SUMMARY
Background and objectives:
Platelet concentrates are frequently transfused to patients with reduced immunity. An exhaustive description of their viral content is needed to prevent unwanted infection.
Material and methods:
To track viral sequences, a shotgun metagenomics approach was used on a bank of 300 platelets concentrates. Sequences were analysed through the diagnostics-oriented pipeline ezVIR.
Results:
We only observed viruses commonly described in healthy individuals.
Conclusion:
Herein is reported the first viral landscape of a platelet concentrates bank.
Keywords: next generation sequencing, platelet concentrates, transfusion-transmissible infections
Using metagenomics, we recently reported on the virome of red blood cell concentrates and fresh frozen plasmas [1]. Platelet concentrates are frequently transfused to immunosuppressed patients. However, due to their limited availability for research, their viral landscape has not been reported. In order to assess the risk of transfusing pathogenic viruses, we performed viral screening of 300 platelet concentrates manufactured at the blood transfusion center of Geneva (Switzerland).
1. Material and methods
Platelet concentrates manufacturing
Three hundred platelet concentrates were collected from single donors in 2016 containing, on average, 39% of donor’s plasma, and 0.23×106 residual leukocytes/unit. Four ml were derived from each concentrate before clinical use. Samples were not pathogen-reduced.
Nucleic acid extraction and Sequencing
Extraction was made using the QIAamp Circulating Nucleic Acid Kit (Qiagen) without carrier RNA. Pools were made of 30 extracts each. Pool libraries were prepared as previously described [1]. As positive controls, two libraries, one DNA (DNA B19V) and one RNA (RNA HRV14), were prepared from one spiked single concentrate with 105 copies/ml of Parvovirus B19 (B19V) and Human Rhinovirus 14 (HRV-14), respectively. Negative controls consisted of empty tubes submitted to the entire process. Libraries were run on the HiSeq 2500, producing paired-end, 100 nt long reads.
Library Analysis
Libraries were analysed with an updated version of ezVIR [2]. Non-human data were assembled using IDBA-UD, and scaffolds were classified with DIAMOND, using NR as the database. Cross-contamination was described as previously reported in [3]. Briefly, for each sequencing lane, a percentage of reads can be misattributed to other libraries. For each lane, we compared the libraries with most abundance of each virus, against the other libraries. If the ratio was below 0.24%, they were considered a contaminants.
2. Results and discussion
Twenty-four libraries were produced: 10 DNA pools, 10 RNA pools, 2 positive controls, and 2 negative controls (DNA and RNA). After removing low quality and complexity reads, DNA libraries were comprised of an average of 91.2% human content and 8.8% non-human content, while RNA libraries had an average of 89.5% human content and 10.5% non-human content. The spiked viruses – Parvovirus B19 (B19V) and Human Rhinovirus 14 (HRV-14) were found with almost complete coverage (99.99% and 99.98% respectively).
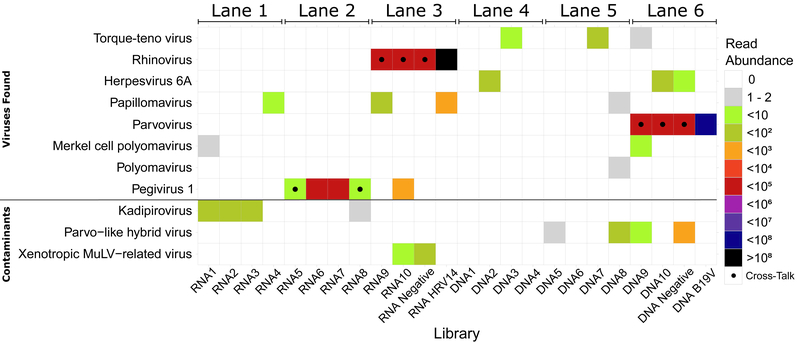
Five virus families were identified (figure 1): Anelloviridae (Torque Teno Virus - TTV), Herpesviridae (Herpesvirus 6 - HHV6), Papillomaviridae (Papillomavirus), Polyomaviridae (Polyomavirus and Merkel Cell Polyomavirus) and Flaviviridae (human Pegivirus - HPgV). The percentage of genome covered and number of reads for each virus are shown in table 1. These signals are consistent with studies from other blood products, namely viruses reported to commonly circulate in the blood of healthy individuals: TTV, Pegivirus, HHV6, Polyomavirus and Papillomavirus. TTV is detected in blood with a prevalence between 1.9% to 62% and a tropism for peripheral mononuclear cells, liver and bone marrow [7]. Pegivirus infects 1/6 of the global population and is known to be transmissible through transfusion [8,9], with a tropism for bone marrow, spleen, and plasma [8]. While Herpesvirus 6 viraemia has been described in the blood virome, the signal may also be due to HHV6 integrated in the genome [10]. While associated with skin flora, Polyomavirus and Papillomavirus have also been reported in the blood of healthy individuals [10]. These observations suggest virus transmission to the product from the donor’s blood. HHV6 reads were also detected in the negative control, mapping to a region common to several HHV6 strains (a repeat region in the first 3000nt of the genome). Since no hits for other species were found for these reads, we discarded the possibility of a reagent contaminant or read misclassification. It could be a case of cross-contamination from sample 10, though the low abundance of reads does not allow us to confirm it. Finally, known reagent contaminants are found: Parvo-like hybrid virus, Kadipiro virus[4,5] and Xenotropic murine leukemia-related virus [6].
Figure 1:
Viral signatures in each individual pool sorted by sequencing lane and sequencing type. The dotted libraries represent cross-talk contaminations.
Table 1:
Quantitative values of virus signals found by ezVIR.
| Library | Virus Family | Genome Name | ID | % covered | Genome length | Total mapped reads |
|---|---|---|---|---|---|---|
| DNA 1 | none | none | ||||
| DNA 2 | Herpesvirus 6A | Human herpesvirus 6A AJ | KP257584.1 | 0.54 | 156714 | 16 |
| DNA 3 | TT virus | Torque teno virus 1 | NC 002076.2 | 5.19 | 3852 | 3 |
| DNA 4 | none | none | ||||
| DNA 5 | none | none | ||||
| DNA 6 | none | none | ||||
| DNA 7 | TT virus | Torque teno midi virus 2 MD2–013 | AB290919.1 | 11.96 | 3’253 | 14 |
| DNA 8 | Polyomavirus | Polyomavirus HPyV6 607b | HM011561.1 | 2.42 | 4’926 | 2 |
| Papillomavirus | Human papillomavirus type 20 | U31778.1 | 1.33 | 7’757 | 2 | |
| DNA 9 | Merkel cell polyomavirus | Merkel cell polyomavirus MCC350 | EU375803.1 | 6.48 | 5’387 | 4 |
| TT virus | Torque teno virus 1 | NC 002076.2 | 2.60 | 3’852 | 2 | |
| DNA 10 | Herpesvirus 6A | Human herpesvirus 6A AJ | KP257584.1 | 0.55 | 156’714 | 12 |
|
DNA negative |
Herpesvirus 6A | Human herpesvirus 6A GS | KJ123690.1 | 0.06 | 156’864 | 3 |
| DNA B19V | Parvovirus | Human parvovirus B19 | NC 000883.2 | 99.98 | 5’596 | 65’895’350 |
| RNA 1 | Merkel cell polyomavirus | Merkel cell polyomavirus MCC350 | EU375803.1 | 3.71 | 5’387 | 2 |
| RNA 2 | none | none | ||||
| RNA 3 | none | none | ||||
| RNA 4 | Papillomavirus | Human papillomavirus type 107 | EF422221.1 | 1.32 | 7’562 | 4 |
| RNA 5 | none | none | ||||
| RNA 6 | Pegivirus 1 | Hepatitis GB virus C GT110 | D90600.1 | 96.66 | 9’395 | 32’787 |
| RNA 7 | Pegivirus 1 | Hepatitis GB virus C GT110 | D90600.1 | 97.79 | 9’395 | 32’322 |
| RNA 8 | none | none | ||||
| RNA 9 | Papillomavirus | Human papillomavirus type 107 | EF422221.1 | 1.32 | 7’562 | 11 |
| RNA 10 | Pegivirus 1 | Hepatitis G virus strain HGV | AF081782.1 | 53.73 | 9’373 | 485 |
|
RNA negative |
none | none | ||||
| RNA HRV 14 | Rhinovirus | Human rhinovirus type 14 | K02121.1 | 99.99 | 7’212 | 102’745’995 |
| Papillomavirus | Human papillomavirus type 18 | X05015.1 | 29.50 | 7’857 | 175 |
Herein is reported a novel insight into the viral landscape of a platelet concentrates bank. It contains less diversity of viral signatures than reported for the blood virome, probably due to the plasma/leukocyte reduction process applied to blood products manufacturing, and the high level of pooling in this study. Despite this, we were able to find several commensal viruses also present in other blood products. Finding signatures of two DNA viruses in RNA libraries suggest a replication phase releasing RNA or incomplete DNase efficacy during library preparation. No uncommon viruses were found, unlike our previous analysis in red blood cells and plasma [1], where we found an astrovirus MLB2 linked to meningitis in immunocompromised patients [11]. As generally observed in high throughput sequencing, unspecific background signal was systematically present, and must be carefully identified and removed. This is achieved by controlling for specific reagents, using double indexing, and complementing the analysis with robust molecular methods, respectively [3–6]. Thus, a metagenomic approach offers an attractive option for the exhaustive screening of platelet concentrate banks, as it allows for the detection of viruses which aren’t part of standard diagnostics, and also can lead to follow up association studies in larger platelet banks and/or defined donor populations [11,12].
Acknowledgements
Production and quality control staff of the blood transfusion center of Geneva, staff of the genomic core facility, faculty of medicine of Geneva, for HTS.
Sources of support: Dubois Ferrière Dinu Lipatti Foundation, unrestricted grant from AstraZeneca (OPS), National Heart, Lung, and Blood Institute (ED, No. R01 HL105770), IGE3 PhD Student Award (FB).
References
- 1.Lau P, Cordey S, Brito F, et al. Metagenomics analysis of red blood cell and fresh-frozen plasma units. Transfusion. 2017;57(7):1787–1800. doi: 10.1111/trf.14148. [DOI] [PubMed] [Google Scholar]
- 2.Petty TJ, Cordey S, Padioleau I, et al. Comprehensive human virus screening using high-throughput sequencing with a user-friendly representation of bioinformatics analysis: a pilot study. J Clin Microbiol. 2014;52(9):3351–3361. doi: 10.1128/JCM.01389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright ES, Vetsigian KH. Quality filtering of Illumina index reads mitigates sample cross-talk. BMC Genomics. 2016;17(1):876. doi: 10.1186/s12864-016-3217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naccache SN, Greninger AL, Lee D, et al. The Perils of Pathogen Discovery: Origin of a Novel Parvovirus-Like Hybrid Genome Traced to Nucleic Acid Extraction Spin Columns. J Virol. 2013;87(22):11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ngoi CN, Siqueira J, Li L, et al. Corrigendum: The plasma virome of febrile adult Kenyans shows frequent parvovirus B19 infections and a novel arbovirus (Kadipiro virus). J Gen Virol. 2017;98(3):517–517. doi: 10.1099/jgv.0.000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simmons G, Glynn SA, Komaroff AL, et al. Failure to confirm XMRV/MLVs in the blood of patients with chronic fatigue syndrome: a multi-laboratory study. Science. 2011;334(6057):814–817. doi: 10.1126/science.1213841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spandole S, Cimponeriu D, Berca LM, Mihăescu G. Human anelloviruses: an update of molecular, epidemiological and clinical aspects. Arch Virol. 2015;160(4):893–908. doi: 10.1007/s00705-015-2363-9. [DOI] [PubMed] [Google Scholar]
- 8.Bailey AL, Lauck M, Mohns M, et al. Durable sequence stability and bone marrow tropism in a macaque model of human pegivirus infection HHS Public Access Together, these results reconcile several paradoxical observations from cross-sectional analyses of HPgV in humans and provide an animal model for studying pegivirus biology. Sci Transl Med. 2015;7(305):305–144. doi: 10.1126/scitranslmed.aab3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernardin F, Operskalski E, Busch M, Delwart E. Transfusion transmission of highly prevalent commensal human viruses. Transfusion. 2010;50(11):2474–2483. doi: 10.1111/j.1537-2995.2010.02699.x. [DOI] [PubMed] [Google Scholar]
- 10.Moustafa A, Xie C, Kirkness E, et al. The blood DNA virome in 8,000 humans. Belshaw R, ed. PLoS Pathog. 2017;13(3):e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordey S, Vu D-L, Schibler M, et al. Astrovirus MLB2, a New Gastroenteric Virus Associated with Meningitis and Disseminated Infection. Emerg Infect Dis. 2016;22(5):846–853. doi: 10.3201/eid2205.151807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson MR, Fedewa G, Stenglein MD, et al. Multiplexed Metagenomic Deep Sequencing To Analyze the Composition of High-Priority Pathogen Reagents. Chia N, ed. mSystems. 2016;1(4):e00058–16. doi: 10.1128/mSystems.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]