Summary
Macroautophagy/autophagy is a key catabolic recycling pathway that requires fine-tuned regulation to prevent pathologies and preserve homeostasis. Here we report a new post-transcriptional pathway regulating autophagy involving the Pat1-Lsm (Lsm1 to Lsm7) mRNA-binding complex. Under nitrogen starvation conditions, Pat1-Lsm binds a specific subset of autophagy-related (ATG) transcripts and prevents their 3' to 5' degradation by the exosome complex, leading to ATG mRNA stabilization and accumulation. This process is regulated through Pat1 dephosphorylation, is necessary for the efficient expression of specific Atg proteins and is required for robust autophagy induction during nitrogen starvation. To the best of our knowledge, this work presents the first example of ATG transcript regulation via 3' binding factors and exosomal degradation.
Keywords: 3'-5' degradation, exosome, Lsm1, mRNA decay, Mrt1, PATL1
Graphical Abstract
In Brief
Autophagy is a catabolic recycling process that must be properly regulated to preserve homeostasis. In this study, Gatica et al. discovered that during nitrogen starvation the Pat1-Lsm complex binds a subset of autophagy-related (ATG) mRNAs, preventing their degradation by the exosome and leading to ATG transcript accumulation and robust autophagy.
Introduction
Macroautophagy (hereafter autophagy) is a highly conserved and key catabolic recycling process for cell survival triggered under stress conditions such as nutrient starvation. During autophagy, a diverse range of cytoplasmic components including proteins, lipids and damaged and/or superfluous organelles are sequestered by phagophores that mature into double-membrane vesicles termed autophagosomes. The autophagosomes subsequently fuse with the vacuole (in yeast and plants) or lysosome (in metazoans) leading to the degradation of the sequestered cargo by resident hydrolytic enzymes. The macromolecules obtained from cargo degradation are then transported back to the cytoplasm for recycling (Feng et al., 2014; He and Klionsky, 2009). During nutrient-rich conditions, autophagy is usually kept at low levels, mostly working as a quality-control mechanism (Hara et al., 2006). However, upon starvation, autophagy activity is dramatically increased to compensate for the lack of nutrients.
Several autophagy-related (ATG) genes and their corresponding proteins have been characterized based on their function(s) in regulating the various stages of autophagy (Feng et al., 2014; He and Klionsky, 2009). Along these lines, multiple studies have highlighted the importance of the post-translational modification of several Atg proteins (Feng et al., 2016; Feng et al., 2015; Kamada et al., 2010), as well as the transcriptional upregulation of ATG genes upon starvation, to increase autophagy activity (Bernard et al., 2015a; Bernard et al., 2015b; Feng et al., 2015). However, less is known about the post-transcriptional regulation of the mRNAs encoding these ATG genes and the mechanisms behind maintaining mRNA stability during autophagy induction. Recently, we reported that following nitrogen starvation the mRNA decapping enzyme Dcp2 is inhibited, which prevents exonuclease Xrn1-mediated 5'-3' degradation of multiple ATG transcripts, and enables ATG mRNA accumulation and efficient autophagy induction (Hu et al., 2015).
Although providing new insights into the post-transcriptional regulation of autophagy, Xrn1-dependent 5'-3' degradation is not the only RNA decay pathway that has been described. The ribonuclease complex known as the exosome also mediates RNA degradation in a 3' to 5' direction (Garneau et al., 2007; Parker, 2012). While both RNA decay mechanisms have been extensively studied, little is known about the crosstalk between the two pathways. The RNA binding protein Pat1/Mrt1 has been proposed as a possible link between 5'-3' and 3'-5' mRNA degradation (Tharun, 2009). Pat1 interacts with the heptameric ring-shaped complex formed by the seven Sm-like proteins Lsm1 to Lsm7, which together form the Pat1-Lsm complex (Bouveret et al., 2000); this complex preferentially binds to the 3' untranslated region (UTR) of oligoadenylated mRNA (Chowdhury et al., 2014; Chowdhury et al., 2007). Initially reported as a decapping enhancer, the Pat1-Lsm complex has also been implicated in protecting the 3' end of oligoadenylated mRNA from shortening by 10-20 nucleotides, also known as trimming (Chowdhury and Tharun, 2009; He and Parker, 2001; Tharun et al., 2005). Furthermore, inhibiting the exosome-dependent 3'-5' mRNA decay pathway leads to the accumulation of trimmed mRNA in pat1Δ and lsm1Δ strains (He and Parker, 2001), providing evidence that the Pat1-Lsm complex can protect the 3' end of mRNA from exosome-dependent 3'-5' degradation. Here we show that upon nitrogen starvation, the Pat1-Lsm complex binds and stabilizes a subset of ATG mRNA by preventing their exosome-mediated 3'-5' degradation. We provide evidence that this process is regulated through Pat1 dephosphorylation, is necessary for the efficient expression of specific Atg proteins, and is required for normal autophagy induction during nitrogen starvation.
Results
The RNA binding protein Pat1 is required for normal ATG mRNA accumulation, Atg protein expression, and autophagy induction during nitrogen starvation.
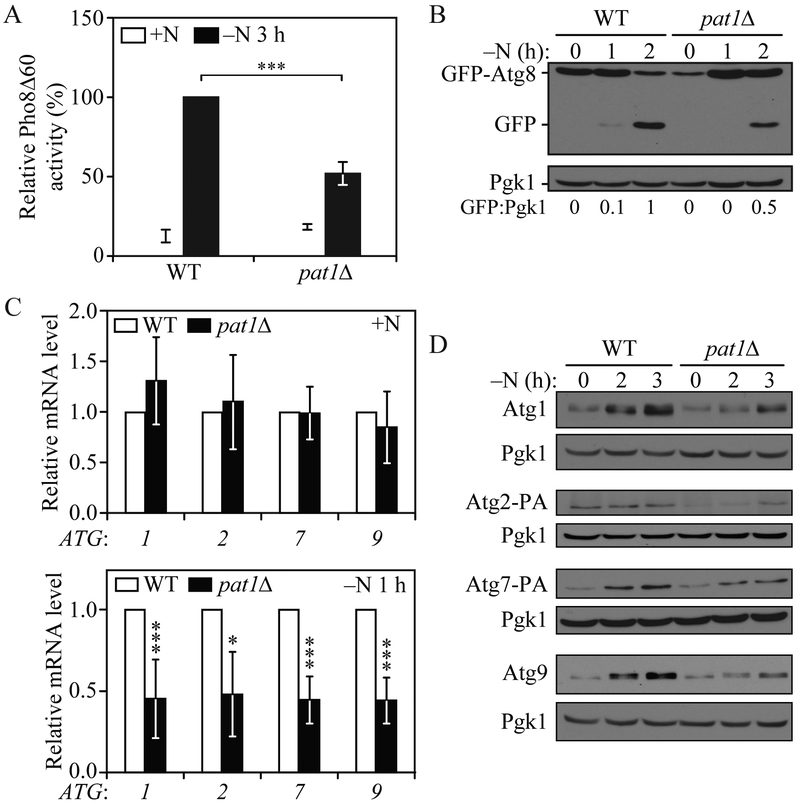
To explore the possibility that Pat1 could have a role in regulating autophagy, we generated pat1Δ cells and measured autophagy activity using the quantitative Pho8Δ60 assay (Noda and Klionsky, 2008). Briefly, we engineered a mutant Pho8 phosphatase that lacks the first 60 amino acids of its N terminus, leaving it unable to reach the vacuole through the secretory pathway. However, by inducing nonselective autophagy through starvation, Pho8Δ60 can be delivered to the vacuole, leading to the cleavage of its propeptide by resident hydrolases and the activation of its phosphatase activity. Thus, by measuring phosphatase activity in Pho8Δ60 cells we can indirectly measure autophagy induction. A significant decrease in autophagy activity was observed in pat1Δ cells after 3 h of nitrogen starvation (Figure 1A). To extend our analysis, we carried out a GFP-Atg8 processing assay (Shintani and Klionsky, 2004). During autophagy induction, Atg8 is covalently conjugated to phosphatidylethanolamine (PE); hence, Atg8–PE can be found on both the inner and outer membrane of the phagophore. When autophagosomes are completed, outer membrane bound Atg8–PE is deconjugated and recycled back into the cytoplasm; however, inner membrane-bound Atg8–PE is trapped inside the autophagosome, leading to its degradation after vacuolar fusion. Unlike Atg8, GFP is relatively resistant to vacuolar hydrolysis; thus by tagging GFP to the N terminus of Atg8, we can measure the amount of Atg8 being delivered to the vacuole during autophagy induction. Similar to the Pho8Δ60 assay, GFP-Atg8 processing showed decreased autophagy induction in pat1Δ cells after 1 and 2 h of nitrogen starvation, as observed by free GFP levels (Figure 1B).
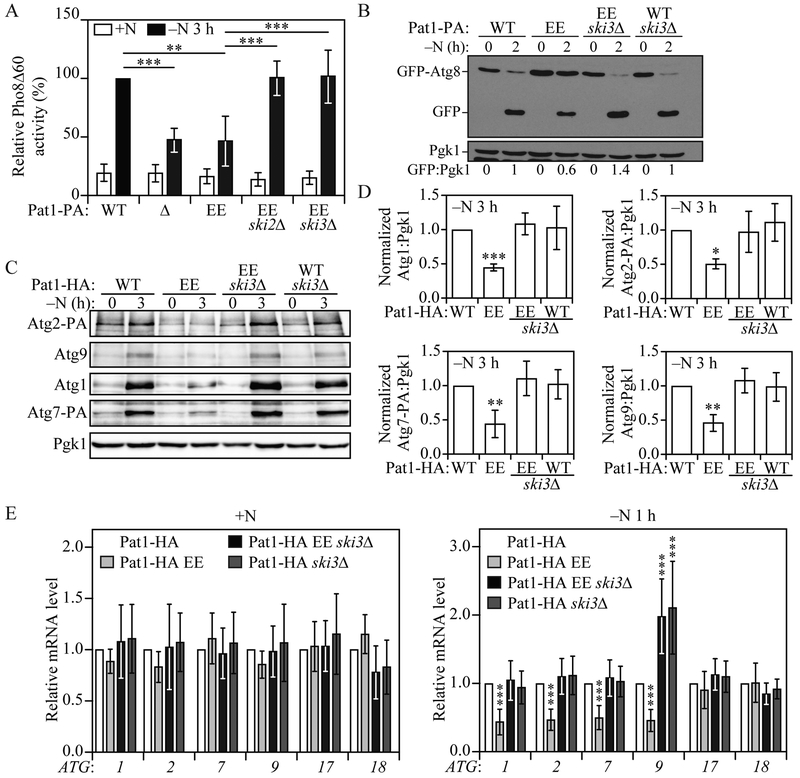
Fig. 1. PAT1 deletion lowers autophagy activity after nitrogen starvation by decreasing ATG mRNA and protein levels.
A) Autophagy activity was measured by the Pho8Δ60 assay in WT and pat1Δ strains under growing conditions (+N) and after 3 h of nitrogen starvation (−N). Error bars indicate the standard deviation of 4 independent experiments. Student’s t-test, ***P <0.001. B) Autophagy was measured by GFP-Atg8 processing in WT and pat1Δ strains under growing conditions and after 1 and 2 h of nitrogen starvation; a representative image is shown. C) ATG1, ATG2, ATG7 and ATG9 mRNA levels were determined in WT and pat1Δ strains under growing conditions and after 1 h of nitrogen starvation by RT-qPCR. Error bars indicate the standard deviation of at least 6 independent experiments. Student’s t-test, * P<0.05 and *** P <0.001. D) Atg1, Atg2-PA, Atg7-PA and Atg9 protein levels were measured by western blot in WT and pat1Δ strains under growing conditions and after 2 and 3 h of nitrogen starvation; representative images are shown. See also Figure S1.
Pat1 interacts with the Lsm1-Lsm7 ring complex, forming the Pat1-Lsm complex (Bouveret et al., 2000). Therefore, we decided to measure autophagy in lsm1Δ cells. Similar to Pat1Δ cells, LSM1 deletion led to a significant decrease in autophagy activity in both the Pho8Δ60 (Figure S1A) and GFP-Atg8 processing assays (Figure S1B). Deleting both PAT1 and LSM1 did not further decrease autophagy activity (Figure S1A), suggesting that the effect of Pat1 and Lsm1 on autophagy involves a similar mechanism. Taken together, these data indicate that the Pat1-Lsm complex is necessary for normal autophagy induction during nitrogen starvation.
Pat1 is an RNA binding protein involved in preventing mRNA degradation (He and Parker, 2001). To determine whether Pat1 plays a similar role with regard to autophagy we measured ATG mRNA levels by RT-qPCR in Pat1Δ cells. Essential autophagy genes ATG1, ATG2, ATG7 and ATG9 were selected as autophagy induction markers due to their high transcriptional induction during nitrogen starvation (Bernard et al., 2015b). In nutrient-replete conditions Pat1Δ cells showed no significant difference relative to the wild-type (WT) control (Figure 1C). Conversely, after 1 h of nitrogen starvation PAT1 deletion caused a significant decrease in ATG1, ATG2, ATG7 and ATG9 transcripts compared to WT cells. Similar results were obtained when checking the levels of the same ATG transcript in lsm1Δ cells (Figure S1C). These results suggest that the presence of the Pat1-Lsm complex is necessary for normal ATG mRNA stabilization and accumulation during nitrogen starvation.
To exclude the possibility that deleting PAT1 could have an effect on ATG mRNA transcription we constructed a Pat1-inducible degradation strain using the auxin-inducible degron (AID) system (Morawska and Ulrich, 2013) and measured ATG mRNA levels after 1 h of nitrogen starvation in the presence or absence of the mRNA synthetic inhibitor 1,10 phenanthroline (Hu et al., 2015). After 30 min of auxin treatment Pat1 levels decreased dramatically in our Pat1-inducible degradation strain (Figure S1D). ATG1, ATG2 and ATG7 mRNA levels decreased significantly after transcriptional inhibition with 1,10 phenanthroline or Pat1 degradation using auxin. Importantly, after both transcriptional inhibition and Pat1-induced degradation, ATG1, ATG2 and ATG7 mRNA levels were further decreased compared to 1,10 phenanthroline- or auxin-treated cells. These results indicate that the PAT1 deletion effect on ATG mRNA levels are not solely due to lower rates of ATG gene transcription, but rather a post-transcriptional regulation of ATG mRNA.
To determine if the decrease in ATG mRNA levels observed in the PAT1 deletion cells during nitrogen starvation translated into lower Atg protein levels, we measured the amounts of the Atg1, Atg2, Atg7 and Atg9 proteins by western blot. After 2 and 3 h of nitrogen starvation, the levels of these protein in Pat1Δ cells failed to increase to the same extent as in WT cells (Figure 1D). Similarly, after nitrogen starvation Atg1 and Atg9 levels in lsm1Δ cells failed to increase to the same level as in WT cells (Figure S1B). Together, these data indicate that the Pat1-Lsm complex is required for normal ATG mRNA accumulation, Atg protein expression and autophagy induction during nitrogen starvation.
Pat1 stabilizes ATG mRNA during nitrogen starvation by preventing efficient 3'-5' degradation.
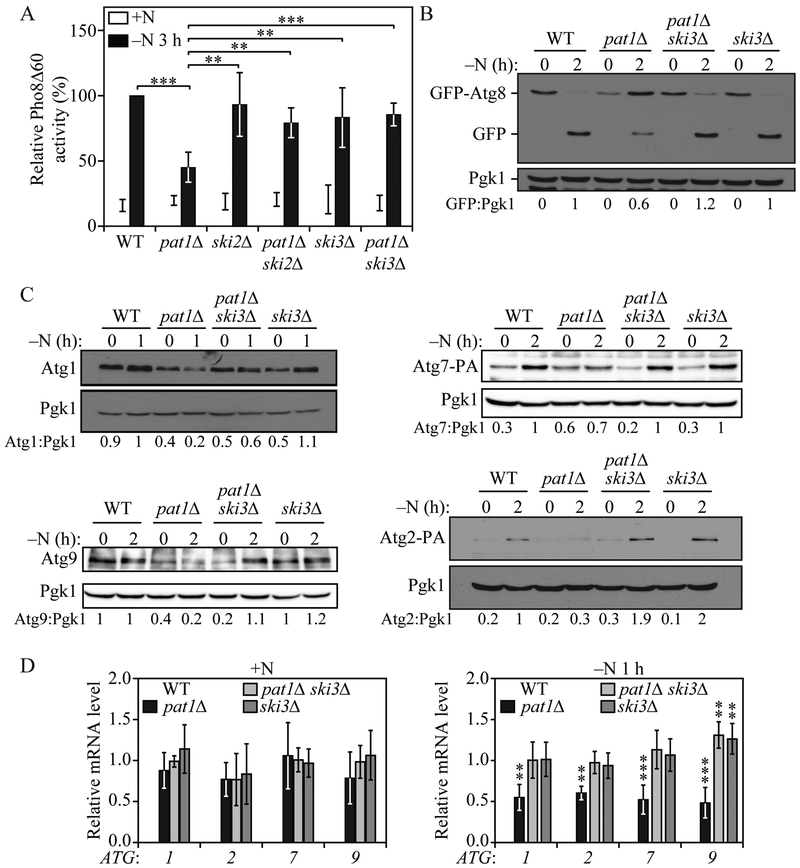
Our data suggest that the Pat1-Lsm complex could be stabilizing ATG mRNAs during nitrogen starvation. The Pat1-Lsm complex protects mRNA from 3’ end trimming and 3'-5' degradation by the exosome (Chowdhury and Tharun, 2009; He and Parker, 2001; Tharun et al., 2005). Efficient 3' to 5' mRNA degradation by the exosome requires the Ski complex (Anderson and Parker, 1998; van Hoof et al., 2000). The Ski complex, formed by Ski2, Ski3 and Ski8 (Brown et al., 2000), together with the adaptor protein Ski7 (Araki et al., 2001), has been proposed to work as a scaffold to modulate the cytoplasmic functions of the exosome without affecting its nuclear role (Anderson and Parker, 1998; van Hoof et al., 2000). Deletion of any members of the Ski complex produce similar phenotypes (Anderson and Parker, 1998; Brown et al., 2000; van Hoof et al., 2000) and prevent efficient 3' to 5' mRNA decay (He and Parker, 2001). To test if the decrease in autophagy observed in Pat1Δ cells during nitrogen starvation could be rescued by inhibiting the exosome-mediated 3' to 5' mRNA decay, we constructed Pat1Δ ski2Δ and Pat1Δ ski3Δ double-deletion strains and measured autophagy by the Pho8Δ60 assay.
Both Pat1Δ ski2Δ and Pat1Δ ski3Δ strains showed a significant increase in autophagy activity after 3 h of nitrogen starvation compared to Pat1Δ cells (Figure 2A). Consistent with this result, the GFP-Atg8 processing assay also showed that autophagy activity could be recovered in Pat1Δ cells by deleting SKI3 (Figure 2B). Similar results were obtained when both LSM1 and SKI3 were deleted (Figure S2A and S2B). Furthermore, Pat1Δ ski3Δ cells showed normal Atg1, Atg2, Atg7 and Atg9 protein expression after nitrogen starvation (Figure 2C) and similar results were seen with lsm1Δ ski3Δ cells compared to lsm1Δ cells (Figure S2B). Correspondingly, ATG1, ATG2, ATG7 and ATG9 mRNA levels after 1 h of nitrogen starvation were significantly higher in the double mutant than in Pat1Δ cells and were similar to WT levels (Figure 2D). Although ATG9 mRNA levels significantly increased after nitrogen starvation in Pat1Δ ski3Δ and ski3Δ cells compared to WT cells, this increase did not lead to a robust increase in Atg9 protein levels. Together, these data indicate that the Pat1-Lsm complex prevents exosome-mediated 3' to 5' ATG mRNA degradation during nitrogen starvation, and this function is necessary for ATG mRNA stabilization, Atg protein expression and normal autophagy induction during starvation.
Fig. 2. SKI3 deletion prevents the pat1Δ-mediated decrease in autophagy.
A) Autophagy activity was measured by the Pho8Δ60 assay in WT, pat1Δ, ski2Δ, sk3Δ pat1Δ ski2Δ, and pat1Δ ski3Δ; strains under growing conditions (+N) and following 3 h of nitrogen starvation (−N). Error bars indicate the standard deviation of 6 independent experiments. ANOVA, **P <0.01 and *** P <0.001. B) Autophagy was measured by GFP-Atg8 processing in WT, pat1Δ, ski3Δ and pat1Δ ski3Δ strains under growing conditions and after 2 h of nitrogen starvation; a representative image is shown. C) Atg1, Atg2-PA, Atg7-PA and Atg9 protein levels were measured by western blot in WT, pat1Δ, ski3Δ and pat1Δ ski3Δ strains under growing conditions and after 1 or 2 h of nitrogen starvation; representative images are shown. D) ATG1, ATG2, ATG7 and ATG9 mRNA levels were determined in WT, pat1Δ, ski3Δ and pat1Δ ski3Δ strains under growing conditions and after 1 h of nitrogen starvation by RT-qPCR. Error bars indicate the standard deviation of 3 independent experiments. ANOVA, ** P<0.01 and *** P <0.001. See also Figure S2.
We decided to extend our analysis on the effect of deleting PAT1 to additional ATG genes by examining Pat1Δ cells with RT-qPCR. Several other ATG genes shared the same mRNA accumulation pattern observed for ATG1, ATG2, ATG7 and ATG9, with no significant differences observed in transcript levels during nutrient-replete conditions and significantly lower mRNA levels after nitrogen starvation (Figure S2C). However, a subset of ATG transcripts including ATG4, ATG17 and ATG18 failed to show any significant difference between Pat1Δ cells and WT cells during both nutrient-replete and nitrogen starvation conditions. Furthermore, ATG3 and ATG8 mRNAs showed distinct patterns of transcript accumulation in Pat1Δ cells. ATG3 mRNA levels were unchanged during nutrient-rich conditions and significantly increased during nitrogen starvation. Conversely, ATG8 transcripts significantly increased during nutrient-replete conditions and remained unchanged compared to WT cells after nitrogen starvation. These data suggest Pat1 is necessary for the accumulation of a specific subset of ATG transcripts.
Pat1 phosphorylation regulates ATG mRNA stabilization, Atg protein expression and autophagy induction during nitrogen starvation.
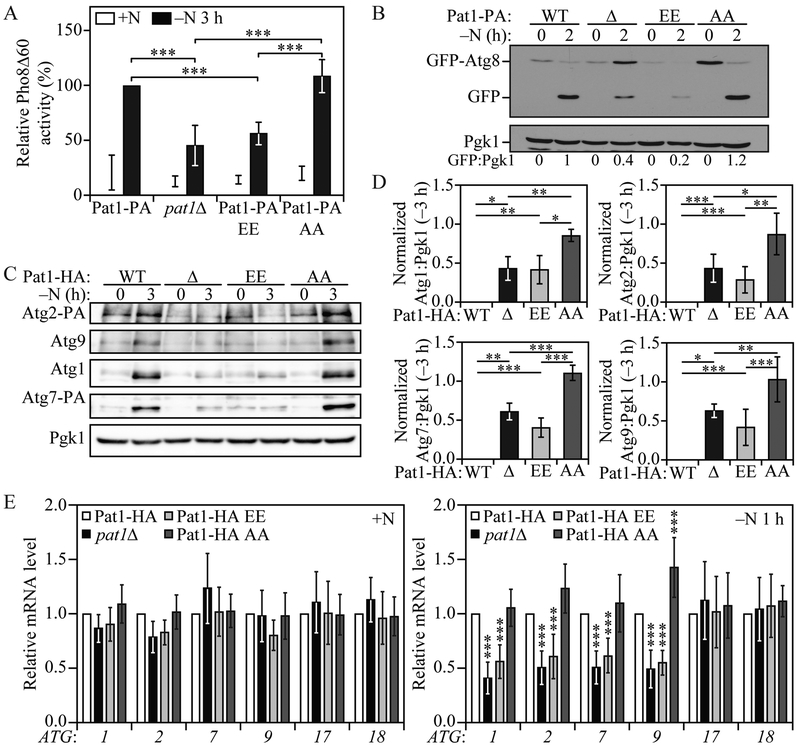
Pat1 has been reported to be phosphorylated in yeast at residues S456 and S457 during nutrient-replete conditions, but to rapidly lose this phosphorylation after glucose starvation. Accordingly, we decided to evaluate if nitrogen starvation could lead to Pat1 dephosphorylation by analyzing the levels of phosphorylated and dephosphorylated Pat1 using SDS-PAGE and a phosphate-binding tag (Phos-tag); Phos-tag decreases the migration speed of phosphorylated proteins when resolved by SDS-PAGE, thus providing better resolution from their dephosphorylated counterparts. Under growing conditions phosphorylated Pat1 levels were higher than that of dephosphorylated Pat1; however, starting at 30 min of nitrogen starvation, a shift in the pool of phosphorylated Pat1 to its dephosphorylated state could be observed (Figure S3A); nitrogen starvation led to a significant decrease in Pat1 dephosphorylation after 60 min and ultimately resulted in a shift to a lower molecular mass form (Figure S3A and S3B). Site-directed mutagenesis of Pat1 S456 and S457 residues to alanine or glutamate have been effectively used to study the effects of nonphosphorylatable and phosphomimetic mutants of Pat1, respectively (Chowdhury et al., 2014; Ramachandran et al., 2011; Shah et al., 2013). We decided to test if Pat1 phosphorylation on these two serine residues could affect autophagy activity by generating Pat1S456,457A (Pat1-AA) and Pat1S456,457E (Pat1-EE) nonphosphorylatable and phosphomimetic mutants, respectively, both containing the PA tag. Pat1-AA migrated similar to Pat1-PA after 30-60 min of nitrogen starvation (Figure S3A), supporting the conclusion that Pat1 was dephosphorylated under these conditions. Whereas Pat1-AA cells failed to show any difference in autophagy activity as measured by both the Pho8Δ60 and GFP-Atg8 processing assays, Pat1-EE cells showed decreased autophagy levels similar to Pat1Δ cells (Figure 3A and 3B). This result suggested that Pat1 dephosphorylation is required for normal autophagy induction during nitrogen starvation.
Fig. 3. Pat1 dephosphorylation on S456 and S457 regulates autophagy by modulating ATG mRNA and protein levels during nitrogen starvation.
A) Autophagy activity was measured by the Pho8Δ60 assay in Pat1-PA, pat1Δ, Pat1S456,457E-PA (Pat1-PA EE), and Pat1S456,457A-PA (Pat1-PA AA) strains under growing conditions (+N) and after 3 h of nitrogen starvation (−N). Error bars indicate the standard deviation of 5 independent experiments. ANOVA, **P <0.01 and *** P <0.001. B) Autophagy was measured by GFP-Atg8 processing in Pat1-PA, pat1Δ, Pat1-PA EE, and Pat1-PA AA strains under growing conditions and after 2 h of nitrogen starvation; a representative image is shown. C) Atg1, Atg2-PA, Atg7-PA and Atg9 protein levels were measured by western blot in Pat1-HA, pat1Δ, Pat1-HA EE, and Pat1-HA AA strains under growing conditions and after 3 h of nitrogen starvation; a representative image is shown. D) Quantification of Atg1, Atg2-PA, Atg7-PA and Atg9 protein levels from (C). Error bars indicate the standard deviation of at least 4 independent experiments. ANOVA, *P <0.05, **P <0.01 and *** P <0.001. E) ATG1, ATG2, ATG7, ATG9, ATG17 and ATG18 mRNA levels were analyzed in Pat1-HA, pat1Δ, Pat1-HA EE, and Pat1-HA AA strains under growing conditions and after 1 h of nitrogen starvation by RT-qPCR. Error bars indicate the standard deviation of at least 8 independent experiments. ANOVA, *** P<0.001. See also Figure S3.
Next, we sought to determine if Pat1 phosphorylation was important for normal Atg protein expression during nitrogen starvation. In agreement with our autophagy activity data, Pat1-AA cells showed no significant difference relative to WT cells, whereas Pat1-EE cells showed a significant decrease in Atg1, Atg2, Atg7 and Atg9 protein expression compared to WT cells, again similar to Pat1Δ cells (Figure 3C and 3D). Finally, to determine if the effects on Atg protein expression in Pat1-EE cells were due to a decreased stabilization/accumulation of ATG mRNA, we measured ATG transcript levels by RT-qPCR. Under nutrient-replete conditions no significant difference in ATG mRNA could be observed. However, after 1 h of nitrogen starvation, Pat1-EE cells showed the same significant decrease in ATG mRNA as Pat1Δ cells (Figure 3E). Conversely, in Pat1-AA cells only ATG9 showed a significant increase in mRNA levels compared to WT cells; however, as shown in figures 3C and 3D, this increase in ATG9 mRNA did not translate into a significant increase in Atg9 protein levels. Neither ATG17 nor ATG18 mRNA, genes for which the transcript levels remained unaffected in Pat1Δ cells (Figure S2C), showed a significant difference in Pat1-EE and Pat1-AA cells (Figure 3E), further indicating that Pat1 and its regulation by phosphorylation only affects a subset of ATG mRNAs. Together, these data indicate that nitrogen starvation leads to Pat1 dephosphorylation, and that dephosphorylation on residues S456 and S457 is required for specific ATG mRNA stabilization, subsequent protein expression and normal autophagy induction.
Pat1 binds specific ATG mRNAs during nitrogen starvation.
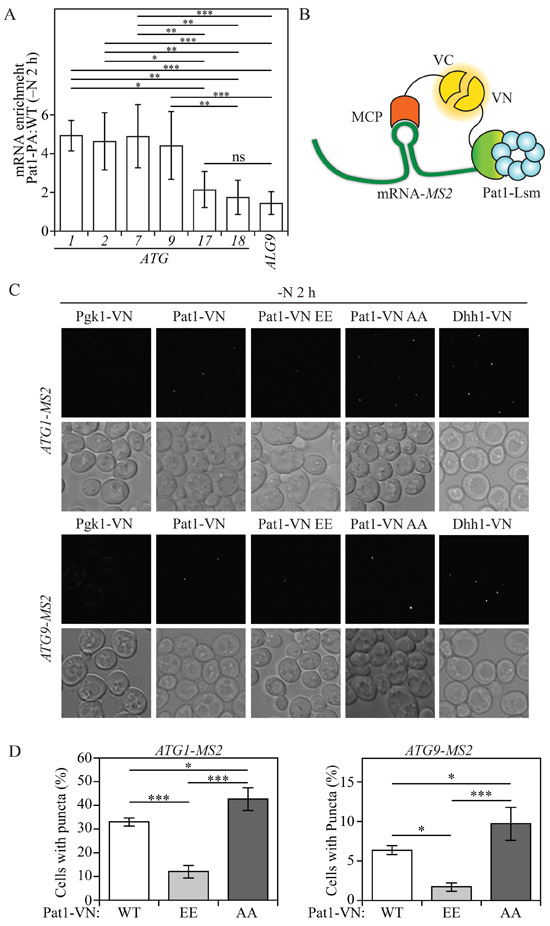
Various reports have indicated that the Pat1-Lsm complex has the ability to bind to the 3' end of oligoadenylated mRNAs (Chowdhury et al., 20l4; Chowdhury et al., 2007; Chowdhury and Tharun, 2008; Tharun et al., 2005), which is proposed to protect the mRNA from exosome-mediated degradation (Chowdhury and Tharun, 2009; He and Parker, 2001; Tharun, 2009). Our data indicate that the Pat1-Lsm complex can prevent the 3' to 5' degradation of certain ATG mRNAs during nitrogen starvation and that this process is regulated by Pat1 phosphorylation on serine residues 456 and 457. To determine if Pat1 binds to ATG mRNA we performed RNA immunoprecipitation (RIP) (Selth et al., 2009). Briefly, Pat1 protein A (PA)-tagged and -untagged cells were starved for 2 h, followed by crosslinking. Lysates were immunoprecipitated and crosslinking was reversed. After DNAse treatment, RNA was extracted and quantified using RT-qPCR.
Our RIP analysis showed that Pat1-PA cells had a significant enrichment in ATG1, ATG2, ATG7, and ATG9 mRNA, but not ATG17 and ATG18 mRNA compared to ALG9 mRNA, which was used as a negative control (Figure 4A). This result indicates that Pat1 preferentially binds ATG1, ATG2, ATG7, and ATG9 mRNA during nitrogen starvation. To further test if Pat1 interaction with ATG mRNA correlated with its effect on ATG mRNA accumulation, we designed a protein-RNA bimolecular fluorescence complementation (protein-RNA BiFC) assay based on the single-molecule analysis of gene expression (Hocine et al., 2013). In brief, an ATG mRNA of interest was tagged with MS2, an RNA hairpin sequence from bacteriophage, without altering the mRNA original 5' UTR and 3' UTR. The MS2 coat protein (MCP) was fused to the C terminus of the Venus variant of the yellow fluorescent protein, vYFP (VC), generating MCP-VC. Similarly, a protein of interest (Pat1, Pgk1 or Dhh1) was tagged with the N terminus of vYFP (VN). By tagging MS2 to the ATG mRNA of interest and MCP to VC, we would expect to observe a vYFP fluorescence signal if the VN-tagged protein of interest interacts with the ATG mRNA of interest (Figure 4B).
Fig. 4. Pat1 binds specific ATG mRNAs.
A) RNA immunoprecipitation was performed in Pat1-PA and WT (untagged) cells. Enrichment (Pat1-PA: WT) of ATG1, ATG2, ATG7, ATG9, ATG17, ATG18 and ALG9 (control) mRNA after 2 h of nitrogen starvation is presented. Error bars indicate the standard deviation of at least 5 independent experiments. ANOVA, * P<0.05, ** P<0.01, *** P<0.001, ns, no statistical significance B) Schematic model for protein-RNA BiFC. Interaction between MS2 coat protein (MCP) tagged with C-terminal vYFP (VC) bound to an MS2 hairpin-tagged ATG mRNA and Pat1-tagged with N-terminal vYFP (VN), leads to a fluorescent signal by a complete vYFP protein. C) Protein-RNA BiFC was used to determine the interaction of Pgk1-VN, Pat1-VN, Pat1-VN AA, Pat1-VN EE and Dhh1-VN with ATG1-MS2- and ATG9-MS2-tagged mRNA after 2 h of nitrogen starvation (−N). D) Quantification of the number of cells showing puncta in Pat1-VN, Pat1-VN EE and Pat1-VN AA cells also expressing either ATG1-MS2- or ATG9-MS2-tagged mRNA. Error bars indicate the standard deviation of at least 3 independent experiments. ANOVA, *P <0.05 and *** P <0.001. See also Figure S4.
During nutrient-rich conditions no fluorescent signal was detected in cells expressing Pat1-VN with either ATG1-MS2, ATG2-MS2 or ATG9-MS2 (Figure S4A and S4B). Conversely, after 2 h of nitrogen starvation, puncta formation could be observed in Pat1-VN cells expressing ATG1-MS2, ATG2-MS2 or ATG9-MS2 (Figure 4C and Figure S4B). This result further indicates that Pat1 binds to these ATG mRNAs during nitrogen starvation. Pgk1-VN cells expressing ATG1-MS2, ATG2-MS2 or ATG9-MS2, which were used as negative controls, failed to form any bright puncta during both nutrient-replete and nitrogen-starved conditions. Conversely, Dhh1-VN cells expressing ATG1-MS2 or ATG9-MS2, which were used as positive controls, formed puncta during both growing and nitrogen-starved conditions, with the latter inducing the formation of a higher number of puncta (Figure 4C and Figure S4A). Dhh1 interaction with ATG1 and ATG9 mRNA during growing conditions is consistent with previous data, highlighting the importance of Dhh1 in regulating ATG1 and ATG9 mRNA decapping and degradation when autophagy is not induced (Hu et al., 2015). Consistently, ATG18 mRNA which failed to show enrichment during our RIP assay, also failed to show Pat1 binding in our protein-RNA BiFC assay, because Pat1-VN cells expressing ATG18-MS2 did not produce any puncta during either growing or nitrogen-starved conditions (Figure S4C). In contrast, our positive control Dhh1-VN cells expressing ATG18-MS2 showed puncta formation after nitrogen starvation (Figure S4C). The fact that Dhh1-VN cells expressing ATG18-MS2 failed to show any puncta during growing conditions, is in line with our previous results showing ATG18 mRNA is not affected by DHH1 deletion (Hu et al., 2015).
To determine the effects of Pat1 phosphorylation on ATG1 and ATG9 mRNA binding we performed our protein-RNA BiFC assay using Pat1-VN EE and Pat1-VN AA, phosphomimetic and nonphosphorylatable mutants, respectively. In both ATG1-MS2- and ATG9-MS2-expressing cells Pat1-VN AA exhibited a significant increase in the number of cells forming puncta after 2 h of nitrogen starvation. Furthermore, cells expressing Pat1-VN EE showed a significant decrease in the number of cells forming puncta with both ATG1-MS2 and ATG9-MS2 after nitrogen starvation (Figure 4C and 4D). Together, these data indicate that Pat1 binds a specific subset of ATG mRNA during nitrogen starvation and that this process is controlled by phosphorylation at residues S456 and S457.
Preventing efficient 3'-5' mRNA degradation recovers autophagy activity in Pat1-EE phosphomimetic cells.
Both PAT1 deletion and the Pat1-EE phosphomimetic point mutation led to a similar decrease in autophagy activity, ATG mRNA accumulation and Atg protein levels under nitrogen starvation conditions. Because the PAT1 deletion phenotype could be rescued by deleting SKI3, which prevents efficient 3’ to 5’ mRNA degradation by the exosome, we hypothesized that the Pat1-EE-mediated decrease in autophagy activity during nitrogen starvation could also be prevented by hindering 3' to 5' mRNA degradation. To this end, we constructed Pat1-EE ski2Δ and Pat1-EE ski3Δ cells and measured autophagy activity using the Pho8Δ60 assay. Deletion of either SKI2 or SKI3 in cells expressing Pat1-EE significantly increased autophagy activity compared to the otherwise WT cells (i.e., Pat1-EE) after 3 h of nitrogen starvation (Figure 5A). Consistent with this finding, the GFP-Atg8 processing assay also showed that the Pat1-EE-mediated decrease in autophagy activity could be prevented by deleting SKI3 (Figure 5B).
Fig. 5. SKI3 deletion prevents the Pat1S456E- and Pat1S457E-mediated decrease in autophagy.
A) Autophagy activity was measured by the Pho8Δ60 assay in Pat1-PA, pat1Δ, Pat1-PA EE, Pat1-PA EE ski2Δ, and Pat1-PA EE ski3Δ strains under growing conditions (+N) and after 3 h of nitrogen starvation (−N). Error bars indicate the standard deviation of 6 independent experiments. ANOVA, **P <0.01 and *** P <0.001. B) Autophagy was measured by GFP-Atg8 processing in Pat1-PA, Pat1-PA EE, Pat1-PA EE ski3Δ and Pat1-PA ski3Δ strains under growing conditions and after 2 h of nitrogen starvation; a representative image is shown. C) Atg1, Atg2-PA, Atg7-PA and Atg9 protein levels were measured by western blot in Pat1-HA, Pat1-HA EE, Pat1-HA EE ski3Δ and Pat1-HA ski3Δ strains under growing conditions and after 3 h of nitrogen starvation; a representative image is shown. D) Quantification of the data from (C). Error bars indicate the standard deviation of at least 4 independent experiments. ANOVA, *P <0.05, **P <0.01 and *** P <0.001. E) ATG1, ATG2, ATG7, ATG9, ATG17 and ATG18 mRNA levels were analyzed in Pat1-HA, Pat1-HA EE, Pat1-HA EE ski3Δ and Pat1-HA ski3Δ strains under growing conditions and after 1 h of nitrogen starvation by RT-qPCR. Error bars indicate the standard deviation of at least 8 independent experiments. ANOVA, *** P <0.001.
We next checked Atg1, Atg2, Atg7 and Atg9 protein levels by western blot during nutrient-rich conditions and after 3 h of nitrogen starvation. As expected, deleting SKI3 in Pat1-EE-expressing cells was able to significantly prevent Atg protein depletion during nitrogen starvation (Figure 5C and 5D). Furthermore, Pat1-EE ski3Δ cells showed a significant ATG1, ATG2 and ATG7 mRNA accumulation after nitrogen starvation compared to Pat1-EE cells. Reminiscent of the Pat1Δ ski3Δ effect on ATG9 mRNA levels, Pat1-EE ski3Δ cells showed a significant increase in ATG9 transcript levels compared to WT cells, which failed to translate into a significant increase in Atg9 protein levels (Figure 5C-E). Once again, ATG17 and ATG18 transcript levels did not show any significant difference among the different strains tested (Figure 5E). Together, these data indicate that when Pat1 residues S456 and S457 are phosphorylated, ATG transcripts fail to accumulate due to 3' to 5' mRNA degradation, which in turn leads to lower Atg protein expression and autophagy induction.
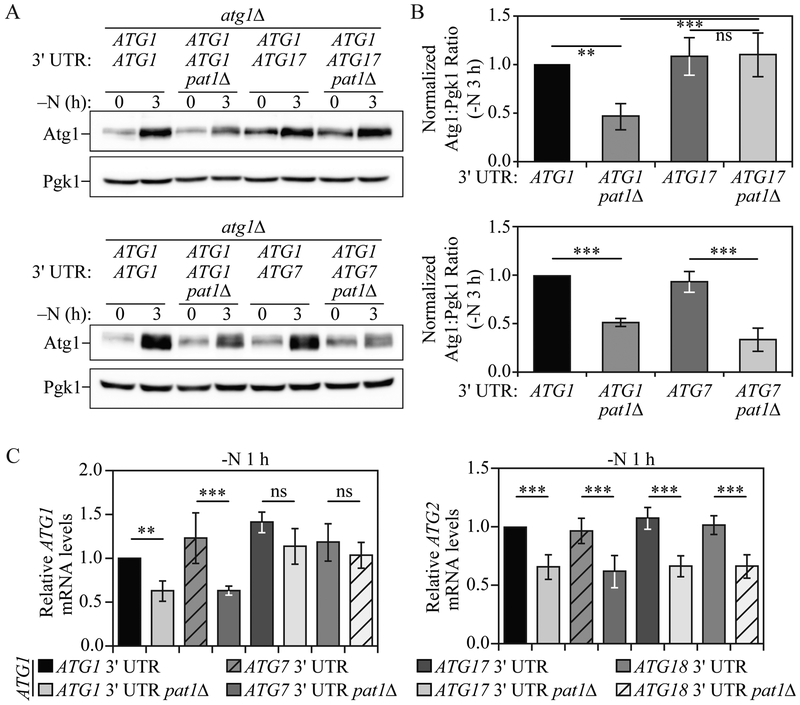
Switching the ATG1 3'UTR with that of ATG17 prevents the PAT1 deletion-mediated decrease in ATG1 mRNA and protein levels.
Our data indicate that during nitrogen starvation Pat1 binds to the 3' UTR of a specific subset of ATG mRNA, such as ATG1, which is necessary for ATG1 transcript stabilization and subsequent protein expression. Because other ATG genes, such as ATG17 and ATG18, were not affected by PAT1 deletion, we hypothesized that switching the ATG1 3' UTR with that of ATG17 or ATG18 could prevent the decrease in ATG1 mRNA and protein levels observed in pat1Δ cells. To this end, we generated atg1Δ strains in which ATG1 was expressed under the control of it endogenous promoter and contained either the ATG1, ATG7, ATG17 or ATG18 3' UTR. The strain where we replaced the ATG1 3' UTR with that of ATG7 was used as a negative control because ATG7/Atg7 mRNA/protein levels were decreased by PAT1 deletion, similar to ATG1/Atg1. Consistent with our previous results (Figure 1), after 3 h of nitrogen starvation atg1Δ pat1Δ cells expressing ATG1 mRNA with its own 3' UTR showed a significant decrease in Atg1 protein levels compared to atg1Δ cells complemented with ATG1 mRNA having its endogenous 3' UTR (Figure 6A and 6B). Conversely, after 3 h of nitrogen starvation PAT1 deletion failed to decrease Atg1 protein levels in atg1Δ cells expressing ATG1 mRNA containing the ATG17 3' UTR. In contrast, switching the ATG1 3' UTR with that of ATG7 did not prevent the decrease in Atg1 protein in pat1Δ cells after 3 h of nitrogen starvation.
Fig. 6. Switching the ATG1 mRNA 3' UTR prevents the PAT1 deletion-mediated decrease in ATG1 mRNA and protein levels.
A) Atg1 protein levels were measured by western blot in atg1Δ; cells and atg1Δ pat1Δ cells expressing ATG1 under its endogenous promoter and with either the ATG1, ATG7 or ATG17 3' UTR during growing conditions and after 3 h of nitrogen starvation; representative images are shown. B) Quantification of the data from (A). Error bars indicate the standard deviation of at least 4 independent experiments. ANOVA, **P <0.01 and *** P <0.001, ns, no statistical significance. C) ATG1 and ATG2 mRNA levels were analyzed by RT-qPCR in atg1Δ; and atg1Δ pat1Δ; cells expressing ATG1 under its endogenous promoter and with either the ATG1, ATG7, ATG17 or ATG18 3' UTR, after 1 h of nitrogen starvation. Error bars indicate the standard deviation of at least 6 independent experiments. ANOVA, **P <0.01, *** P <0.001, ns, no statistical significance.
Similarly, after 1 h of nitrogen starvation, ATG1 mRNA levels, measured by RT-qPCR, were significantly decreased in atg1Δ pat1Δ cells expressing ATG1 mRNA having either the ATG1 or ATG7 3' UTR (Figure 6C). Conversely, switching the ATG1 3' UTR with that of either ATG17 or ATG18 prevented the PAT1 deletion-mediated decrease in ATG1 mRNA levels. Consistent with our previous data (Figure 1C), deleting PAT1 still led to a significant decrease in the mRNA levels of ATG2 with its endogenous 3' UTR in these same strains, indicating that suppression of mRNA loss only occurred in cis. These results further indicate Pat1 is necessary for normal ATG1 mRNA accumulation and protein expression during nitrogen starvation and highlight the importance of the ATG1 3' UTR in its post-transcriptional regulation.
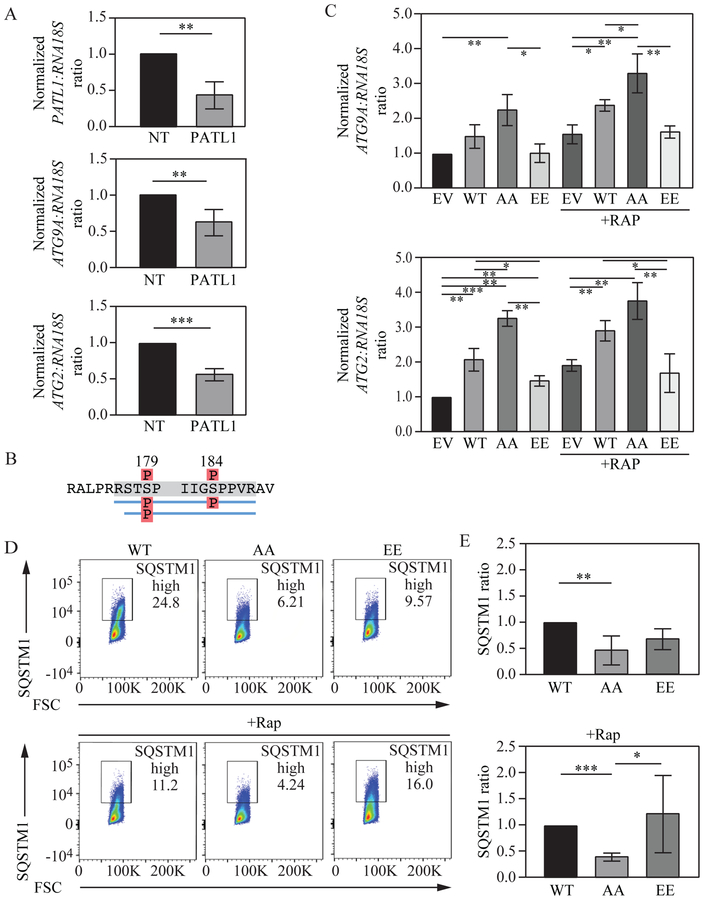
The Pat1 mammalian homolog, PATL1 is required for normal ATG2 and ATG9 mRNA accumulation and autophagy induction.
To determine if the effect of yeast Pat1 on ATG mRNA and autophagy activity is conserved in mammalian cells, we sought to study PATL1, one of the two human homologs of yeast Pat1 (Scheller et al., 2007). The role of PATL1 in mRNA deadenylation and decapping via its interacting protein partners makes it more similar in function to yeast Pat1 in comparison to PATL2, for which a role in mRNA regulation is still not completely understood (Ozgur et al., 2010). To determine the effects of PATL1 on ATG mRNA, PATL1 mRNA levels were depleted using siRNA (Figure 7A). Similar to our data in yeast, both ATG2 and ATG9 mRNA were significantly reduced after PATL1 depletion, suggesting that PATL1 could play a role in ATG mRNA stabilization in mammalian cells.
Fig. 7. PATL1 levels and phosphorylation state regulate ATG2 and ATG9A mRNA levels and autophagy activity.
A) PATL1, ATG2 and ATG9A mRNA levels were measured in Jurkat cells after non-target or PATL1 siRNA treatment. B) Cell lysate from actively growing Jurkat cells was subjected to PATL1 immunoprecipitation followed by mass spectrometry. Identified phosphorylated residues are indicated. C) Jurkat cells were transfected with constructs expressing PATL1 wild-type, AA, EE or empty vector and ATG2 and ATG9A mRNA levels were determined after control and rapamycin treatment. D) Jurkat cells were transfected with constructs expressing PATL1 wild-type, AA or EE and SQSTM1 protein levels were measured through flow cytometry. E) Quantification of the data from (D). Error bars indicate the standard deviation of at least 3 independent experiments. Student’s t-test, ANOVA, *P <0.05, **P <0.01, *** P <0.001.
Yeast Pat1 was dephosphorylated at serine residues 456 and 457 upon nitrogen starvation (Figure S3A and S3B). Previous whole phosphoproteome studies (Hsu et al., PMID: 21659604) had identified two possible phosphorylatable serine residues, S179 and S184 in PATL1 under nutrient-rich conditions, which were confirmed in the present studies by mass spectrometry (Figure 7B). To study the effects of S179 and S184 phosphorylation on ATG mRNA levels, plasmids coding for wild-type, phosphomimetic (PATL1S179,184E; PATL1-EE) or unphosphorylatable (PATL1S179,184A; PATL1-AA) PATL1 were transfected into Jurkat cells, and ATG2 and ATG9A mRNA levels were measured. In nutrient-rich conditions cells expressing PATL1-AA showed a significant increase in ATG9A and ATG2 mRNA levels (Figure 7C) compared to cells expressing exogenous WT PATL1. Conversely, under the same nutrient-rich conditions PATL1-EE-expressing cells showed a significant decrease in both ATG2 and ATG9A mRNA levels. Consistent with our previous yeast data, when autophagy was induced through rapamycin treatment, both ATG2 and ATG9A mRNA levels showed a significant increase in PATL1-AA-expressing cells compared to WT cells. Conversely, PATL1-EE cells showed a significant decrease in ATG2 and ATG9A mRNA levels under autophagy-inducing conditions. To determine if PATL1 unphosphorylatable or phosphomimetic constructs could affect autophagy activity, SQSTM1/p62 levels were determined using a flow cytometry-based assay. Briefly, SQSTM1 is a receptor protein that links poly-ubiquitinated substrates to the Atg8 mammalian homolog LC3B. During autophagy SQSTM1 and the ubiquitinated cargo are sequestered inside autophagosomes and degraded once lysosomal fusion occurs (Pankiv et al., 2007). Thus, by measuring SQSTM1 degradation we can monitor autophagy activity. In both nutrient-rich and autophagy-inducing conditions PATL1-AA-expressing cells showed a significant decrease in SQSTM1 levels indicating increased autophagy flux (Figure 7D and Figure 7E). These results suggest a conserved mechanism in more complex eukaryotes where PATL1 is an important regulatory factor for normal ATG mRNA accumulation and autophagy induction.
Discussion
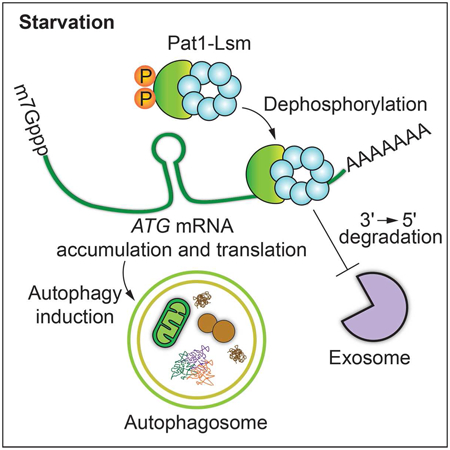
Autophagy is a rapid and robust cellular response to cope with nutrient starvation and other types of stress. To this end, rapid induction of ATG mRNA synthesis through transcription factor activation and inhibition of transcriptional and post-transcriptional repressors is essential for quick autophagy induction. Thus, simultaneous induction of ATG mRNA transcription in addition to reducing degradation proves an efficient strategy for triggering robust autophagy induction. Conversely, during nutrient-rich conditions autophagy is kept at basal levels in part by inhibiting ATG mRNA synthesis and inducing transcript degradation. In a previous study we described a similar mechanism, in which the decapping enzyme Dcp2 was phosphorylated during nutrient-rich conditions, but rapidly dephosphorylated during starvation leading to its inactivation and the accumulation of ATG mRNA (Hu et al., 2015). Here we propose an additional mechanism, which furthers our understanding of how ATG mRNA accumulates under conditions of nutrient starvation. In our model, we propose that during nutrient-replete conditions, many of the ATG mRNAs are continuously degraded by Dcp2-dependent decapping and subsequent 5' to 3' Xrn1-mediated degradation (Delorme-Axford et al., 2018). This prevents ATG mRNA accumulation and autophagy induction, keeping autophagy at basal levels. However, when cells go through periods of nutrient deprivation, Dcp2-dependent decapping is inhibited, preventing 5' to 3' mRNA degradation. Simultaneously, Pat1 is dephosphorylated leading to Pat1-Lsm complex binding to specific ATG mRNA 3' UTRs, which prevents exosome-mediated 3' to 5' degradation; the result is the accumulation of ATG transcripts and robust autophagy induction (Figure S5).
We identified several ATG mRNA targets whose accumulation patterns during nitrogen starvation were affected by PAT1 deletion. Four of these ATG mRNA targets, ATG1, ATG2, ATG7 and ATG9 were further examined, and showed increased Pat1 binding after nitrogen starvation. However, several other ATG mRNAs, including ATG17 and ATG18 transcripts, were not affected by PAT1 deletion and did not show increased Pat1 binding during nitrogen starvation. Further analysis of ATG1 mRNA after replacing its 3' UTR with that of genes not affected by PAT1 deletion, ATG17 or ATG18, further highlighted the importance of the ATG1 3' UTR in Pat1-mediated regulation of ATG mRNA accumulation and subsequent protein expression during nitrogen starvation. While no specific Pat1-Lsm complex mRNA binding sequence has been found, several studies have reported Pat1-Lsm complex affinity for the 3' UTR of oligoadenylated mRNAs carrying U-tracts (6 or more uracils) on or near their 3' end (Chowdhury et al., 2014; Chowdhury et al., 2007; Song and Kiledjian, 2007; Tharun, 2009). Whereas all ATG mRNAs affected by PAT1 deletion contain uracil repeats near their 3' UTR, other non-affected ATG transcripts also have uracil repeats close to their 3' end. Thus, U-tracts may be important for Pat1-Lsm binding, but are not the only required feature. It is possible that the Pat1-Lsm complex binds not a specific sequence, but rather a secondary structure formed in oligoadenylated mRNA containing U-tracts. While this publication was under revision a recent study proposed that the Pat1-Lsm complex can bind to stress-activated mRNA during hyperosmotic shock (Garre et al., 2018). Further supporting the idea that the Pat1-Lsm complex can selectively bind and regulate specific mRNAs. Further studies will be required to determine the exact mechanism by which the Pat1-Lsm complex discriminates between different subsets of ATG and other stress-related mRNAs.
Two specific mRNAs that we evaluated, ATG3 and ATG8, showed distinct patterns when PAT1 was deleted. ATG8 transcript levels were high during growing conditions and remained similar to the WT during nitrogen starvation. In contrast, ATG3 mRNA levels were similar to WT in growing conditions and increased during nitrogen starvation. In particular, the ATG8 mRNA accumulation pattern was similar to the one observed when DHH1 was deleted (Hu et al., 2015). Pat1 is a key scaffold protein in the recruitment of different components involved in RNA decapping and translation inhibition, including Dhh1 (Nissan et al., 2010; Ramachandran et al., 2011). Thus, we cannot exclude the possibility that the effects of PAT1 deletion on ATG8 mRNA levels may be an indirect consequence of not recruiting Dhh1. This possibility would explain the similarities in ATG8 transcript accumulation between DHH1 and PAT1 deletion.
PKA has been reported as the kinase responsible for Pat1 phosphorylation on residues Ser456 and Ser457 (Ramachandran et al., 2011). The same study provided evidence that glucose starvation, a known inactivator of PKA signaling, triggers rapid Pat1 dephosphorylation. Our current study indicates that Pat1 dephosphorylation on these residues also occurs during nitrogen starvation, a condition that inactivates TOR signaling. We observed that nitrogen starvation shifted the pool of phosphorylated Pat1 to its dephosphorylated state, suggesting that the TOR signaling pathway could regulate Pat1 phosphorylation. Furthermore, Pat1 phosphorylation was predicted to decrease following treatment with rapamycin, a well characterized TOR inhibitor (Huber et al., 2009), and was found to decrease in the mammalian PATL1 (Hsu et al). The roles of both PKA and TOR in negatively regulating autophagy have been extensively studied (Stephan et al., 2010), and possible cross-talk between the TOR and PKA pathways has been proposed (Schmelzle et al., 2004; Soulard et al., 2010). Thus, nitrogen starvation could lead to Pat1 dephosphorylation by TOR inactivation and subsequent PKA inhibition.
Multiple studies have highlighted Pat1-Lsm as a decapping enhancer complex (Bonnerot et al., 2000; Bouveret et al., 2000; Tharun et al., 2000). However, Pat1-Lsm has also been proposed to stabilize the 3' end of oligoadenylated mRNA by preventing trimming and 3' to 5' degradation by the exosome (He and Parker, 2001; Tharun, 2009). In this study, we have uncovered a specific role for the mRNA stabilizing function of Pat1-Lsm. During nitrogen starvation, the Pat1-Lsm complex facilitates the accumulation of a specific subset of ATG transcripts by preventing their 3' to 5' degradation by the exosome. While not all ATG transcripts are stabilized by Pat1-Lsm, the critical role in autophagy of most of those affected by Pat1-Lsm accounts for the drastic decrease in autophagy induction when PAT1 or LSM1 are deleted. Furthermore, key aspects were repeated with the mammalian homolog, PATL1, which indicated a similar role in autophagy flux and nutrient-dependent PATL1 phosphorylation, pointing to this function as being conserved through evolution. Autophagy is a key recycling process involved in a diverse variety of physiological conditions, ranging from development to disease prevention. The present data further expand our knowledge surrounding the post-transcriptional regulation governing autophagy.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the Lead Contact, Daniel J. Klionsky (klionsky@umich.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAIL
All yeast Saccharomyces cerevisiae strains used in this study were constructed in the SEY6210 genetic background (see Table S1). Deletions and double-deletions were performed using PCR-based methods (Gueldener et al., 2002) (see Table S2). Strains carrying PA-tagged Atg2 and Atg7 maintaining their original 3' UTRs were generated by PCR-amplifying plasmid pMJ160 and, following transformation, selection and Cre-loxP marker removal as described previously (Gueldener et al., 2002). Pat1-PA, and the Pat1S456,457E-HA and Pat1S456,457A-HA genomic point mutations were generated as previously described (Longtine et al., 1998; Toulmay and Schneiter, 2006).
Strains were grown in YPD medium (1% [w:v] yeast extract, 2% [w:v] peptone, and 2% [w:v] glucose) at 30°C to mid-log phase and then collected. Strains grown in YPD medium were shifted into medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% [w:v] glucose) cultured at 30°C for the indicated time points and then collected. Pho8Δ60 and western blot experiments and analyses were performed as previously described (Noda and Klionsky, 2008; Shintani and Klionsky, 2004). Plasmid pRS-ATG1 (406) expressing ATG1 under its endogenous promoter was used to switch the ATG1 3' UTR with that of ATG7, ATG17 or ATG18 through fast cloning (Li et al., 2011). Strains expressing ATG1 with different 3' UTR sequences were generated by digesting the corresponding plasmids with StuI and transforming in a atg1Δ strain.
Human Jurkat cells were cultured in RPMI 1640 (10% FBS and 1% penicillin and streptomycin) at 37°C in the presence of 5% CO2. Cell transfections were carried out using an Amaxa Cell line Optimization Nucleofector Kit (LONZA). PATL1 siRNA (E-015591-00-0005) and non-target siRNA pools (D-001919-10-05) were purchased from Dharmacon.
METHOD DETAILS
RNA and RT-qPCR
The RNA extraction protocol and qPCR primers were published previously (Hu et al., 2015). cDNA samples were analyzed using a Bio-Rad CFX Connect Real-Time System. Samples were tested in Hard-Shell® 96-clear well black shell plates (Bio-Rad). The reaction mix (15 μl final volume) consisted of 7.5 μl Radiant™ Green Lo-ROX qPCR kit (Radiant), 0.6 μl each primer (400 nM final concentration), 1.3 μl H2O, and 5 μl of a 1:5 dilution of the cDNA preparation. The thermocycling program consisted of one hold at 95°C for 3 min, followed by 40 cycles of 5 sec at 95°C and 25 sec at 62°C. After completion, a melting curve was generated to verify PCR specificity, as well as the absence of contamination and primer dimers. The transcript abundance in samples was determined using the CFX Manager™ Software regression method. Relative abundance of reference mRNAs and normalization for different total RNA amounts was done as described previously (Hu et al., 2015).
Auxin-inducible degron (AID) system and transcriptional inhibition
Saccharomyces cerevisiae SEY6210 cells were first transformed with the plasmid pNHK53 (ADH1p-OsTIR1-9MYC). PAT1 was then tagged with AID-9MYC by homologous recombination (see Table S2). The DNA fragments used for transformation were amplified with pHIS3-AID*-9MYC (Addgene, 99524; deposited by Dr. Helle Ulrich) as the template DNA. The auxin-inducible degron refers to the 71-116 amino acids of the AT1G04250/ATIAA17 protein in plants.
To deplete Pat1 protein levels, the cells were treated with 300 μM 3-indoleacetic acid (auxin; Sigma) or DMSO (vehicle) during mid-log phase growth in YPD medium for 30 min to induce degradation of Pat1. Subsequently samples were collected for western blot. For RNA extraction and RT-qPCR, cells were grown in YPD medium to mid-log phase, washed and shifted to SD-N for nitrogen starvation. Auxin (300 μM), DMSO and/or 1,10 phenathroline (200 μg/ml) was added to the SD-N medium. After 1 h of starvation and treatment, samples were collected as described above.
Protein-RNA bimolecular fluorescence complementation assay
We constructed ATG(X)-MS2 MCP-VC strains through several steps. First, ATG genes of interest were tagged with 24 MS2 hairpins. pDZ415 (24MS2SL-loxP-KANMX6-loxP; Addgene, 45162, deposited by Dr. Robert Singer and Dr. Daniel Zenklusen) was used as a template to PCR amplify the DNA fragments containing 24 MS2 hairpins, homologous regions to the 3' UTR of ATG genes and the KANMX6 marker flanked by loxP sites. Yeast strain SEY6210 was transformed with the PCR products, and positive colonies were selected for by growth on YPD plates containing G418, then confirmed by PCR. Next, the KANMX6 marker was removed by introducing Cre recombinase into the cells, resulting in ATG(X)-MS2 strains. Next, we made the pCu-MCP-2yeGFP (405) plasmid by two-step cloning. The CUP1 promoter was amplified by PCR and ligated into the pRS405 vector between the XmaI and SpeI sites to make pCu405. Then MCP-2yeGFP together with the CYC1 terminator sequence was PCR amplified from pDZ274 (MET25p-MCP-2yEGFP; Addgene, 45929, deposited by Dr. Robert Singer and Dr. Daniel Zenklusen) (Hocine et al., 2013) and ligated into pCu405 between the SpeI and SacII sites to make pCu-MCP-2yeGFP(405); this was linearized by AflII digestion and integrated into the genome of ATG(X)-MS2 strains to make the ATG(X)-MS2 MCP-2yeGFP strains.
Finally, to make the ATG(X)-MS2 CUP1p-MCP-VC strains, we exchanged the 2yeGFP fragment in the genome of the ATG(X)-MS2 CUP1p-MCP-2yeGFP strains with the VC fragment through homologous recombination. To do this, a DNA fragment with the VC peptide-coding sequence and homologous regions from the MCP-coding sequence was amplified by PCR using pFA6a-VC-His3MX6 (Sung and Huh, 2007) as template. ATG(X)-MS2 CUP1p-MCP-VC strains were generated from ATG(X)-MS2 CUP1p-MCP-2yeGFP strains by transforming the VC-encoding DNA fragment, selecting colonies and confirming correct integration by PCR. In the background of ATG(X)-MS2 CUP1p-MCP-VC strains, C-terminal tagging of VN at the PAT1, PGK1, and DHH1 loci in the genome was carried out as described previously (Sung and Huh, 2007); for primers specifics see Table S2. The BiFC strains were examined by fluorescence microscopy under the indicated conditions using softWoRx software (GE Healthcare).
RNA immunoprecipitation
The RNA immunoprecipitation protocol was adapted from a previously published procedure (Selth et al., 2009). A Pat1-PA tagged strain and an untagged strain were cultured to mid-log phase and nitrogen starved for 2 h. Cross-linking was performed by adding formaldehyde to 0.8% and shaking slowly for 10 min at room temperature. To stop cross-linking, glycine was added to a final concentration of 0.2 M and cultures were slowly shaken for 5 min. Cultures were then harvested, washed in PBS and resuspended in FA lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1 % SDS), containing 5 mM PMSF, 1 tablet of complete protease inhibitor cocktail (Roche) and RNasin® PLUS RNAse inhibitor (Promega). Cultures were vortexed at 4°C using glass beads to lyse the cells, centrifuged (1000 x g, 1 min) and the supernatant was collected. Samples were sonicated at 4°C using three 15-sec pulses of 45% amplitude, with a minute break in between when samples were held on ice. Sonicated samples were collected by centrifugation, and the supernatant was collected and then divided into input and IP fractions. IP fractions were incubated with IgG Sepharose™ 6 Fast Flow beads (GE healthcare Life Sciences), overnight at 4°C, while input fractions were frozen in liquid nitrogen and left at −80°C. IP fractions were washed with FA lysis buffer several times, resuspended in RIP elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1% SDS) and incubated at 70°C for 10 min. IP supernatant and input samples were collected and reverse cross-linking was performed by addition of proteinase K and incubation for 1 h at 42°C, followed by 1 h at 65°C. Next, acid-phenol:chloroform was added to the samples, which were mixed and centrifuged. The aqueous layer of each sample was recovered and 25 μl 3M sodium acetate, 20 μg glycogen, and 625 μl ice-cold 100% ethanol were added to precipitate RNA for 1-2 h at −80°C. Samples were centrifuged, washed with 70% ethanol and dried for 15 min. Pellets were resuspended in 90 μl of RNA-free water after which 10 μl of TURBO Dnase buffer, 2 μl of TURBO™ DNase (TURBO DNA-free™ kit, Invitrogen) and 0.5 μl of RNasin® PLUS RNAse inhibitor were added. Samples were incubated for 45 min at 37°C to eliminate DNA. Following incubation, DNAse was inhibited using the DNAse inactivation reagent that came with the kit. Samples were then subjected to RT-qPCR as described above.
Western blot imaging
Differences in western blot images between panels are due to two different imaging techniques being used. For some images, photographic film was used to developed membranes, whereas imaging was also performed through the ChemiDoc™ Touch imaging system (Bio-Rad).
PATL1 DNA constructs and site-directed mutagenesis
The cDNA encoding full-length human PATL1 was amplified by RT-PCR from total RNA extracted from Jurkat cells using primers PATL1-S-KPN1 and PATL1-A-NOT1 (see Table S2). and cloned into compatible sites of a pCMV-GFP vector (Addgene). Mutations were introduced by site-directed mutagenesis and all expression constructs were confirmed by DNA sequencing. Primers used to generate expression constructs for the phosphomimic were PATL1-EE1 and PATL1-EE2, and phosphodeficient mutations used PATL1-AA1 and PATL1-AA2 (see Table S2).
SQSTM1 flow cytometry
Jurkat cells were grown in RPMI 1640 media with 10% FBS and 1% penicillin and streptomycin at 37°C with 5% CO2 until cell counts were 3×105/ml. Cells were treated with rapamycin (200 ng/ml) overnight, and non-treated cells were used as a control. Cells were harvested and washed with 0.2% BSA in PBS twice, and then fixed in fixation buffer (Invitrogen) 1 h at room temperature. After fixation, cells were washed with 1X permeabilization buffer for 30 min at room temperature. Goat serum (10 μl) was added for blocking and incubated for another 30 min at room temperature. SQSTM1/p62 monoclonal antibody (1 μl; Abcam, ab56416) was added, and mouse monoclonal IgG2a (Abcam, ab170191) was used as an isotype control. After incubation for 1 h at room temperature, cells were washed once with 1X permeabilization buffer, and then 1 μl secondary antibody (Alexa Fluor 594 goat anti-mouse) was added. Cells were incubated at room temperature for 1 h and analyzed with a BD Fortessa flow cytometer.
Mass spectrometry and protein identification
Jurkat cells were grown in RPMI, cells were lysed and PATL1 immunoprecipitated using an anti-PATL1 antibody (Abcam, ab124257); immunoprecipitated proteins were resolved by to SDS-PAGE. Identification of all proteins was performed on reduced and alkylated, trypsin-digested samples prepared by standard mass spectrometry protocols. The supernatant and two washes (5% formic acid in 50% acetonitrile) of the gel digests were pooled and concentrated by SpeedVac (Labconco) to dryness directly in 200 μl polypropylene auto-sampler vials (Sun Sri). The recovered peptides were resuspended in 5 μl of solvent A (0.1% formic acid, 99.9% water).
Mass spectrometry analysis was performed on an Orbitrap Fusion Tribrid Mass Spectrometer with in-line chromatography with a PepMap 100 C18 pre-column (3-μm particle size, 75-μm ID, 2-cm length). Nano LC-MS (LC-MS/MS) was performed with a ProXeon Easy-nLC 1000 multi-dimensional liquid chromatograph and temperature controlled Nanospray Flex Ion Source (ThermoFisher Scientific). Peptides were separated at 200 nl/min using a PepMap 100 C18 column (2-μm particle size, 75-μm ID, 50-cm length). The mobile phase consisted of a linear gradient prepared from solvent A and solvent B (0.1% formic acid, 99.9% acetonitrile) at room temperature. Acquisitions followed a decision tree EThcD/CID methodology with a combination of data-dependent acquisition and an inclusion list (Swaney et al., 2008). The targeted list was formulated to cover all potential m/z for phosphorylated PATL1 peptides and were subjected to fragmentation by EThcD.
Data processing and databank searching were performed with Peaks Studio 8.5 (Bioinformatics Solution Inc, Waterloo, ON, Canada). The data was searched against the human, pig, and cow proteins deposited in the Uniprot KB (9/2017) and the common Repository of Adventitious Proteins (theGPM.org). Carbamidomethylation [C] was set as a fixed modification, while oxidation [M], phosphorylation [S/T/Y] and the SILAC labels 13C(6)15N(2) [K] and 13C(6)15N(4) [R] were used as variable modifications. Peptides were filtered at a 0.5% false discovery rate calculated using a decoy sequence approach with a 2 peptides per protein minimum.
QUANTIFICATION AND STATISTICAL ANALYSES
Microscopy imaging data analysis and image processing was carried out in softWoRx software (GE Healthcare). Cellular imaging sample sizing were chosen to be the minimum number of independent experiments required for statistically significant results. Western blot images were quantified by ImageJ software. Statistical analyses were performed using GraphPad Prism 6. Statistical significance was determined in all cases from at least 3 independent experiments using either Student’s t-test or ANOVA. Differences with a P value < 0.05 or lower were considered significant. *p<0.05, **p<0.01, ***p<0.001. Number of independent experiments (n), statistical test utilized, dispersion of measurements and significance is described in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-YFP | Clontech | 632381 |
| Anti-peroxidase (anti-protein A) | Jackson Immunoresearch | 323-005-024 |
| Anti-Pgk1 | Dr. Jeremy Thorner | N/A |
| Anti-Atg1 | In house | N/A |
| Anti-Atg9 | In house | N/A |
| Goat anti Rabbit IgG, HRP | Fisher Scientific | ICN55676 |
| Rabbit anti Mouse IgG, HRP | Jackson Immunoresearch | 315-035-003 |
| Mouse monoclonal IgG2a | Abcam | ab170191 |
| Alexa Fluor 594 goat anti-mouse | Thermo Fisher scientific | A-11005 |
| Anti-PATL1 antibody | Abcam | ab124257 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1,10 Phenanthroline | Sigma-Aldrich | 320056 |
| 3-Indole acetic acid | Sigma-Aldrich | I2886 |
| p-Nitrophenyl phosphate | Sigma-Aldrich | N4645 |
| Radiant™ Green 2x qPCR Mix Lo-ROX | Alkali Scientific | QS1020 |
| Complete EDTA-free protease inhibitor cocktail | Roche | 05056489001 |
| IgG Sepharose™ 6 Fast Flow beads | Fisher Scientific | 45-000-173 |
| TURBO DNA-free™ kit | Fisher Scientific | AM1907 |
| RNasin® PLUS RNAse inhibitor | Fisher Scientific | PRN2615 |
| Critical Commercial Assays | ||
| Nucleospin® RNA | Clontech | 740955.250 |
| BCA Protein assay | Fisher Scientific | PI23223, PI23224 |
| Amaxa Cell line Optimization Nucleofector Kit | LONZA | VC0-1001 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Jurkat cells | ATCC | ATCC TIB-152 |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: strain background: SEY6210 | See Supp Table 1. | |
| Oligonucleotides | ||
| Primers for RT-qPCR | Hu et al., 2015 | |
| Plasmid Cloning and strain construction primers | See Supp Table 2. | |
| PATL1 siRNA | Dharmacon | E-015591-00-0005 |
| non-target siRNA | Dharmacon | D-001919-10-05 |
| Recombinant DNA | ||
| pMJ160 | This study | |
| pRS-ATG1 (406) | This study | |
| pRS-ATG1 ATG7-3’UTR (406) | This study | |
| pRS-ATG1 ATG17-3’UTR (406) | This study | |
| pRS-ATG1 ATG18-3’UTR (406) | This study | |
| pNHK53 | Morawska and Ulrich. 2013 | |
| pHIS3-AID*-9MYC | Morawska and Ulrich. 2013 | Addgene, 99524 |
| pDZ415 | Hocine et al. 2013 | Addgene, 45162 |
| pCu-MCP-2yeGFP (405) | This study | |
| pDZ274 | Hocine et al. 2013 | Addgene, 45929 |
| pFA6a-VC-His3MX6 | Sung and Huh. 2007 | |
| pCMV-GFP vector | Matsuda and Cepko 2004 | Addgene, 11153 |
| Software and Algorithms | ||
| CFX Manager™ Software | Bio-Rad | |
| Other | ||
Highlights.
The Pat1-Lsm complex binds to a subset of ATG mRNAs during nitrogen starvation.
Pat1-Lsm binding to ATG mRNAs prevents their 3' to 5' degradation by the exosome.
Dephosphorylation of Pat1 during nitrogen starvation regulates binding to ATG mRNA.
Pat1-dependent accumulation of ATG mRNA allows robust autophagy induction.
Acknowledgements
This work was supported by NIH grant GM053396 to DJK and in part by the Intramural Research Program of the NIH, NIAID to PRW.
Abbreviations:
- Atg
autophagy related
- BiFC
bimolecular fluorescence complementation
- MCP
MS2 coat protein
- Pat1-AA
Pat1S456,457A
- Pat1-EE
Pat1S456,457E
- RIP
RNA immunoprecipitation
- UTR
untranslated region
- vYFP
venus variant of the yellow fluorescent protein
- WT
wild type
Footnotes
Declaration of Interests
The authors declare no competing interests.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JS, and Parker RP (1998). The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. EMBO J. 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, and Katada T (2001). Ski7p G protein interacts with the exosome and the Ski complex for 3'-to-5' mRNA decay in yeast. EMBO J. 20, 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Jin M, Gonzalez-Rodriguez P, Fullgrabe J, Delorme-Axford E, Backues SK, Joseph B, and Klionsky DJ (2015a). Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 25, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Jin M, Xu Z, and Klionsky DJ (2015b). A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy 11, 2114–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, and Lapeyre B (2000). The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20, 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, and Seraphin B (2000). A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JT, Bai X, and Johnson AW (2000). The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Kalurupalle S, and Tharun S (2014). Pat1 contributes to the RNA binding activity of the Lsm1-7-Pat1 complex. RNA 20, 1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, and Tharun S (2007). The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13, 998–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, and Tharun S (2008). lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA 14, 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A, and Tharun S (2009). Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7-Pat1 complex. RNA 15, 1837–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E, Abernathy E, Lennemann NJ, Bernard A, Ariosa A, Coyne CB, Kirkegaard K, and Klionsky DJ (2018). The exoribonuclease Xrn1 is a post-transcriptional regulator of autophagy. Autophagy 14, 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Backues SK, Baba M, Heo JM, Harper JW, and Klionsky DJ (2016). Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy 12, 648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, and Klionsky DJ (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yao Z, and Klionsky DJ (2015). How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 25, 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, and Wilusz CJ (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 8, 113–126. [DOI] [PubMed] [Google Scholar]
- Garre E, Pelechano V, Sanchez Del Pino M, Alepuz P, and Sunnerhagen P (2018). The Lsm1-7/Pat1 complex binds to stress-activated mRNAs and modulates the response to hyperosmotic shock. PLoS Genet. 14, e1007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, and Hegemann JH (2002). A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H et al. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. [DOI] [PubMed] [Google Scholar]
- He C, and Klionsky DJ (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, and Parker R (2001). The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3' termini from partial degradation. Genetics 158, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocine S, Raymond P, Zenklusen D, Chao JA, and Singer RH (2013). Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods 10, 119–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, McQuiston T, Bernard A, Park YD, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, et al. (2015). A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 17, 930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, and Loewith R (2009). Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23, 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, and Ohsumi Y (2010). Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wen A, Shen B, Lu J, Huang Y, and Chang Y (2011). FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Morawska M, and Ulrich HD (2013). An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, and Parker R (2010). Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39, 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, and Klionsky DJ (2008). The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 451, 33–42. [DOI] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, and Stoecklin G (2010). Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell. Biol. 30, 4308–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, and Johansen T (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- Parker R (2012). RNA degradation in Saccharomyces cerevisae. Genetics 191, 671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Shah KH, and Herman PK (2011). The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43, 973–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Resa-Infante P, de la Luna S, Galao RP, Albrecht M, Kaestner L, Lipp P, Lengauer T, Meyerhans A, and Diez J (2007). Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim. Biophys. Acta 1773, 1786–1792. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, and Hall MN (2004). Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24, 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Gilbert C, and Svejstrup JQ (2009). RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb. Protoc. 2009, pdb prot5234. [DOI] [PubMed] [Google Scholar]
- Shah KH, Zhang B, Ramachandran V, and Herman PK (2013). Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, and Klionsky DJ (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, and Kiledjian M (2007). 3' Terminal oligo U-tract-mediated stimulation of decapping. RNA 13, 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, and Hall MN (2010). The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell. 21, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, and Herman PK (2010). The Tor and cAMP-dependent protein kinase signaling pathways coordinately control autophagy in Saccharomyces cerevisiae. Autophagy 6, 294–295. [DOI] [PubMed] [Google Scholar]
- Sung MK, and Huh WK (2007). Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 24, 767–775. [DOI] [PubMed] [Google Scholar]
- Swaney DL, McAlister GC, and Coon JJ (2008). Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S (2009). Lsm1-7-Pat1 complex: a link between 3' and 5'-ends in mRNA decay? RNA Biol. 6, 228–232. [DOI] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, and Parker R (2000). Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404, 515–518. [DOI] [PubMed] [Google Scholar]
- Tharun S, Muhlrad D, Chowdhury A, and Parker R (2005). Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3' end protection. Genetics 170, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, and Schneiter R (2006). A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 23, 825–831. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Staples RR, Baker RE, and Parker R (2000). Function of the ski4p (Csl4p) and Ski7p proteins in 3'-to-5' degradation of mRNA. Mol. Cell. Biol. 20, 8230–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.