Abstract
Background and study aims Detection of polyps during colonoscopy is essential for screening colorectal cancer and computer-aided-diagnosis (CAD) could be helpful for this objective. The goal of this study was to assess the efficacy of CAD in detection of polyps in video colonoscopy by using three methods we have proposed and applied for diagnosis of polyps in wireless capsule colonoscopy.
Patients and methods Forty-two patients were included in the study, each one bearing one polyp. A dataset was generated with a total of 1680 polyp instances and 1360 frames of normal mucosa. We used three methods, that are all binary classifiers, labelling a frame as either containing a polyp or not. Two of the methods (Methods 1 and 2) are threshold-based and address the problem of polyp detection (i. e. separation between normal mucosa frames and polyp frames) and the problem of polyp localization (i. e. the ability to locate the polyp in a frame). The third method (Method 3) belongs to the class of machine learning methods and only addresses the polyp detection problem. The mathematical techniques underlying these three methods rely on appropriate fusion of information about the shape, color and texture content of the objects presented in the medical images.
Results Regarding polyp localization, the best method is Method 1 with a sensitivity of 71.8 %. Comparing the performance of the three methods in the detection of polyps, independently of the precision in the location of the lesions, Method 3 stands out, achieving a sensitivity of 99.7 %, an accuracy of 91.1 %, and a specificity of 84.9 %.
Conclusion CAD, using the three studied methods, showed good accuracy in the detection of polyps with white light colonoscopy.
Introduction
Colorectal cancer (CRC) is one of the major health issues 1 . It is reasonable to expect that colonoscopy will continue to play an important role in CRC screening programs 2 , for CRC prevention.
The ability of screening colonoscopy to reduce CRC mortality and incidence is mainly due to the capacity for detecting polyps/adenomas 3 . This strategy implies that endoscopic detection of the polyps must be highly efficient. Nevertheless, it is a well-known fact that adenoma detection rate (ADR) varies widely among gastroenterologists 4 . Many aspects should be considered when trying to improve the ADR, and computer-aided-diagnosis (CAD) for colonoscopy is certainly one of them 5 .
The purpose of this study was to assess the efficacy of CAD in detection of colonic polyps in video colonoscopy by using three methods we have proposed and applied in the diagnosis of polyps in capsule colonoscopy 6 7 8 . For convenience of the reader we give a brief description of these three methods.
Methods 1 6 and 2 7 are binary classifiers and threshold-based methods. Each one of these methods assigns a numerical value to a given frame. If this value is larger than a predefined threshold the frame is classified as a polyp frame, otherwise it is a normal mucosa frame. In Method 1 the numerical value mainly represents the strength of the protrusion produced by the polyp, and it also incorporates fused information about shape and color content of the image. In Method 2 the numerical value essentially represents the radius of the circle that best fits the (candidate) polyp region. As in the case of Method 1 this numerical value also involves implicit fused shape, color and texture information. Moreover, in both methods, the numerical value is also associated to a pixel location in the image which corresponds the position of the detected polyp. Thus Methods 1 and 2 provide the binary classification of a frame (as polyp or normal mucosa frame), as well as the location of the polyp in the image.
Method 3 8 belongs to the class of machine learning methods. It consists of two steps. First, by using a training dataset, where each frame is a priori labelled either as polyp or normal mucosa frame, Method 3 generates a “separation mathematical object” that separates the frames of this training dataset into two classes: the class of polyp frames and the class of normal mucosa frames. Secondly, Method 3 uses this “separation mathematical object” as a binary classifier for determining for a given new frame (that is not contained in the training dataset) whether it is as polyp or normal mucosa frame. Method 3 is thus a binary classifier, but as opposed to Methods 1 and 2 it does not indicate the location of the polyp in the image.
In the “Methods” section, we give further information about the different mathematical techniques involved in these three methods and in Section “Results” we discuss the selection of the thresholds for Methods 1 and 2.
Patients and methods
Study cohort
The study included 42 patients, 28 male (66.7 %), with a mean age of 57 years (standard deviation 10.23) submitted to colonoscopy, using an Olympus colonoscope (Q165 L). Each patient had one polyp, with a mean dimension of 9.6 mm (standard deviation 5.2 mm), 32 (76.2 %) measuring less than 10 mm, all of them protruding (Paris classification 0-Ip and 0-Is). Of the 35 polyps recovered for histology, three were hyperplastic (7.1 %) and the remaining 32 were adenomas.
Written Informed consent was obtained from all patients before colonoscopy. Ethical board approval was granted on January 29, 2018 by the institutional review board of the Faculty of Medicine, University of Coimbra, Coimbra, Portugal (registry number 020-CE-2018). The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a prior approval by the institution’s human research committee.
Dataset
A dataset was generated from different colonoscopy short videos of the 42 different patients. In this collection, the 42 videos correspond to sequences of 42 different polyps recorded with a white light video colonoscope. For each video, a total of 40 frames were extracted by sampling the video every 10 frames, aiming to exclude very similar images. Thus, there is a total of 1680 polyp instances from the recorded 42 different polyps.
We also had 1360 frames of normal mucosa. We note that the videos we have used did not contain the whole examination, but mainly the parts of the colonoscopy that exhibit polyps. From those videos, we extracted not only the frames with polyps but also frames of normal mucosa. These mucosa frames were lower in number because the fragments of the videos with normal mucosa were smaller than the ones with polyps. In addition, some of these normal mucosa frames were excluded because they were blurred.
In what concerns preparation, those frames that exhibited poor preparation, with Boston score under 2 9 , were excluded.
Methods
We studied the performance, in optical colonoscopy images, of three automated polyp identification methods that we have proposed before for wireless colon capsule endoscopy images 6 7 8 . The methods described in references 6 and 7 act as binary classifiers, labelling a frame as either containing polyps or not based essentially on the geometrical analysis and texture content of the frame. The method in reference 8 uses a support vector machine (SVM) technique: for discriminating between polyp and non-polyp images, several main visible and crucial features related to colonic polyps (such as high-level shape information, color information and local texture information involving binary decision pixel intensity operators) are used in the SVM model classifier. We note that all three methods 6 7 8 are binary classifiers, but methods described in references 6 and 7 also provide information about the location of the polyp (this is not the case for the method of reference 8 ).
Method 1
This method is a binary classifier and a threshold-based method. It is based on the definition of a polyp detector function we have proposed before in 6 10 , herein called P . On a given video frame, each pixel is given a P value. One may think of the P function associated to a given frame as an image whose intensity estimates the level of protrusion at each pixel of the given frame. The P function relies on the hypothesis that polyps are protrusions in the colonic mucosa that are mostly round in shape. Therefore, this function P incorporates shape information about the image content. But, in addition, P also incorporates color information. In fact, since in colonoscopy images the polyps are characterized by a more pronounced reddish color than the surrounding mucosa, we use the a-channel of the CIE Lab color space 11 as input channel for computing this function P . As this a-channel of CIE Lab represents the colors between magenta/red and green, the choice of this a-channel potentially enhances the contrast between polyps and normal mucosa in the computation of P . However, it is also known that the blue channel (B-channel), the third component of the RGB color space, provides a better enhancement of the polyp. Therefore, in the experiments, we have also used as input channel, for computing the function P , the product of the a-channel with the B-channel (hereafter denoted by a & B channels).
To each medical image, Method 1 assigns a numerical value, which corresponds to the highest value of function P , and the possible polyp location corresponds the pixel location of this highest P value ( Fig. 1 ).
Fig. 1 .

Left, original frame of the dataset displaying a polyp. Middle, the graphic of the corresponding P function, with highest value in the polyp region. Right, the blue curve is the ground truth segmentation of the polyp and the yellow curve is a circle centered at the highest value of the P function.
Method 2
In 7 , we proposed an algorithm that is a binary classifier that labels a frame as either containing a polyp or not, based on geometrical analysis and a pre-selection textural criterion. For optical colonoscopy images, we have slightly modified this method. Here, our Method 2 corresponds to the method described in 7 , but without this pre-selection criterion, because in our dataset the frames display overall a similar textural content and this criterion was not useful to discriminate between polyp and non-polyp frames. Thus, for each frame we have only used the geometrical analysis of the corresponding a-channel of the CIE Lab color space (as opposed to the grayscale channel adopted in the method described in reference 7 ), and an extra texture discriminant. The geometrical analysis relies, again, on the fact that the polyps are more round in shape and protrusions on a flatter surrounding tissue ( Fig. 2 ). Similar to Method 1, to each medical image Method 2 assigns a numerical value that corresponds to the best fit ball radius of the polyp region.
Fig. 2.

Left, image from dataset exhibiting a colonic polyp; middle: selected regions (red) with Method 2 with the corresponding ellipses of inertia superimposed (yellow curve). Right, best fit ball (yellow curve) obtained with the method overlapping the polyp (the blue curve is the segmentation of the polyp obtained from the polyp mask).
Method 3
In 8 we employ SVM binary classifiers, involving different shape/color/texture features, for distinguishing images containing colonic polyps from images displaying normal colonic mucosa, in wireless capsule endoscopy images. In the present paper, our Method 3, relies on the application of 8 to optical colonoscopy images.
The shape features are extracted based on the P function, which is described in Method 1. The texture features are obtained by using the traditional local binary pattern (LBP) operator 12 13 , which relies on binary decision pixel-based-intensity operators (using thresholds on pixel intensities for a decision on the local structure of the image) and also by using a combined monogenic local binary pattern (M-LBP) operator 14 . The latter gives information about the local lines, orientations and edges patterns in the image.
In the tests, three different SVM binary classifiers were built depending on three chosen features: LBP or LBP + P or M-LBP. Therefore, Method 3 has these three variants: LBP or LBP + P or M-LBP. A training set with half of the frames was used for building the binary classifiers (for each variant, LBP or LBP + P or M-LBP), and the remaining half of the frames, hereafter called the testing set, was used to assess the performance of these three variants of Method 3.
We recall that Method 3 generates, in a first step, a “separation mathematical object” that separates the training set into two classes: frames with polyps and frames without polyps. Then in a second step, Method 3 uses its “separation mathematical object” as a binary classifier for checking any new frame, of the testing set, either as polyp or normal mucosa frame. Method 3 is also a binary classifier, but as opposed to Methods 1 and 2 it does not indicate the location of the polyp in the image.
Pre-processing
Aiming to detect the specular highlights, white dots originated by reflections from the endoscope light that should be removed as they can affect the performance of the methods, we used a threshold technique based on the sum of the three colors components in the RGB color space, followed by an inpainting technique 15 ( Fig. 3 ).
Fig. 3 .
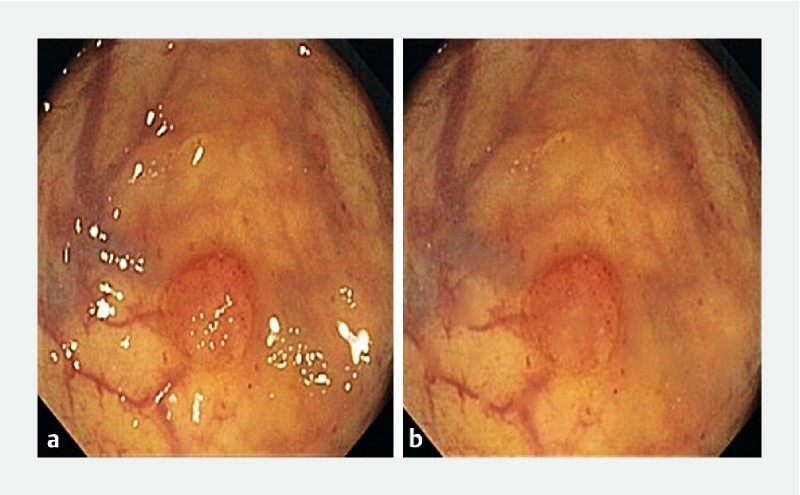
One example of a frame of our dataset displaying specular highlights (left) and the corresponding corrected frame obtained after this preprocessing step (right).
Performance comparison with the public dataset CVC-Clinic DB
A polyp detection comparison of Methods 1 and 2, to a recent automated polyp detection method 16 is also presented, at the end to the “Results” Section. The comparison is made in terms of the correct location of the polyp in a polyp frame. We used the public dataset CVC-Clinic DB 16 , consisting of 612 frames of colonic polyps built by gathering 29 sequences of polyps in 25 videos, with one sequence per polyp.
Statistical analysis
All continuous variables are expressed using means with standard deviation (SD). Accuracy was calculated by dividing the number of true positives plus the number of true negatives by the number of frames with polyps plus the number of frames without polyps. When comparing sensitivity, specificity and accuracy between different methods, McNemar’s test was used. The calculations were done with SPSS 24 statistical software (SPSS Inc., Chicago, Illinois, United States).
The receiver operator characteristic (ROC) curve is obtained by plotting sensitivity versus false acceptance rate (100-specificity) for different decision thresholds, i. e., each point on the ROC curve represents the sensitivity and the false acceptance rate obtained with a specific decision threshold. For Method 1, the decision threshold varies from 0 to the maximum value of all the functions P . For Method 2, the decision threshold varies from 0 to the maximum radius of all the best fit balls.
All tests were two-sided and probability values (hereafter denoted by “p-value”) < 0.05 were considered statistically significant.
Results
The intention of the first experiment was to assess the ability of Methods 1 and 2 to locate correctly lesions in the 1680 polyp frames. In this context, the frame was considered a true positive if the region corresponding to the result of the method intersected the polyp mask in the binary ground truth image, previously identified and manually segmented ( Fig. 4 ). On the other hand, a frame was considered false negative frame if the region corresponding to the result of the method did not intersect the polyp mask of the associated binary ground truth image.
Fig. 4 .
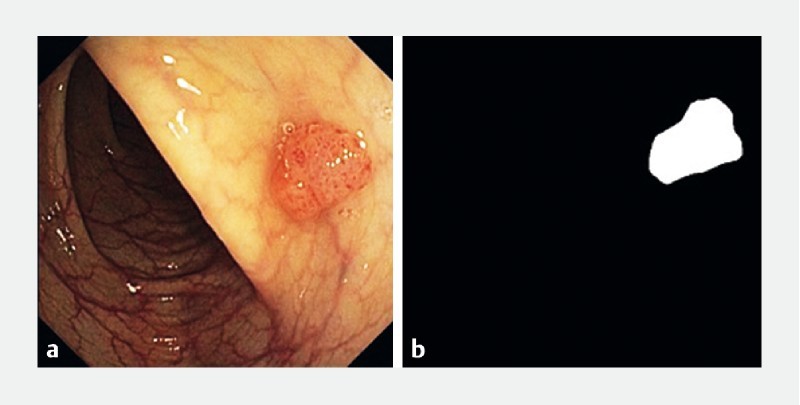
Original image displaying a polyp (left) and the corresponding binary image exhibiting the ground truth for the polyp, i. e. the polyp mask (right).
The results are summarized in Table 1 , which shows the sensitivity of the methods in localizing, per frame, the polyps in 1680 polyp images.
Table 1. Sensitivity of Methods 1 and 2 (CI – confidence interval).
| Method | Sensitivity |
| Method 1 (a &B channels) | 71.8 % (95 %CI [9.5 %-74.0 %]) |
| Method 1 (a-channel) | 68.8 % (95 %CI [66.7 %-71.0 %]) |
| Method 2 (a-channel) | 69.8 % (95 %CI [67.7 %-72.2 %]) |
Method 1 (a & B channels), with a sensitivity of 71.8 % [95 %CI (69.5 % – 74.0 %)], performed significantly better than Method 1 (a channel), which showed a sensitivity of 68.8 % [95 %CI (66.7 % – 71.0 %)] (“p-value” = 0.019). The difference between Method 1 (a and B channels) and Method 2 (a channel) that presented a sensitivity of 69.8 % [95 %CI (67.7 % – 72.2 %)] is not statistically significant.
The second experiment concerned the performance of the three methods in the detection of polyps independent of precision in the location of the lesions.
The outputs obtained with Method 3 are shown in Table 2 (testing set). Here a positive frame for the method was considered a true positive if it contained a lesion and a false positive if it did not, and vice-versa a negative frame for the method was a true negative if it did not contain a lesion, and a false negative if it contained a lesion. For implementation of SVM, we considered the library LIBSVM 17 .
Table 2. Results of Method 3 (CI- confidence intervals).
| Method | Sensitivity | Specificity | Accuracy |
| LBP | 99.6 % [95 %CI (99.0 %-100 %)] | 78.4 % [95 %CI (75.3 %-81.5 %)] | 89.5 % [95 %CI (87.8 %-91.0 %)] |
| LBP + P | 99.7 % [95 %CI (99.3 %-100 %)] | 79.6 % [95 %CI (76.5 %-82.5 %)] | 90.1 % [95 %CI (88.60 %-91.6 %)] |
| M-LBP | 97.2 % [95 %CI (95.9 %-98.4 %)] | 84.9 % [95 %CI (82.2 %-87.5 %)] | 91.1 % [95 %CI (89.70 %-92.50 %)] |
| LBP (without inpaiting) | 99.6 % [95 %CI (99.0 %-100 %)] | 72.6 % [95 %CI (69.4 %-76.0 %)] | 86.8 % [95 %CI (85.0 %-88.6 %)] |
| M-LBP (without inpaiting) | 98.4 % [95 %CI (97.4 %-99.3 %)] | 75.4 % [95 %CI (72.2 %-78.7 %)] | 87.3 % [95 %CI (85.5 %-89.0 %)] |
LBP, local binary pattern; LBP + P , local binary pattern + polyp detection function; M-LBP, monogenic local binary pattern
Aiming to emphasize the efficacy of the pre-processing step (removal of specular highlights followed by the inpainting technique) we also show in Table 2 the results of Method 3 with and without this pre-processing step (referred to as “with and without inpainting”).
Considering sensitivity, LBP + P performed significantly better than M-LBP (“p-value” = 0.001) and M-LBP without inpainting (“p-value” = 0.009). Considering specificity, M-LBP performed significantly better than LBP (“p-value” < 0.001), LBP + P (“p-value” = 0.009), LBP without inpainting and M-LBP without inpainting (“p-value” < 0.001). The methods that did not use inpainting were significantly less accurate than the methods that used this technique (“p-value” < 0.001). M-LBP did not perform significantly better than LBP and LBP + P in terms of accuracy (“p-value” = 0.084 and “p-value” = 0.309 respectively).
The results obtained with Methods 1 and 2 are shown in Fig. 5 , and exhibit the ROC curves 18 , computed in the training set previously defined (using the a-channel, as input channel). Each ROC curve is the graphical plot of the sensitivity versus the specificity (or equivalently the FAR = 100 % specificity), for the maximum of the P function defined in Method 1, and for the maximum of the fit ball radii in Method 2. The optimal threshold (indicated as a red circle in each curve of Fig. 5 ) is selected as one that maximizes the accuracy.
Fig. 5.
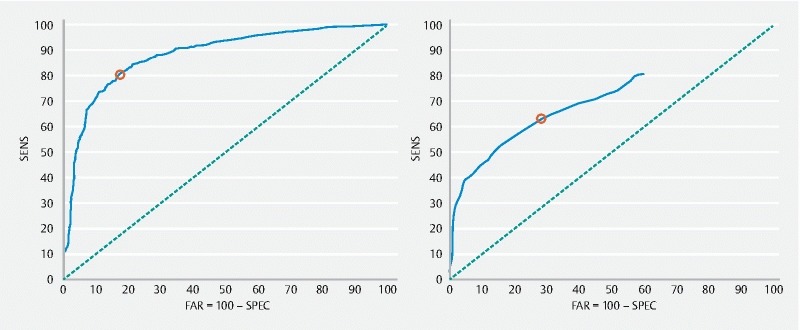
ROC curves obtained with Method 1 (left) and 2 (right) using the training set (a – channel) for the polyp detection (840 polyp frames and 680 normal frames). Each red circle corresponds to the threshold that maximizes the accuracy.
The optimal thresholds found – 1233.9105 for Method 1 and 47 for Method 2 – were then adopted and the results for the testing set are shown in Table 3 .
Table 3. Sensitivity, specificity and accuracy of Methods 1 and 2 in detecting the polyp (CI – confidence intervals).
| Method | Sensitivity | Specificity | Accuracy |
| Method 1 (a-channel) | 83.7 % [95 %CI (80.9 %-86.3 %)] | 66.6 % [95 %CI (63.1 %-70.3 %)] | 74.3 % [95 %CI (72.0 %-76.5 %)] |
| Method 2 (a-channel) | 61.6 % [95 %CI (57.8 %-65.4 %)] | 61.3 % [95 %CI (57.8 %-64.9 %)] | 63.2 % [95 %CI (60.8 %-65.7 %)] |
Sensitivity, specificity and accuracy for Method 1 were significantly better than for Method 2 (“p-value” < 0.001, “p-value” = 0.016, “p-value” < 0.001 respectively) ( Table 3 ).
Method 3 performed significantly better than Method 1 and 2 in detecting the polyps, considering sensitivity, specificity and accuracy (“p-value” < 0.001).
Finally, sensitivity of Methods 1 and 2 in detection of polyps was compared with sensitivity of the method described in 16 and applied in a recently published paper 19 , using for that purpose the public dataset CVC-ClinicDB. Methods 1 and 2, with a sensitivity of 78.5 % and 74.5 %, respectively, outperformed the method described in 16 , which reached a sensitivity of 70.3 %. We note that Method 3 was not tested in this public dataset because there are no frames without polyps and those are needed to build the SVM binary classifier.
Discussion
Our results can be discussed in terms of the methods’ ability to localize the polyps in the frames and their ability to detect the polyp, independent of the position of the lesion in the frame. Concerning localization of the polyp in the frame, Method 1 performed better than Method 2. For this purpose, we only have sensitivity because only frames with polyps were used. In terms of detection, the best results were obtained with Method 3, probably because it involved different shape, color and texture features for distinguishing images containing polyps from images displaying normal colonic mucosa. It is interesting to note that, when applied to the public dataset CVC-ClinicDB, Methods 1 and 2 achieved better results than those reported by G. Fernandez-Esparrach, et al 19 . One possible explanation is that the method works better with zenithal views of a polyp, somehow different from the technique used in Methods 1 and 2, which do not need this condition, but rely essentially on the fact that the polyps are protrusions on a flat surrounding tissue.
Although the question of the dimension of a polyp is, of course, very important, in this kind of study it was difficult to evaluate this parameter because the dimension of the polyp in the frame may have varied a lot depending on the lens’ relative position, and independent from the dimension of the lesion. The only way to overcome this problem would be to execute the algorithm in real time and compare its performance with the accuracy of the gastroenterologist.
Another important question is bowel preparation and how well the algorithm worked when bowel preparation was less than 3 by the Boston classification system in one segment 9 . G. Fernandez-Esparrach 19 found that the applied method was not influenced by poor preparation, but fewer than 10 % of the frames had a Boston score of 1. We deliberately excluded frames presenting Boston score of 1 because when that happens, the colonoscopy should be repeated.
An interesting pre-processing technique involving removal of specular highlights followed by the inpainting technique 15 was used for all three methods, allowing amelioration of the input image to be processed and achieving better accuracy. By way of illustration, this is clearly demonstrated in Table 2 .
The issue of measuring the performance in colonoscopy is very important, and one of the domains, “identification of pathology,” includes ADR and polyp detection rate, those being considered a surrogate for meticulous inspection of the colorectal mucosa 20 . Even though the accuracy is not optimal, we must not forget that these methods are designed to help the gastroenterologist in the detection of polyps undetected during colonoscopy, meaning that the ADR and polyp detection rate will probably improve with the help of CAD.
Apart from detection of polyps, the other indication for CAD may be classification of colorectal polyps 21 . This is also an issue of major importance when dealing with diminutive colorectal polyps 22 . In fact, CAD may help in performing optical biopsy 23 . In the future, CAD that includes both detection and classification of polyps seems to be essential. In fact, it does not make sense to have a CAD fully operating in real time that only detects lesions and does not immediately classify them as adenomatous or hyperplastic.
Conclusion
In conclusion, our results show that the methods used can detect polyps with a reasonable accuracy. Further work is necessary, namely by applying the algorithms in real time.
Acknowledgements
This work was partially supported by the FCT research project POCI-01-0145-FEDER-028960. The authors Isabel N. Figueiredo and Luís Pinto also acknowledge some support from CMUC – UID/MAT/00324/2013, funded by the Portuguese Government through FCT/MEC and co-funded by the European Regional Development Fund through the Partnership Agreement PT2020. Luís Pinto was also supported by FCT scholarship SFRH/BPD/112687/2015.
For their work in the manual identification and segmentation of the polyps, followed by the building of the masks, the authors would like to thank the following physicians from the Department of Gastroenterology, Centro Hospitalar e Universitário de Coimbra, Coimbra, Portugal: Sara Campos, Elisa Soares, Marta Soares, Ana Rita Alves, Diogo Branquinho and Filipe Taveira. The authors also thank the mathematician Carlos Tenreiro, (CMUC, Department of Mathematics, University of Coimbra, Coimbra, Portugal) for the valuable statistical input.
Footnotes
Competing interests None
References
- 1.Globocan. Estimated cancer incidence, mortality and prevalence worldwide in 2012 2012. Available from:http://globocan.iarc.fr/Default.aspx
- 2.Rex D K. Colonoscopy: the current king of the hill in the USA. Dig Dis Sci. 2015;60:639–646. doi: 10.1007/s10620-014-3448-0. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Chang-Claude J, Jansen L et al. Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology. 2014;146:709–717. doi: 10.1053/j.gastro.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Shaukat A, Oancea C, Bond J H et al. Variation in detection of adenomas and polyps by colonoscopy and change over time with a performance improvement program. Clin Gastroenterol Hepatol. 2009;7:1335–1340. doi: 10.1016/j.cgh.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Mori Y, Kudo S E, Berzin T M et al. Computer-aided diagnosis for colonoscopy. Endoscopy. 2017;49:813–819. doi: 10.1055/s-0043-109430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueiredo P N, Figueiredo I N, Prasath S et al. Automatic polyp detection in pillcam colon 2 capsule images and videos: Preliminary feasibility report. Diagn Ther Endosc. 2011:182435. doi: 10.1155/2011/182435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mamonov A V, Figueiredo I N, Figueiredo P N et al. Automated polyp detection in colon capsule endoscopy. IEEE Trans Medical Imaging. 2014;33:1488–1502. doi: 10.1109/TMI.2014.2314959. [DOI] [PubMed] [Google Scholar]
- 8.Figueiredo I N, Kumar S, Figueiredo P N.An intelligent system for polyp detection in wireless capsule endoscopy imagesIn: Computational Vision and Medical Image Processing IV, João Tavares & Natal Jorge (eds)Taylor & Francis Group; London: 2014235–239.ISBN 978-1-138-00081-0
- 9.Calderwood A H, Jacobson B C. Comprehensive validation of the Boston Bowel Preparation Scale. Gastrointest Endosc. 2010;72:686–692. doi: 10.1016/j.gie.2010.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueiredo I N, Prasath S, Tsai Ret al. Automatic detection and segmentation of colonic polyps in wireless capsule images. UCLA-CAM Report 201010–65.. Accessed April 8, 2018. Available from:http://www.math.ucla.edu/applied/cam/
- Schanda J. John Wiley & Sons; 2007. Colorimetry: understanding the CIE system. [Google Scholar]
- 12.Ojala T, Pietikainen M, Harwood D. A comparative study of texture measures with classification based on feature distributions. Pattern Recognition. 1996;29:51–59. [Google Scholar]
- 13.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Patt Anal Machine Intell. 2002;24:971–987. [Google Scholar]
- 14.Zhang L, Zhang L, Guo Z 17th IEEE International Conference on Image Processing (ICIP); 2010. Monogenic-LBP: A new approach for rotation invariant texture classification. pp. 2677–2680.
- 15.DʼErrico J. inpaint_nans, MATLAB Central File Exchange, MathWorks, Natick, Massachusetts, United States. 2012. https://www.mathworks.com/matlabcentral/fileexchange/4551-inpaint-nans https://www.mathworks.com/matlabcentral/fileexchange/4551-inpaint-nans
- 16.Bernal J, Sanchez F J, Fernandez-Esparrach G et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comp Med Imaging Graphics. 2015;43:99–111. doi: 10.1016/j.compmedimag.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Chang C C, Lin C J. LIBSVM: A library for support vector machines. ACM Trans Intell Systems Tech. 2011;27:1–27. [Google Scholar]
- 18.Fawcett T. An introduction to ROC analysis. Patt Recog Lett. 2006;27:861–874. [Google Scholar]
- 19.Fernandez-Esparrach G, Bernal J, López-Cerón M et al. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837–842. doi: 10.1055/s-0042-108434. [DOI] [PubMed] [Google Scholar]
- 20.Kaminski M F, Thomas-Gibson S, Bugajski M et al. Performance measures for lower gastrointestinal endoscopy: a European Society of Gastrointestinal Endoscopy (ESGE) Quality Improvement Initiative. Endoscopy. 2017;49:378–397. doi: 10.1055/s-0043-103411. [DOI] [PubMed] [Google Scholar]
- 21.Mori Y, Kudo S E, Berzin T M et al. Computer-aided diagnosis for colonoscopy. Endoscopy. 2017;49:813–819. doi: 10.1055/s-0043-109430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu Dayyeh B K, Thosani N, Konda V et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015;81:502. doi: 10.1016/j.gie.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Kudo S E, Chiu P W et al. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110–1118. doi: 10.1055/s-0042-113609. [DOI] [PubMed] [Google Scholar]


