM Jr., Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress, Proc. Natl. Acad. Sci. U. S. A 108 (2000) 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Machwe A, Ganunis R, Bohr VA, Orren DK, Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA, Nucleic Acids Res. 28 (2000) 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tadokoro T, Kulikowicz T, Dawut L, Croteau DL, Bohr VA, DNA binding residues in the RQC domain of Werner protein are critical for its catalytic activities, Aging (Albany NY) 4 (2012) 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Turaga RV, Paquet ER, Sild M, Vignard J, Garand C, Johnson FB, Masson JY, Lebel M, The Werner syndrome protein affects the expression of genes involved in adipogenesis and inflammation in addition to cell cycle and DNA damage responses, ABBV Cell Cycle 8 (2009) 2080–2092. [DOI] [PubMed] [Google Scholar]
- [20].Lachaud AA, Auclair-Vincent S, Massip L, Audet-Walsh E, Lebel M, Anderson A, Werner’s syndrome helicase participates in transcription of phenobarbital-inducible CYP2B genes in rat and mouse liver, Biochem. Pharmacol 79 (2010) 463–470. [DOI] [PubMed] [Google Scholar]
- [21].Klionsky DJ, Emr SD, Autophagy as a regulated pathway of cellular degradation, Science 290 (2000) 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rubinsztein DC, Marin G, Kroemer G, Autophagy and aging, Cell 146 (2011) 682–695. [DOI] [PubMed] [Google Scholar]
- [23].Mizushima N, Autophagy: process and function, Genes Dev. 21 (2007) 2861–2873. [DOI] [PubMed] [Google Scholar]
- [24].Cuervo AM, Autophagy and aging: keeping that old broom working, Trends Genet. 12 (2008) 604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rubinsztein DC, Marino G, Autophagy and aging, Cell 146 (2011) 682–695. [DOI] [PubMed] [Google Scholar]
- [26].Toth ML, Sigmond T, Borsos E, Barna J, Erdélyi P, Taka cs-Vellai K, Orosz L, Kovacs L, Csikos G, Sass M, Vellai T, Longevity pathways converge on autop-hagy genes to regulate life span in Caenorhabditis elegans, Autophagy 4 (2008) 330–338. [DOI] [PubMed] [Google Scholar]
- [27].Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, Karin M, Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies, Science 327 (2010) 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Martinez-Vicente M, Cuervo AM, Autophagy and neurodegeneration: when the cleaning crew goes on strike, Lancet Neurol. 6 (2007) 352–361. [DOI] [PubMed] [Google Scholar]
- [29].Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T, Theautophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice, J. Clin. Invest 118 (2008) 2190–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Onofre I, Mendonça N, Lopes S, Nobre R, de Melo JB, Carreira IM, Januario C, Gonçalves F, Almeida LPD, Fibroblasts of Machado Joseph Disease patients reveal autophagy impairment, Sci. Rep 6 (2016) 28220–28230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Costa Mdo C, Paulson HL, Toward understanding Machado-Joseph disease, Prog. Neurobiol 97 (2012) 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kieling C, Prestes PR, Saraiva-Pereira ML, Jardim LB, Survival estimates for patients with Machado-Joseph disease (SCA3), Clin. Genet 72 (2007) 543–545. [DOI] [PubMed] [Google Scholar]
- [33].Mizushima N, Levine B, Cuervo AM, Klionsky DJ, Autophagy fights disease through cellular self-digestion, Nature 451 (2008) 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey C, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood R, Winslow AR, Rubinsztein DC, Regulation of mammalian autophagy in physiology and pathophysiology, Physiol. Rev 90 (2010) 1383–1435. [DOI] [PubMed] [Google Scholar]
- [35].Shiratori M, Suzuki T, Itoh C, Goto M, Furuichi Y, Matsumoto T, WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex, Oncogene 21 (2002) 2447–2454. [DOI] [PubMed] [Google Scholar]
- [36].Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B, Induction of autophagy and inhibition of tumorigenesis by beclin 1, Nature 402 (1999) 672–676. [DOI] [PubMed] [Google Scholar]
- [37].He C, Levine B, The beclin 1 interactome, Curr. Opin. Cell Biol. 2 (2014) 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen L, Mizushima N, Ohsumi Y, Cattoretti G, Levine B, Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene, J. Clin. Invest 122 (2003) 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Maity J, Bhor VA, Laskar A, Karmakar P, Transient overexpression of Werner protein rescues starvation induced autophagy in Werner syndrome cells, BBA Mol. Basis Dis. 1842 (2014) 2387–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D, An integrated stress response regulates amino acid metabolism andresistance to oxidative stress, Mol. Cell 11 (2003) 619–633. [DOI] [PubMed] [Google Scholar]
- [41].Zhang XJ, Chen S, Huang KX, Le WD, Why should autophagic flux be assessed? Acta Pharmacol. Sin 34 (2013) 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sliwinska MA, The role of lamins and mutations of LMNA gene in physiological and premature aging, Postepy Biochem. 53 (2007) 46–52. [PubMed] [Google Scholar]
- [43].Ju YJ, Lee KH, Park JE, Yi YS, Yun MY, Ham YH, Kim TJ, Hyun MC, Gwi JH, Lee JH, Lee J, Jong SH, Lee KM, Park GH, Decreased expression of DNA repair proteins Ku70 and Mre11 is associated with aging and may contribute to the cellular senescence, Exp. Mol. Med 38 (2006) 686–693. [DOI] [PubMed] [Google Scholar]
- [44].Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK, Elevated histone expression promotes life span extension, Mol. Cell 39 (2010) 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Piazzesi A, Papic D, Bertan F, Salomoni P, Nicotera P, Bano D, Replication-independent histone variant H3.3 controls animal lifespan through the regulation of pro-longevity transcriptional programs, Cell Rep. 17 (2016) 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Peleg S, Feller C, Ladurner AG, Imhof A, The metabolic impact on histone acetylation and transcription in ageing, Trends Biochem. Sci 41 (2016) 700–711. [DOI] [PubMed] [Google Scholar]
- [47].Pernodet N, Dong K, Pelle E, Autophagy in human skin fibroblasts: comparison between young and aged cells and evaluation of its cellular rhythm and response to ultraviolet A radiation, J. Cosmet. Sci 67 (2016) 13–20. [PubMed] [Google Scholar]
- [48].Kusumoto R, Muftuoglu M, Bohr VA, The role of WRN in DNA repair is affected by post-translational modifications, Mech. Ageing Dev. 128 (2007) 50–57. [DOI] [PubMed] [Google Scholar]
- [49].Lozada E, Yi J, Luo J, Orren DK, Acetylation of Werner syndrome protein (WRN): relationships with DNA damage, DNA replication and DNA metabolic activities, Biogerontology 15 (2014) 347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li K, Wang R, Lozada E, Fan W, Orren DK, Luo J, Acetylation of WRN protein regulates its stability by inhibiting ubiquitination, PLoS One 5 (2010) e10341–10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Karmakar P, Piotrowski J, Brosh RM Jr., Sommers JA, Miller SP, Cheng WH, Snowden M, Ramsden DA, Bohr VA, Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation, J. Biol. Chem 277 (2002) 18291–18302. [DOI] [PubMed] [Google Scholar]
- [52].Pichierri P, Rosselli F, Franchitto A, Werner’s syndrome protein is phosphorylated in an ATR/ATMdependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle, Oncogene 22 (2003) 1491–1500. [DOI] [PubMed] [Google Scholar]
- [53].Harrigan JA, Piotrowski J, Di Noto L, Levine RL, Bohr VA, Metal-catalyzed oxidation of the Werner syndrome protein causes loss of catalytic activities and impaired protein-protein interactions, J. Biol. Chem 282 (2007) 36403–36411. [DOI] [PubMed] [Google Scholar]
- [54].Aumailley L, Garand C, Dubois MJ, Johnson FB, Marette A, Lebel M, Metabolic and phenotypic differences between mice producing a werner syndrome helicase mutant protein and wrn null mice, PLoS One 10 (2015) e0140292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cox JS, Walter P, A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response, Cell 87 (1996) 391–404. [DOI] [PubMed] [Google Scholar]
- [56].Yorimitsu T, Nair U, Yang Z, Klionsky DJ, Endoplasmic reticulum stress triggers autophagy, J. Biol. Chem 281 (2006) 30299–30304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Spear ED, Ng DTW, Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways, Mol. Biol. Cell 14 (2003) 2756–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Meusser B, Meusser B, Hirsch C, Jarosch E, Sommer T, ERAD: the long road to destruction, Nat. Cell Biol. 7 (2005) 766–772. [DOI] [PubMed] [Google Scholar]
- [59].Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T, A regulatory link between ER-associated protein degradation and the unfolded-protein response, Nat. Cell Biol. 2 (2000) 379–384. [DOI] [PubMed] [Google Scholar]
- [60].Kawabe Y, Seki M, Seki T, Wang WS, Imamura O, Furuichi Y, Saitoh H, Enomoto T, Covalent modification of the Werner’s syndrome gene product with the ubiquitin-related protein, SUMO-1, J. Biol. Chem 275 (2000) 20963–20966. [DOI] [PubMed] [Google Scholar]
- [61].Gupta MK, McLendon PM, Gulick J, James J, Khalili K, Robbins J, UBC9-Mediated sumoylation favorably impacts cardiac function in compromised hearts, Circ. Res 118 (2016) 1894–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Motizuki M, Yokota S, Tsurugi K, Autophagic death after cell cycle arrest at the restrictive temperature in temperature-sensitive cell division cycle and secretory mutants of the yeast Saccharomyces cerevisiae, Eur. J. Cell Biol. 68 (1995) 275–287. [PubMed] [Google Scholar]
- [63].Tilstra JS, Clauson CL, Niedernhofer LJ, Robbins PD, kNF B in aging and disease, Aging Dis. 2 (2011) 449–465. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.