Abstract
Mechanical force plays pivotal roles in vascular development during tissue growth and regeneration. Nevertheless, the process by which mechanical force controls the vascular architecture remains poorly understood. Using a systems bioengineering approach, we show that intercellular tension negatively regulates tip cell formation via Notch1-Dll4 signaling in mouse retinal angiogenesis in vivo, sprouting embryoid bodies, and human endothelial cell networks in vitro. Reducing the intercellular tension pharmacologically by a Rho-associated protein kinase inhibitor or physically by single cell photothermal ablation of the capillary networks promotes the expression of Dll4, enhances angiogenic sprouting of tip cells and increases the vascular density. Computational biomechanics, RNA interference, and single cell gene expression analysis reveal that a reduction of intercellular tension attenuates the inhibitory effect of Notch signaling on tip cell formation and induces angiogenic sprouting. Taken together, our results reveal a mechanoregulation scheme for the control of vascular architecture by modulating angiogenic tip cell formation via Notch1-Dll4 signaling.
Keywords: angiogenesis, tip cells, nanoparticles, Notch signaling, mechanoregulation
Graphical abstract
A systems bioengineering approach enabled by nanoparticle based single cell photothermal ablation and dynamic gene expression analysis is presented. Using the systems bioengineering approach, this study reveals the mechanoregulation of angiogenic sprouting is mediated by Notch1-Dll4 signaling angiogenic sprouting of endothelial tip cells and the resulting vascular architecture are regulated mechanically via Notch1-Dll4 signaling in mouse retinal angiogenesis in vivo, sprouting embryoid bodies, and human endothelial cell networks in vitro.
1. Introduction
Angiogenesis is the outgrowth of new capillaries from existing vasculature. The multicellular morphogenic process plays essential roles in tissue engineering and regenerative medicine. The formation of new blood vessels also contributes to numerous malignant, ischemic, inflammatory, infectious and immune disorders[1]. Identification of the regulatory mechanisms of angiogenesis will have direct implications in repairing/replacing damaged tissues and inhibiting pathological neovascularization. Biochemical factors (e.g., VEGF) and physical signals (e.g., fluid shear stress) in the microenvironment, for instance, are well-known regulators of capillary morphogenesis[2]. The cell-matrix mechanical interaction has been demonstrated to modulate vascular development[3]. In particular, cell traction force is shown to control the expression of VEGF receptor 2 (VEGFR2) in vitro and in vivo by applying an inhibitor of Rho signaling and modulating the ECM elasticity[4]. Tissue deformation modulates VEGF gradients and endothelial cell proliferation in deformable tissue constructs[5]. Endothelial cell traction and ECM density influence the formation and maintenance of capillary via a cell traction force associated mechanism[6]. Geometric control by microfabrication has also been demonstrated to control the capillary network topology via cell-matrix mechanical interactions[7]. Nevertheless, the mechanistic pathway by which mechanical forces regulate the capillary architecture remains poorly understood.
A key mechanism in the regulation of capillary architecture is angiogenic sprouting with a tip-stalk organization[8]. During capillary morphogenesis, endothelial cells adapt distinct tip cell and stalk cell phenotypes: tip cells in sprouting vessels extend active filopodia and lead the sprout outgrowth while stalk cells proliferate and elongate to support angiogenic sprouting. The density of tip cells is regulated via Notch1-Dll4 lateral inhibition[9]. The interaction between Notch1 and Dll4 limits the formation of tip cells and dynamically controls the proper ratio between tip cells and stalk cells for regulating the branching morphology and network architectures[10]. Notably, Notch signaling is shown to be force dependent using atomic force microscopy and optical tweezers[11]. Mechanical force is required to pull on the Notch receptor and expose the ADAM protease cleavage site for proteolysis and downstream signaling. The involvement of Notch signaling in the mechanoregulation of vascular development, however, has not been explored.
In this study, we hypothesize that intercellular tension regulates capillary architectures by controlling tip cell formation via Notch1-Dll4 signaling. Using a systems bioengineered approach enabled by single cell photothermal ablation and dynamic single cell analysis, we investigate the effects of mechanical force on vascular development in mouse retinal angiogenesis in vivo, sprouting embryoid bodies, and human endothelial cell networks in vitro. The intercellular tension is perturbed pharmacologically by applying cell traction force modulating drugs. We also physically disrupt the intercellular tension by establishing a single cell laser ablation technique. Single cell gene expression analysis and computational biomechanics are applied to elucidate the interplay between mechanical force and Notch1-Dll4 signaling in the regulation of tip cell formation and vascular development.
2. Results
2.1. Reducing intercellular tension by ROCK inhibition enhances tip cell formation
To explore the roles of intercellular tension on vascular development in vivo, we inhibited Rho-associated protein kinase (ROCK) by systemic administration of Y-27632 and investigated its effects on neonatal mouse retinal angiogenesis. ROCK signaling regulates the cell contractility by phosphorylating myosin light chain[12]. Y-27632 treatment led to hyperbranching and abnormalities of the vascular plexus (Figure 1A-B). The number of vascular branch points and vessel density increased significantly in the developing (but not the formed) region of the plexus (Figure 1C-D). At the developing front of the vascular plexus, Y-27632 treatment resulted in a hypersprouting phenotype. The number of sprouting tip cells with active filopodia increased significantly (Figure 1E). N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT), a γ-secretase inhibitor that blocks Notch receptor cleavage and signaling, was also included in the study. Notch1-Dll4 signaling is known to inhibit the formation of tip cells and control the vessel density during angiogenic development. Similar hypersprouting and vessel density increase were observed in DAPT treatment, which is consistent with the function of Notch signaling in restricting tip cell formation. Co-treatment of DAPT and Y-27632, however, did not further enhance hypersprouting, as indicated by the vessel density, branch points and tip cells (Figure 1C-E).
Figure 1. Inhibition of ROCK signaling enhances tip cell formation in vivo.
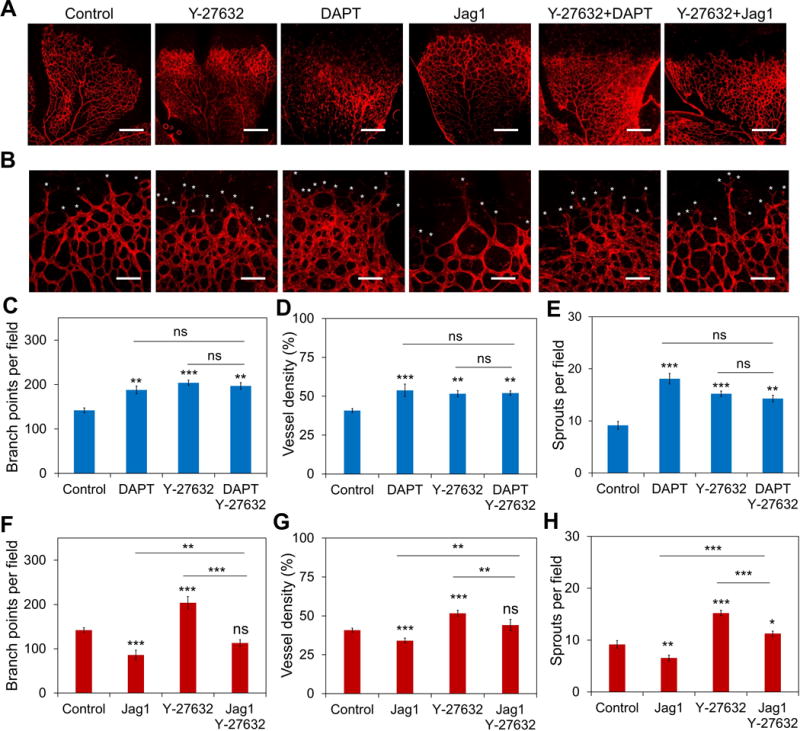
(A-B) Whole-mounted retinas stained with Isolectin B4. Mouse pups were treated with Y-27632 (10 μg/g), DAPT (50 μg/g), Jag1 (50 μg/g), Y-27632 + DAPT, and Y-27632 + Jag1 at P3, P4, P5, and P6 and sacrificed humanely at P7. Retinas were collected for staining. White asterisks indicate filopodial protrusions. Images are representative from three independent experiments. Scale bars, 400 μm in (a) and 100 μm in (b). (C-E) Quantification of the (C) branch point density, (D) vessel density, and (E) sprouts density with the treatment of Y-27632, DAPT, and Y-27632 + DAPT. (F-H) Quantification of (F) branch point density, (G) vessel density, and (H) sprouts density with the treatment of Y-27632, Jag1, and Y-27632 + DAPT. Data are expressed as mean ± s.e.m. (n = 5; ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
To further evaluate the effects of intercellular tension and Notch signaling on tip cell formation, the experiment was performed in human umbilical vein endothelial cell (HUVEC) networks in vitro and HUVEC spheroids in 3D gel culture (supplementary Figure S1-2). Hypersprouting, as indicated by the mean cord length of the network and the density of angiogenic sprouts, was observed in both models, supporting the notion that reducing intercellular tension by ROCK inhibition enhances tip cell formation (supplementary Figure S1A and S2A). Similar to the in vivo model, DAPT did not further increase the Y-27632 mediated tip cell enhancement in the in vitro models (supplementary Figure S1C and S2B). These results suggest DAPT and Y-27632 may modulate tip cell formation via similar mechanistic pathways.
Jag1 peptide, a Notch ligand that activates Notch signaling in vivo[9a, 9c], was administered to mouse pups from P3 to P6 to investigate the influence of Notch signaling in Y-27632 mediated hypersprouting (Figure 1A-B). In agreement with previous studies[9a, 9c], Jag1 peptide administration decreased the number of branch point and vessel density of the vascular plexus significantly (Figure 1F-G). The number of tip cells, as characterized by sprouting cells with active filopodia, also decreased significantly at the development front of the vascular plexus (Figure 1H). Co-treatment of Jag1 with Y-27632 reduced the effect of Y-27632 on the vascular architecture (Figure 1F-H). The Y-27632 mediated hyperbranching was attenuated by Jag1. The branch point density and vessel density were similar to control (Figure 1F-G). Furthermore, Jag1 attenuated the Y-27632 mediated tip cell enhancement (Figure 1H). The combined effects of Y-27632 and Jag1 were also examined in HUVEC networks in vitro and 3D HUVEC spheroids (supplementary Figure S1B,D and S3). Jag1 attenuated the effects of Y-27632 and partially restored the normal mean cord length of HUVEC networks in a dose dependent manner (supplementary Figure S1D-F). Similar inhibitory effects of Jag1 on Y-27632 mediated tip cell formation were also observed in 3D HUVEC spheroids (supplementary Figure S3B).
2.2. Reducing intercellular tension by ROCK inhibition increases Dll4 expression
The ability to attenuate the effects of ROCK inhibition on vascular development with a Notch ligand suggests that Y-27632 mediates its effect on tip cell formation via Notch1-Dll4 signaling. Immunostaining with confocal microscopy was performed to evaluate the expression of Dll4 in vascular plexuses (Figure 2A-B). The formation of mosaic or checker box patterns with alternating high and low Dll4 expressions were observed between tip and stalk cells in the sprouting front of vascular plexuses. With DAPT administration, the expression of Dll4 increased in both tip and stalk cells (Figure 2C-E). The Dll4 pattern became relative uniform near the sprouting front due to the attenuation of Notch inhibition by DAPT. Remarkably, reducing intercellular tension by Y-27623 had a similar effect on the Dll4 expression compared to DAPT treatment. Y-27632 disrupted the mosaic pattern and resulted in a uniform distribution of Dll4 near the sprouting front. Close examination of the sprouts confirmed the Dll4 expression was upregulated in both tip cells and stalk cells. In contrast, Jag1 treatment significantly reduced the expression of Dll4 in stalk cells.
Figure 2. Inhibition of ROCK signaling modulates Dll4 expression in vivo.
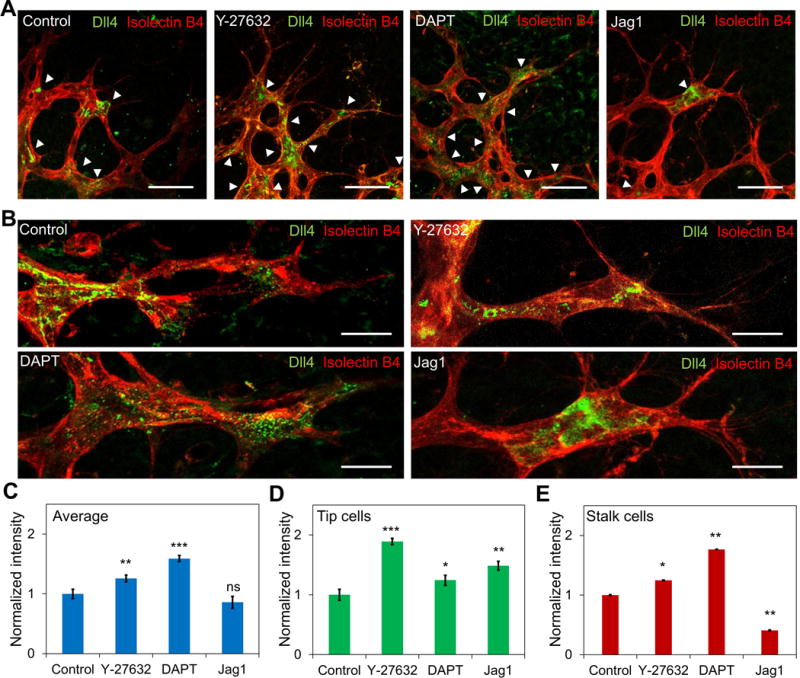
(A) Dll4 distribution at the sprouting plexus margin. Whole-mounted retinas were stained with Isolectin B4 (red) and Dll4 (green). Mice were treated with Y-27632, DAPT, and Jag1 at P3, P4, P5, and P6. White arrow heads indicate Dll4 expressing cells. Scale bars, 50 μm. (B) Confocal images of Dll4 distribution in retinal sprouts with different treatments. Images are representative from three independent experiments. Scale bars, 20 μm. (C-E) Quantification of the average Dll4 expression in (C) the sprouts, (D) tip cells, and (E) stalk cells under different treatments. Data are expressed as mean ± s.e.m. (n = 5; ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
2.3. Cell contractility negatively regulates tip cell formation in sprouting embryoid bodies
Y-27632 reduced the intercellular tension by decreasing the contractility of cells. To study the effects of cell contractility on tip cell formation and vascular architectures, several traction force modulating drugs, including Y-27632, blebbistatin, nocodazole and calyculin A, were applied to sprouting embryoid bodies (Figure 3). Y-27632, and myosin II inhibitor, blebbistatin that reduced the cell traction force enhanced angiogenic sprouting significantly (Figure 3A-B). Consistent with the in vivo experiments, reducing the cell traction force by Y-27632 or blebbistatin led to a hypersprouting phenotype. The sprouts branched out and formed dense interconnected networks with short sprouting tips at the front. The branching density was increased significantly compared to control (Figure 3B). To rule out the uncertainty due to the size of the embryoid body, the sprouting density was also estimated and confirmed the hypersprouting effect of Y-27632 and blebbistatin. In contrast, nocodazole and calyculin A, which enhanced the traction force, reduced the size of the embryoid bodies (Figure 3A-B). The number of sprouts and sprout density decreased significantly compared to control (Figure 3C). Unlike the interconnecting sprouts, distinct sprouts with very few branching were observed for embryoid bodies treated with nocodazole and calyculin A. Similar effects of the traction force modulating drugs were observed in HUVEC networks in vitro (supplementary Figure S4). Y-27632 and blebbistatin reduced the mean cord length while nocodazole and calyculin A increased the mean cord length. These results further support the notion that intercellular tension originated from the cell traction negatively regulates the formation of tip cells.
Figure 3. Mechanical force regulates tip cell formation in sprouting embryoid bodies.
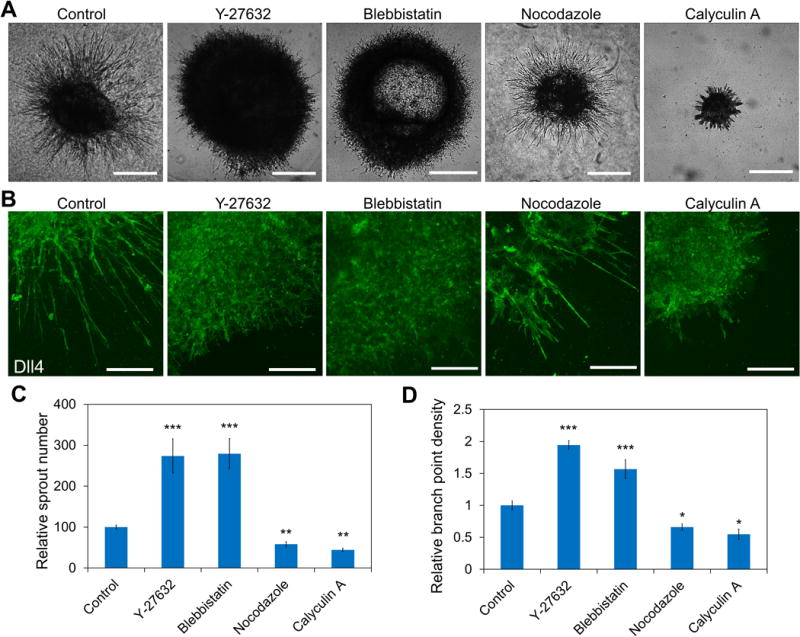
(A) Representative phase-contrast images of embryoid bodies with treatment of traction force modulating drugs. Scale bars, 500 μm. (B) Effects of traction force modulating drugs on the morphology of the sprouts. Cells were stained for Dll4. Confocal images are representative of five independent experiments. Scale bars, 200 μm. (C-D) Quantification of the sprout (C) densities and (D) lengths under the treatment of traction force modulating drugs. Data are expressed as mean ± s.e.m. (n = 5; ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
2.4. Mechanical force modulates Notch1-Dll4 signaling
The effects of intercellular tension on Notch1-Dll4 signaling were examined in sprouting embryoid bodies. Dll4 expression was monitored near the surface of the embryoid body (Figure 4A and supplementary Figure S5A). High levels of Dll4 were observed in the developing sprouts during the initial sprouting phase. Examining Dll4 distributions in the developed sprouts also revealed alternating levels of Dll4, similar to the checker box pattern in mouse retinal angiogenesis (Figure 4B and supplementary Figure S5b). With Y-27632 treatment, the level of Dll4 increased in the sprout in general. Close examination of the sprouts revealed Y-27632 treatment disrupted the alternating Dll4 patterns in the sprout and led to an even Dll4 distribution in the sprout (Figure 4B-C). Western blot analysis supported the negative effect of mechanical force on Notch1-Dll4 signaling (Figure 4D). Y-27632 treatment increased the expressions of Dll4 in embryoid bodies by 36.1±5.6% (Figure 4E). In contrast, nocodazole decreased Notch1 and Dll4 levels at the sprouts and downregulated the overall expression of Notch1 by 11.4±2.1% in the embryoid body.
Figure 4. Mechanical force modulates Dll4 expression in sprouting embryoid bodies.
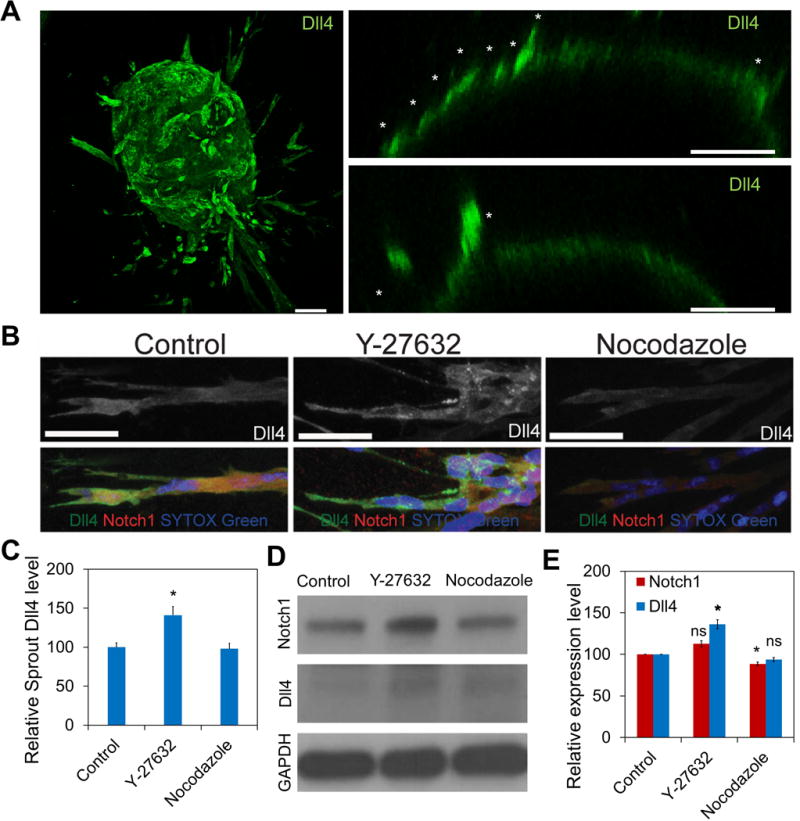
(A) Confocal image of a sprouting embryoid body (left). Sprouting embryoid bodies with Dll4-positive tip cells (white asterisks) near the surface of the embryoid body are indicated (right). Images are representative of five independent experiments. Scale bars, 100 μm. (B) Confocal images of Dll4 (red) and Notch1 (green) distributions in sprouts of embryoid bodies treated with Y-27632 and nocodazole. White asterisks indicate Dll4-positive cells in the sprout. Images are representative of three independent experiments. Scale bars, 50 μm. (C) Quantification of Dll4 expression in the sprout. (n = 6; ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test). (D-E) Western blot analysis of Notch1 and Dll4 expression in embryoid bodies at day 5. Gene expression was normalized to GAPDH. Data are representative from three independent experiments and are expressed as mean ± s.e.m. (n = 3; ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
The influences of intercellular tension on Notch1 and Dll4 expressions were further examined using HUVEC network in vitro (Figure 5A-B). Y-27632 treatment upregulated the expression of both Notch1 (46.4±11.6%) and Dll4 (179.7±10.5%) in HUVEC network. In contrast, nocodazole treatment downregulated the expression for Notch1 (22.5±4.4%) and Dll4 (30.7±8.9%). The negative effects of mechanical force on Notch1-Dll4 signaling were also confirmed at the transcription level using a live cell biosensor (Figure 5C and supplementary Figure S6)[13]. In particular, gold nanorod based single cell biosensors with specific locked nucleic acid probes were applied to detect the expressions of Notch1 and Dll4 mRNA. Single cell analysis minimized the uncertainty due to the pro-angiogenic and anti-angiogenic effects of the drugs on the morphology of the networks. As indicated by the single cell analysis, both Notch1 and Dll4 mRNA expressions were upregulated by Y-27632 and blebbistatin. Nocodazole and calyculin A, in contrast, downregulated the expressions of Notch1 and Dll4.
Figure 5. Mechanical force modulates Dll4 expression in HUVEC network in vitro.
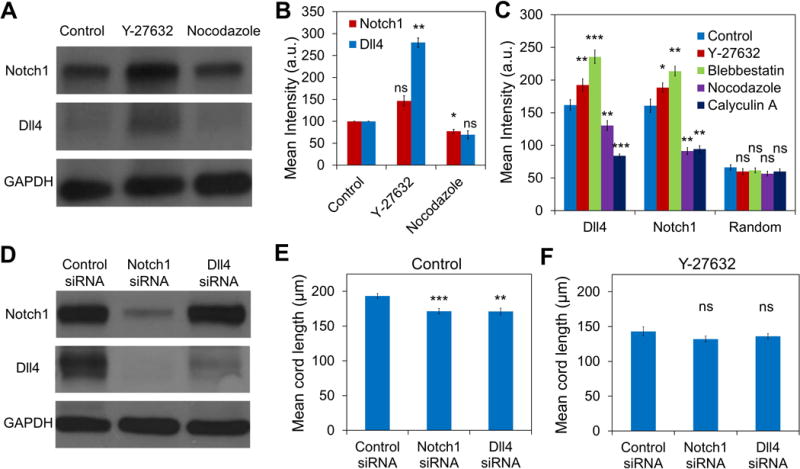
(A-B) Western blot analysis of Notch1 and Dll4 expressions in HUVEC networks treated with Y-27632 and nocodazole. Data are representative from three independent experiments and are expressed as mean ± s.e.m. (n = 3; ns, not significant; *P<0.05, **P<0.01; unpaired Student’s t-test). (C) Notch1 and Dll4 mRNA expressions in HUVEC networks in vitro. Gene expressions were measured by a gold nanorod biosensor with locked nucleic acid probes targeting Notch1 and Dll4 mRNA. Data are representative from three independent experiments and each data point represents at least 150 cells (ns, not significant; *P<0.05, **P<0.01; unpaired Student’s t-test). (D) Western blot analysis of Notch1 and Dll4 expression in HUVECs pretreated with Notch 1 siRNA or Dll4 siRNA. Data are representative from three independent experiments. (E-F) Mean cord lengths of siRNA-treated HUVEC networks without (E, n = 10, 9 hours after cell seeding) or with (F, n = 12, 9 hours after cell seeding) Y-27632. Data are expressed as mean ± s.e.m. (ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
To gain insight into the roles of Notch1-Dll4 signaling on the mechanoregulation of tip cell formation, Notch1 and Dll4 siRNAs were applied with and without Y-27632 (Figure 5D and supplementary Figure S7). The effects of siRNA were first verified (Figure 5D). Immunoblotting showed that the efficiency for Notch1 siRNA and Dll4 siRNA was 77.3±4.5% and 50.7±4.9% respectively. Notch1 siRNA reduced both Notch1 and Dll4 expressions while Dll4 siRNA did not have a significant effect on the expression of Notch1. Notch1 and Dll4 siRNA treatments resulted in hyperbranching and formed dense HUVEC networks compared to cells treated with control siRNA (Figure 5E). Importantly, Notch1 and Dll4 siRNA treatments decreased the mean cord length of the HUVEC networks with and without Y-27632 (Figure 5E-F and supplementary Figure S7).
2.5. Single cell laser ablation perturbed intercellular tension and induced angiogenic sprouting
Our results using traction force modulating drugs supported that intercellular tension negatively regulates Dll4 expression and tip cell formation. To investigate the mechanoregulation of Notch1-Dll4 signaling and tip cell formation using a non-pharmacological approach, we established a single cell laser ablation technique to reduce the intercellular tension of the HUVEC networks (Figure 6A-B). The technique takes advantage of the absorption of near infrared laser by gold nanorods to create a local hyperthermic effect for single cell ablation and rupture the HUVEC network in selective locations. The laser ablation experiment was performed by focusing the near infrared laser to a desired location of the network for less than two seconds (Figure 6C). The photothermal effect was previously optimized for single cell ablation[13a]. Retraction of the cells was observed immediately after ablation of the network, indicating that the cells were under intercellular tension. The mechanical interaction, as indicated by the retraction length, could be perturbed by Y-27632 and nocodazole (Figure 6B). The results verified the HUVEC network is under tension and the biomechanical effects of the pharmacological reagents. As a control, the Notch ligand, Jag1, did not have any observable effect on the retraction length indicating that Notch signaling did not directly regulate the intercellular tension.
Figure 6. Laser ablation of the HUVEC network disrupts the intercellular tension and induces tip cell formation.
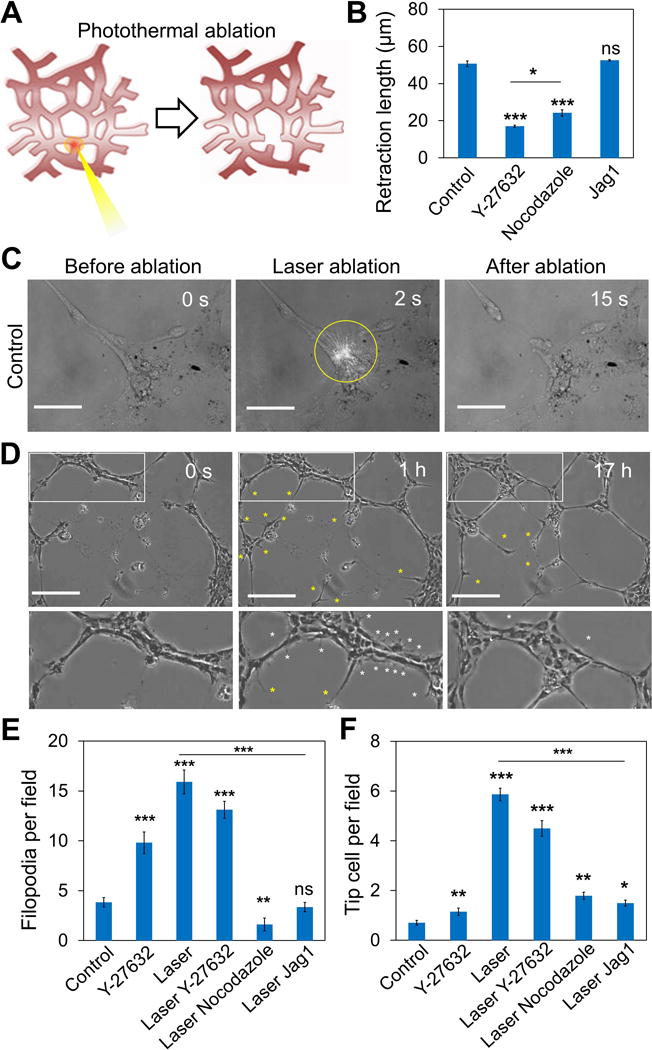
(A) Schematic of the laser ablation experiment using a near infrared laser coupled with gold nanorods. The gold nanorods were incubated and internalized in the cells before the experiment. (B) Retraction length of the cells after laser ablation under different treatments. (C) A representative laser ablation experiment by irradiating a near infrared laser to a single cell for 2 s. Yellow circle indicates the location of irradiation. The network retracted after cell ablation. The gap distance between the retracting ends of the network is defined as the retraction length in this study. Scale bars, 50 μm. (D) Time-lapse images illustrating capillary anastomosis after laser ablation. After ablation, tip cells (yellow asterisks) were observed to sprout out from the network. White rectangles indicated the zoom-in regions at the bottom. Cells with active filopodial protrusions (white asterisks) were observed in the undamaged region of the network. Scale bars, 200 μm. (E-F) Formation of (E) sprouting tip cells and (F) cells with active filopodial protrusions under different conditions. Data are expressed as mean ± s.e.m. (ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
Laser ablation on the network redistributed the intercellular tension in the networks and reduced intracellular stress near the ablated end. Computational biomechanical analysis was performed to illustrate the reduction of intercellular tension by laser ablation (supplementary Figure S8). Ablation of the network resulted in a hyperactive phenotype with active filopodia that protrude and retract (Figure 6D). Tip cells sprouted out from existing cell structures and anastomosed the network (supplementary Video S1). The formation of filopodial protrusion decreased during network anastomosis and negatively correlated with the intercellular tension (supplementary Figure S9-10). Cells with active filopodia were not observed after the HUVEC network reconnected and restored the intercellular tension. The laser ablation experiments were performed with Y-27632 and nocodazole to further study the mechanism of ablation mediated tip cell formation (Figure 6E-F). The formation of tip cells and cells with filopodial protrusion after laser ablation were similar with and without Y-27632 treatment. In contrast, enhancing mechanical force by nocodazole significantly attenuated the effects of laser ablation. Jag1 treatment also diminished the effects of laser ablation on the numbers of tip cells and filopodia observed.
Our results suggested reducing intercellular tension by laser ablation induced tip cell formation via Notch1-Dll4 signaling mechanically. To study the involvement of Notch1-Dll4 signaling, single cell gene expression analysis was performed to monitor the dynamic gene expression profiles in the ablated HUVEC networks (Figure 7A and C). Laser ablation enhanced Notch1-Dll4 expression near the ablated region. Upregulation of Dll4 and Notch1 mRNA was observed in cells along the cord. The cells, therefore, appeared to communicate the ablation signal and the upregulation of Since Notch1 and Dll4 expressions were upregulated in many cells away from the ablated end, the observation cannot be explained solely by the release of contact inhibition. In contrast, the intensity of the random probe control was not affected, supporting that the result was not due to the cell injury and morphology change. The experiment was also performed under Y-27632, nocodazole and Jag1 (Figure 7B and D). Laser ablation increased the levels of Dll4 in Y-27632 and Jag1 treated cells. The laser ablation mediated Dll4 increase was completely abolished by nocodazole, further supporting the notion that the laser ablation induced Dll4 upregulation is mediated mechanically.
Figure 7. Laser ablation of HUVEC networks enhances Notch1-Dll4 expression in vitro.

(A) Dll4 and Notch1 mRNA expression of endothelial cells before and after laser ablation. A random probe sequence is included as control. Scale bars, 50 μm. Yellow circles indicate locations of laser ablation. Endothelial cells were incubated with GNR-LNA probes for 4 hours before seeding on Matrigel for network formation. After 4 hours, HUVEC networks were ablated with a near infrared laser (0.85 mW/μm2). (B) Dll4 mRNA expression before and after injury for cell networks treated with Y-27632, nocodazole, and Jag1, respectively. Yellow circles indicate locations of laser ablation. (C) Quantification of Dll4 mRNA, Notch1 mRNA, and random expression before and after laser ablation. (D) Comparison of Dll4 mRNA expression before and after laser ablation with the treatments of Y-27632, nocodazole, and Jag1. Data are expressed as mean ± s.e.m. (ns, not significant; *P<0.05, **P<0.01, ***P<0.001; unpaired Student’s t-test).
3. Discussion
In this study, we demonstrate the inhibitory role of intercellular tension on tip cell formation during vascular development using a systems bioengineering approach. Reducing the traction force by ROCK inhibition modified the tip-stalk organization and led to vascular abnormality. The density of tip cells and branch density of the vascular structures negatively correlated with the intercellular tension. The inhibitory effects of intercellular tension on tip cell formation and angiogenic sprouting were consistently observed in mice retinal angiogenesis in vivo, mouse embryonic stem cells, HUVEC spheroids and HUVEC networks in vitro. These results suggest that mechanical force modulates the vascular architecture by inhibiting tip cell formation during angiogenesis. The mechanoregulation of tip cell formation was confirmed using pharmacological reagents targeting different mechanisms, including ROCK signaling, myosin II activity, microtubules polymerization, and myosin-light-chain phosphatase activity[12a]. In addition, we established a laser ablation assay to physically perturb the intercellular tension to complement the pharmacological experiments. The intercellular tension of the networks and the mechanical effects of laser ablation were verified by experimental measurement of the retraction length and computational biomechanical analysis. The reduction of intercellular tension by laser ablation was sufficient to mediate aggressive phenotypes of endothelial cells in the networks and to induce angiogenic sprouting that repaired the ablated network. The observation of aggressive tip cells with the active filopodial protrusions in the undamaged region of the network cannot be explained solely using Notch1-Dll4 lateral inhibition, supporting the inhibitory role of intercellular tension on tip cell formation. After capillary anastomosis, the networks were stabilized and the aggressive behaviors in endothelial cells disappeared. Furthermore, reducing the cell traction force by ROCK inhibition did not further increase tip cell formation while enhancing the cell traction force by nocodazole attenuated the effects of laser ablation. These observations support that intercellular tension inhibits the formation of tip cell formation during vascular development.
Our study reveals that intercellular tension negatively regulates tip cell formation via Notch1-Dll4 signaling. Dll4 is known to negatively regulate the formation of tip cells through Notch1 signaling[9b, 9c]. Notch lateral inhibition forms alternating Dll4 patterns near the sprouting front and regulates network density by preventing hypersprouting[9b]. In our experiment, reduction of the cell traction force by ROCK inhibition with Y-27632 and myosin II inhibition with blebbistatin modulated Notch1-Dll4 signaling and enhancing hypersprouting. Y-27632 treatment disrupted the alternating Dll4 patterns resulting in a uniform Dll4 distribution in the sprouting front. The effect of ROCK inhibition by Y-27632 on Dll4 distribution was similar to Notch inhibition with DAPT[9b], suggesting that the reduction of intercellular tension mediates its effect by interfering Notch signaling. In contrast, enhancing the cell traction force by nocodazole and calyculin A reduced the level of Dll4 in embryonic bodies and HUVEC networks. The inhibitory roles of intercellular tension on Dll4 expression and tip cell formation were consistently observed when targeting different mechanoregulation pathways. The mechanoregulation of Notch1-Dll4 signaling was also observed in the single cell laser ablation assay. Reducing the intercellular tension by laser ablation, as confirmed in cell retraction and computational biomechanical analysis, enhanced Dll4 expression and tip cell formation in ablated HUVEC networks. Single cell analysis revealed upregulation of Notch-Dll4 signaling in laser ablated HUVEC networks and Dll4 expressing tip cells sprouted out from the network after ablation. The effects of laser ablation on Notch1-Dll4 signaling and tip cell formation were eliminated by nocodazole treatment. The notion that intercellular tension regulates tip cell formation by perturbing Notch lateral inhibition was further supported by modulating traction force and Notch activity simultaneously. If the reduction of mechanical force mediates tip cell by inhibiting Notch signaling, Notch inhibition should not further increase Y-27632 mediated tip cell formation enhancement. In fact, DAPT treatments did not further enhance the Y-27632-mediated tip cell formation in mouse retina, embryonic bodies and HUVEC networks. In contrast, activating Notch activity by Jag1 restored the normal topology of the vascular structures. Furthermore, the importance of Notch signaling in the mechanoregulation of tip cell formation was evaluated by Notch1 and Dll4 siRNA. The effects of Notch1 and Dll4 siRNAs on the density of HUVEC networks were attenuated by Y-27632, supporting the negative regulation of Notch1-Dll4 signaling by mechanical force.
An interesting finding in our study is the mechanosensitivity of Notch1-Dll4 signaling for regulating tip cell formation during angiogenesis. Mechanistically, mechanical force controls the expression of VEGFR[4], which modulates the VEGFR-Dll4-Notch signaling circuit[10a]. Notch signaling is also known to be mechanosensitive using laser tweezer trapped microbeads coated with Notch ligands[11]. Mechanical pulling is required to expose the ADAM cleavage site of Notch for proper proteolysis and release of the intracellular domain. The mechanosensitivity of Notch signaling provides a mechanistic explanation for the regulatory effects of Rho and ROCK signaling on capillary morphogenesis[14]. From a systems perspective, our finding clarifies the spatial control of angiogenic sprouting during capillary morphogenesis. In developing vascular plexuses, the intercellular tension is lower near the migrating front (supplementary Figure S11). Recent computational modeling also revealed the creation of a mechanical tension in the angiogenic sprout by tip cells[15]. The intercellular tension, therefore, serves as a mechanoregulation signal in the sprouting front to communicate among endothelial cells for proper spatial response of VEGF-mediated angiogenesis. Interestingly, the mechanosensitivity of Notch signaling has also been identified for regulating leader cell formation during epithelial wound healing[16]. The reduction of mechanical force near the wound boundary promotes Notch1-Dll4 signaling to regulate the density of epithelial leader cells. In contrast, Dll4 regulates the formation of angiogenic tip cells through Notch1 signaling negatively, suggesting distinct regulatory mechanisms may be involved effects in the regulation of epithelial cells and endothelial tip cells. Despite the difference in cell types and mechanisms involved, intercellular tension appears to serve as a general mechanical communication mechanism to initiate or stop the anastomosis and wound healing processes. The results also clarify why Dll4 expression is confined near the sprouting front of the vascular plexus and how ectopic vascular sprouting is avoided under normal physiological condition.
4. Materials and Methods
Animal procedures
The University of Arizona Institutional Animal Care and Use Committee approved all animal protocols. The C57BL/6J mice were fed on laboratory food and tap water ad libitum in a regular 12 hours dark/light cycle. Postnatal day (P) 3 newborn pups were injected daily with DAPT (Sigma, 50μg/g), Jag1 peptide (AnaSpec, 50 μg/g), Y-27632 (Sigma, 10μg/g), the combination of DAPT and Y-27632, or the combination of Jag1 and Y27632 daily for 4 days. The control pups were injected with 1× PBS. The mouse pups were humanely sacrificed at P7. The eyelids were cut and the eyes were enucleated using a pair of curved forceps to pinch the optic nerve and ocular muscles. The eyes were transferred to a petri dish containing 4% paraformaldehyde in artificial cerebrospinal fluid solution for 5-10 minutes. The eyes were transferred to a petri dish containing Dulbecco’s phosphate-buffered saline placed under a dissection microscope to dissect the retina.
Mouse retina immunostaining
The fixed retina tissues were rinsed in 1× PBS for 3 times and covered with 100 μL of block solution (PBS + 0.5% Triton +1% BSA+ 5% donkey serum) for 1 hour. After removing the block solution, the retina tissues were incubated with 100 μL primary antibody (diluted 1:50 in PBSTX, rabbit polyclonal to Dll4, Abcam) at 4°C overnight. The samples were then incubated with secondary antibody (diluted 1:500 in PBSTX, Goat anti-rabbit IgG H&L (Alexa Fluor 488, Abcam) and TO-PRO-3 (1 μM, diluted 1/1000 in PBSTX, Invitrogen) at room temperature for 4 hours. The staining specificity was verified by substituting primary antibodies with non-immune serum. The samples were mounted onto coverslips with 50 μl of Prolong Gold antifade reagent (Invitrogen).
Embryoid body formation
R1/SVJ 129 murine embryonic stem cells were obtained from Dr. Andras Nagy (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, ON, Canada) and cultured as previously described[17]. Embryoid bodies were formed using the hanging drop methods. Briefly, 20 μl of droplets containing 103 embryonic stem cells were suspended from the lid of a petri dish for 4 days. The spheroids were then embedded in 1.5 mg/mL collagen type I and cultured with addition of 30 ng/mL VEGF-A164 (Peprotech) for another 5 days.
Cell culture and reagents
Human Umbilical Vein Endothelial Cells (HUVECs) were purchased from Corning Inc. HUVECs were cultured in endothelial basal medium-2 (EBM-2) culture medium (Lonza Inc.) with 2% fetal bovine serum. Cells were grown at 37°C and 5% CO2 on tissue culture dishes in a humidified incubator with media changes every two days. Cells were passaged using 0.25% Trypsin (Invitrogen), and passages 2-7 were used in the experiments. DAPT, Y-27632, blebbistatin, nocodazole and calyculin A were purchased from Sigma Aldrich. Jag1 peptide (188-204) was acquired from Anaspec. HUVECs were treated with DAPT (20 μM), Jag1 peptide (20 μM), blebbistatin (20 μM), nocodazole (1 μM), and calyculin A (5 nM).
HUVEC network formation in vitro
Matrigel (growth factor reduced, BD) was thawed overnight with ice in a refrigerator, added into 48-well plates, and incubated at 37°C for 30 minutes to allow complete gelation. Cells were seeded (~400 cells/mm2) on top of the gel, cultured and imaged at 9 hours and 22 hours after cell seeding with a CCD camera (Cooke SensiCam).
GNR-LNA for intracellular mRNA detection
Analysis of single cell gene expression was performed using gold nanorods (GNR) with locked nucleic acid (LNA) probes targeting specific mRNA sequences[13a, 13b]. Mercaptoundecyltrimethylammonnium bromide coated GNR with 10 nm diameter and 67 nm length (Nanopartz) were used. The 20-base LNA probes with alternating DNA/LNA monomers and a fluorophore (6-FAM) labeled at the 5′ end were synthesized by Integrated DNA Technologies Inc. When in close proximity, the fluorophore in the LNA probe was quenched by the GNR due to its intrinsic quenching ability. The target mRNA thermodynamically displaced the LNA probe from the GNR, allowing it to fluoresce. Two specific LNA probes targeting Dll4 and Notch1 mRNA were designed. A random probe was included as the positive and negative control, respectively. The LNA sequences and synthetic targets for calibration are listed in supplementary Table S1.
Single cell gene expression analysis and laser ablation in HUVEC networks
In the GNR-LNA assay, 2 × 104 HUVEC were seeded on the 35 mm tissue culture dish. GNR-LNA complex with a density of 2 × 1011 particles per ml were added to the tissue culture dish when the cells reached 70-80% confluency, and incubated with HUVECs for 8 hours at 37°C for endocytic uptake. HUVECs were washed with 1×PBS for 3 times and were harvested. The cells were seeded on matrigel-precoated glass-bottom 24-well tissue culture plates with a density of 2 × 104 cells per well. HUVEC networks were self-organized after 3 hours of culture. For laser ablation, a 1064 nm fiber laser with TEM00 beam was utilized. Upon laser illumination, individual cells in the capillary networks were ablated within 2 seconds with an irradiation intensity of 0.85 mW/μm2.
Imaging and data analysis
All bright field images and fluorescence images of HUVECs were captured using an inverted fluorescence microscope (Nikon TE2000-U) with a HQ2 CCD camera. Fluorescence images were taken with the same settings for comparison. All the animal tissue images were captured using a confocal microscope (Zeiss LSM 510 Meta). 3D images of embryoid bodies were obtained with a laser scanning confocal microscope (Leica TCS SP8) equipped with a 25× objective lens. Data collection and imaging analysis were performed using NIH ImageJ software.
Western blot and immunofluorescence
Embryoid bodies grown in collagen were fixed on day 5 by 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. After blocking with 5% BSA in PBS for 60 min, the samples were incubated with primary antibodies against Dll4 (1:100, R&D Systems, Minneapolis, MN, USA) and Notch (1:100, Abcam, Cambridge, MA, USA) overnight at 4°C and conjugated with appropriate Alexa Fluor 647 and 555 secondary antibodies at 4°C overnight. Control experiment was performed to verify the specificity of the secondary antibody. Nuclei were stained with Sytox green (1:1000, Life Technologies, Grand Island, NY, USA).
To isolate HUVEC networks or embryoid bodies from matrigel, PBS-EDTA, which contains 5 mM EDTA, 1 mM NaVO4, 1.5 mM NaF and 1× Protease Inhibitor Cocktail (Sigma), was applied[18]. HUVEC networks or embryoid bodies were lysed by RIPA buffer (Santa Cruz Biotechnology). After protein quantifications with BCA assay (Santa Cruz Biotechnology), proteins (20 μg per well) were subjected to SDS–PAGE electrophoresis (10%, Bio-Rad) and transfer to a PVDF membrane (Santa Cruz Biotechnology, CA). The PVDF membrane was incubated with 5% BSA and 5% non-fat milk in Tris-buffered saline with Tween 20 (TBST) at room temperature for 1 hour, and further incubated with rabbit monoclonal GAPDH antibody (1:1,000), a rabbit monoclonal Notch1 antibody (1:800) and a rabbit polyclonal Dll4 antibody (1:400) at 4°C overnight, followed by three washes with TBST and incubation with horseradish peroxidase-conjugated anti-rabbit antibody (1:1,000) at room temperature for 1 hour. Autoradiography films and western blotting luminol reagent (Santa Cruz Biotechnology, CA) were employed for the detection of protein bands. All the antibodies were purchased from Cell Signaling Technology (Danvers, MA).
Gene silence with siRNA
Notch1 or Dll4 siRNAs (0.6 μL, 20 μM, Qiagen) were added into 50 μL serum-free medium (Opti-MEM, Life Technologies) with 0.5 μL PLUS™ Reagent. The siRNAs were verified and used in our previous study[16]. Lipofectamine® LTX reagent (2 μL, Life Technologies) was added to another 50 μL Opti-MEM. The reagents were mixed and incubated at room temperature for 20 min. The mixture was then added to HUVEC (~80% confluent) in 500 μL medium in one well of a 24-well plate. After 36 hours of culture, HUVECs were ready for the network formation.
Computational biomedical analysis
A 3D finite-element model of the cell monolayer was constructed with Comsol Multiphysics 5.0 and with two components, a traction layer and a passive layer. During the simulation, contractility was introduced by prescribing a thermal strain[19]. The traction layer (20 μm height; other dimensions prescribed as indicated) was treated as an isotropic elastic material with a Young’s modulus of 500 Pa, a Poisson’s ratio of 0.499 (incompressible), a thermal conductivity of 10 W·m-1·K-1, and a coefficient of expansion of 0.05 K-1. The passive layer (4 μm height) was treated as an isotropic elastic material with values of 100 Pa and 0.499 for the Young’s modulus and Poisson’s ratio, respectively.
Statistical analysis
Data are presented as mean ± s.e.m. Experiments were conducted in triplicate, and repeated at least three independent times. Student’s t-tests were performed to analyze statistical significance between experimental groups. For comparing multiple groups, a one-way analysis of variance and Tukey’s post hoc test were used. Statistically significant P values were assigned as follows: *, P<0.05; **, P<0.01 or ***, P<0.001.
Supplementary Material
Acknowledgments
We thank Dr. Erika Eggers and Dr. Min Long for assistance on the mouse retina experiment. The authors also thank Dr. Reza Riahi for suggestions on single cell gene expression analysis and laser ablation. This work was supported by National Institutes of Health Director’s New Innovator Award (DP2OD007161).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Shue Wang, Department of Aerospace and Mechanical Engineering, The University of Arizona, Tucson, AZ 85721 USA.
Dr. Jian Sun, Department of Aerospace and Mechanical Engineering, The University of Arizona, Tucson, AZ 85721 USA.
Dr. Yuan Xiao, Department of Aerospace and Mechanical Engineering, The University of Arizona, Tucson, AZ 85721 USADepartment of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802 USA.
Dr. Yi Lu, Department of Aerospace and Mechanical Engineering, The University of Arizona, Tucson, AZ 85721 USADepartment of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802 USA
Prof. Donna D. Zhang, Department of Pharmacology and Toxicology, The University of Arizona, Tucson, AZ 85721 USA
Prof. Pak Kin Wong, Department of Aerospace and Mechanical Engineering, The University of Arizona, Tucson, AZ 85721 USADepartment of Biomedical Engineering, The Pennsylvania State University, University Park, PA 16802 USA.
References
- 1.a) Carmeliet P, Jain RK. Nature. 2011;473:298. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Carmeliet P, Jain RK. Nature. 2000;407:249. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]; c) Warren CM, Ziyad S, Briot A, Der A, Iruela-Arispe ML. Sci Signal. 2014;7:ra1. doi: 10.1126/scisignal.2004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Song JW, Munn LL. Proc Natl Acad Sci U S A. 2011;108:15342. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Indrasekara ASDS, Meyers S, Shubeita S, Feldman LC, Gustafsson T, Fabris L. Nanoscale. 2014;6:8891. doi: 10.1039/c4nr02513j. [DOI] [PubMed] [Google Scholar]; c) Rivron NC, Vrij EJ, Rouwkema J, Le Gac S, van den Berg A, Truckenmuller RK, van Blitterswijk CA. P Natl Acad Sci USA. 2012;109:6886. doi: 10.1073/pnas.1201626109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Tornavaca O, Chia M, Dufton N, Almagro LO, Conway DE, Randi AM, Schwartz MA, Matter K, Balda MS. The Journal of cell biology. 2015;208:821. doi: 10.1083/jcb.201404140. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Edgar LT, Hoying JB, Utzinger U, Underwood CJ, Krishnan L, Baggett BK, Maas SA, Guilkey JE, Weiss JA. Journal of biomechanical engineering. 2014;136:021001. doi: 10.1115/1.4026471. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ingber DE. Circ Res. 2002;91:877. doi: 10.1161/01.res.0000039537.73816.e5. [DOI] [PubMed] [Google Scholar]
- 4.Mammoto A, Connor KM, Mammoto T, Yung CW, Huh D, Aderman CM, Mostoslavsky G, Smith LEH, Ingber DE. Nature. 2009;457:1103. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivron NC, Vrij EJ, Rouwkema J, Le Gac S, van den Berg A, Truckenmuller RK, van Blitterswijk CA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6886. doi: 10.1073/pnas.1201626109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kniazeva E, Putnam AJ. American Journal of Physiology-Cell Physiology. 2009;297:C179. doi: 10.1152/ajpcell.00018.2009. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Jamilpour N, Wang FY, Wong PK. Biomaterials. 2014;35:3273. doi: 10.1016/j.biomaterials.2013.12.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Phng LK, Gerhardt H. Developmental Cell. 2009;16:196. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]; b) Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. The Journal of cell biology. 2003;161:1163. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Seano G, Chiaverina G, Gagliardi PA, di Blasio L, Puliafito A, Bouvard C, Sessa R, Tarone G, Sorokin L, Helley D, Jain RK, Serini G, Bussolino F, Primo L. Nature cell biology. 2014;16:931. doi: 10.1038/ncb3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. Cell. 2009;137:1124. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]; b) Suchting S, Freitas C, le Noble F, Benedito R, Breant C, Duarte A, Eichmann A. Proc Natl Acad Sci U S A. 2007;104:3225. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalen M, Gerhardt H, Betsholtz C. Nature. 2007;445:776. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]; d) Stenzel D, Franco CA, Estrach S, Mettouchi A, Sauvaget D, Rosewell I, Schertel A, Armer H, Domogatskaya A, Rodin S, Tryggvason K, Collinson L, Sorokin L, Gerhardt H. EMBO reports. 2011;12:1135. doi: 10.1038/embor.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, Schulte-Merker S, Gerhardt H. Nature cell biology. 2010;12:943. doi: 10.1038/ncb2103. [DOI] [PubMed] [Google Scholar]; b) Bentley K, Franco CA, Philippides A, Blanco R, Dierkes M, Gebala V, Stanchi F, Jones M, Aspalter IM, Cagna G, Westrom S, Claesson-Welsh L, Vestweber D, Gerhardt H. Nature cell biology. 2014;16:309. doi: 10.1038/ncb2926. [DOI] [PubMed] [Google Scholar]
- 11.a) Ahimou F, Mok LP, Bardot B, Wesley C. Journal of Cell Biology. 2004;167:1217. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shergill B, Meloty-Kapella L, Musse AA, Weinmaster G, Botvinick E. Developmental Cell. 2012;22:1313. doi: 10.1016/j.devcel.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Meloty-Kapella L, Shergill B, Kuon J, Botvinick E, Weinmaster G. Developmental Cell. 2012;22:1299. doi: 10.1016/j.devcel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Wang X, Ha T. Science. 2013;340:991. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Sun J, Xiao Y, Wang S, Slepian MJ, Wong PK. J Lab Autom. 2015;20:127. doi: 10.1177/2211068214554802. [DOI] [PubMed] [Google Scholar]; b) Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Science. 1999;285:895. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 13.a) Riahi R, Wang S, Long M, Li N, Chiou PY, Zhang DD, Wong PK. Acs Nano. 2014;8:3597. doi: 10.1021/nn500107g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang S, Riahi R, Li N, Zhang DD, Wong PK. Adv Mater. 2015;27:6034. doi: 10.1002/adma.201502814. [DOI] [PubMed] [Google Scholar]; c) Wang S, Sun J, Zhang DD, Wong PK. Nanoscale. 2016;8:16894. doi: 10.1039/c6nr03907c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) van der Meel R, Symons MH, Kudernatsch R, Kok RJ, Schiffelers RM, Storm G, Gallagher WM, Byrne AT. Drug discovery today. 2011;16:219. doi: 10.1016/j.drudis.2011.01.005. [DOI] [PubMed] [Google Scholar]; b) Bryan BA, Dennstedt E, Mitchell DC, Walshe TE, Noma K, Loureiro R, Saint-Geniez M, Campaigniac JP, Liao JK, D’Amore PA. Faseb J. 2010;24:3186. doi: 10.1096/fj.09-145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos-Oliveira P, Correia A, Rodrigues T, Ribeiro-Rodrigues TM, Matafome P, Rodriguez-Manzaneque JC, Seica R, Girao H, Travasso RDM. Plos Comput Biol. 2015:11. doi: 10.1371/journal.pcbi.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riahi R, Sun J, Wang S, Long M, Zhang DD, Wong PK. Nature communications. 2015;6:6556. doi: 10.1038/ncomms7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Rossant J, Nagy R, Abramownewerly W, Roder JC. P Natl Acad Sci USA. 1993;90:8424. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee GY, Kenny PA, Lee EH, Bissell MJ. Nature methods. 2007;4:359. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Nam KH, Jamilpour N, Mfoumou E, Wang FY, Zhang DD, Wong PK. Scientific reports. 2014;4:6965. doi: 10.1038/srep06965. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. PNAS. 2005;102:11594. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


