Abstract
Purpose
The purpose of this study was to develop a feasible approach for single sperm isolation and chromosome analysis by next-generation sequencing (NGS).
Methods
Single sperm cells were isolated from semen samples of normozoospermic male and an infertile reciprocal translocation (RcT) carrier with the 46,XY,t(7;13)(p12;q12.1) karyotype using the optimized fluorescence-activated cell sorting (FACS) technique. Genome profiling was performed using NGS.
Results
Following whole-genome amplification, NGS, and quality control, the final chromosome analysis was performed on 31 and 6 single cell samples derived from the RcT carrier and normozoospermic male, respectively. All sperm cells from normozoospermic male showed a normal haploid 23-chromosome profile. For the RcT carrier, the sequencing data revealed that 64.5% of sperm cells harbored different variants of chromosome aberrations, involving deletion of 7p or 7q, duplication of 7p, and duplication of 13q, which is concordant with the expected chromosome segregation patterns observed in balanced translocation carriers. In one sample, a duplication of 9q was also detected.
Conclusions
We optimized FACS protocol for simple and efficient isolation of single human sperm cells that subsequently enabled a successful genome-wide chromosome profiling and identification of segmental aneuploidies from these individual cells, following NGS analysis. This approach may be useful for analyzing semen samples of infertile men or chromosomal aberration carriers to facilitate the reproductive risk assessment.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1340-0) contains supplementary material, which is available to authorized users.
Keywords: Single sperm genomic analysis, Reciprocal translocation, Fluorescence-activated cell sorting, Whole-genome amplification, Next-generation sequencing
Introduction
Chromosomally derived male infertility is estimated to affect 14% of azoospermic and 5% of oligozoospermic men [1]. In azoospermic patients, sex chromosome abnormalities predominate, while in oligozoospermic men, autosomal structural abnormalities (reciprocal and Robertsonian translocations) are most frequent [2]. Balanced reciprocal translocations (RcT) are caused by the mutual exchange of chromosomal segments between two non-homologous chromosomes which results in balanced karyotype with two reorganized derivative chromosomes, being phenotypically neutral to the carriers. However, in RcT carrier males, the aberrant meiotic behavior of affected chromosomes is rather common, resulting in unbalanced spermatozoa in frequency of 20–80%, depending upon the chromosomes, positions of breaks, and the technique used for chromosomal analysis [3]. Therefore, the genetic counseling of RcT carriers for reproductive risk estimation and family planning purposes needs more personalized approaches and determination of meiotic behavior for each particular translocation.
Some conventional methods of cytogenetic sperm segregation analysis are available, including the zona-free hamster oocyte penetration test by human spermatozoa [4] and the non-radioactive in situ hybridization technique on the nuclei of spermatozoa [5]. However, sperm karyotyping through a fusion assay is laborious and technically demanding, and enables only to investigate the sperms that have fused with hamster oocytes, while in situ hybridization allows only the screening of a restricted number of chromosomes. In recent years, the array comparative genomic hybridization and next-generation sequencing (NGS) have provided the valuable tool for genome-wide chromosome screening in single sperm cells [6–9].
Furthermore, the development of human single sperm cell-isolation techniques, such as micromanipulation [7, 10] and microfluidics approaches [8], has facilitated the use of NGS in single sperm studies. Micromanipulation is the most cost-effective method to isolate small numbers of single sperm cells. It also provides direct visual control, allowing selection of morphologically normal spermatozoa. Nevertheless, manual handling of single cells requires experienced personnel and becomes challenging when the number of cells necessary for subsequent analysis increases [11]. Alternately, various microfluidics systems have been developed that allow automated single cell isolation and processing with controlled management of nanoliters of reactions [12]. However, microfluidic devices are usually specifically designed for certain applications and exhibit only little flexibility regarding upstream sample preparation and downstream analysis methods [13]. Conversely, flow cytometry (FC) is a fast, sensitive, and high-throughput technique for isolating single cells from heterogeneous cell mixtures which is also suitable for any downstream applications, including NGS [14]. Therefore, cell sorting by FC, mainly using fluorescence-activated cell sorting (FACS) systems, is currently the method of choice to separate single cells both in basic and in clinical research [13]. However, there are no studies reporting the use of FC in human single sperm cell genomic studies in combination with NGS.
In this study, we developed an optimized experimental workflow including isolation of single sperm cells by FACS, followed by whole-genome analysis by NGS. Our pipeline allows a comprehensive whole-genome chromosomal copy number profiling and represents a powerful tool for analyzing sperm chromosomal composition for personalized family planning purposes in reproductive medicine.
Materials and methods
Study participants and sample collection
The study was approved by the Research Ethics Committee of the University of Tartu, Estonia (approval no. 267/T-2), and each participant provided a written informed consent. Semen samples were obtained from a normozoospermic man (sperm concentration 156 × 106/mL, progressive motility 58%) and from a RcT carrier with the 46,XY,t(7;13)(p12;q12.1) karyotype. The patient was 35 years old (body mass index 20.4), diagnosed with oligoasthenoteratozoospermia, and suffered from secondary infertility of a 7-year duration. The karyotype of the patient was generated from peripheral blood lymphocyte cultures using the GTG (G bands by trypsin using Giemsa) banding technique. Semen collection was conducted after 3 days of abstinence by masturbation, and the semen was liquefied at 37 °C for 30 min. After liquefaction, a semen analysis was performed within 1 h of collection, according to the World Health Organization (2010) recommendations [15]. The total volume of the ejaculate was 1 mL, sperm concentration was 1 × 106/mL, and progressive motility was 8%, and no sperm with normal morphology was present. For cryopreservation, semen samples were diluted slowly and dropwise with an equal volume of TNE (10 mM Tris-HCl, pH = 7.4, 140 mM NaCl, and 1 mM EDTA). The samples were placed into cryovials and kept in a vertical position with the bottom edge 5 cm above the surface of liquid nitrogen bath for 30 min. Finally, the tubes were plunged into liquid nitrogen for a long-term storage.
Isolation of single sperm cells
Prior to processing, cryopreserved specimens were thawed by placing them in an incubator at 37 °C for 30 min. Semen aliquots were diluted in 2 mL PBS buffer to contain 1–2 × 106 spermatozoa for the normozoospermic male sample and 0.5 × 106 spermatozoa for the RcT carrier sample. The samples were centrifuged for 10 min at 460×g, and the pellet was resuspended in 3 mL of PBS. The samples were kept on ice until the cells were labeled with a double-strand DNA binding dye SYBR Green I (Invitrogen, USA) to a final concentration of 1:10,000 for 20 to 30 min. A BD FACSMelody™ (BD Biosciences, USA) cell sorter equipped with 100-μm nozzles was used for single sperm separation. BD FACSFlow solution (BD Biosciences) was used as sheath fluid for all flow cytometry experiments. Gating and data analysis were done with BD FACSChorus™ (BD Biosciences) software. SYBR Green I was excited by the blue laser with a wavelength of 488 nm and nominal power of 16 mW. A signal was detected using 507 LP mirrors and a 527/32-nm optical filter. “Single cell” precision mode was chosen for sorting. Single sperms were collected into 96-well PCR plates (Thermo Fisher Scientific, USA) containing 3 μL PBS per well. Samples were immediately stored at − 80 °C for further analysis.
Whole-genome amplification
For whole-genome amplification (WGA) of single sperm cells, SurePlex DNA Amplification System (BlueGnome, UK) was used with minor modifications. Because of the tightly packaged DNA in sperm cells, a more aggressive method for cell lysis is needed prior to WGA. Therefore, cell lysis was optimized for sperm samples and was performed under the following conditions: 6 μL of lysis buffer (0.1 mg/mL proteinase K [Qiagen, Germany] and 5 mM DTT [Merck, Germany]) was added to sperm cells, and the mixture was incubated at 56 °C for 40 min, followed by proteinase K inactivation at 95 °C for 5 min. Subsequent pre-amplification and amplification steps of SurePlex protocol were performed according to the manufacturer’s instructions without any modifications.
To analyze WGA efficiency, 10× SolisGreen Detection mix (Solis BioDyne, Estonia) was added at 0.125× final concentration in the amplification cocktail and amplification was performed using the ABI 7500 Fast Real-time PCR system (Applied Biosystems, USA). Samples showing a threshold cycle value < 10 were determined to contain a cell and were suitable for further analysis.
Sex chromosome validation by PCR
To examine the number of sperm cells in the samples, PCR amplification using sex chromosome-specific primers (Supplementary File 1) was performed. PCR was carried out in a total volume of 20 μL containing 1 μL of 10× diluted WGA product, 1× HOT FIREPol Master Mix Ready (Solis BioDyne), 10 pmol of both primers (Tag Copenhagen A/S, Denmark), and PCR-grade water. Prior to PCR cycling, reaction mixtures were preheated for 15 min at 95 °C, followed by 35 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s. PCR products were subsequently loaded on 1.5% agarose gel and assessed for target fragments.
Next-generation sequencing
WGA products were processed using the commercially available VeriSeq™ PGS kit (Illumina Inc., USA), according to the manufacturer’s instructions and sequenced on Illumina MiSeq® platform (Illumina Inc.). BlueFuse Multi software (V4.4) (Illumina, Inc.) was utilized to analyze the data with default settings. For every single sperm, roughly 0.8–1.0-M raw reads were generated. The rate of reads mapped to the genome was approximately 72.0%. The average sequencing depth per genome was 0.02.
Results
A semen sample of normozoospermic male was used to determine the efficiency of single sperm isolation by FACS. The workflow of the used protocol is shown in Fig. 1. After FACS, WGA was used on 51 individual sperm cells, resulting in 47 (92.2%) successfully amplified samples (Supplementary File 2). Afterwards, PCR was performed to verify the presence of sex chromosomes and to confirm that the WGA product was derived from a single cell with the haploid genome and either X or Y chromosome. The results showed that 18 (38.3%) and 21 (44.7%) sperms were positive either for X or Y chromosome. Both sex chromosomes were present in four out of 47 (8.5%) sperms, indicating that the samples most likely contained at least two sperms. In four cells (8.5%), no X or Y chromosome-specific PCR products were obtained, which could be a result of allelic dropout (ADO), occurring during WGA. Six WGA products of single sperms from a normozoospermic male were randomly chosen for NGS and showed a normal 23-chromosome profile. Four samples had an X chromosome, and the other two cells had a Y chromosome, which was in accordance with the PCR results (Supplementary File 3).
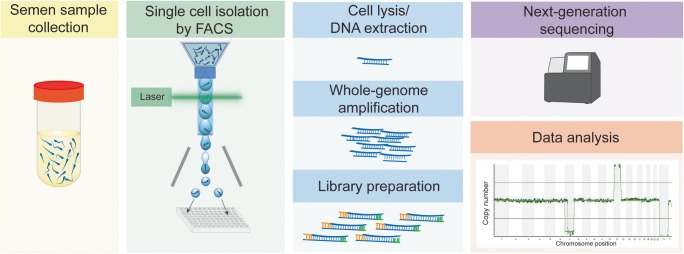
Fig. 1.
Workflow outlining the major steps of the study. SYBR Green I-stained single sperm cells were isolated by fluorescence-activated cell sorting (FACS) to 96-well PCR plates; DNA was amplified and used for next-generation sequencing
The same FACS sorting and WGA protocol with subsequent sequencing and whole-genome analysis was then applied to a sperm sample from a patient with balanced translocation 46,XY,t(7;13)(p12;q12.1). The overall success rate of WGA was 78.1% (50/64 sperm cells), and PCR validation showed that 18 (36.0%) and 26 (52.0%) of the examined sperms carried an X or Y chromosome, respectively (Supplementary File 2). In three samples (6.0%, 3/50), PCR showed the presence of both sex chromosomes and amplification has failed for three (6.0%) samples. Subsequently, NGS was applied on 40 successfully amplified and PCR-validated single sperm cells. Analysis of NGS data revealed inconclusive sequencing results for eight (20%) sperm cells, likely due to the damaged or fragmented DNA, as infertile patients might have higher sperm DNA fragmentation rates [16]. Surprisingly, one sample was positive for both sex chromosomes and was therefore excluded from the final analysis as it may contain genomes from two sperm cells or a single disomic sperm. As such, the final analysis was made on the remaining 31 (77.5%) samples with good-quality NGS data and showing either X or Y chromosome-specific sequencing reads (Supplementary File 3). The sex chromosomes revealed by NGS (15 and 16 for X and Y chromosomes, respectively) coincided with the results of PCR analysis.
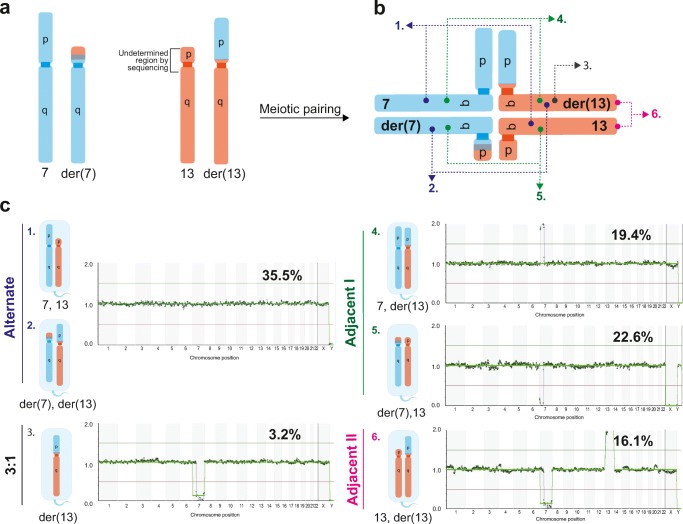
The BlueFuse analysis of sequenced data provided more precise information about the approximate location of translocation breakpoint on chromosomal region 7p12.1, indicating that almost entire 7p was involved in reciprocal translocation. Due to the heterochromatic region of the short arm of the acrocentric chromosome 13 with repetitive sequences, the short arm of chromosome 13 is not annotated by the BlueFuse software [17]. As such, the potential breakpoint, present either at the centromeric region or at the short arm of chromosome 13, was not detected (Fig. 2a). Based on this assumption, the analysis described further is specified only for the long arm of chromosome 13, starting from region 13q12.11.
Fig. 2.
The meiotic segregation patterns of affected chromosomes in single sperm cells from reciprocal translocation carrier with the 46,XY,t(7;13)(p12;q12.1) karyotype. a Schematic representation of affected chromosomes. b Affected chromosomes form a quadrivalent structure upon meiotic pairing. c Segregation of a quadrivalent can occur in alternate mode that produces spermatozoa with balanced normal and balanced translocated karyotypes (1 and 2) or produce unbalanced post-meiotic products according to 3:1 (3), adjacent I (4 and 5), and II (6) segregation modes
The proportion of sperms with normal chromosomal profile was found to be 11 out of 31 (35.5%). However, the low-coverage sequencing used in the current study does not allow distinguishing euploid sperm cells with normal chromosomes 7 and 13 from the sperm cells having a balanced karyotype with the pair of derivative chromosomes. The sequencing analysis revealed that 20 out of 31 (64.5%) sperm cells showed different variants of segmental aneuploidies. The sperm cells with segmental aneuploidies for chromosomes 7 and 13 represented the meiotic products of adjacent I and II segregations, and also from the 3:1 segregation pattern (Fig. 2b, c). Seven of the analyzed sperms (22.6%, 7/31) exhibited the deletion of 7p22.3p12.2, being the most frequent genomic aberration. These sperm cells were generated by adjacent I meiotic segregation, leading to the cells with normal chromosome 13 and derivative chromosome 7 with missing 7p (Fig. 2c). In six sperm cells (19.4%, 6/31) duplication of 7p22.3p12.2 was found (adjacent I segregation, with chromosome 7 and derivative chromosome 13, with extra 7p), while in five sperm cells (16.1%, 5/31), double chromosomal abnormalities were identified, consisting of loss of 7p12.1q36.3 and simultaneous duplication of 13q (adjacent II segregation, with normal chromosome 13 and derivative chromosome 13). In addition, we found one sperm (3.2%) with deletion of 7p12.1q36.3 (3:1 segregation, with derivative chromosome 13) and one with chromosomal aneuploidy not involving chromosomes 7 or 13 but showing the partial duplication of the long arm of chromosome 9 (Supplementary File 3, sample 25). The latter is likely representing the interchromosomal effect, a common phenomenon described in reciprocal translocation carriers [18]. In our study, we did not observe any sperm cells originating from the 4:0 meiotic segregation pattern, which should present only full chromosomal aneuploidies of both affected chromosomes. However, this is probably caused by the limited number of analyzed sperm cells of the RcT carrier in the current study.
Discussion
In this study, we optimized a FC-based protocol for single sperm cell sorting and lysis, and combined it with Illumina VeriSeq PGS workflow for comprehensive chromosome screening. To allow a quick isolation of large numbers of single sperm cells, we optimized the cell sorting conditions and used fluorescent dye to label cells. As the commonly used Hoechst 33342 is not compatible with the FACS instrument used in this study, we utilized SYBR Green I dye. Analogously to Hoechst 33342, SYBR Green I penetrates the live cell’s membrane and binds to double-stranded DNA of spermatozoa, thus discriminating live cells from cellular debris and increasing the yield of sorted cells. Stained sperm cells of a healthy man appeared as clearly distinguishable populations on light scatter and SYBR Green I fluorescence intensity plots (Supplementary File 4). Appearance of the RcT carrier’s semen sample on SYBR Green I fluorescence intensity dot plot was very similar to the one of normal sperm sample; however, its light scatter profile was different. No clearly distinguishable populations appeared, and the vast majority of events looked like a debris when compared to normal sperm patterns (Supplementary File 4). This result is in good correlation with determined semen parameters of the RcT carrier, showing morphological and motility defects. In addition, it has been shown that because of the protein aggregates, samples from azoospermic men contain large amounts of low-intensity light scatter particles [19]. It is commonly agreed that two light scattering parameters—forward and side scatter—resemble a relative cell size and granularity, respectively. Setting the sorting gate around the normal sperm population on a light scatter dot plot and applying the same gate for the RcT carrier sperm sample allowed us to sort out cells with the same relative size and granularity for both semen samples.
To further evaluate the efficiency of our cell isolation workflow, we whole-genome profiled gametes of the RcT patient with the 46,XY,t(7;13)(p12;q12.1) karyotype, as RcT carriers are at high risk of having genomic aberrations in the germ cells due to defective chromosome pairing during meiosis, affecting both quantitative as well as qualitative sperm production [20, 21]. By whole-genome analysis of single sperm cells, we identified a high number of chromosomally abnormal spermatozoa in the RcT patient, which is a common characteristic of balanced translocation carriers [3]. Namely, we detected 35.5% genetically normal/balanced sperm cells, resulting from the alternate pattern of quadrivalent segregation (Fig. 2). The frequencies of adjacent I and adjacent II segregations were 42.0% and 16.1%, respectively, and the occurrence of 3:1 segregation was the lowest, occurring only in 3.2% of the sperm cells. This result is consistent with the majority of previous observations made by FISH studies in RcT patients [20, 22].
The risk of an overall chromosomal aneuploidy in spermatozoa is inversely correlated to sperm concentration and total progressive motility [23, 24]. Unfortunately, using FACS for single sperm sorting, we did not distinguish between motile and immotile sperms; however, the study examining motile and immotile sperms’ aneuploidy rate on a single sperm level indicated that morphologically normal, immotile but viable spermatozoa have an aneuploidy rate similar to that of normal motile spermatozoa [25]. This observation suggests that the detected aneuploidy rate and types in our study are not affected by the motility status of the studied sperms. Hence, the protocol developed in the current study holds promise for RcT patient counseling and can be beneficial for male infertility diagnostics, as sperm meiotic studies have also unveiled more frequent errors in chromosomal segregation in infertile men [26]. Furthermore, it has been proposed that the combined semen analysis and molecular assessment of chromosome meiotic segregation are essential for a complex counseling to assist infertile couples in making their reproductive choices. If there is a high risk of having chromosomally unbalanced spermatozoa, an intracytoplasmic sperm injection with a preimplantation genetic diagnosis is considered as the first relevant reproductive approach [27]; however, if the percentage of abnormal spermatozoa is very high, the number of balanced embryos that can be implanted would be very low, and another reproductive choices, i.e., using donor sperms for insemination should be considered [28].
The limitation of our study is the small number of patients and samples processed and analyzed. In addition, the sorting efficiency of the RcT carrier’s semen sample was still considerably lower compared to normozoospermic man sample (78.1% vs. 92.2%), which might also be associated with a lower proportion of sperm cells containing intact genomic DNA, which is important both for cell sorting applications and for whole-genome amplification. As a consequence, low quality of initial DNA material (fragmented or degraded) will result in uneven amplification and ADO with following poor genome representation. In our case, from the total of 40 sequenced single-sperm samples of the RcT carrier, 20% (8/40) had bad-quality sequencing data, most likely because of the poor-quality WGA. Although in our protocol we used SYBR Green I DNA dye to discriminate the live sperm cells from cellular debris, the apoptotic cells may still have remained unnoticed. Also, the sperm DNA is tightly packed with protamines, making DNA extraction complicated. Furthermore, significant variations in protamine-1 and protamine-2 levels exist on the single sperm level within sperm populations that associate with the susceptibility of sperm cells to DNA damage induction [29].
Conclusion
In conclusion, we optimized a fast and high-throughput FACS protocol for single sperm cell isolation. We have successfully validated the presence of individual cells after sorting and identified segmental aneuploidies in single sperms. Our protocol offers robust laboratory procedure, which can be implemented into clinics to support the personalized family planning in several patients’ groups with reproductive failure, such as men with chromosomal aberrations and/or male infertility and in couples with recurrent miscarriages. With the fast development of NGS, we expect that the sample preparation and sequencing cost will be further reduced in the future, making the developed method more accessible for routine clinical use.
Electronic supplementary material
(PDF 405 kb)
(PDF 288 kb)
(PDF 951 kb)
(PNG 1120 kb)
Acknowledgments
We would like to thank the study participants who enrolled in the study.
Funding information
This study was funded by the Estonian Ministry of Education and Research (IUT34-16), Enterprise Estonia (EU48695), and the Horizon 2020 innovation program (WIDENLIFE, 692065).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by the Research Ethics Committee of the University of Tartu, Estonia (approval no. 267/T-2), and written informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Yatsenko AN, Yatsenko SA, Weedin JW, Lawrence AE, Patel A, Peacock S, Matzuk MM, Lamb DJ, Cheung SW, Lipshultz LI. Comprehensive 5-year study of cytogenetic aberrations in 668 infertile men. J Urol. 2010;183:1636–1642. doi: 10.1016/j.juro.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elghezal H, Hidar S, Braham R, Denguezli W, Ajina M, Saâd A. Chromosome abnormalities in one thousand infertile males with nonobstructive sperm disorders. Fertil Steril. 2006;86:1792–1795. doi: 10.1016/j.fertnstert.2006.04.041. [DOI] [PubMed] [Google Scholar]
- 3.Benet J, Oliver-Bonet M, Cifuentes P, Templado C, Navarro J. Segregation of chromosomes in sperm of reciprocal translocation carriers: a review. Cytogenet Genome Res. 2005;111:281–290. doi: 10.1159/000086901. [DOI] [PubMed] [Google Scholar]
- 4.Rudak E, Jacobs PA, Yanagimachi R. Direct analysis of the chromosome constitution of human spermatozoa. Nature. 1978;274:911–913. doi: 10.1038/274911a0. [DOI] [PubMed] [Google Scholar]
- 5.Godo A, Blanco J, Vidal F, Anton E. Accumulation of numerical and structural chromosome imbalances in spermatozoa from reciprocal translocation carriers. Hum Reprod. 2013;28:840–849. doi: 10.1093/humrep/des431. [DOI] [PubMed] [Google Scholar]
- 6.Patassini C, Garolla A, Bottacin A, Menegazzo M, Speltra E, Foresta C, Ferlin A. Molecular karyotyping of human single sperm by array—comparative genomic hybridization. PLoS One. 2013;8:e60922. doi: 10.1371/journal.pone.0060922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sha Y, Sha Y, Ji Z, Ding L, Zhang Q, Ouyang H, Lin S, Wang X, Shao L, Shi C, Li P, Song Y. Comprehensive genome profiling of single sperm cells by multiple annealing and looping-based amplification cycles and next-generation sequencing from carriers of Robertsonian translocation. Ann Hum Genet. 2017;81:91–97. doi: 10.1111/ahg.12187. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, Bai F, Qiao J, Tang F, Li R, Xie XS. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkness EF, Grindberg RV, Yee-Greenbaum J, Marshall CR, Scherer SW, Lasken RS, Venter JC. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–832. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valihrach L, Androvic P, Kubista M. Platforms for single-cell collection and analysis. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed]
- 12.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 13.Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P. Technologies for single-cell isolation. Int J Mol Sci. 2015;16:16897–16919. doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, Wigler M, Navin N, Hicks J. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO laboratory manual for the examination and processing of human semen. World Health Organization; 2010.
- 16.Saleh RA, Agarwal A, Nada EA, El-Tonsy MH, Sharma RK, Meyer A, et al. Negative effects of increased sperm DNA damage in relation to seminal oxidative stress in men with idiopathic and male factor infertility. Fertil Steril. 2003;79(Suppl 3):1597–1605. doi: 10.1016/S0015-0282(03)00337-6. [DOI] [PubMed] [Google Scholar]
- 17.Dunham A, Matthews LH, Burton J, Ashurst JL, Howe KL, Ashcroft KJ, Beare DM, Burford DC, Hunt SE, Griffiths-Jones S, Jones MC, Keenan SJ, Oliver K, Scott CE, Ainscough R, Almeida JP, Ambrose KD, Andrews DT, Ashwell RIS, Babbage AK, Bagguley CL, Bailey J, Bannerjee R, Barlow KF, Bates K, Beasley H, Bird CP, Bray-Allen S, Brown AJ, Brown JY, Burrill W, Carder C, Carter NP, Chapman JC, Clamp ME, Clark SY, Clarke G, Clee CM, Clegg SCM, Cobley V, Collins JE, Corby N, Coville GJ, Deloukas P, Dhami P, Dunham I, Dunn M, Earthrowl ME, Ellington AG, Faulkner L, Frankish AG, Frankland J, French L, Garner P, Garnett J, Gilbert JGR, Gilson CJ, Ghori J, Grafham DV, Gribble SM, Griffiths C, Hall RE, Hammond S, Harley JL, Hart EA, Heath PD, Howden PJ, Huckle EJ, Hunt PJ, Hunt AR, Johnson C, Johnson D, Kay M, Kimberley AM, King A, Laird GK, Langford CJ, Lawlor S, Leongamornlert DA, Lloyd DM, Lloyd C, Loveland JE, Lovell J, Martin S, Mashreghi-Mohammadi M, McLaren SJ, McMurray A, Milne S, Moore MJF, Nickerson T, Palmer SA, Pearce AV, Peck AI, Pelan S, Phillimore B, Porter KM, Rice CM, Searle S, Sehra HK, Shownkeen R, Skuce CD, Smith M, Steward CA, Sycamore N, Tester J, Thomas DW, Tracey A, Tromans A, Tubby B, Wall M, Wallis JM, West AP, Whitehead SL, Willey DL, Wilming L, Wray PW, Wright MW, Young L, Coulson A, Durbin R, Hubbard T, Sulston JE, Beck S, Bentley DR, Rogers J, Ross MT. The DNA sequence and analysis of human chromosome 13. Nature. 2004;428:522–528. doi: 10.1038/nature02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anton E, Vidal F, Blanco J. Interchromosomal effect analyses by sperm FISH: incidence and distribution among reorganization carriers. Syst Biol Reprod Med. 2011;57:268–278. doi: 10.3109/19396368.2011.633682. [DOI] [PubMed] [Google Scholar]
- 19.Ferrara F, Daverio R, Mazzini G, Bonini P, Banfi G. Automation of human sperm cell analysis by flow cytometry. Clin Chem. 1997;43:801–807. [PubMed] [Google Scholar]
- 20.Shi Q, Martin RH. Aneuploidy in human spermatozoa: FISH analysis in men with constitutional chromosomal abnormalities, and in infertile men. Reproduction. 2001;121:655–666. doi: 10.1530/rep.0.1210655. [DOI] [PubMed] [Google Scholar]
- 21.Oliver-Bonet M, Ko E, Martin RH. Male infertility in reciprocal translocation carriers: the sex body affair. Cytogenet Genome Res. 2005;111:343–346. doi: 10.1159/000086908. [DOI] [PubMed] [Google Scholar]
- 22.Guttenbach M, Engel W, Schmid M. Analysis of structural and numerical chromosome abnormalities in sperm of normal men and carriers of constitutional chromosome aberrations. A review. Hum Genet. 1997;100:1–21. doi: 10.1007/s004390050459. [DOI] [PubMed] [Google Scholar]
- 23.Collodel G, Capitani S, Baccetti B, Pammolli A, Moretti E. Sperm aneuploidies and low progressive motility. Hum Reprod. 2007;22:1893–1898. doi: 10.1093/humrep/dem099. [DOI] [PubMed] [Google Scholar]
- 24.Vegetti W, Van Assche E, Frias A, Verheyen G, Bianchi MM, Bonduelle M, et al. Correlation between semen parameters and sperm aneuploidy rates investigated by fluorescence in-situ hybridization in infertile men. Hum Reprod. 2000;15:351–365. doi: 10.1093/humrep/15.2.351. [DOI] [PubMed] [Google Scholar]
- 25.Zeyneloglu HB, Baltaci V, Ege S, Haberal A, Batioglu S. Detection of chromosomal abnormalities by fluorescent in-situ hybridization in immotile viable spermatozoa determined by hypo-osmotic sperm swelling test. Hum Reprod. 2000;15(4):853–856. doi: 10.1093/humrep/15.4.853. [DOI] [PubMed] [Google Scholar]
- 26.Martin RH. Cytogenetic determinants of male fertility. Hum Reprod Update. 2008;14:379–390. doi: 10.1093/humupd/dmn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munné S. Preimplantation genetic diagnosis of numerical and structural chromosome abnormalities. Reprod BioMed Online. 2002;4:183–196. doi: 10.1016/S1472-6483(10)61938-4. [DOI] [PubMed] [Google Scholar]
- 28.Leclercq S, Auger J, Dupont C, Le Tessier D, Lebbar A, Baverel F, et al. Sperm FISH analysis in two healthy infertile brothers with t(15;18) unbalanced translocation: implications for genetic counselling and reproductive management. Eur J Med Genet. 2010;53:127–132. doi: 10.1016/j.ejmg.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Aoki VW, Emery BR, Liu L, Carrell DT. Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl. 2006;27:890–898. doi: 10.2164/jandrol.106.000703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 405 kb)
(PDF 288 kb)
(PDF 951 kb)
(PNG 1120 kb)