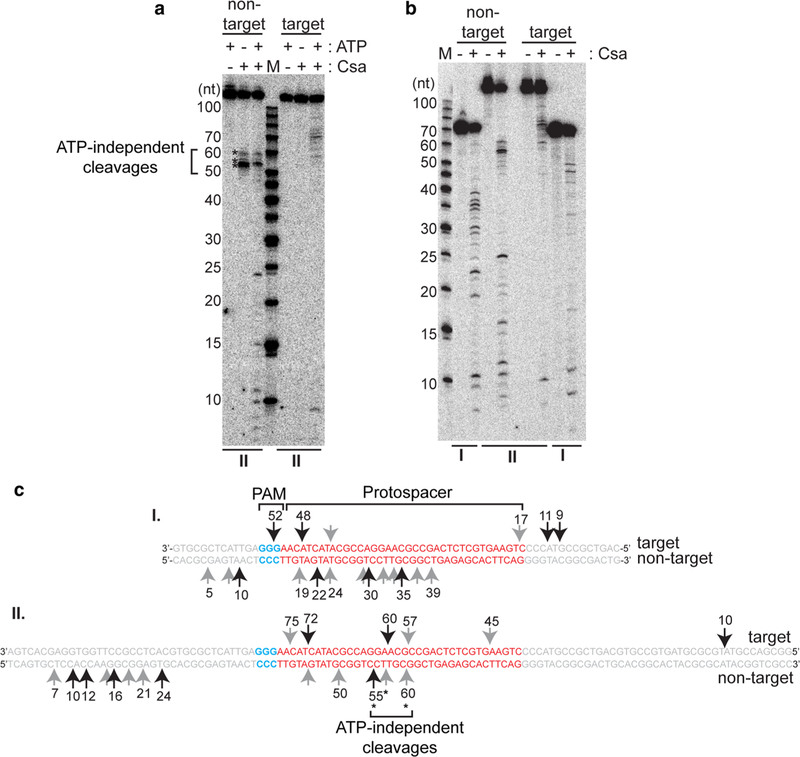
Fig. 6.
Type I-A crRNP-induced cleavages map to multiple sites in both strands of DNA. a Effect of ATP (∓) on cleavage of double- stranded DNA (117 base pairs, same as Figs. 1, 2, 3, 4, 5 and II in Fig. 6) and b Cleavage profiles of two different sized double-stranded DNA [67 base pairs (I) and 117 base pairs (II)] with no protein (−) or Csa crRNP [(+) assembled with crRNA, Csa4–1, Cas3″, Cas3′, Cas5a, Csa2 and Csa5]. The upper label on each gel indicates location of 5′ radiolabel (32P) on the non-target or target strand of double-stranded DNA. 5′ radiolabeled DNA size-standard (Affymetrix, 10–100 bases) is labeled (M). c Major (long black arrows) and minor (short gray arrows) cleavage products (in a and b) mapped to target and non-target strand sequences of double-stranded DNA substrates (I and II) tested in b. Protospacer (red, labeled), PAM (blue, labeled) and flanking (gray) sequences are indicated. Numbers correspond to size of cleavage products (counting from 5′ end of each strand). ATP- independent cleavages (on non-target strand) are denoted (asterisks and bracket) and labeled (in a and c)