Abstract
Comparative studies of microbiome variation in world populations and different developmental stages of organisms are essential to decipher the linkages among microbiome, health, and disease. Notably, the gut microbiota are believed to mature in early life. In this context, we compared the gut microbiota diversity in Korean adolescent healthy samples (KAHSs) to healthy Korean adults (HKAs) as well as the Human Microbiome Project healthy samples (HMPHSs), the latter being one of the largest adult cohorts, based on organismal composition, alpha- and beta-diversities, function/pathway prediction analysis, and co-occurrence networks. We found that the gut microbiota compositions, including the ratios of firmicutes to bacteroidetes, between KAHSs and HMPHSs were different, and the diversities of KAHSs were less than those of HMPHSs. The predicted functions, for example, secondary bile acid synthesis and insulin signaling of KAHSs and HMPHSs, were also significantly different. Genus-level networks showed that co-occurrences among different taxa more frequently happened in HMPHSs than in KAHSs. Even though both KAHSs and HMPHSs represent healthy microbiomes, comparisons showed substantial differences, likely implicating different diets, environments, and demographics. Interestingly, we observed lower microbial diversities and less frequent co-occurrences among different taxa in KAHSs than adult HMPHSs and HKAs. These new findings collectively suggest that the adolescent gut microbiota in the present Korean sample did not reach the extent of maturity of adult microbiota diversity. In all, further population studies of microbiome variation across geographies and developmental stages are warranted, and should usefully inform future diagnostics and therapeutics innovation targeting the microbiome.
Keywords: population microbiome science, Korean adolescents, co-occurrence network, Human Microbiome Project, developmental biology
Introduction
The human gut microbiota facilitates numerous vital biological processes, including energy harvesting, metabolism of short-chain fatty acids (SCFAs), and inflammation, and its dyshomeostasis contributes to diverse diseases such as obesity, neurodegeneration, and cancer (Bonfili et al., 2017; Cani, 2018; Dimitrov, 2011; Houser and Tansey, 2017; Human Microbiome Project Consortium, 2012; Jiao et al., 2018; Kau et al., 2011; Koliada et al., 2017; Malan-Muller et al., 2018; Sonnenburg and Backhed, 2016; Zeller et al., 2014).
Recently, high-throughput sequencing technology has enabled determination of the gut microbiome composition (Laudadio et al., 2018; Nishijima et al., 2016; Tsai et al., 2015), allowing for analysis of individual organismal proportions, as well as microbiota diversity, which associates with good health (Sonnenburg and Backhed, 2016). These techniques have been applied to gut microbiota studies for diverse large cohorts of healthy subjects, and those suffering from rheumatoid arthritis, diabetes, and colorectal cancer, in Europe, United States, Russia, and Asia (Backhed et al., 2015; Forslund et al., 2015; Karlsson et al., 2013; Nakayama et al., 2015; Nishijima et al., 2016; Qin et al., 2010, 2012, 2014; Turnbaugh et al., 2009; Tyakht et al., 2013; Zeller et al., 2014; Zhang et al., 2015).
In Korean cohorts, gut microbiome characterization is now underway, primarily of healthy adults or those with various disease states (Hu et al., 2015; Kim et al., 2013, 2014; Nam et al., 2011; Song et al., 2014; Yun et al., 2017).
Notably, the adolescents experience rapid changes, physically and physiologically, in human development (Christie and Viner, 2005). The changes during adolescence are often affected by individual factors (e.g., sex) and environmental factors (e.g., undernutrition, substance use) (Christie and Viner, 2005). Adolescent development changes the disease aspects in reproductive health problems, injuries, and mental illness (World Health Organization, 2006). Also, health-related behaviors often affect long-term outcomes on present and future health (World Health Organization, 2006). Hence, comparative studies of microbiome variation in world populations and developmental stages of organisms are essential to decipher the linkages among microbiome, health, and disease.
In this study, Korean adolescents (Hu et al., 2015) were compared with adult cohorts, including the Human Microbiome Project (HMP), one of the largest healthy adult samples (Human Microbiome Project Consortium, 2012; Turnbaugh et al., 2007), and Korean adult samples (Yun et al., 2017), in terms of gut microbiota. Through the comparison, we have not only examined the gut microbiota differences, but also observed whether or not gut microbial diversity of the adolescents reached to the extent of diversity of healthy adults. By comparing microbiome composition, microbiota diversities, function/pathway analysis, and co-occurrence networks, we developed a deeper understanding of the Korean healthy adolescents' gut microbiota specifically, and with a view to developmental biology broadly.
Materials and Methods
Datasets and preprocessing
The Korean adolescent healthy samples (KAHSs) (Hu et al., 2015), Human Microbiome Project healthy samples (HMPHSs) (Human Microbiome Project Consortium, 2012), and healthy Korean adults (HKAs) (Yun et al., 2017) are described in Table 1. An overview of the data analysis workflow is shown in Supplementary Figure S1. The HMPHSs are available at https://portal.hmpdacc.org, and the KAHSs (Hu et al., 2015) were downloaded from http://metagenome.cafeomics.com/public/download.php
Table 1.
Description of the Datasets in the Present Study
| Dataset | Raw reads (median) | Reads after filtering low quality and short reads | Cleaned reads | Sequencing platform |
|---|---|---|---|---|
| KAHSs (n = 67) | 9958 | 3761 | 2861 | GS Junior |
| HMPHSs (n = 325) (processed data used) | — | 13974 | 10884 | GS FLX |
| HKAs (n = 1463) | 42,379 | 10,940 | 12,405 | Illumina Miseq |
| IAHSs (n = 10) | 14,285 | 5606 | 5398 | GS FLX |
HKAs, healthy Korean adults; HMPHSs, Human Microbiome Project healthy samples; IAHSs, Italian adolescent healthy samples; KAHSs, Korean adolescent healthy samples.
Low-quality KAHS reads (average quality score below 30, and <250 bp) were filtered out using fastx_toolkit 0.014 (hannonlab.cshl.edu/fastx_toolkit). Chimera detection and removal were performed with vsearch (v2.4.3) (Rognes et al., 2016) and the Ribosomal Database Project (RDP) training dataset (release 16_022016) (Cole et al., 2014) guided by the Microbiome_helper (Comeau et al., 2017).
The HKAs (Yun et al., 2017) were downloaded from CODA (Clinical & Omics Data Archive) available at coda.nih.go.kr/coda We merged raw reads of HKAs using Paired-End reAd mergeR (PEAR; version 0.9.6) (Zhang et al., 2014) and filtered low-quality reads (less than average quality score 25, and <400 bp) using fastx_toolkit 0.014. The same procedure above was applied to chimera removal for the reads in the HKAs.
Italian adolescent healthy samples (IAHSs) (Del Chierico et al., 2018) were obtained from the NCBI BioProject accession PRJNA280490 (www.ncbi.nlm.nih.gov/bioproject). KAHSs were age 13–16 years (13.8 ± 3), and IAHSs 13–19 years (16.9 ± 1.1).
The data used in the present study were drawn from four public datasets (Table 1) that have been approved by their own institutional Ethics Committees (Del Chierico et al., 2018; Hu et al., 2015; Human Microbiome Project Consortium, 2012; Yun et al., 2017).
16S ribosomal RNA sequence analysis
Operational Taxonomic Unit (OTU) clustering was done by SortMeRNA (version 2.1b) (Kopylova et al., 2012), contained in Quantitative Insights into Microbial Ecology (QIIME) (Caporaso et al., 2010) in a closed reference approach (97% identity threshold). OTU sequences were assigned to the Greengenes database (release 13_8) (DeSantis et al., 2006) with UCLUST (version 1.2.22q) (Edgar, 2010), inside QIIME. We subsampled 3000 reads from each sample, to rarify for diversity analysis in QIIME (Caporaso et al., 2010). Subsequently, we estimated alpha-diversity and beta-diversity in QIIME (Caporaso et al., 2010).
Functional analysis
Functional profiling prediction was performed with Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt; version1.1.3) (Langille et al., 2013), and was visualized by STAMP (version 2.1.3) (Parks et al., 2014). Taxonomy composition plot and rarefaction curves were made using R (version 3.4.3) and QIIME.
Network analysis
For phylum-level network analysis, SPIEC-EAST (Kurtz et al., 2015) was used, and CoNet (Faust and Raes, 2016) was used to generate genus-level networks for the two populations. Low abundant taxa, with <20 reads across the samples, were removed. The thresholds for Pearson correlation were set to 0.7 for both positive and negative values. Other than those, we went with the default options.
Results
Comparisons of KAHSs and HMPHSs stool samples
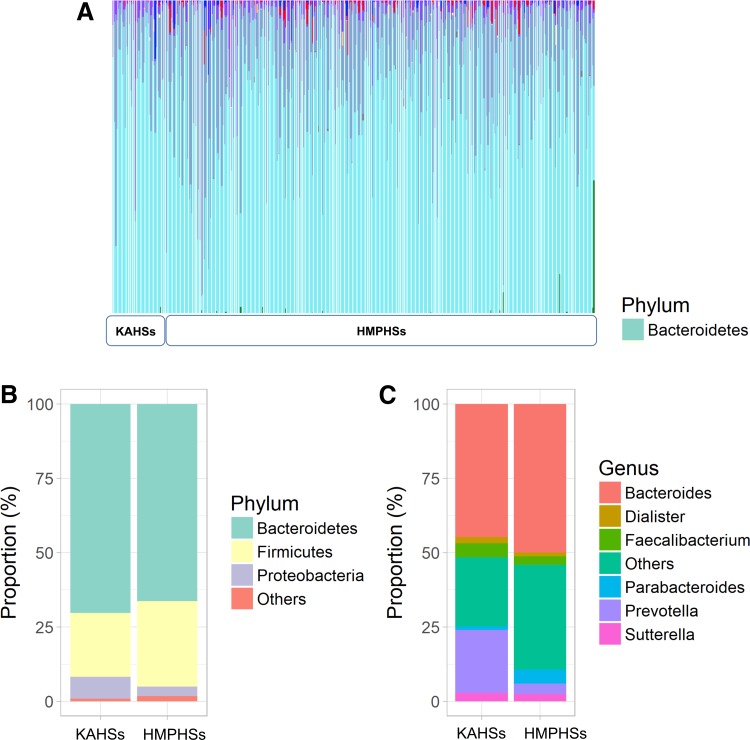
To characterize the stool microbiota of KAHSs, we compared samples to those of adult HMPHSs, showing both to possess prevalent cyan-colored species (bacteriodetes) (Fig. 1A). At the phylum level, bacteroidetes amounted to 70.3% and 66.4% of KAHSs and HMPHSs, respectively (Fig. 1B), whereas firmicutes amounted to 21.5% and 28.7% in KAHSs and HMPHSs, respectively. At the genus level of KAHSs and HMPHSs, respectively, bacteroidetes amounted to 44.7% and 49.9%, prevotella 21.1% versus 3.5%, and faecalibacterium 4.8% versus 2.9% (Fig. 1C).
FIG. 1.
Gut microbiome composition of KAHSs and HMPHSs, at the phylum and genus levels. (A) Phylum-level taxonomy proportion plot for all samples, including KAHSs and HMPHSs. (B) Taxonomy proportion plots for the phylum level. (C) Taxonomy proportion plots for the genus level. HMPHSs, Human Microbiome Project healthy samples; KAHSs, Korean adolescent healthy samples.
α- and β-diversities in the gut microbiome of KAHSs and HMPHSs
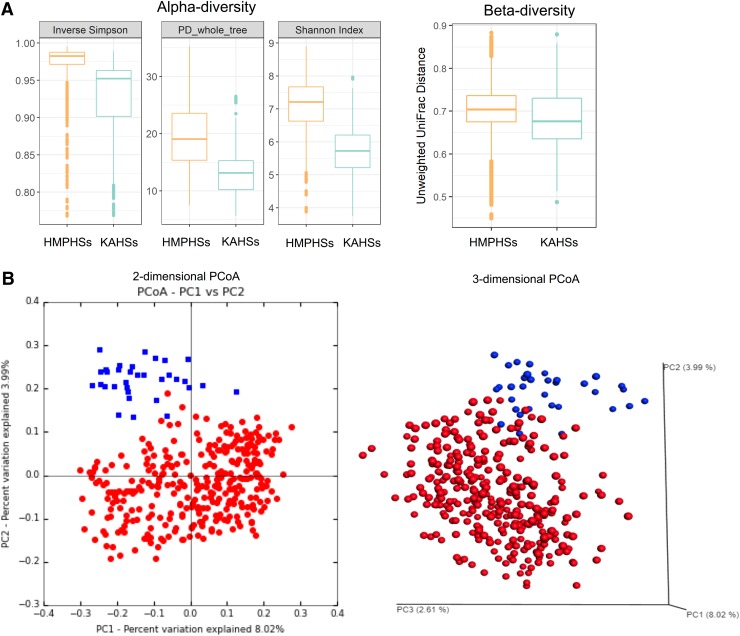
The relative abundance of gut taxa was measured for KAHSs and HMPHSs, in the terms of alpha- and beta-diversities. Alpha-diversities of KAHSs were less than those of HMPHSs (Fig. 2A) in the three measurements [inverse Simpson, phylogenetic diversity (PD) whole trees, and Shannon index], implying higher within-sample organismal diversity in HMPHSs. By using unweighted UniFrac distances (Morgan and Huttenhower, 2012) in QIIME (Caporaso et al., 2010), the beta-diversities of KAHSs (average of 0.685) were significantly (p-value 3.51e-26) less than those of HMPHSs (average of 0.707).
FIG. 2.
Microbiota diversity estimation of KAHSs and HMPHSs. (A) Alpha-diversity estimation showing that the inverse Simpson index for KAHSs was 0.927; for HMPHSs, 0.973. The PD whole_tree index for KAHSs was 13.31; for HMPHSs, it was 19.38; and the Shannon indices were 5.714 for KAHSs and 7.091 for HMPHSs, respectively. Beta-diversity indicated that HMPHSs were higher than KAHSs. (B) Two- and three-dimensional PCoA plots for KAHSs and HMPHSs were provided (blue: KAHSs; red: HMPHSs). Distance metric for the PCoA plots was calculated through unweighted UniFrac distance. PCoA, Principal Coordinate Analysis.
Moreover, Principal Coordinate Analysis (PCoA) (Caporaso et al., 2010) of the beta-diversities of KAHSs (Caporaso et al., 2010) showed them to separate from HMPHSs (Fig. 2B). Analysis of similarities tests (in QIIME) between KAHSs and HMPHSs were performed using the unweighted UniFrac distance measurement and the weighted UniFrac distance measurement, yielding p-values of 0.0099 and 0.0021, respectively. We performed statistical analyses for the OTU differences between KAHSs and HMPHSs. Under false discovery rate cutoff 0.05, 293 out of 21348 OTUs were statistically significant. Twenty out of the 293 are summarized in Supplementary Table S1. Overall, our results indicate that, even in healthy controls, the gut microbiomes of Korean adolescents and HMP populations were different.
Functional differences between the gut microbiome of KAHSs and HMPHSs
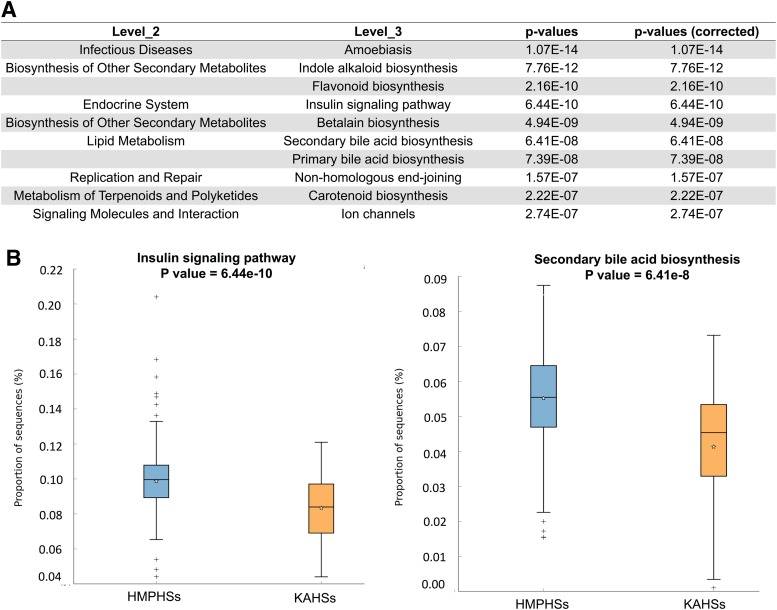
Using PICRUSt (Langille et al., 2013), functional differences between KAHSs and HMPHSs were predicted. In the first-level classification Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000), pathways of PICRUSt, cellular processes, genetic information processing, and organismal systems were significantly different between KAHSs and HMPHSs (Supplementary Fig. S2) Also, 37 second-level classification KEGG pathways, by PICRUSt, were reported to be significant, and 163 significant third-level classification KEGG pathways, based on the structure of the gastrointestinal microbiota between the two populations. The ten most significant third-level classification KEGG pathways are summarized in Figure 3A. Two of the 10, insulin signaling pathway and secondary bile acid synthesis, are depicted in boxplots in Figure 3B.
FIG. 3.
Functional prediction of PICRUSt. (A) One hundred and sixty-three significant third-level classification KEGG pathways/terms were reported, based on gastrointestinal microbiota compositions between the two populations (KAHSs and HMPHSs). The 10 significant terms of the 163 are summarized in tabular format. (B) Two significant third-level terms (insulin signaling, secondary bile acid synthesis) in (A) are drawn as boxplots. KEGG, Kyoto Encyclopedia of Genes and Genomes; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
Moreover, the abundances of the two pathways of KAHSs were lower than those of HMPHSs. Both bile acids (Ridlon et al., 2014) and insulin signaling (Carvalho et al., 2012) have been recognized as important mediators of health status and well-being of gut microbiota.
Network comparisons between gut microbiome of KAHSs and HMPHSs
Based on the OTU tables, we constructed microbial correlation networks representing pair-wise associations (equivalently, co-occurrences) among different microbial communities. At a phylum-level network of KAHSs, the largest cluster showed pair-wise associations between bacteroidetes and firmicutes (Supplementary Fig. S3). Also, a HMPHS phylum-level network (Supplementary Fig. S3) showed its largest cluster to link bacteroidetes and firmicutes. Relatedly, recent studies have reported that the relative ratio between bacteroidetes and firmicutes in gut microbiota clinically associates with healthy status and inflammatory diseases (Johnson and Versalovic, 2012; Koenig et al., 2011).
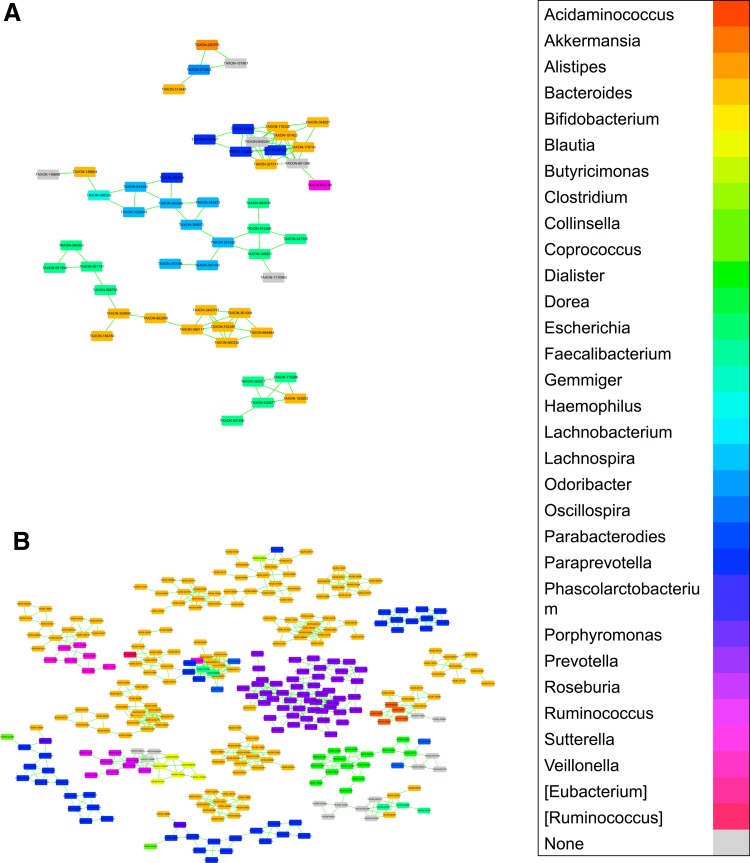
Moreover, the abundance ratio of firmicutes/bacteriodetes (0.43) of HMPHSs was higher than that (0.31) of KAHSs, in agreement with another study showing increasing ratios of firmicutes to bacteriodetes during growth into adulthood (Mariat et al., 2009). The second largest clusters of both KAHSs and HMPHSs also indicated that actinobacteria associate with bacteroidetes and firmicutes, respectively (black arrow in Supplementary Fig. S3). Interestingly, in genus-level networks (Fig. 4 and Supplementary Fig. S4), HMPHSs showed more associations among different taxa than KAHSs.
FIG. 4.
Co-occurrence network among microbial communities of KAHSs and HMPHSs at genus level. (A) Co-occurrence networks in KAHSs. (B) Co-occurrence networks in HMPHSs. Full networks in genus level for both populations are in Supplementary Figure S4.
Looking into the different taxa associations (co-occurrence) in Supplementary Figure S4, commonality and difference of the associations between KAHSs and HMPHSs were revealed (Supplementary Fig. S5). HMPHSs had more different taxa associations than KAHSs, indicating that HMPHS microbial communities likely interacted with each other.
Gut microbiota comparisons of KAHSs with HKAs and IAHSs
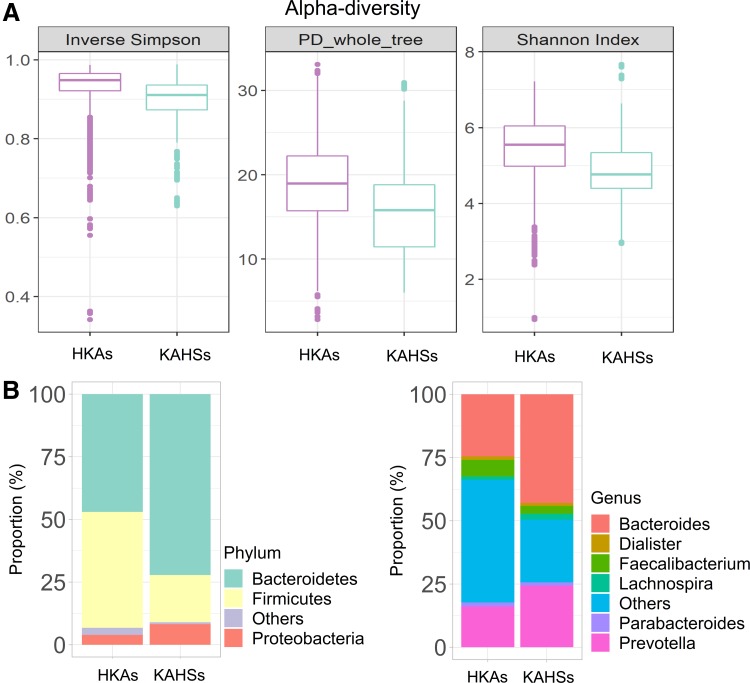
For considering the same population, we compared KAHSs and HKAs (Yun et al., 2017) (Fig. 5A and Supplementary Fig. S6A). Even though two- and three-dimensional PCoA plots indicate that KAHSs and HKAs were not separate (Supplementary Fig. S6A), microbiota diversity was not reached to mature (Fig. 5A).
FIG. 5.
Microbiota comparisons of KAHSs and HKAs. (A) Alpha-diversity of KAHSs was lower than that of HKAs. (B) Taxonomy composition plots were presented for HKAs and KAHSs. HKAs, healthy Korean adults.
We further inspected microbiota proportional changes between HKAs and healthy Korean adolescents (KAHSs). At phylum level, from the adolescents (KAHSs) to the adults (HKAs), bacteroidetes were decreased from 72% to 47%, and firmicutes increased from 19% to 46% (Fig. 5B). At genus level, from the adolescents (KAHSs) to the adults (HKAs), bacteroides were changed 43–25%; prevotella 24–16%; faecalibacterium 3–7%; and lachnospira 2–1% (Fig. 5B). These changes from adolescents to adults may demonstrate value for future clinical contexts, not to mention developmental biology.
In addition, to consider an age-matching factor, IAHSs (Del Chierico et al., 2018) were compared with KAHSs. Despite similar ages, the two adolescent cohorts were separate in two- and three-dimensional PCoA plots (Supplementary Fig. S6B). We performed microbiota composition comparison of KAHSs and IAHSs, and found microbiota differences (Supplementary Fig. S7). At phylum level, KAHSs and IAHSs were 71% and 29% for bacteroidetes, respectively; and 20% and 67% for firmicutes, respectively (Supplementary Fig. S7). At genus level, bacteroides were KAHSs 46% and IAHSs 15%; prevotella KAHSs 22% and IAHSs 0.5%; faecalibacterium KAHSs 5% and IAHSs 12%; and lachnospira KAHSs 2% and IAHSs 0.2% (Supplementary Fig. S7).
Discussion
In this study, we compared gut microbiota between KAHSs and HMPHSs, in terms of microbiota composition (Fig. 1), diversities (Fig. 2), functions/pathways (Fig. 3), and co-occurrence networks (Fig. 4).
Higher ratios of prevotella to bacteroidetes are preferably observed in agrarian diets (i.e., those high in fiber and agricultural products) (Simpson and Campbell, 2015). Of interest, we found the prevotella to bacteroide ratio (0.471) of KAHSs to be higher than that of HMPHSs (0.070) in our study. Such a striking ratio difference likely results from high-fiber ingredients in the Korean diet (Kim et al., 2016).
Recent studies report that the abundance ratio between bacteroidetes and firmicutes, in gut microbiota, clinically associates with both healthy well-being and inflammatory disease (Koliada et al., 2017; Lopez et al., 2016; Verdam et al., 2013). As mentioned earlier, the ratio of firmicutes/bacteroidetes in Korean adolescents (KAHSs) was lower than that in HMP adults (HMPHSs). This may likely indicate that KAHSs have not fully reached mature microbiota composition.
In our PICRUSt analysis (Fig. 3B), we found different abundances of secondary bile acid synthesis between adolescent and adult populations. In this synthetic pathway, secondary bile acids are generated from primary fecal acids (Wong et al., 2006). Recently, this conversion process (i.e., primary to secondary bile acids in fecal samples) was associated with firmicutes and bacteroidetes (Kakiyama et al., 2013). Thus, the significant abundance differences of firmicutes and bacteroidetes between KAHSs and HMPHSs may affect activation of the secondary bile acid synthesis pathway. Also, considering the role of the gut microbiome as a regulator of bile acids (Ridlon et al., 2014; Sayin et al., 2013; Wahlstrom et al., 2016), cholesterol and bile acid pools may be different between KAHSs and HMPHSs.
Insulin signaling is also related to microbiota diversity (Sonnenburg and Backhed, 2016), since diverse SCFAs (e.g., butyrate, acetate), mediated by gut microbiota, associate with increases in insulin sensitivity and AMPK activity (Jiao et al., 2018; Jones, 2016). In that regard, we found that the functional abundance of insulin signaling (Fig. 3B) was lower in KAHSs, than in HMPHSs, again suggesting that microbiota diversity (i.e., alpha- and beta-diversity) of KAHSs are likely still progressing toward the maturity of adult-like microbiota diversity found in HMPHSs. This finding was supported by another comparison of KAHs and HKAs, in that alpha-diversity of Korean adolescents was also lower than that of HKAs (Fig. 5).
Another recent report (Hollister et al., 2015) indicated that microbiota development likely continues over the first 3 years of life, during which gut microbiota are believed to mature, and it is presumed that continuous development of microbiota may make rich associations among different taxa. Consequently, associations among different taxa in KAHSs may not reach the extent of diversities of HMPHSs. In fact, our visual inspection of the two genus-level networks (Supplementary Fig. S4) of KAHSs and HMPHSs showed that adult-like microbiota (HMPHSs) had more associations, among different taxa, than KAHSs.
Since we also targeted the 16S ribosomal RNA (rRNA) regions of KAHSs and HMPHSs, within V1–V3 and V1–V3/V3–V5, respectively (Hu et al., 2015; Human Microbiome Project Consortium, 2012), we concede that different target regions may affect the results, thus warranting careful interpretation of these findings. Also, our study should be carefully interpreted, since microbiota comparison of adolescents and adults from different populations would have unaccounted demographic and other confounding factors.
Conclusions
In this study, we report the microbiota composition, diversities, functions/pathways, and co-occurrence networks that characterize gut microbiota in Korean healthy adolescents (KAHSs), compared with the large healthy adult cohort, HMPHSs. These differences represent unique characteristics of Korean healthy adolescent gut microbiota. In particular, we noted that KAHSs had a higher ratio of prevotella to bacteroidetes than HMPHSs, with lower microbiota diversity and 163 significantly different KEGG pathways (third level of PICRUSt). Co-occurrences among different taxa happened in HMPHSs rather than in KAHSs. While these differences await further computational and laboratory analysis for robust validation, such approaches will provide insight into the differences between maturing and adult microbial physiology and pathology.
In all, further population studies of microbiome variation across geographies and developmental stages are warranted, and should usefully inform future diagnostics and therapeutics innovation targeting the microbiome.
Supplementary Material
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2016R1D1A1B03933145 to S.N.). HKAs used in this study were provided by the CODA (http://coda.nih.go.kr), CODA accession number R000635.
Abbreviations Used
- CODA
Clinical & Omics Data Archive
- HKAs
healthy Korean adults
- HMP
Human Microbiome Project
- HMPHSs
Human Microbiome Project healthy samples
- IAHSs
Italian adolescent healthy samples
- KAHSs
Korean adolescent healthy samples
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OTU
operational taxonomic unit
- PCoA
principal coordinate analysis
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- QIIME
Quantitative Insights Into Microbial Ecology
- SCFAs
short-chain fatty acids
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Backhed F, Roswall J, Peng Y, et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 [DOI] [PubMed] [Google Scholar]
- Bonfili L, Cecarini V, Berardi S, et al. (2017). Microbiota modulation counteracts Alzheimer's disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci Rep 7, 2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD. (2018). Human gut microbiome: Hopes, threats and promises. Gut 67, 1716–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BM, Guadagnini D, Tsukumo DML, et al. (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55, 2823–2834 [DOI] [PubMed] [Google Scholar]
- Christie D, and Viner R. (2005). Adolescent development. BMJ 330, 301–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, et al. (2014). Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res 42, D633–D642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Douglas GM, and Langille MG. (2017). Microbiome helper: A custom and streamlined workflow for microbiome research. mSystems 2 e00127–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Chierico F, Abbatini F, Russo A, et al. (2018). Gut microbiota markers in obese adolescent and adult patients: Age-dependent differential patterns. Front Microbiol 9, 1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72, 5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov DV. (2011). The human gutome: Nutrigenomics of the host-microbiome interactions. OMICS 15, 419–430 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 [DOI] [PubMed] [Google Scholar]
- Faust K, and Raes J. (2016). CoNet app: Inference of biological association networks using Cytoscape. F1000Res 5, 1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister EB, Riehle K, Luna RA, et al. (2015). Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 3, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser MC, and Tansey MG. (2017). The gut-brain axis: Is intestinal inflammation a silent driver of Parkinson's disease pathogenesis? NPJ Parkinsons Dis 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HJ, Park SG, Jang HB, et al. (2015). Obesity alters the microbial community profile in Korean adolescents. PLoS One 10, e0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao N, Baker SS, Nugent CA, et al. (2018). Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: A meta-analysis. Physiol Genomics 50, 244–254 [DOI] [PubMed] [Google Scholar]
- Johnson CL, and Versalovic J. (2012). The human microbiome and its potential importance to pediatrics. Pediatrics 129, 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM. (2016). The Influence of the gut microbiota on host physiology: In pursuit of mechanisms. Yale J Biol Med 89, 285–297 [PMC free article] [PubMed] [Google Scholar]
- Kakiyama G, Pandak WM, Gillevet PM, et al. (2013). Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol 58, 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 [DOI] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, and Gordon JI. (2011). Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BS, Bae HS, Lim CY, et al. (2013). Comparison of gut microbiota between Sasang constitutions. Evid Based Complement Alternat Med 2013, 171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BS, Song MY, and Kim H. (2014). The anti-obesity effect of Ephedra sinica through modulation of gut microbiota in obese Korean women. J Ethnopharmacol 152, 532–539 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim MS, Lee MS, et al. (2016). Korean diet: Characteristics and historical background. J Ethnic Foods 3, 26–31 [Google Scholar]
- Koenig JE, Spor A, Scalfone N, et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl 1), 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliada A, Syzenko G, Moseiko V, et al. (2017). Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol 17, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E, Noe L, and Touzet H. (2012). SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 [DOI] [PubMed] [Google Scholar]
- Kurtz ZD, Muller CL, Miraldi ER, Littman DR, Blaser MJ, and Bonneau RA. (2015). Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol 11, e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudadio I, Fulci V, Palone F, Stronati L, Cucchiara S, and Carissimi C. (2018). Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS 22, 248–254 [DOI] [PubMed] [Google Scholar]
- Lopez P, de Paz B, Rodriguez-Carrio J, et al. (2016). Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep 6, 24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan-Muller S, Valles-Colomer M, Raes J, Lowry CA, Seedat S, and Hemmings SMJ. (2018). The gut microbiome and mental health: Implications for anxiety- and trauma-related disorders. OMICS 22, 90–107 [DOI] [PubMed] [Google Scholar]
- Mariat D, Firmesse O, Levenez F, et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, and Huttenhower C. (2012). Chapter 12: Human microbiome analysis. PLoS Comput Biol 8, e1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Watanabe K, Jiang J, et al. (2015). Diversity in gut bacterial community of school-age children in Asia. Sci Rep 5, 8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YD, Jung MJ, Roh SW, Kim MS, and Bae JW. (2011). Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One 6, e22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima S, Suda W, Oshima K, et al. (2016). The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res 23, 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, and Beiko RG. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 [DOI] [PubMed] [Google Scholar]
- Qin N, Yang F, Li A, et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB, and Bajaj JS. (2014). Bile acids and the gut microbiome. Curr Opin Gastroenterol 30, 332–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, and Mahe F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin SI, Wahlstrom A, Felin J, et al. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17, 225–235 [DOI] [PubMed] [Google Scholar]
- Simpson HL, and Campbell BJ. (2015). Review article: Dietary fibre-microbiota interactions. Aliment Pharmacol Ther 42, 158–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MY, Kim BS, and Kim H. (2014). Influence of Panax ginseng on obesity and gut microbiota in obese middle-aged Korean women. J Ginseng Res 38, 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, and Backhed F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KN, Lin SH, Liu WC, and Wang D. (2015). Inferring microbial interaction network from microbiome data using RMN algorithm. BMC Syst Biol 9, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, and Gordon JI. (2007). The Human Microbiome Project. Nature 449, 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyakht AV, Kostryukova ES, Popenko AS, et al. (2013). Human gut microbiota community structures in urban and rural populations in Russia. Nat Commun 4, 2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdam FJ, Fuentes S, de Jonge C, et al. (2013). Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 21, E607–E615 [DOI] [PubMed] [Google Scholar]
- Wahlstrom A, Sayin SI, Marschall HU, and Backhed F. (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24, 41–50 [DOI] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, and Jenkins DJ. (2006). Colonic health: Fermentation and short chain fatty acids. J Clin Gastroenterol 40, 235–243 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2006). Orientation Programme on Adolescent Health for Health-care Providers. Geneva, Switzerland: Department of Child and Adolescent Health and Development, World Health Organization [Google Scholar]
- Yun Y, Kim HN, Kim SE, et al. (2017). Comparative analysis of gut microbiota associated with body mass index in a large Korean cohort. BMC Microbiol 17, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G, Tap J, Voigt AY, et al. (2014). Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol 10, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, and Stamatakis A. (2014). PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21, 895–905 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.