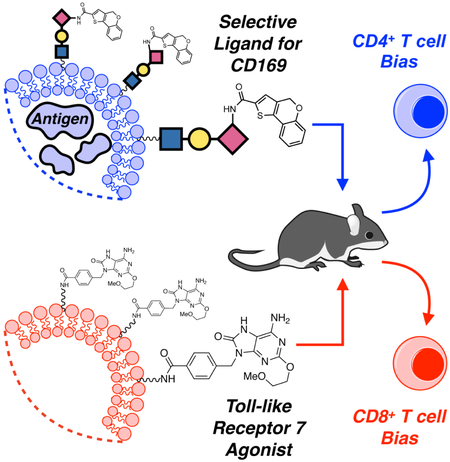
SUMMARY
Macrophages (MØs) expressing the endocytic sialic acid-binding immunoglobulin-like lectin 1 (siglec-1, CD169, sialoadhesin) are known to be adept at antigen capture — primarily due to their strategic location within lymphatic tissues. Antigen concentrated in these cells can be harnessed to induce potent anti-tumor/anti-pathogen cytotoxic (CD8+) T cell responses. Here, we describe a chemical platform that exploits the CD169-mediated antigen capture pathway for biased priming of antigen-specific CD4+ or CD8+ T cells in vivo. In the absence of a toll-like receptor (TLR) agonist, antigen delivery through CD169 produced robust CD4+ T cell priming only. However, simultaneous treatment with targeted antigen and a TLR7 agonist induced CD8+ T cell priming, with concomitant suppression of the CD4+ T cell response. We exploited these observations to manipulate the activation ratio of CD4+/CD8+ T cells in the same animal. These findings represent a unique chemical strategy for targeting CD169+ macrophages to modulate antigen-specific T cell immunity.
Graphical Abstract
eTOC Paragraph:
Macrophages expressing the sialic acid-binding lectin CD169 are involved in mounting antigen specific T cell responses. Here, Edgar et al. describe a CD169-targeted liposome-based platform able to selectively activate and deliver antigen to these macrophages. This modular platform enabled control over antigen-specific CD4+:CD8+ T cell ratios in vivo.
INTRODUCTION
Macrophages (MØs) are key mediators of innate immunity, possessing phenotypes that are plastic in response to environmental cues such as pathogen associated molecular patterns and cytokines produced by other leukocytes (Sica et al., 2012; Chávez-Gálan et al. 2015). Beyond their role as innate immune sentinels, recent work has underscored the importance of MØs in the adaptive immune response (Varol et al., 2015; Varol et al., 2010; Junt et al., 2007). Specifically, MØs expressing sialoadhesin, also known as sialic acid-binding immunoglobulin-like lectin 1 (siglec-1), or CD169 (CD169MØs), have generated significant attention as they have been shown to be adept at capture of antigen in circulating lymph, mainly due to their strategic location in the marginal zone of the spleen (Mebius et al., 2005). Antigen capture by these cells can result in potent cytotoxic (CD8+) T cell responses, either through direct crosspresentation of antigen, or indirectly, through antigen transfer to cross-presenting dendritic cells (DCs) (Junt et al. 2007; Backer et al., 2010; Asano et al., 2011; Bernhard et al., 2015; van Dinther et al., 2018). Due to the importance of CD8+ T cell responses in anti-tumor immunity and infection, CD169MØs have become an attractive target for selective delivery of antigen. We previously demonstrated that IFN-α-stimulated MØs cultured from primary bone marrow progenitors (BMMØs) express CD169 and can induce CD4+ T cell expansion in vitro when an appropriate antigen is delivered via CD169-targeted liposomal nanoparticles; however, these BMMØs do not necessarily represent a comparable surrogate for native splenic CD169MØs (Chen et al., 2012). Here, we extend the liposome platform to delivery of antigen to native CD169MØs in vivo and interrogate the downstream consequences of this delivery on both CD4+ and CD8+ T cell priming. We discovered that in the absence of adjuvant, selective delivery of antigen to CD169MØs induced robust CD4+ T cell priming; however, no CD8+ T cell responses were observed. To promote CD8+ T cell priming, we coupled targeted antigen delivery to TLR7-induced activation of CD169MØs to measure the effect of TLR7 ligation on cross-presentation bias to CD8+ T cells and discovered that conditions favorable for expansion of CD8+ T cells were less favorable for CD4+ T cells. This reciprocal T cell behavior enabled us manipulate CD4+/CD8+ T cell ratios in a single organism through strategic liposome dosing regimes.
RESULTS AND DISCUSSION
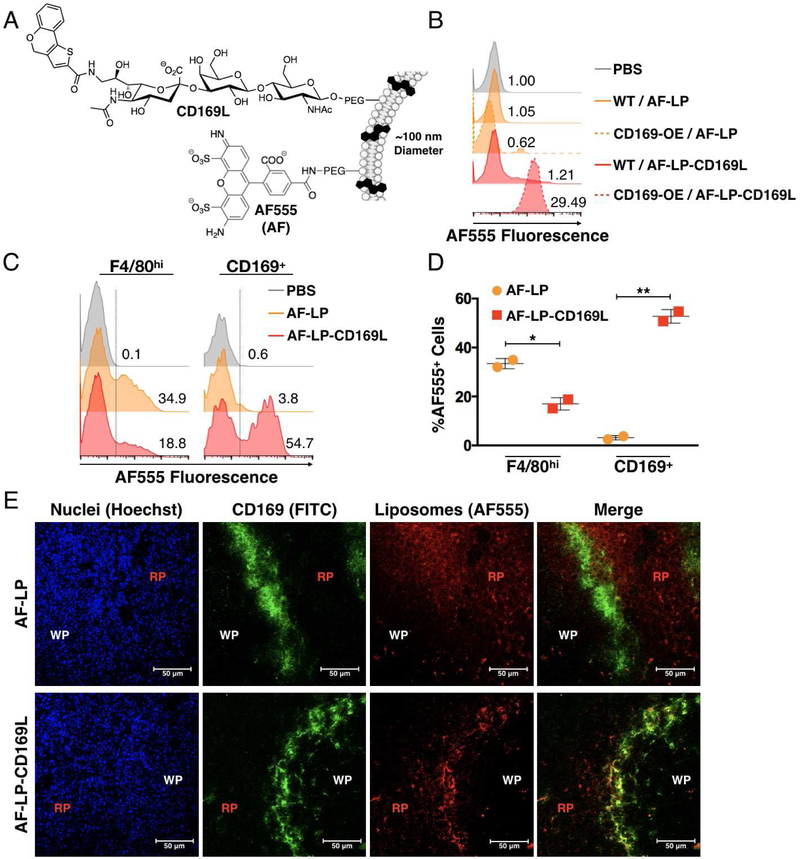
Liposomes decorated with a synthetic sialylated glycan possessing high affinity for CD169 (9-N-(4H-thieno[3,2-c]chromene-2-carbamoyl)-Neu5Acα2–3Galβ1–4GlcNAc (CD169L), Figure 1A) have been shown to enable delivery of lipid antigen to CD169MØs in vivo, resulting in enhanced innate immune activation of invariant natural killer T cells through a CDld-related pathway (Nycholat et al., 2012; Kawasaki et al. 2013). We reasoned that this platform could be adapted for delivery of soluble protein antigen able to influence adaptive, rather than innate, immunity in vivo. As a foundation for our CD169-targeted liposome platform, we first investigated the selectivity of the liposomes for CD169MØs over other phagocyte subsets — specifically, conventional F4/80hi MØs (F4/80MØs). We formulated liposomes with CD169L and a fluorescent reporter group (AF-LP-CD169L, Figure 1A). These liposomes exhibited ~30-fold selectivity for THP-1 cells expressing high levels of CD169 as compared to untargeted fluorescent liposomes in vitro (AF-LP, Figure 1B).
Figure 1. Targeting CD169 via Liposomal Nanoparticles.
(A) Schematic of AF-LP-CD169L (B) Performance of CD169-targeted liposomes in vitro using CD169 knock-in THP-1 cells. Solid and dashed histograms represent WT and CD169 knock-in THP-1 cells respectively. AF-LP-CD169L and AF-LP correspond to liposomes with and without CD169L respectively. Numbers represent median fluorescence intensity normalized to the PBS control. (C) Competitive uptake of liposomes between subpopulations of splenic macrophages in vivo. Numbers represent the percentage of cells within the AF555+ region (right side of dashed line). (D) Quantification of data in B. Each symbol corresponds to an individual animal. Error bars represent SD. *P<0.05, **P<0.01. (E) Confocal microscopy images of murine spleen from mice injected with either AF-LP or AF-LP-CD169L. AF555 (red) / α-CD169 (green) / Hoechst nuclear dye (blue). WP and RP represent white pulp and red pulp respectively.
In vivo, AF-LP-CD169L demonstrated excellent selectivity for CD169MØs as compared to AF-LP, with the former concentrating primarily in the F4/80MØ population (Figure 1C and 1D, gating strategy in Figure S2). Competition between the two MØ populations was observed in the AF-LP-CD169L-treated group, suggesting that even though F4/80MØs represent the majority of macrophages (> 98%) in the spleen, CD169L provides sufficient affinity/selectivity to significantly bias uptake to the CD169MØs (< 2%) (Figures S1, 1C, and 1D). It has been reported that some F4/80MØs express low levels of CD169 and this may account for some of the uptake observed in the F4/80+ population when liposomes are decorated with CD169L (Phan et al., 2009). Confocal microscopy data was consistent with results from FC; AF-LP-CD169L clearly concentrated in CD169+ cells within the marginal zone, whereas AF-LP localized mainly in the red pulp — a region rich in F4/80MØs (Figure 1E) (Oldenborg et al., 2000).
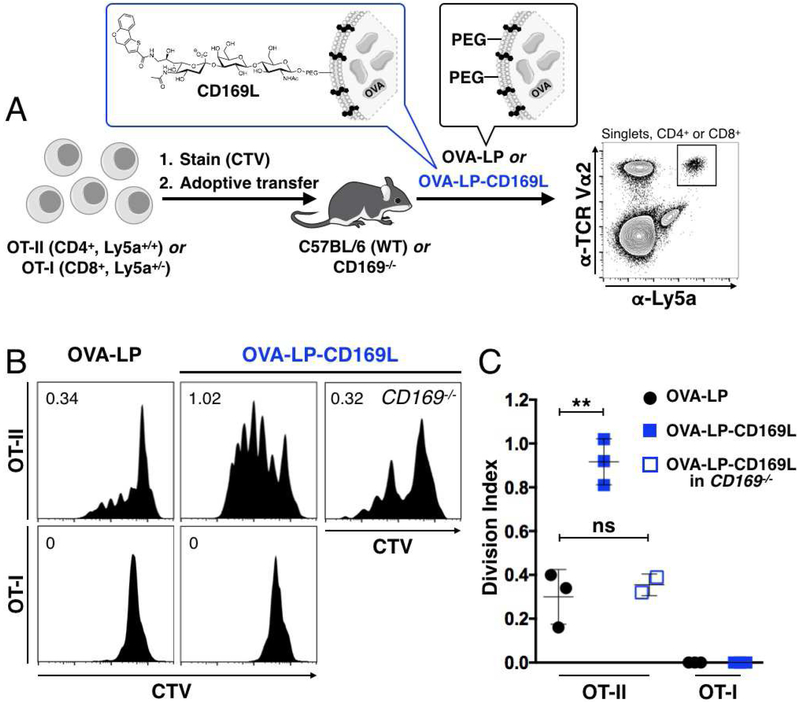
Given the robustness of CD169MØ targeting in vivo, we next investigated if a protein antigen, ovalbumin (OVA), could be selectively delivered to CD169MØs for activation of OVA specific CD4+ (OT-II) or CD8+ (OT-I) T cells. Accordingly, we encapsulated OVA in liposomes with or without CD169L, OVA-LP-CD169L and OVA-LP respectively (Table S1). Cell trace violet (CTV)-stained CD4+ (OT-II, Ly5a+/+) or CD8+ (OT-I, Ly5a+/−) T cells were adoptively transferred into Ly5b+/+ host mice, allowing facile identification of the adoptively transferred cells with the Ly5a surface marker. Mice were injected with liposomal OVA 24 h later (Figure 2A), and after 3 days, host spleens were analyzed for CTV dilution in the Ly5a+ adoptively transferred T cells. Every cell division dilutes the CTV dye two-fold, allowing assessment of the degree of proliferation by flow cytometry. In the presence of OVA-LP, minimal CD4+ T cell activation was observed; however, inclusion of CD169L (2 mol %) greatly enhanced CD4+ T cell expansion, suggesting that selective delivery of antigen to CD169MØs bolsters antigen specific CD4+ T cell priming in vivo, despite the small percentage of CD169+ MØs (Figures 2B and2C). CD4+ T cells from CD169−/− mice did not display significantly enhanced expansion, confirming the role of CD169 in the enhanced proliferation (Figures 2B and2C). In contrast, neither liposome formulation was able to mount a detectable CD8+ T cell response, suggesting that CD169-targeting of antigen alone is insufficient to induce cross-presentation.
Figure 2. Delivery of antigen to CD169MØs enhances CD4+ T cell priming.
(A) WT C57BL/6 or CD169−/− mice were injected with ~3×105 OT-I or OT-II cells stained with CTV. After 24 h, liposomes were injected. Proliferation of adoptively transferred cells, identified as CD4+ or CD8+TCR Vα2+Ly5a+ (bold gate), was measured 72 h later via CTV dilution. (B) Proliferation histograms for adoptively transferred CTV stained T cells in Ly5b+/+ mice injected with OVA-LP or OVA-LP-CD169L. Numbers represent cell division indices. (C) Quantification of data in B. ns = not statistically significant (P≥0.05).
Toll-like receptor (TLR) stimulation has been reported to induce efficient cross-presentation of antigen to CD8+ T cells in conventional F4/80MØs and DCs (Datta et al., 2005). In MØs, the ability to crosspresent antigen is often coupled with a phenotypic shift towards a classically activated (inflammatory) state. The potential for classical activation of CD169MØs through TLR stimulation has not been directly investigated. Classical MØ activation involves upregulation of key proteins involved in CD8+ T cell priming, such as CD80/CD86 and major histocompatibility complex I (MHC I) (Sica et al., 2012; Chávez-Gálan et al., 2015). Increased levels of these proteins on antigen presenting cells can drive enhanced antigen-specific CD8+ T cell priming, providing stimulatory signals through CD28 and a T cell receptor recognizing the antigen bound to MHC I (Kurts et al., 2010). We reasoned that classical activation of CD169MØs may enable cross presentation, even if the CD169MØs themselves are not directly involved in CD8+ T cell priming (Backer et al., 2010; van Dinther et al., 2018). Accordingly, we first investigated administration of lipopolysaccharide (LPS) for TLR4-mediated classical activation of CD169MØs. Flow cytometry (FC) analysis of CD169MØs from the spleens of LPS-treated mice revealed that these MØs adopted a classically activated phenotype closely related to that of conventional MØs (Figures S2A–S2E) (Sica et al., 2012; Chávez-Gálan et al., 2015). Indeed, both MØ populations produced increased levels of inducible nitric oxide synthase (iNOS), a hallmark of classical MØ activation (Figures S2B and S2C), and upregulated CD80/86 (Figures S2D and S2E) (Chávez-Gálan et al., 2015).
Having demonstrated that CD169MØs could be classically activated, we next examined the impact of this activation on T cell proliferation via simultaneous injection of both liposomal OVA and LPS into mice with adoptively transferred CD4+ or CD8+ T cells. After 3 days, robust proliferation of CD4+ T cells was observed for both OVA-LP and OVA-LP-CDL169L treatment groups. In contrast, while CD8+ T cell proliferation was observed with untargeted liposomes (OVA-LP), proliferation was dramatically enhanced in mice given CD169-targeted antigen (OVA-LP-CD169L) (Figures S2F and S2G). These data show that CD8+ T cell priming is maximized when antigen is delivered directly to CD169MØs in combination with TLR-stimulation.
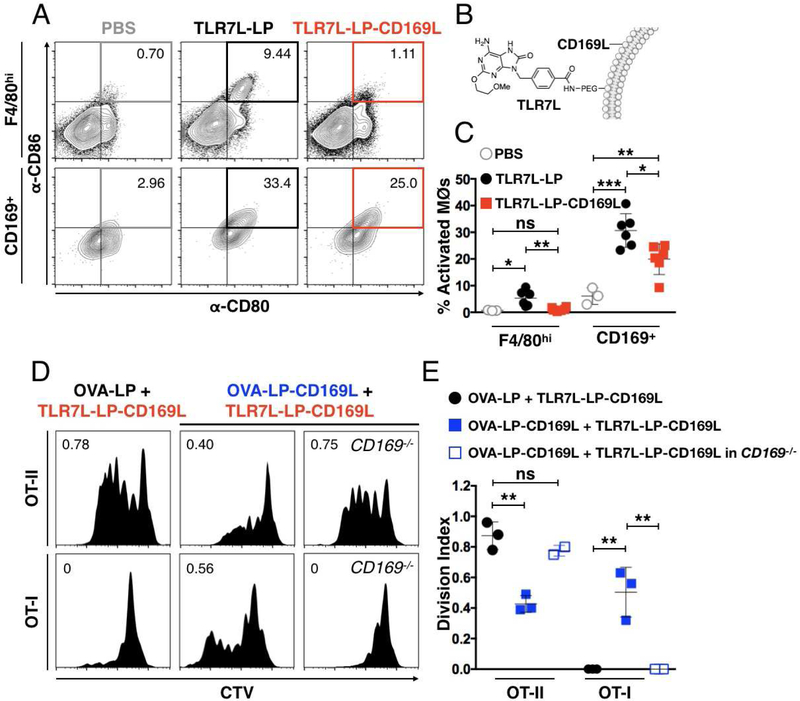
With the knowledge that classical activation of CD169MØs enabled CD8+ T cell priming, we next investigated selective delivery of both antigen and a TLR ligand to this MØ subset. Liposomes were formulated that contained a synthetic agonist against TLR7 (TLR7L) for selective activation of CD169MØs (Figure 3B) (Chan et al., 2009). We chose TLR7L since this receptor is known to be restricted to the endosome, ensuring delivery by liposomes targeting the endocytic receptor CD169 (Jiménez-Dalmaroni et al. 2016; Macauley et al., 2014). We observed robust activation of both MØsubsets upon treatment with untargeted TLR7L (TLR7L-LP). In contrast, the CD169-targeted TLRL liposomes (TLR7L-LP-CD169L) provided more selective activation of CD169MØs (Figures 3A and3C). In both treatment groups, a larger percentage of CD169MØs were activated relative to F4/80MØs, as observed with LPS treatment (Figures S2E and 3C).
Figure 3. Delivery of antigen to TLR7-activated CD169MØs enables CD8+ T cell priming.
. (A) Classical activation of F4/80hi and CD169+ macrophages through stimulation with liposomes bearing TLR7L in vivo. Cells populating the CD80+CD86+ quadrant are considered activated (grey, orange, and red squares). Numbers represent the percentage of activated cells. (B) Schematic of CD169-targeted liposomes bearing CD169L and TLR7L (TLR7L-LP-CD169L). (C) Quantification of data in A. (D) Proliferation histograms for adoptively transferred CTV stained OT-II or OT-I cells in Ly5b+/+ mice injected with both OVA-LP(-CD169L) and TLR7L-LP-CD169L. Numbers represent cell division indexes. (E) Quantification of data in D. ***P<0.001.
The impact of targeted TLR7L (TLR7L-LP-CD169L) on T cell activation with both untargeted and targeted OVA is shown in Figures 3D and3E. With OVA-LP, there was robust proliferation of CD4+ T cells, but no significant expansion of CD8+ T cells. Surprisingly, with OVA-LP-CD169L, CD4+ T cell proliferation was suppressed, while enhanced expansion of CD8+ T cells was observed (Figures 3D and3E). In CD169-deficient mice, CD4+ T cells expanded, but not CD8+ T cells. The results suggest that selective delivery of antigen coupled with TLR7L stimulation causes activation of CD169MØs and promotes bias for induction of CD8+ T cell priming, while suppressing a CD4+ T cell response (Figures 3D and3E).
To further interrogate the relationship between specific MØactivation and CD4+ T cell responses, we treated mice with TLR7L-LP — liposomes that activate both MØsubsets (Figures 3A and3C). Here, we observed that global classical activation combined with CD169-targeted antigen suppressed CD4+ T cell expansion in a manner similar to polarization of CD169MØs alone (Figure S3). In contrast, when neither TLR7L or antigen were targeted, CD4+ T cell priming was robust (Figure S3). Taken together, this data suggests that CD4+ T cell priming is enhanced by the presence of TLR7L-bearing liposomes when antigen is provided by non-targeted liposomes, which are primary sequestered in the F4/80hi population (Fig. 1C and Fig. 3D, left panels). Conversely, when antigen and TLR7L are both targeted to CD169MØs, CD4+ T cell expansion is impaired (Fig. 3D, middle panels).
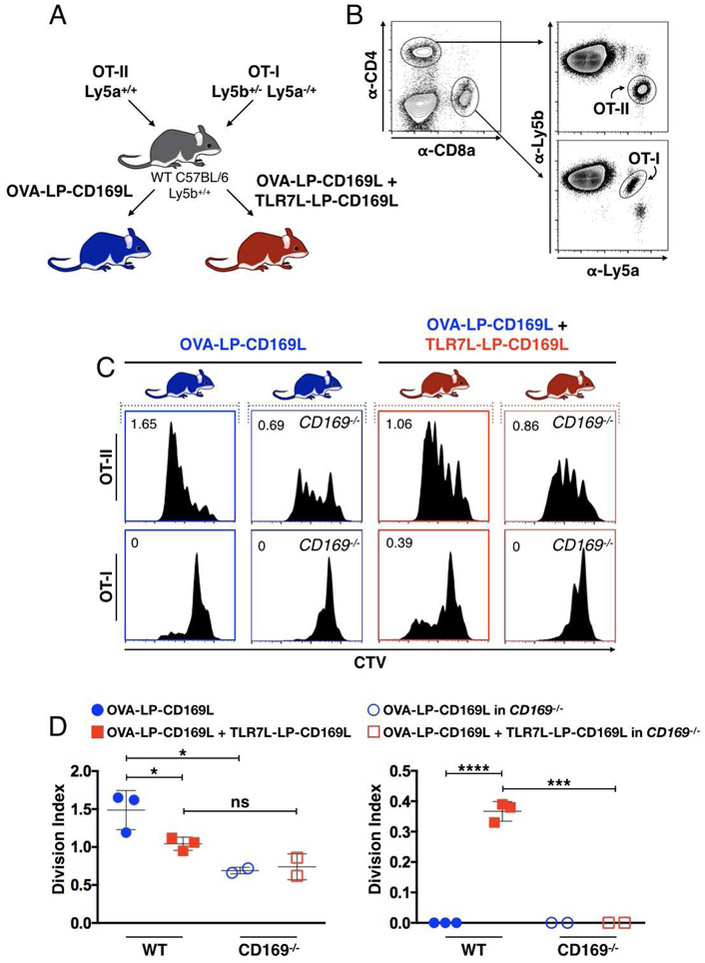
Our observations of TLR7-induced modulation of CD4+ and CD8+ T cell priming through antigen delivery to CD169MØs suggested that the OVA/TLR7L liposome platform may be useful for modulating the relative expansion of antigen specific T cell subsets in a single organism. To test this, we adoptively transferred equal numbers of CD4+ and CD8+ T cells, from Ly5a homozygotes and heterozygotes respectively, followed by treatment with either OVA-LP-CD169L or OVA-LP-CD169L + TLR7L-LP-CD169L (Figure 4A). We were able to deconvolute adoptively transferred T cells via their Ly5a/Ly5b expression profiles and observed the predicted CTV dilution curves, consistent with the trends in our previous results (Figures 4B-4D). CD169−/− host mice exhibited markedly reduced T cell proliferation under both treatment groups, further supporting a CD169-dependence for this system (Figures 4C and4D).
Figure 4. CD169-targeted antigen delivery coupled with TLR7-induced activation provides control over CD4+:CD8+ T cell ratios in a single organism.
(A) Schematic of co-adoptive transfer experiments. Equal numbers of CTV stained OT-II (Ly5a+/+) and OT-I (Ly5b+/−Ly5a−/+) cells were adoptively transferred into WT Ly5b+/+ host mice. Host mice were then injected with OVA-LP-CD169L (blue) or OVA-LP-CD169L + TLR7L-LP-CD169L (red). (B) Gating strategy for identifying adoptively transferred OT-II and OT-I cells from the same host. (C) OT-II and OT-I cell proliferation histograms from individual mice. Vertical plots represent OT-II and OT-I proliferation from the same mouse. Numbers represent division indexes. (D) Quantification of data in C.
Taken together, these data demonstrate that antigen delivery to CD169MØs, coupled with TLR7-induced activation, can introduce a bias towards activation of CD8+ T cells in vivo. This work represents a rare example of both chemically-controlled activation of T cell subsets in vivo and targeting of specific macrophage subsets through judicious liposome design. The modular nature of this platform makes it an attractive alternative to strategies that rely on chimeric antibody-antigen constructs for antigen delivery (van Dinther et al., 2018). Future work will focus on extension of this platform to delivery of diseaserelevant antigens and explore differential T cell priming via involvement of additional endosomal TLRs.
STAR★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. James C. Paulson (jpaulson@scripps.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mice used in this study mice were 6–8-week-old males, housed in a pathogen-free facility at The Scripps Research Institute (La Jolla, California) and used in compliance with the guidelines of the Institutional Animal Care Committee at the National Institutes of Health.
Cell Culture
The monocyte cell line (derived from a 1-year-old human male), THP-1, was maintained in complete glutamine-supplemented RPMI-1640 containing 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified incubator at 37 °C. THP-1 cells over-expressing CD169 were a generous gift from Dr. Hans Rempel and Dr. Lynn Pulliam (The University of California, San Francisco, USA).
METHOD DETAILS
Instrumentation
Flow cytometry data was collected using the following instruments: 5 laser (355, 405, 488, 561, and 640 nm) Bio-Rad ZE5 cell analyzer, 4 laser (405, 488, 561, and 640 nm) Bio-Rad ZE5 cell analyzer, 4 laser (405, 488, 561, 640) Beckman Coulter Cytoflex cell analyzer. FACS was performed using a 3-laser (405, 488, and 633 nm) BD Biosciences Aria fluorescence-activated cell sorter.
Confocal microscopy data was collected using a Zeiss LSM 710 laser scanning confocal microscope with a 40x oil-immersion objective lens.
Syntheses
9-N-(4H-thieno[3,2-c]chromene-2-carbamoyl)-Neu5Acα2–3Galβl–4GlcNAc-PEG-DSPE (CD169L) was synthesized as described previously (Nycholat et al., 2012). 4-((6-amino-2-(2-methoxyethoxy)-8-oxo-7,8-dihydro-9H-purin-9-yl)methyl)benzoic acid was synthesized as described previously (Chan et al., 2009). For coupling to PEG-DSPE, the above compound (5.3 mg, 0.015 mmol, 1.1 eq.) was dissolved in anhydrous DMF (0.25 ml) under argon at room temperature. HATU (5.6 mg, 0.014 mmol, 1.1 eq.) and triethylamine (3.8 μl, 0.023 mmol, 2 eq.) were then added. A solution of NHS-PEG-DSPE (38.4 mg, 0.013 mmol, 1 eq.) in CH2Cl2-DMF (1:1, 0.25 ml each) was added dropwise to the above mixture. The reaction was stirred at room temperature under argon atmosphere overnight. The reaction mixture was concentrated under reduced pressure. The remaining residue was resuspended in deionized distilled H2O (10 ml) and the product purified by dialysis. The aqueous solution was transferred to a Thermo Scientific Slide-A-Lyzer dialysis cassette (10,000 MWCO, 3-12 ml capacity) then dialyzed against deionized distilled H2O (4 l × 3). The aqueous solution was removed from the cassette and lyophilized to a white amorphous solid (quant.). The coupling efficiency (>90%) was estimated by 1H-NMR by comparing the aromatic resonances of the TLR7 ligand with the resonances for the methyl groups of DSPE chains.
Preparation of Liposomes
Lipids were prepared as a solution in DMSO in the ratios displayed in Table S1 as described previously (Chen et al., 2012; Kawasaki et al., 2013). Following removal of DMSO via lyophilization, lipids were hydrated in 1 mL PBS (for AF-LP(-CD169L) and TLR7L-LP (-CD169L)) or 1 mL of a 10 mg/mL solution of OVA (for OVA-LP(-CD169L)). The final concentration of total lipid was 1 mM for (AF-LP-CD169L and TLR7L-LP(-CD169L) and 5 mM for OVA-LP(-CD169L). Hydrated lipids were sonicated in a water bath for 60 s and then extruded through membranes of decreasing pore size: 0.8 μm, 0.2 μm, and finally, 0.1 μm. The resultant preparations of AF-LP(-CD169) and TLR7L-LP(-CD169) were then used without further processing. For preparations of OVA-LP(-CD169L), liposomes were separated from free OVA protein via size-exclusion chromatography using Sepharose CL-4B resin. Fractions containing liposomes were identified via absorbance at 220–280 nm and the protein content of each fraction was measured using a LavaPep protein quantification kit.
Fluorescent Liposome Uptake In Vitro
THP-1 cells (1×106) were exposed to 250 μM liposomes for 45 min in complete RPMI-1640 medium (see cell culture section). Following incubation, cells were centrifuged at 350 rcf and washed 3 times with cold FACS buffer (see flow cytometry section). Cells were then analyzed via flow cytometry.
Fluorescent Liposome Uptake In Vivo
Six- to eight-week old male C57BL/6J mice were injected with 200 μL of 250 μM AF-LP or AF-LP-CD169L (concentration based on total lipid content—see liposome preparation sections) via the tail vein. After 90 min., mice were sacrificed, their spleens harvested, and leukocytes prepared for analysis via flow cytometry (see flow cytometry section). Spleens for analysis via confocal microscopy (see immunohistochemical staining and analysis section) were obtained from mice treated under the same conditions.
Immunohistochemical staining and analysis
Immediately after harvesting, spleens were rinsed with cold PBS, dried on absorbent towel, immersed in O.C.T. medium, flash-frozen, and stored at −80 °C. Spleens were then cut into 10 μm thick sections and placed on glass slides. Tissue sections were then rehydrated in PBS (10 min., room temperature), blocked with 1% goat serum in PBS (30 min. room temperature), and stained with FITC-conjugated antimouse CD169 in PBS (MOMA-1 clone) overnight at 4 °C. Slides were then washed 3x with PBS and stained with Hoechst dye (3 min., room temperature). Slides were again washed 3x with PBS, dried, and mounted with anti-fade mounting media. Glass cover slips were adhered using clear lacquer. Slides were then analyzed on a confocal microscope (see instrumentation section).
Flow Cytometry
Single cell suspensions of murine splenocytes were obtained via mechanical digestion of spleens followed by erythrocyte lysis (see adoptive transfer experiments for details). Cells were filtered through 100 μm nylon mesh, stained with a fixative-compatible viability dye (where appropriate, according to the manufacturer’s instructions), blocked with anti-mouse CD16/32 in FACS buffer (1% BSA, 2 mM EDTA in HBSS), and stained with appropriate fluorophore-tagged antibodies. Cells were then washed twice with FACS buffer, and for experiments analyzing macrophages, fixed in a 4% solution of paraformaldehyde, followed by analysis by flow cytometry.
Adoptive Transfer Experiments
Six- to eight-week old Ly5a+/+ B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II, CD4+) or Ly5a+/− C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I, CD8+) mice were sacrificed and their spleens harvested. Spleens were mechanically digested into a single cell suspension and erythrocytes were lysed via exposure to a solution of 150 mM NH4Cl, 1 mM NaHCO3, and 1.27 μM EDTA for 3 min. at room temperature. After quenching with FACS buffer (1% BSA, 2 mM EDTA in HBSS), leukocytes were centrifuged at 350 rcf for 5 min., washed with PBS, filtered through 40 μm nylon mesh, centrifuged (350 rcf, 5 min.), and then resuspended in PBS to a final concentration of 1×107 cells/mL. A solution of cell trace violet in DMSO, prepared according the manufacturer’s instructions, was then added (1:1000 dilution). Cells were incubated in the dark at room temperature for 10 min., after which the solution was quenched via addition of pre-warmed complete media (RPMI-1640 containing 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin). Following centrifugation, cells were resuspended in PBS to a final concentration of 1.5×106 TCR Vα2+ CD4+ (for OT-II donors) or TCR Vα2+ CD8+ (for OT-I donors) cells/mL, as assessed by flow cytometry. Six- to eight-week old WT male C57BL6/J mice then each received 3×105 CTV-stained TCR Vα2+ CD4+ or TCR Va2+ CD8+ cells via the tail vein. For co-transfer experiments, 3×105 of both CD4+ (OT-II) and CD8+ (OT-I) cells were adoptively transferred into the same host animal. After 24 h, mice received either PBS, LPS, and/or liposomes via the tail vein. In all treatment groups involving OVA-LP(-CD169L), mice were given 5 ng total OVA (see liposome preparation for protein quantification details). For treatment groups involving TLR7L-LP(-CD169L), mice were given 5 ng total TLR7L. For experiments involving LPS, 230 ng LPS was administered. For control experiments involving CD169−/− host mice, CTV-stained OT cells were purified via FACS prior to adoptive transfer to ensure that no CD169+ cells from donor mice would be transferred. After 72 h, mice were sacrificed, their spleens harvested, and leukocytes prepared for analysis via flow cytometry.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical parameters were calculated via student’s t test in Prism v. 7 (GraphPad). Division indices were calculated using FlowJo v.10 (BD).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies (all monoclonal rat anti-mouse except where noted) | ||
| α-CD3 (clone: 17A2) | Biolegend | Cat.# 100234 |
| α-CD4 (clone: GK1.5) | BD Biosciences | Cat.# 561828 |
| α-CD8a (clone: 53-6.7) | Biolegend | Cat.# 100705 |
| α-CD11b (clone: M1/70) | Biolegend | Cat.# 101225 |
| α-CD16/32 (Fc receptor block) (clone: 93) | Biolegend | Cat.# 101319 |
| α-CD19 (clone: 6D5) | Biolegend | Cat.# 115545 |
| α-CD80 (clone: 16-10A1, hamster anti-mouse) | BD Biosciences | Cat.# 563687 |
| α-CD86 (clone: GL-1) | Biolegend | Cat.# 105011 |
| α-CD45.1/Ly5a (clone: A20, mouse anti-mouse) | Biolegend | Cat.# 110713 |
| α-CD45.2/Ly5b (clone: 104, mouse anti-mouse) | Biolegend | Cat.# 109805 |
| α-CD169 (clone: MOMA-1) | Bio-Rad | Cat.# MCA947F |
| α-F4/80 (clone: T45-2342 | BD Biosciences | Cat.# 565411 |
| α-iNOS (clone: CXNFT) | eBioscience | Cat.# 12–5920-80 |
| α-Ly6C (clone: HK1.4) | Biolegend | Cat.# 128011 |
| α-Ly6G (clone: 1A8) | Biolegend | Cat.# 127633 |
| α-NK1.1 (clone: PK136, mouse anti-mouse) | Biolegend | Cat.# 108737 |
| α-TCR Vα2 (clone: B20.1) | Biolegend | Cat.# 127807 |
| IgG2a, k isotype control | Biolegend | Cat.# 400507 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1,2-distearoyl-sn-glycero-3-phosphocoline (DSPC) | Avanti Polar Lipids Inc. | SKU 850365P |
| 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG) | Avanti Polar Lipids Inc. | SKU 880128P |
| 3-(N-succinimidyloxyglutaryl)aminopropyl polyethyleneglycol-carbamyl distearoylphosphatidyl-ethanolamine (NHS-PEG-DSPE) | NOF America Corp. | DSPE-034GS |
| 9-N-(4H-thieno[3,2-c]chromene-2-carbamoyl)-Neu5Acα2–3Galβ1–4GlcNAc-PEG-DSPE (CD169L) | This study | N/A |
| AF555-PEG-DSPE | Prof. Matthew Macauley | N/A |
| Cholesterol | MilliporeSigma | SKU C8667 |
| Lipopolysaccharide (LPS) (E. coli 0127:B8) | MilliporeSigma | SKU L3129 |
| Chicken ovalbumin (OVA) | Worthington Biochemical Corp. | Product Code OAEF |
| O.C.T. medium | Tissue-Tek (Sakura®) | Item # 4583 |
| Anti-fade mounting medium | Vector Laboratories Inc. | Cat.# H-1000 |
| Cell Trace Violet (CTV) | Invitrogen | Cat.# C34557 |
| Critical Commercial Assays | ||
| LavaPep protein quantification kit | Gel Company Inc. | LP022010 |
| Experimental Models: Cell Lines | ||
| THP-1 (wild type) | ATCC | TIB-202 |
| THP-1 (CD169 over-expressers) | Hans Rempel and Lynn Pulliam | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | Scripps rodent breeding colony | N/A |
| CD57BL/6J CD169−/− mice | Paul Crocker | N/A |
| C57BL/6-Tg(TcraTcrb)1100Mjb/J, Ly5a+/− (OT-I, CD8+) | The Jackson Laboratory (bred to Ly5a+/+ in house) | Stock#: 003831 |
| B6.Cg-Tg(TcraTcrb)425Cbn/J, Ly5a+/+ (OT-II, CD4+) | The Jackson Laboratory (bred to Ly5a+/− in house) | Stock#: 004194 |
| Software and Algorithms | ||
| FlowJo v. 10 | BD | https://www.flowjo.com |
| Prism v. 7 | Graph Pad Software | https://www.graphpad.com/scientific-software/prism/ |
| Image Pro Premier v 9.2 | Media Cybernetics Inc. | http://www.mediacy.com/imagepro |
| Zen Black & Blue | Zeiss Inc. | https://www.zeiss.com/microscopy/int/downloads/zen.html |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Other | ||
| Membrane supports for liposome extrusion | Whatman plc./Fisher | Cat.# NC9759169 |
| Membrane (0.8 μm) for liposome extrusion | Whatman plc./Fisher | Cat.# 09-800-945 |
| Membrane (0.2 μm) for liposome extrusion | Whatman plc./Fisher | Cat.# 05-715-115 |
| Membrane (0.1 μm) for liposome extrusion | Whatman plc./Fisher | Cat.# 09-805-346 |
SIGNIFICANCE.
Chemical tools that enable the manipulation of antigen-specific immune responses can enhance our understanding of adaptive immunity. Here, we describe a liposome-based system for targeting soluble protein antigen and/or a Toll-like receptor agonist to macrophages expressing CD169 - cells known to be involved in mounting antigen-specific T cell responses. This platform enabled us to expand CD4+ T cells selectively without initiating a CD8+ T cell response in a manner dependent on CD169. By adapting the platform to delivery of a TLR7 agonist, we demonstrated that CD169MØs can be classically activated in a manner similar to other macrophage subsets, and that this activation presented a pathway for expansion of antigen-specific CD8+ T cells through delivery of liposome-encapsulated antigen. Further, we discovered that classical activation of CD169MØs coupled with antigen targeted to those cells impaired expansion of CD4+ T cells. We exploited this phenomenon to influence CD4+/CD8+ T cell ratios in the same organism. Manipulation of T cell ratios is important in diseases such as HIV infection, with recent work suggesting that the CD4+/CD8+ ratio is a more important biomarker for disease progression as compared to CD4+ T cell levels alone (McBride et al., 2017). Furthermore, large CD4+/CD8+ ratios have been associated with enhanced patient survival in non-small cell lung cancer, but can also contribute to progression of autoimmune disorders (Jackute et al., 2015; Janahi et. al., 2018). This platform will be of use to investigators interested in studying the effects of varying T cell ratios in models of disease.
Highlights.
Antigen or activator was targeted to CD169+ MØs via a synthetic glycan on liposomes
Targeting of antigen alone promoted expansion of CD4+ T cells
CD169+ MØs were selectively activated by targeted liposomes with a TLR7 agonist
Targeting of antigen to activated CD169+ macrophages biased priming of CD8+ T cells
ACKNOWLEDGMENTS
The authors acknowledge the following staff in the flow cytometry core at The Scripps Research Institute for assistance in experiments: Alan Saluk, Matthew Haynes, and Brian Seegers. We thank: Britni Arlian Cruz for assistance with animal experiments, Matthew S. Macauley for providing lipid-conjugated fluorophore, Kevin Worrell for assistance with synthesizing the carbohydrate core of CD169L, Dennis A. Carson and Tomoko Hayashi for supplying TLR7L (without PEG-DSPE), Hans Rempel and Lynn Pulliam for THP-1 cells expressing CD169, and Paul R. Crocker for CD169−/− mice. We also thank Namir Shaabani and Vincent Vartabedian for productive discussions. Open source graphics for liposome lipid bilayers and protein (OVA) were provided by Servier Medical Art. This work was supported by grants AI150143 and HL107151 from the NIH (to J.C.P). L.J.E. is supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (# 502448 - 2017).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing financial interest.
SUPPLEMENTAL INFORMATION
Supporting figures S1-S3, table S1, and the 1H NMR spectrum for TLR7L-PEG-DSPE may be found in the Supplemental Information document.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Asano K, Nabeyama A, Miyake Y, Qiu C, Kurita A, Tomura M, Kanagawa O, Fujii S, and Tanaka M (2011). CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity. 34, 85–95. [DOI] [PubMed] [Google Scholar]
- Backer R, Schwandt T, Greuter M, Oosting M, Jüngerkes F, Tüting T, Boon L, O’Toole T, Kraal G, Limmer A, and den Haan JMM (2010). Effective collaboration between marginal metallophilic macrophages and CD8+ dendritic cells in the generation of cytotoxic T cells. Proc. Natl. Acad. Sci. 107(1) 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard CA, Ried C, Kochanek S, and Brocker T (2015). CD169+ macrophages are sufficient for priming of CTLs with specificities left out by cross-priming dendritic cells. Proc. Natl. Acad. Sci 112(17) 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Gálan L, Olleros ML, Vesin D, and Garcia I (2015). Much more than M1 and M2 macrophages, there are also CD169(+) and TCR(+) macrophages. Front. Immunol, 6, article 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M, Hayashi T, Kuy CS, Gray CS, Wu CCN, Corr M, Wrasidlo W, Cottam HB, and Carson DA (2009). Synthesis and immunological characterization of Toll-like receptor 7 agonistic conjugates. Bioconjugate Chem. 20, 1194–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WC, Kawasaki N, Nycholat CM, Han S, Pilotte J, Crocker PR, and Paulson JC (2012). Antigen delivery to macrophages using liposomal nanoparticles targeting sialoadhesin/CD169. PLoS ONE. 7(6) e39039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SK, and Raz E (2005). Induction of antigen cross-presentation by Toll-like receptors. Springer Sem. Immun 26, 247–255. [DOI] [PubMed] [Google Scholar]
- Jackute J, Zemaitis M, Pranys D, Sitkauskiene B, Miliauskas S, Bajoriunas V, Lavinskiene S, Sakalauskas R (2015). The prognostic influence of tumor infiltrating Foxp3+CD4+, CD4+ and CD8+ T cells in resected non-small cell lung cancer. J. Inflamm 12, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janahi EMA, Das S, Bhattacharya SN, Haque S, Akhter N, Jawed A, Wahid M, Mandal RK, Lohani M, Areeshi MY, Ramachandran VG, Almalki S (2018). Cytomegalovirus aggravates the autoimmune phenomenon in systemic autoimmune diseases. Microb. Pathog 120, 132–139. [DOI] [PubMed] [Google Scholar]
- Jiménez-Dalmaroni M, Gerswhin ME, and Adamopoulos IE (2016). The critical role of toll-like receptors – From microbial recognition to autoimmunity: A comprehensive review. Autoimmunity Rev. 15, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, Henrickson SE, Shayakhmetov DM, Di Paolo NC, van Rooijen N, Mempel TR, Whelan SP, and von Andrian UH (2007). Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 450, 110–114. [DOI] [PubMed] [Google Scholar]
- Kawasaki N, Vela JL, Nycholat CM, Rademacher C, Khurana A, van Rooijen N, Crocker PR, Kronenberg M, and Paulson JC (2013). Targeted delivery of lipid antigen to macrophages via the CD169/sialoadhesin endocytic pathway induces robust invariant natural killer T cell activation. Proc. Natl. Acad. Sci 110(19) 7826–7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurts C, Robinson BWS, and Knolle PA (2010). Cross-priming in health and disease. Nat. Rev. Immunol. 10, 403–414. [DOI] [PubMed] [Google Scholar]
- Macauley MS, Crocker PR, and Paulson JC. (2014). Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol 14, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JA, Striker R (2017). Imbalance in the game of T cells: What can the CD4/CD8 T-cell ratio tell us about HIV and health?. PLoS Pathog 13(11) e1006624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebius RE, and Kraal G (2005). Structure and function of the spleen. Nat. Rev. Immunol. 5, 606–616. [DOI] [PubMed] [Google Scholar]
- Nycholat CM, Rademacher C, Kawasaki N, and Paulson JC (2012). In silico-aided design of a glycan ligand of sialoadhesin for in vivo targeting of macrophages. J. Am. Chem. Soc 134, 15696–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg P, Zheleznyak A, Fang Y, Lagenaur CF, Gresham HD, and Lindberg FP (2000). Role of CD47 as a marker of self on red blood cells. Science. 288, 2051–2054. [DOI] [PubMed] [Google Scholar]
- Phan TG, Green JA, Gray EE, Xu Y, Cyster JG (2009). Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol 10(7) 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, and Mantovani A (2012). Macrophage plasticity and poliarization: in vivo veritas. J. Clin. Invest. 122(3) 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dinther D, Veninga H, Oborra S, Borg EGF, Hoogterp L, Olesek K, Beijer MR, Schetters STT, Kalay H, Garcia-Vallejo JJ, Franken KL, Cham LB, Lang KS, van Kooyk Y, Sancho D, Crocker PR, and den Haan JMM (2018). Functional CD169 on macrophages mediates interaction with dendritic cells for CD8+ T cell cross-priming. Cell Reports. 22, 1484–1495. [DOI] [PubMed] [Google Scholar]
- Varol C, Mildner A, and Jung S (2015) Macrophages: development and tissue specialization. Annu. Rev. Immunol 33, 643–675. [DOI] [PubMed] [Google Scholar]
- Varol C, Zigmond E, and Jung S (2010). Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat. Rev. Immunol 10, 415–426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.