SUMMARY
Clostridium scindens, biotransforms primary bile acids into secondary bile acids, is correlated with inhibition of Clostridium difficile growth in vivo. The aim of the current study was to determine how C. scindens regulates C. difficile growth in vitro and if these interactions might relate to the regulation of gut microbiome structure in vivo. The bile acid 7α-dehydroxylating gut bacteria, C. scindens and C. sordellii, were found to the secrete tryptophan-derived antibiotics, 1-acetyl-β-carboline and turbomycin A, respectively. Both antibiotics inhibited growth of C. difficile and other gut bacteria. The secondary bile acids, deoxycholic acid (DCA) and lithocholic acid (LCA), but not cholic acid (CA), enhanced the inhibitory activity of these antibiotics. These antibiotics appear to inhibit cell division of C. difficile. The results help explain how endogenously synthesized antibiotics and secondary bile acids may regulate C. difficile growth and the structure of the gut microbiome in health and disease.
Keywords: Turbomycin A, 1-acetyl-β-carboline, cyclicdipeptides, Clostridium difficile, Clostridium scindens, Clostridium sordellii, gut microbiome, dysbiosis
Graphical Abstract
ETOC Blurb
Kang et al. describes the discovery of tryptophan-derived antibiotics secreted by specific gut bacteria that inhibit C. difficile, a bacterial pathogen causing diarrhea and colitis. These results help explain why antibiotic treatment, which disrupts the normal protective gut microbiota, allows C. difficile to colonize the colon and cause disease.
INTRODUCTION
Clostridium difficile, the cause of antibiotic associated diarrhea and colitis, is a growing health threat for patients taking broad spectrum antibiotics (Rupnik et al., 2009). Antibiotics markedly disrupt the gut microbiome homeostasis by decreasing the levels of protective gut microbiota, allowing colonization and proliferation of C. difficile normally found in low levels in some individuals (Loo et al., 2011). Patients treated with antibiotics, especially in hospitals, are also at risk for colonization by C. difficile spores, which germinate in the gastrointestinal tract (stimulated by taurocholate) producing vegetative cells that secrete toxins A and B which cause diarrhea and colitis (Kuehner et al., 2010; Sorg and Sonenshein, 2010). Patients with antibiotic associated diarrhea and/or colitis are routinely treated with either metronidazole or vancomycin to kill C. difficile vegetative cells colonizing the colon; however, up to 35% of all patients successfully treated with these antibiotics will relapse following cessation of antibiotic therapy.
Fecal transplants using gut microbiota from healthy donors have been highly successful in treating relapsing patients (van Nood et al., 2013; Weingarden et al., 2014). A previous study attempted to elucidate the members of the human gut microbiota most responsible for resistance to C. difficile infection (Buffie et al., 2015). This paper reported that C. scindens, a human gut bacterium that converts the primary bile acids, CA and chenodeoxycholic acid (CDCA), to secondary bile acids, DCA and LCA, respectively, is strongly associated with inhibition of C. difficile infections in animal models and in human patients (Buffie et al., 2015). Moreover, several clinical studies have support this association, the most recent study reported that the abundance of the baiCD gene cluster, which is a measurement of the levels of bile acid 7α-dehydroxylating gut bacteria, are negatively correlated with C. difficile infection (Solbach et al., 2018). The aim of the current study was to try and elucidate the interactions between C. difficile and bile acid 7α-dehydroxylating gut bacteria to help understand the possible mechanisms of regulation of C. difficile in vivo and the gut microbiome structure. The results of our investigation offer new insights into the how bile acid 7α-dehydroxylating gut bacteria, which secrete both antibiotics and secondary bile acids, regulate C. difficile growth in the intestine and homeostasis of the gut microbiome structure.
RESULTS
C. difficile Growth is Inhibited by C. scindens in the Presence but Not Absence of Cholic Acid
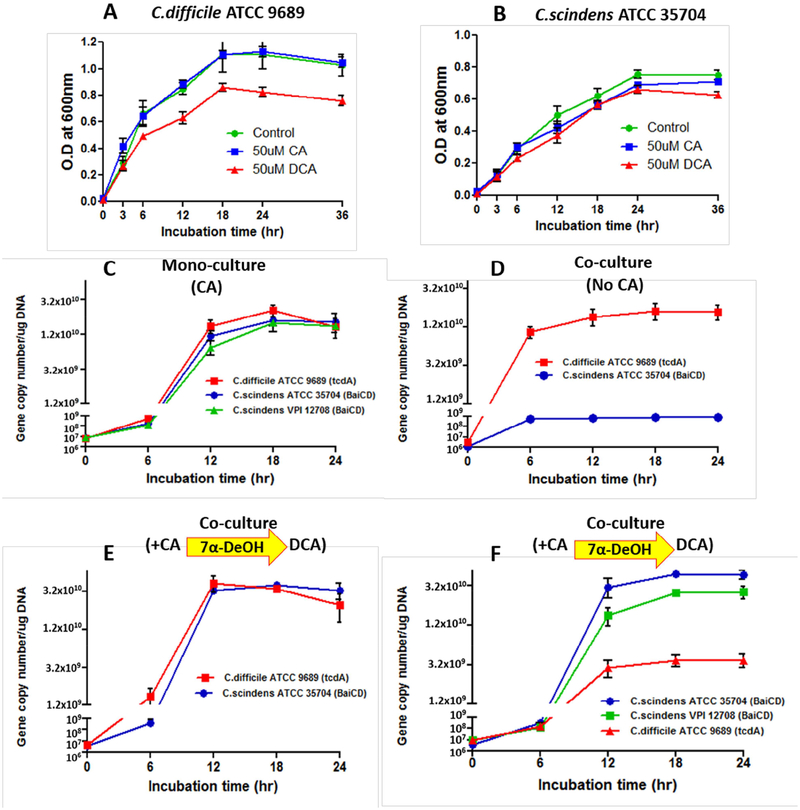
It has been reported that DCA inhibits C. difficile growth in vitro (Sorg and Sonenshein, 2008, 2010) and is hypothesized to inhibit the growth of this bacterium in vivo (Buffie et al., 2015).Bile acid 7α-dehydroxylating gut bacteria, such as C. scindens, convert cholic acid into deoxycholic acid which increases the hydrophobicity and toxicity of the bile acid. We grew C. diffiicle and C. scindens in PYF medium with either CA or DCA. CA (50 μM) had no significant effect on the growth of either bacterium. DCA (50 μM) inhibited (~25%) C. difficile but had no significant effect on C. scindens growth (Figure 1 A and B). Next, we decided to grow C. difficile ATCC 9689 (type strain) and C. scindens ATCC 35704 in PYF medium in coculture with and without added CA (50 μM). In order to measure the growth of each bacterium, we developed specific PCR assays to quantify a gene unique to C. scindens (bai CD gene encoding bile acid 3-oxo-A4–7α-hydroxysteroid oxidoreductase) and the gene encoding Toxin A in C. difficile (STAR Methods). Surprisingly, C. difficile strongly inhibited C. scindens growth under co-culture conditions in the absence (Figure 1 D) but not presence of CA (Figure 1 E). The addition of a second strain of C. scindens ATCC 12708 further inhibited C. difficile growth (Figure 1 F).
Figure 1. C. difficile Growth is Inhibited by C. scindens in the Presence but Not Absence of Cholic Acid.
PYF medium containing 50 μM cholic acid (CA) or 50 μM deoxycholic acid (DCA) was inoculated with 106 vegetative cells/ml of C. difficile (Panel A) or C. scindens (Panel B), incubated for a 24 hr. time course, and growth determined at indicated time points by measuring optical density (OD) at 600 nm. PYF medium was inoculated with 106 vegetative cells/ml of C. difficile and C. scindens grown in either mono culture with 50 μM CA (Panel C) or co-culture without CA (Panel D) or with 50 μM CA (Panels E and F), incubated over a 24 hour time course and levels of bacteria estimated at indicated time points by measuring gene dosage of Toxin A gene for C. difficile and baiCD gene for C. scindens (STAR Methods).
Clostridium difficile Secretes Proline-based Cyclic Dipeptides
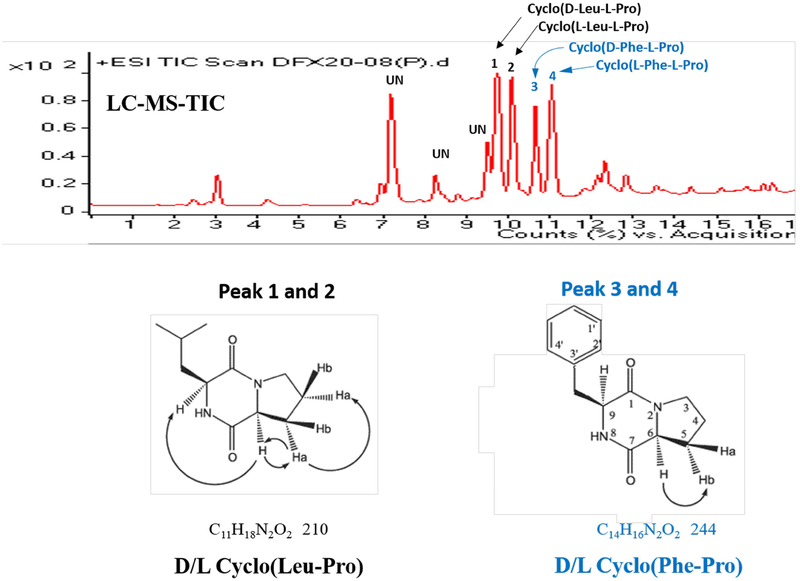
How does C. difficile inhibit C. scindens growth? After filter sterilizing the spent growth medium from a pure culture of C. difficile ATCC 9689 grown in PYF medium and adjusting the pH to 7.2, we discovered that the spent medium inhibited several other common human gut bacteria as well as Staphylococcus aureus MRSA (Table 1). Moreover, three clinical isolates of C. difficile (BBA-1870, BBA-1801, BBA-1814) also secrete antibacterial compound(s) that inhibit C.scindens ATCC 35704 and Staphylococcus aureus MRSA under the same culture conditions (Table 1, Table S1). Next, we decided to purify and character the major antibacterial compound(s) secreted into the culture medium by C. difficile ATCC 9689. Initial experiments showed that the antibacterial compound(s) were less than 3kDa in size (by ultrafiltration), hydrophobic (extracted into ethyl acetate), heat stable (autoclaving), lack a charge (did not bind to anion or cation exchange resins), and resistant to proteolytic enzymes (trypsin). Based on these traits, a protocol was developed to purify the major inhibitory compound(s) from C. difficile for characterization and identification. The purification started with 16 liters of PYF spent culture medium (24 hrs. incubation) from C. difficile ATCC 9689 and sequentially involved: molecular size ultrafiltration, extraction with ethyl acetate, Sephadex LH-20 column chromatography, and 2 different C-18 reverse-phase HPLC elution solvent systems (Figure 2, Figure S1). Two major antibacterial compounds were purified and identified by liquid chromatography-electrospray ionization mass spectrometry (LC-ESI/MS) and 1H and 13C NMR chemical shifts as mixed chiral isomers of cyclo (Phe-Pro) and cyclo(Leu-Pro) (Figure 2). Proline-base cyclic dipeptides are known to be secreted by a number of bacteria (Giessen et al., 2014) but not members of the genus Clostridium. Proline-based cyclodipeptides have been reported to be quorum-sensing molecules for certain bacteria and to inhibit the growth of both bacteria and fungi (Belin et al., 2012; Gonzalez et al., 2017). The synthesis and secretion of these molecules by Clostridium difficile raises the possibility that these cyclodipeptides may enhance the ability of this bacterium to colonize the colon following antibiotic treatment by inhibiting the remaining members of the gut microbiota. In this regard, broad spectrum antibiotics also reduce the levels of bile acid 7α-dehydroxylating gut bacteria.
Table 1.
Inhibition of different gut bacteria by compounds secreted by C. difficile ATCC 9689, C. scindens ATCC 35704, and C. sordellii ATCC 9714
| Test bacteria strains | Cell growth of bacteria(O.D at 600nm) in PYF for 24hr | |||||||
|---|---|---|---|---|---|---|---|---|
| C.diffiicle ATCC 9689 | C.scindens ATCC 35704 | C.sordellii ATCC 9714 | Staph.aureus (MRSA) | Ent.faecium ATCC 2316 | E.coli ATCC 2340 | B.vulgatus ATCC 8482 | ||
| 0.81 | 0.78 | 0.85 | 0.58 | 0.69 | 0.71 | 0.98 | ||
| C.scindens ATCC 35704 spent culture medium | UN† | 0.54±0.07* | 0.31±0.11** | 0.34±0.23* | 0.30±0.01** | 0.41±0.03* | 0.47±0.04* | |
| IN‡ | 0.26±0.05** | 0.10±0.02** | 0.08±0.03** | 0.25±0.03** | 0.38±0.04* | 0.24±0.11** | ||
| C.sordellii ATCC 9714 spent culture medium | UN | 0.08±0.01** | 0.08±0.01** | 0.08±0.03** | 0.10±0.02** | 0.07±0.03** | 0.10±0.03** | |
| C.difficile ATCC 9689 spent culture medium | - | 0.12±0.04** | 0.32±0.03** | 0.24±0.14* | 0.33±0.05** | 0.28±0.03** | 0.53±0.05* | |
: No addition of CA,
: Induced by 100μM CA.
: Significant growth inhibition p<0.05 and p< 0.01, respectively.
Figure 2. Identification of antibacterial compounds secreted by C. difficile.
The purification protocol for antibacterial compounds secreted by C. difficile is described in Figure S1. The mass spectra and 1H and 13C NMR chemical shifts of peaks 1 through 4 were used to identify compounds. Antibacterial compounds were identified as chiral isomers of cyclo(Leu-Pro) for peaks 1 and 2 and chiral isomers of cyclo (Phe-Pro) for peaks 3 and 4.
Bile Acid 7α-Dehydroxylating Bacteria Secrete Tryptophan-based Antibiotics
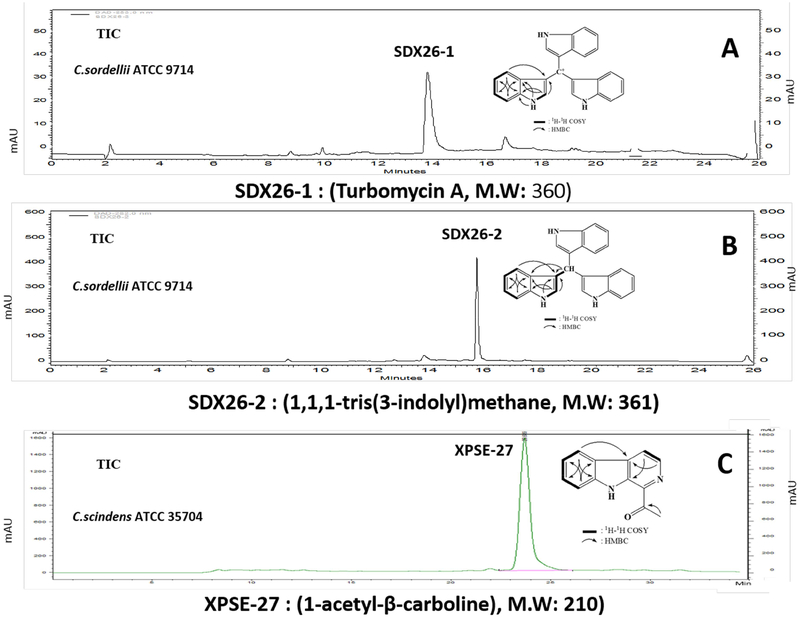
Because C. scindens ATCC 35704 was able to grow much better in co-culture with C. difficile ATCC 9689 in the presence, but not absence of CA, we hypothesized that CA may be inducing antibacterial compound(s) in C. scindens ATCC 35704 and/or enhancing their activity (Figure 1D, E). Next, we screened ten different strains and species of bile acid 7α-dehydroxylating human gut bacteria for the presence of antibacterial compound(s) secreted into the culture medium that inhibit C. difficile growth. Interestingly, all species and strains of human bile acid 7α-dehydroxylating gut bacteria tested were positive for inhibition of C. difficile ATCC 9689 growth to varying degrees. C. scindens ATCC 35704 and C. sordellii ATCC 9714 secrete antibacterial compound(s) that strongly inhibited C. difficile ATCC 9689 and a number of other bacteria, including several antibiotic resistant species (Table 1). The addition of CA to the culture medium enhanced the antibacterial activity of compounds secreted by C. scindens. We decided to purify and characterize the antibacterial compound(s) secreted by C. sordellii ATCC 9714 and C. scindens ATCC 35704 as these two bacteria are in different phylogenetic groups of bile acid 7α-dehydroxylating gut bacteria. Protocols were developed to purify the major antibacterial compound(s) secreted by C. sordellii ATCC 9714 (red) and C. scindens ATCC 35704 (blue) under our culture conditions (Figure 3, Figure S2). The results show that C. sordellii ATCC 9714 secretes two potent antibacterial compounds that absorb at 205–215 nm and strongly at 280 nm. These compounds were identified by LC-ESI/MS and H 1 and C13 NMR chemical shifts as turbomycin A (Figure 3 A) and 1,1,1-tris(3-indolyl)-methane [TIM] (Figure 3B). These antibiotics are derived from indole and indole-3-carboxyaldehyde and have been previously reported to be synthesized by soil microorganisms (Gillespie et al., 2002) but not anaerobic gut bacteria.
Figure 3. Identification of antibacterial compounds secreted by C. sordellii ATCC 9714 and C. scindens ATCC 35704.
The purification protocols for antibacterial compounds secreted by C. sordellii and C. scindens are described in Figure S2. The LC-ESI/MS and 1H and 13C NMR chemical shifts of SDX26–1 and SDX26–2 secreted by C. sordellii were positively identified as turbomycin A and 1,1,1-tris(3-indolyl)-methane, respectively. The LC-ESI/MS and 1H and 13C NMR chemical shift assignments of antibacterial compound (XPSE-27) was positively identified as 1-acetyl-β-carboline.
The structure of the major antibacterial compound secreted by C. scindens ATCC 35704 was determined by LC-ESI/MS and H1and C13 NMR chemical shifts to be 1-acetyl-β-carboline (Figure 3 C). 1-Acetyl-β-carboline is an alkaloid synthesized from L-tryptophan and oxaloacetaldehyde and reported to be synthesized by a Gram-positive marine bacterium with algicide activity (Kim et al., 2015) and by a marine Streptomyces albus (Shin et al.,2010). β-Carboline alkaloids are widely distributed in nature and are known to have a variety of biological activities. However, 1-acetyl-β-carboline has not been reported to be synthesized by gut bacteria. It is of interest that antibiotics synthesized by both species of bile acid 7α-dehydroxylating bacteria studied are derived from L-tryptophan or indole, a bacterial metabolite of L-tryptophan. Indole is found in mammalian feces in high concentrations (0.250 to 1.1 mM), is an inter-kingdom signaling molecule, and regulates the physiology of colonic epithelial cells (Lee et al., 2015).
Secondary Bile Acids Enhance the Activity of Antibiotics Secreted by Gut Bacteria
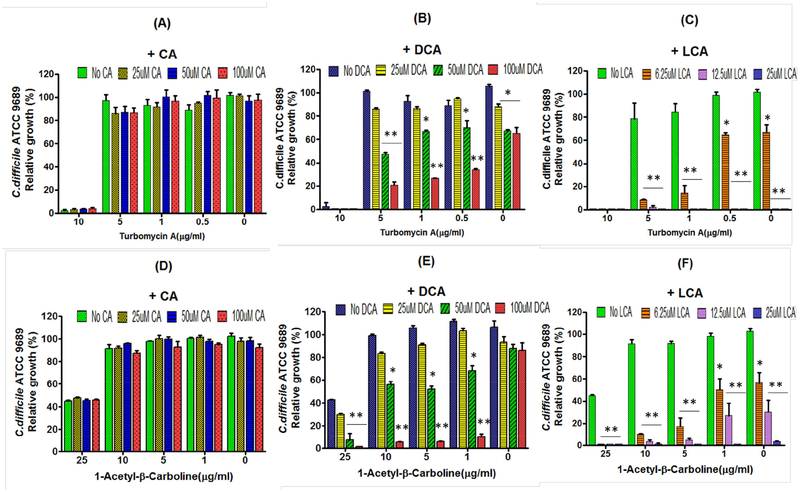
Why does bile acid 7α-dehydroxylating gut bacteria secrete antibiotics and secondary bile acids? We decided to determine if varying concentrations of different bile acids would enhance the antibacterial effects of turbomycin A and 1-acetyl-β-carboline using C. difficile ATCC 9689 as the test bacterium. First, we diluted filtered sterilized and pH 7.2 adjusted spent culture medium from C. sordellii ATCC 9714 and added varying amounts of fresh PYF medium until growth of C. difficile grew to about 40% to 60% maximal. Varying concentrations of different bile acids, similar to ranges found in human fecal water (Ditscheid et al., 2009), were added to the culture medium, inoculated with 106 C. difficile vegetative cells/ml, incubated for 24 hours, and optical density readings at 600 nm measured (STAR Methods). The results showed a strong concentration-dependent enhancement of inhibition of the growth of C. difficile by spent culture medium of C. sordellii by DCA and LCA, but not CA (Figure 4, Figure S3 A–C).
Figure 4. Effects of different bile acids on inhibition of growth of C. difficile by turbomycin A and 1-acetyl- β-carboline.
Varying concentrations of Turbomycin A (0, 0.5, 1, 5 and 10 μg/ml) or 1-acetyl-β-carboline (0, 1, 5, 10 and 25 μg/ml) and either cholic acid (CA) (0 to 100 μM), deoxycholic acid (DCA), (0 to 100 μM) or lithocholic acid (LCA), (0 to 25 μM) were added to PYF medium, inoculated with 106 vegetative cells/ml of C. difficile, incubated for 24 hrs. and optical density reading determined at 600 nm. * p< 0.05, ** p< 0.01
Because turobmycin A and TIM are not commercially available, we chemically synthesized turbomycin A and TIM. The chemical synthesis of these antibiotics was carried out under anaerobic conditions and optimal substrate concentrations which allowed higher yields and fewer side-products compared to the original method of chemical synthesis (Gillespie et al., 2002). LC-ESI/MS was used to demonstrate that the new chemical synthesis protocol for turbomycin A and TIM resulted in the expected product. Chemically synthesized turbomycin A inhibited the growth of C. difficile in a manner similar to the biologically synthesized compound (Figure 4, Figure S4).
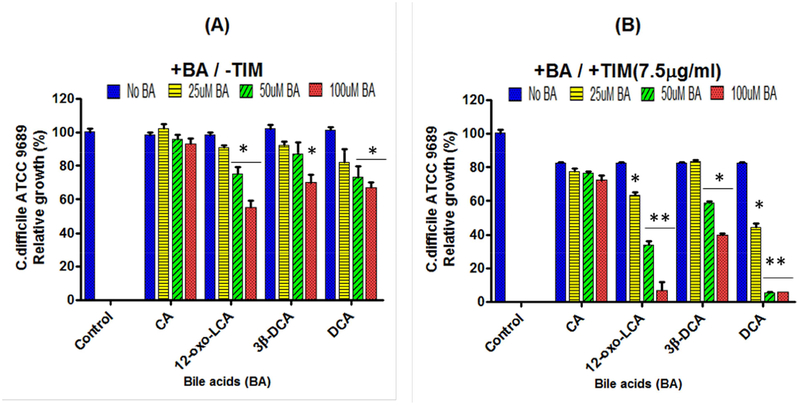
Next, we carried out similar experiments with the spent culture medium from C. scindens ATCC 35704. The results also showed enhancement of inhibition of C. difficile by DCA and LCA, but not CA (Figure S3 D–F). We next tested the ability of different bile acids to enhance the antibacterial activity of commercially available 1-acetyl-β-carboline (Figure 4 D–F). The results showed a concentration dependent enhancement of inhibition of C. difficile by DCA and LCA, but not CA. We next tested the effect of different bile acids found in human fecal water on inhibition of C. difficile by TIM (Ditscheid, 2009). Interestingly, iso-deoxycholic acid (30-DCA) and 12-oxo-lithocholic acid (12-oxo-LCA) were less effective than DCA in inhibiting C. difficile growth (Figure 5). The mechanism of enhancement of inhibition of these antibiotics by DCA and LCA is unknown, but DCA has been reported to be a better solubilizer of various hydrophobic drugs than CA (Enright et al., 2017).
Figure 5. Effect of 1,1,1-tris(3-indolyl)-methane (TIM) and different bile acids (BA) on inhibition of C. difficile ATCC 9689 growth.
A. Varying concentrations of cholic acid (CA) (0 to 100 μM), 12-oxo-lithocholic acid (12-oxo-LCA) (0 to 100 μM), iso-deoxycholic acid (3β-DCA) (0 to 100 μM) or deoxycholic acid (DCA) (0 to 100 μM) were added to PYF medium, inoculated with 106 vegetative cell/ml of C. difficile, incubated for 24 hrs, and O.D. readings at 600 nm determined. B. Experiments were repeated with the addition of TIM (7.5 μg/ml) added to the culture medium. *p<0.05, **p<0.01.
Turbomycin A and TIM Inhibit C. difficile Cell Division
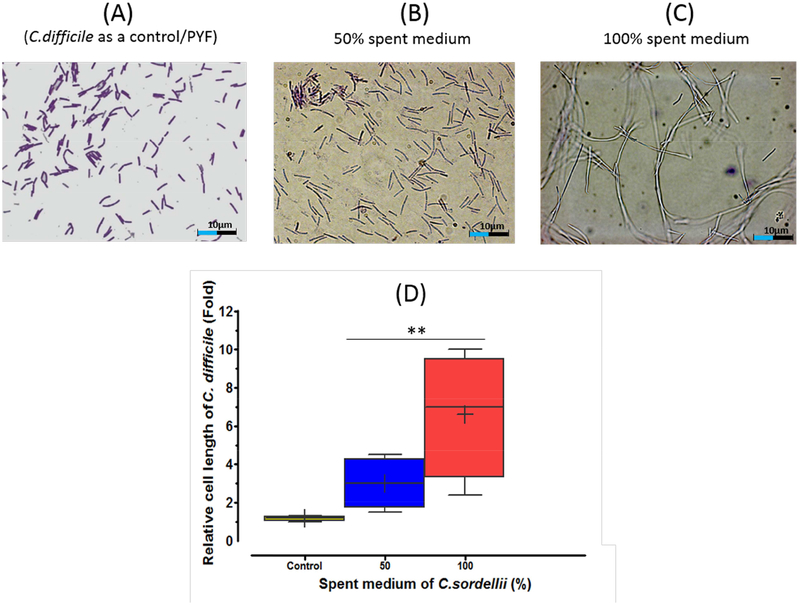
How does antibiotics secreted by C. sordellii and C. scindens inhibit the growth of C. difficile? We observed that the growth of C. difficile in spent culture medium of C. sordellii resulted in the formation of highly elongated C. difficile cells (Figure 6). Higher concentrations of spent culture medium increased the cell length of individual cells. Moreover, the addition of chemically synthesized turbomycin A or 1-acetyl-β-carboline to fresh PYF growth medium also resulted in inhibition of cell growth and elongated cell morphology of C. difficile (Figure 6, Figure S5). The type of morphological changes (2 to 7-fold longer cells in antibiotic treated cells vs controls) may be due to inhibition of the bacterial division septum (Xiao and Goley, 2015). However, additional experiments will be required to determine the mechanism(s) of growth inhibition of these antibiotics and their enhancement by DCA and LCA.
Figure 6. Changes in C. difficile ATCC 9689 cell morphology induced by antibacterial compounds secreted by C. sordelli ATCC 9714.
A) PYF medium control; B) C. difficile grown in 50% C. sordellii spent culture medium; C) C. diffiicle incubated in 100% C. sordellii spent culture medium. Each culture (1ml) was concentrated to 100 μl by centrifugation, 5 μl of cell suspension loaded on to a slide for Gram staining and photo images taken (STAR Methods). D) Cell length was determined using a scale bar generated by Neurolucia 1027 software in several microscopic fields. (Magnification 1000x). ** P<0.01
DISCUSSION
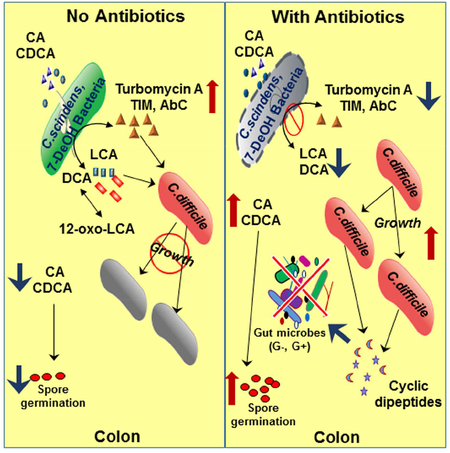
The surprising results of the current study was the discovery that human bile acid 7αdehydroxylating gut bacteria secrete tryptophan-derived antibiotics that are enhanced by secondary bile acids and that C. difficile secretes two proline-based cyclodipeptides that may function as quorum sensing molecules and/or colonization factors. The colonic ecosystem may select for bacteria that secrete antibacterial compounds giving them a competitive growth advantage in an ecosystem with 100 to 150 operational taxonomic units (OTU) at any given time and greater than 1011 bacterial cells/gram wet weight feces. Interestingly, the effectiveness of these hydrophobic antibiotics to inhibit gut bacteria is enhanced by hydrophobic secondary bile acids produced by bile acid 7α-dehydroxylating gut bacteria (Figure 4, Figure S3). The secretion of antibiotics by bile acid 7α-dehydroxylating gut bacteria and their concentration-dependent enhancement by DCA and LCA, but not by CA, is hypothesize to be a major regulator of the structure of the gut microbiome (Graphical Abstract). The minimal inhibitory concentrations (MICs) of turbomycin A and 1-acetyl-β-carboline to inhibit C. difficile are in the range of 7.5 μg/ml and 25 μg/ml, respectively, in PYF medium (Figure 4). These are relatively high MICs to be effective antibiotics; however, if combined with DCA and/or LCA then the mixture appears to become much more potent than either alone. Because all bile acid 7α-dehydroxylating bacteria tested are capable of secreting both secondary bile acids and antibacterial compounds, then this mixture may be very effective in controlling gut microbiome homeostasis under normal physiological conditions. Moreover, it is also possible that the antibiotics may enhance the toxicity of secondary bile acids especially if both are inhibiting the formation of the division septum. However, this possibility will have to be tested in an appropriate in vivo model where each antibiotic can be used alone and in combination with a secondary bile acid.
Human gut bacteria are capable of the formation of more than 20 different metabolites of cholic acid and chenodeoxycholic acid (Ridlon et al., 2006). What is the physiological purpose of these bile acid biotransformations? Bile acid hydroxyl group oxidations and epimerizations (α> β) increase the hydrophilicity of the bile acid molecule making them less toxic to bacterial cells and possibly decreasing their ability to enhance hydrophobic antibiotics (Devlin and Fischbach, 2015). In this regard, iso-deoxycholic acid (3β-hydroxyl derivative) and 12-oxoLCA, which are found in human fecal water (Ditscheid et al., 2009), were less effective in enhancing TIM than deoxycholic acid in inhibiting C. difficile growth (Figure 5). Therefore, the hydrophilic-hydrophobic balance of the colonic bile acid pool may be a key factor in determining the structure of the gut microbiome
Bile acids have been hypothesized to play an important role in the regulation of the gut microbiome structure by a variety of mechanisms. As detergents, bile acids directly inhibit gut bacteria growth by membrane-damage related on their hydrophobicity (Devlin and Fischbach, 2015; Ridlon et al., 2006). Unconjugated secondary bile acids are generally more hydrophobic than corresponding primary bile acids. During their enterohepatic circulation, bile acids can activate the bile acid activated nuclear receptor, farnesoid X receptor (FXR), to induce genes that are enteroprotective by inhibiting bacterial overgrowth (Inagaki et al., 2006). However, these FXR-dependent antibacterial effects may be more relevant to mucosal associated bacteria than those in the lumen of the intestine.
There is evidence that bile acids may be important regulators of the human gut microbiome in cirrhotic liver disease. In this regard, cirrhotic patients have a decreased bile acid pool size due to loss of bile acid synthesis in the liver, resulting in a marked reduction in bile acid 7α-dehydroxylating gut bacteria, and a shift to a more “toxic” Gram-negative gut microbiota (Bajaj et al. 2012; Chen et al. 2011; Kakiyama et al., 2014). In contract, when cirrhotic patients are transplanted with a new liver, there is increased primary bile acids synthesis, increased fecal secondary bile acid levels and a return to a more diverse fecal microbiome (Bajaj et al., 2018). Moreover, in animal model studies, feeding CA to rats markedly shifts the bacterial gut composition from a ~1:1 ratio of Firmicutes/Bacteroidetes to greater than 98% Firmicutes (Islam et al., 2011) while feeding CA to mice has been reported to increase the levels of bile acid 7αdehydroxylating bacteria by ~1000-fold as these bacteria use primary bile acids as an electron acceptor (Ridlon et al., 2013). In addition, animals fed high-fat diets increase the Firmicutes/Bacteroidetes ratio possibily because of increased enterohepatic cycling of bile acids and increased loss into feces (Yokata et al., 2012; Turnbaugh et al. 2008; Hildebrandt et al. 2009). The concentration of various secondary bile acids found in human fecal water samples (unbound bile acids) appear not to adequately explain the marked shift in the microbiome structure with changes in fecal bile acid pool composition and concentration (Ditscheid et al., 2009). However, the combination of secondary bile acids and tryptophan-derived antibiotics may work as a powerful “cocktail” to regulate the structure of the gut microbiome.
STAR METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
“Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Phillip Hylemon (Phillip.hylemon@vcuhealth.org)”
EXPERIMENTAL MODE AND SUBJECT DETAILS
Bacterial Strains and Culture Conditions
The following bacterial strains were obtained from the American Type Culture Collection (ATCC): Clostridium difficile ATCC 9689; Clostridium difficile clinical isolates from the ATCC include: BAA-1870 (ribotype 027); BAA-1801 (ribotype 010); BAA-1814 (ribotype 251); Clostridium scindens ATCC 35704; Clostridium sordelli ATCC 9714; Bacteroides vulgatus ATCC 8482. Clostridium scindens VPI 12708 was obtained from the Virginia Tech Anaerobe Laboratory. Bacteria were cultured in peptone-yeast extract-fructose (PYF) medium and growth monitored at 600 nm.
METHODS DETAILS
Light Microscopy
Clostridium difficile cultures were smeared, fixed on glass slides, stained using Remel Gram Staining Kit according to manufacturer’s directions and allowed to air dry. Bacteria were covered with DPX Mountant and a cover glass. Bacteria were examined under bright field microscopy using a Zeiss AxioImager microscope with computer controlled stage using the Neurolucida 2017 software. Images were obtained using x63 and x100 oil-immersion objectives using a Zeiss MRC camera as .jpg files.
DNA Purification
One ml of a pure culture of C. difficile ATCC 9689, C. scindens ATCC 35704 and C. scindens VPI 12708, respectively, were subjected to genomic DNA isolation using a Fast DNA Spin Kit, according to the manufacturer’s instructions. The final elution volume was 100 [l and the nucleic acid concentration was measured fluorimetrically using a Nano Spectrophotometer.
PCR Quantitation of Bacterial Strains
Target DNA (1 μg) was added and the copy number of the tcdA and baiCD genes were determined based on the molecular weight of 3.8- to 4.18-mbp-sized genome of C. difficile ATCC 9689, C. scindens ATCC 35704, and C. scindens VPI 12708, respectively. 1 ng of DNA was calculated to equal to 6 × 105 times the entire genomes and that the tcd A and baiCD are single-copy genes. Gene specific primers were synthesized for C. difficile ATCC 9689 toxin A(tcdA) gene as follows: (CDF9689T cdA-1F, Forward: T GGGCT GAT ATT AAT GCAGAA; CDF9689TcdA-1R, Reverse: TTCCCAACGGTCTAGTCCAA) and baiCDs of C. scindens ATCC 35704 and C. scindens VPI 12708 (CS357N127BaiCDWhole-F, Forward: T GGCT GTT CAGGCT GGATTT GA; CS357N127VaiCDWhole-R, Reverse: TCGTAGCATCCCTGGTCACA). The PCR assay was performed at 95°C for 4 min., followed by 35 cycles of 95°C for 30 sec., 52°C for 1 min, 72°C for 2 min with a final 10 min extension step 72°C. PCR products from conventional PCR assays were detected on 1% Agarose gels and the size of the PCR products (588 bp for Toxin A and 595 bp for baiCD genes) were confirmed.
Quantitative PCR of Species Specific Genes
Gene specific primers were used to quantitate C. difficile and C. scindens in co-culture using a designed target region of the toxin A gene (tcdA) and baiCD genes that are highly conserved in C. difficile ATCC 9689, C. scindens ATCC 35704, and C.scindens VPI 12708. The primers sets specific for the individual bacteria are as follows: C.difficile ATCC 9689 toxin A gene (CDF9689NtxA-1F, Forward: TCTACCACTGAAGCATTAC; CDF9689NtxA-1R, Reverse, TAGGTACTGTAGGTTTATTGj, baiCD for C. scindens ATCC 35704 (CS35704baiCD-1F, Forward: CTT CATTCGCAACT GT AT AT GA, CS35704baiCD-1R, Reverse: T AACCGGGAT ATT GAT CT GCTT) and for C. scindens VPI 12708 (CS1270 8baiCD-1F, Forward: CAACCGT AT AT GAAGTTCC, CS12708baiCD-1R, Reverse: ATCCCGGTTATGGGAGTTGG). Amplification and detection of DNA by real-time PCR was performed with the IQ5 Realtime PCR system (BioRad, CA) using optical grade 96-well plates. Triplicate samples were routinely used for the determination of DNA by real-time PCR, and the mean values calculated. The PCR reaction was performed in a total volume of 25 μl. C. difficile ATCC 9689, C. scindens ATCC 35704, and C. scindens VPI 12708 were detected by using the Sso Advanced Universal SYBR Green PCR Master Mix (Bio-Rad, CA), with 100 nmol l−1 of each of the forward and reverse primers and 1 ng DNA for each reaction. The PCR reaction conditions for amplification of DNA were 50°C for 2 min, 95°C for 10 sec. and 40 cycles of 95°C for 15 sec. and 60°C for 1 min. A melting curve analysis was done after amplification. The threshold cycle (CT) values and baseline settings were determined by automatic analysis settings. Data analysis was performed using iQ5 Optical System Software supplied by Bio-Rad. The relative abundance of C. difficile ATCC 9689 (toxin A gene copies), C. scindens ATCC 35704 (baiCD genes), and C. scindens VPI 12708 (baiCD genes) were related to the cycle threshold (Ct) which is the number of PCR cycles needed to obtain a detectable fluorescent signal above background and typically was the point on the amplification curve where it becomes linear. Ct was experimentally confirmed and determined to be related to copy number of the strains by amplifying a series of serial 10-fold dilutions of the genomic DNA from each bacterium.
Purification of Antibacterial Compounds from C. difficile, C. scindens. and C. sordellii
The purification protocols for the cyclic dipeptides from C. difficile, turbomycin A and TIM from C. sordellii, and 1-acetyl-β-carboline from C. scindens are described in Figures S1 and S2, respectively.
Identification of the Antibacterial Compounds Secreted by C. difficile, C. sordellii and C. scindens
Characterization and identification of turbomycin A, 1,1,1-tris(3-indolyl)-methane and cyclic dipeptides was carried out by Drs. In-Kyoung Lee and Bong-Sik Yun at Chonbuk National University, Republic of Korea. Antibacterial compounds were characterized and identified by spectroscopic methods. ESI/MS data were obtained on a QTRAP-3200 mass spectrometer (Applied Biosystems, USA) in positive and negative modes. NMR spectra were obtained on a JNM-ECA600 600MHz FT-NMR spectrometer (JEOL, Japan) with 1H NMR at 600 MHz and 13C NMR at 150 MHz in CD3OD, CDCl3 or DMSO-d6. Chemical shifts are given in ppm (δ) using tetramethylsilane as internal standard. Chemical structures were determined by one- and two-dimensional NMR experiments including 1H−1H COSY, HMQC, HMBC, and NOESY.
Chemical Synthesis of Turbomycin A and 1,1,1-tris(3-indolyl)-methane
Because turbomycin A and 1,1,1-tris(3-indolyl)-methane are not commercially available, we chemically synthesized these two antibiotics using a modification of the protocol reported by Gillespie D.E. et al. 2002. Briefly, we optimized the concentration of reactants, incubation times, and carried out synthesis under anaerobic conditions. The reaction was carried out in 100 ml stoppered bottles flushed with N2 gas. Two hundred μmoles of indole-3-carboxyaldehyde and 600 μmoles of indole were dissolved in 450 μL ethanol and 50 μL acetic acid under N2 gas. In order to maintain anaerobic condition, the bottle was flushed with N2 gas, immediately sealed and placed at 65°C water bath for 12 hours or overnight with mixing. Then, 600 μl of 10 N NaOH was added to achieve a pH of ~7.2. After which 4 mmoles of tetrachloro-1,4-benzoquinone was added and the bottle was immediately resealed and contents mixed. The bottle was placed in a water bath at 65°C for 2 hrs with mixing. Then, 50 ml of distilled water was added and the crude product, extracted 3 times with an equal volume of ethyl acetate and allowed to air dry. The crude product was then dissolved in 100% acetonitrile, run through a Sep-Pak column, and fractions collected. Five μl of each fraction was loaded onto TLC plates and reaction products separated using (chloroform:methanol:acetone, 80:15:2, vol/vol/vol) for turbomycin A and (hexane:chloroform:acetone, 20:60:10, vol/vol/vol) for 1,1,1-tris-(3-indolyl)-methane as solvents, respectively (Figure S4). TLC spray regent was used for the detection of the products on the TLC (Ehmann, 1977). Active fractions were assayed for bacteria inhibition activity and positive fractions purified using a preparative reversed-phase HPLC column (Kinetex C18, 5 μm, 21.2 mm inner diameter and × 150 mm length) with linear gradient elution from 50% to 70% acetonitrile and a flow rate (3.0 ml/min) for 35 min for the separation of turbomycin A and 1,1,1-tris-(3-indolyl)-methane. Active fractions were subjected to an analytical C18 HPLC column (Agilent, Eclipse XDB-C18, 4.6 mm inner diameter × 150 mm length) to confirm identification of compound by mass spectrometry analysis.
STATISTICAL ANALYSIS
All experiments were performed in triplicate and repeated at least three times. The values were expressed as Means ± Standard deviation (SD) in all graphs. The statistical analysis of the results was performed by one-way analysis of variance (ANOVA) to compare tests and control using Graphpad Prism (Graphpad software, San Diego, CA, U.S). Symbols * and ** indicate statistical significance, p<0.05 and p<0.01, respectively.
DATA AVAILABILITY
The software used in this study are listed in the Key Resources Table. Additional experimental data are provided as supplementary information and are available from the corresponding author upon request.
Supplementary Material
Highlights.
Bile acid 7α-dehydroxylating gut bacteria secrete tryptophan-derived antibiotics
Secondary bile acid enhanced the activity of these antibiotics
Tryptophan-derived antibiotics appear to inhibit the division septum of bacteria
Clostridium difficile secretes proline-based cyclic dipeptides
SIGNIFICANCE.
This work reports that bile acid 7α-dehydroxylating bacteria secrete tryptophan-derived antibiotics and their antibacterial activity enhanced by secondary bile acids. These discoveries appear relevant to how the structure of the gut microbiome is regulated by bile acids and bile acid 7α-dehydroxylating gut bacteria and how this ecosystem may be disrupted by antibiotics or in liver diseases where there is a decrease in secondary bile acids. Moreover, we also report that Clostridium difficile secretes proline-based cyclic dipeptides which are known quorum sensing molecules for some bacteria. They have been reported to have antibacterial activity and may be important colonization factors in C. difficile infection in the absence of bile acid 7α-dehydroxylating bacteria.
ACKNOWLEDGEMENTS
This work was supported by NIH grant R01-DK104893 and R01-DK115377 to P.B.H and H. Z and VA Merit Grant BX0013828 to P.B.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes six figures and one table and can be found with this article online at https://
DECLARATION OF INTEREST
A patent application has been filed based on this study.
REFERENCES
- Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM Linkage of gut microbiome with cognition in hepatic encephalopathy (2012) Am J Physiol Gastrointest Liver Physiol 302, G168–G175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj JS, Kakiyama G, Cox IJ, Nitono H, Takei H, White M, Fagan A, Gavis EA Heuman DM, Gilles HC et al. Alterations in gut microbial function following liver transplant (2018). Liver Transpl. 24(6), 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin P, Moutiez M, Lautru S, Seguin J, Pernodet JL, Gondry M The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthease pathways (2012). Nat Prod Rep. 29, 961–79. [DOI] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A et al. (2015) Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang F, Lu H, Wang B, Chen Y Lei D, Wang Y, Zhu B, Li L (2011) Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology 54, 562–572. [DOI] [PubMed] [Google Scholar]
- Devlin AS and Fischbach MA (2015) A biosynthetic pathway for a prominent class of microbiota- derived bile acids. Nat Chem Biol 11, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditscheid B, Keller S, Jahreis G (2009) Faecal steroid excretion in humans is affected by calcium supplementation and shows gender-specific differences. Eur J Nutr. 48, 22–30. [DOI] [PubMed] [Google Scholar]
- Ehmann A The Van Urk-Salkowski reagent-A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification for indole derivatives (1977). J Chromatography 132, 267–276. [DOI] [PubMed] [Google Scholar]
- Enright EF, Joyce SA, Gahan CG, Griffin BT (2017) Impact of gut microbiota-mediated bile acid metabolism on the solubilization capacity of bile salt micelles and drug solubility. Mol Pharm 14, 1251–1263. [DOI] [PubMed] [Google Scholar]
- Giessen TW and Marahiel MA (2014) The tRNA-dependent biosynthesis of modified cyclic dipeptides. Int J Mol Sci. 15, 14610–14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR Rondon MR, Clardy J, Goodman RM, Handelsman J (2002) Isolation of antibiotics turbomycin A and B from a metagenomics library of soil microbial DNA. Appl Environ Microbiol 68, 4301–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez O, Ortlz-Castro R, Diaz-Perez C, Diaz-Perez AL, Maganas V, Lopez-Bucio J, Campos-Garcia J (2017) Non-ribosomal peptide synthases from Pseudomonas aeruginosa play a role in cyclodipeptide biosynthesis, quorum-sensing regulation, and root development in a plant host. Microb. Ecol 73, 616–629. [DOI] [PubMed] [Google Scholar]
- Heuman DM (1989) Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J Lipid Res 30, 719–730. [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann CS, Sherrill-Mix A, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima R, Bushman SF, Wu GD (2009) High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 237, 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee Y-K, Peng L, Zhao P, Downes M, Yu RT, Shelton JM, Richardson JH, Repa JJ et al. (2006) Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci (USA) 103, 3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam KB, Fukiya S, Hagio M, Fujii MN, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A (2011) Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141, 1773–1781. [DOI] [PubMed] [Google Scholar]
- Kakiyama G, Hylemon PB, Zhou H, Pankak WM, Heuman DM, Kang DJ, Takei H, Nittono H, Ridlon JM, Fuchs M et al. (2014) Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastroentest Liver Physiol 306, G929–G937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Son HJ, Jeong S (2015) Isolation of an algicide from a marine bacterium and its effects against the toxic dinoflagellate Alexandrium catenella and other harmful algal blooms species. J Microbiol 53, 511–517. [DOI] [PubMed] [Google Scholar]
- Kuehner SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP (2010). The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713. [DOI] [PubMed] [Google Scholar]
- Lee JH, Wood TK, Lee J (2015) Role of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 23, 707–718. [DOI] [PubMed] [Google Scholar]
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, et al. (2015) Burden of Clostridium difficile infection in the United States. N Engl J Med. 372, 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo VG, Bourgault AM, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R, et al. (2011) Host and pathogen factors for Clostridium difficile Infection and colonization. N Engl J Med. 365, 1693–1703. [DOI] [PubMed] [Google Scholar]
- Nelson RL, Suda KJ, Evans CT (2017) Antibiotic treatment of Clostridium difficile-associated diarrhea in adults. Cochrane Database Syst Rev 3, CD004610. doi: 10.1002/14651858.CD004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB (2006) Bile salt biotransformations by human intestinal bacteria. J. Lipid Res 47, 241–259. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Alves JM, Hylemon PB, J. Bajaj JS (2013) Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes 4, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nature Rev. Microbiol 7, 526–536. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Lee H-S, Lee D-S (2010) The synergistic antibacterial activity of 1-acetyl- β-carboline and P-lactams against methicillin-resistant Staphylococcus aureus (MRSA). J Microbiol Biotechnol 20, 501–505. [PubMed] [Google Scholar]
- Solbach P, Chhatwal P, Woltemate S, Tacconelli E, Buhl M, Gerhard M, Thoeringer CK, Vehreschild MJGT, Jazmati N, Rupp J et al. (2018) BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PloS One 13(5): e0196977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg JA and Sonenshein AL (2008) Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 190, 2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg JA, Sonenshein AL (2010) Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol. 192, 4983–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JL (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 10271031. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JL (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. (2013) Duodenal infusion of feces for recurrent Clostridium difficile. N Engl J Med. 368, 407–415. [DOI] [PubMed] [Google Scholar]
- Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A (2014) Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 306, G310–G319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Goley ED (2015) Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr Opin Microbiol. 34, 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota A, Fukiya S, Islam KB, Ooka T, Ogura Y, Hayashi T, Hagio M, Ishizuka S (2012) Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes 3, 455–459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The software used in this study are listed in the Key Resources Table. Additional experimental data are provided as supplementary information and are available from the corresponding author upon request.