Abstract
AMPK is a crucial regulator of energy homeostasis that acts downstream of its upstream kinase liver kinase B1 (LKB1) and calcium/calmodulin-dependent protein kinase 2 (CaMKK2). LKB1 primarily phosphorylates AMPK after energy stress, whereas calcium-mediated activation of AMPK requires CaMKK2, although the regulatory mechanisms of calcium-mediated AMPK activation remain unclear. Using biochemical, microscopic, and genetic approaches, we demonstrate that the stromal interaction molecule (STIM)2, a calcium sensor, acts as a novel regulator of CaMKK2-AMPK signaling. We reveal that STIM2 interacts with AMPK and CaMKK2 and that the increase in intracellular calcium levels promotes AMPK colocalization and interaction with STIM2. We further show that STIM2 deficiency attenuates calcium-induced but not energy stress–induced AMPK activation, possibly by regulating the CaMKK2-AMPK interaction. Together, our results identify a previously unappreciated mechanism that modulates calcium-mediated AMPK activation.—Chauhan, A. S., Liu, X., Jing, J., Lee, H., Yadav, R. K., Liu, J., Zhou, Y., Gan B. STIM2 interacts with AMPK and regulates calcium-induced AMPK activation.
Keywords: CaMKK2, energy stress, LKB1, STIM1
Metazoans have evolved adaptive mechanisms that help maintain their survival under altered physiologic conditions. To achieve this, an intricate signaling network has evolved to sense, respond, and adapt to changes in intra- and extracellular environments. AMPK is one such master regulator of cellular homeostasis. AMPK plays a key role in regulating energy homeostasis in response to energy stress (1, 2). Energy deprivation results in depletion of cellular ATP levels with concomitant increases of cellular AMP levels, inducing energy stress. AMPK can sense cellular energy status by detecting fluctuations of cellular AMP levels. AMPK exists as a heterotrimeric complex, consisting of 3 subunits: AMPK-α, AMPK-β, and AMPK-γ (3). The catalytic kinase activity of AMPK resides in its α subunit, whereas the β and γ subunits function as regulatory subunits (4). AMP can directly bind to the γ subunit of AMPK, resulting in conformational change and allosteric activation of the AMPK complex (5, 6). Energy stress–induced AMPK activation also requires the phosphorylation of Thr172 in its α catalytic subunit by the upstream kinase liver kinase B1 (LKB1) (7–9). Energy stress–induced AMPK activation involves other regulatory mechanisms. For example, in response to energy stress, long noncoding RNA neighbor of BRCA1 gene 2 (NBR2) interacts with the AMPK-α subunit, promotes AMPK kinase activity, and potentiates AMPK activation during energy stress (10–12). In addition, the scaffolding axis inhibition protein (AXIN) regulates the complex formation between LKB1 and AMPK and thus promotes energy stress–induced AMPK activation (13, 14). These regulatory mechanisms serve to fine-tune AMPK activity in response to energy stress.
Once activated, AMPK phosphorylates a number of downstream substrates, which shift from ATP-consuming anabolic processes to ATP-generating catabolic processes, restoring energy homeostasis under energy stress (2). For example, in response to energy stress, AMPK suppresses mechanistic target of rapamycin complex 1 (mTORC1, also called mammalian target of rapamycin complex 1)-mediated protein synthesis and cell growth, an anabolic process that is highly energy consuming. AMPK inhibits mTORC1 by directly phosphorylating mTORC1 component Raptor and its upstream regulator tuberous sclerosis complex (TSC)1–TSC2 complex (15, 16). Dysregulation of AMPK, its upstream regulators such as LKB1, or its downstream effectors such as TSC1 and TSC2 results in defective cellular response to energy stress and leads to diseases such as cancer and diabetes (17, 18).
In addition to LKB1, calcium/calmodulin-dependent protein kinase 2 (CaMKK2) can phosphorylate and activate AMPK. The basal phosphorylation of AMPK Thr172 is significantly reduced but not completely abolished in LKB1-deficient cells, leading to the identification of CaMKK2 as another upstream kinase of AMPK (19–21). Unlike LKB1, CaMKK2 does not appear to mediate energy stress–induced AMPK phosphorylation but phosphorylates AMPK at Thr172 in response to increased intracellular calcium levels (19–21). It has been proposed that increases of intracellular calcium levels upon calcium ionophore treatment cause a conformational change in CaMKK2, resulting in AMPK activation (22). Although energy stress–mediated and LKB1–mediated AMPK activation has been extensively characterized, the regulatory mechanism of calcium- and CaMKK2-mediated AMPK activation is still poorly understood.
Calcium plays essential roles in regulating many signaling pathways and cellular processes, such as cell growth and differentiation (23). To maintain appropriate levels of intracellular calcium, cells have evolved elegant mechanisms to sense and adjust intracellular calcium levels, prominent among which is the stromal interaction molecule (STIM)1 and ORAI1-mediated store-operated calcium entry (SOCE) (24). When calcium levels in the endoplasmic reticulum (ER) lumen are high, calcium can bind to the luminal EF hand motif of STIM1, a single-transmembrane protein that mainly localizes on ER. As calcium in the ER lumen is depleted, calcium dissociates from the luminal side of STIM1, leading to STIM1 conformational change, oligomerization, and translocation to ER–plasma membrane junctions, where STIM1 couples with and gates ORAI1 and subsequently triggers ORAI1-mediated extracellular calcium influx (24, 25). Thus, similar to AMPK serving as an energy sensor to maintain energy homeostasis, STIM1 functions as a calcium sensor to maintain cellular calcium homeostasis. STIM2 is a homolog of STIM1 (26). Despite extensive sequence homology and predominant localization on ER, STIM1 and STIM2 appear to have very different functions in SOCE. Previous studies have shown that, whereas STIM1 knockdown largely eliminated SOCE, STIM2 knockdown had only a minimal effect on SOCE, and its overexpression even inhibited SOCE (27–32). It has been suggested that STIM2 is a much weaker activator of SOCE, with poor ORAI-coupling efficacy compared with STIM1 (26). Whether STIM2 play any additional role in mediating calcium signaling remains unknown.
In the present study, we identified STIM2, but not STIM1, as a novel AMPK-binding protein. Our results demonstrated that STIM2 regulates calcium-mediated AMPK signaling in an LKB1-independent manner. Our data further suggested that STIM2 might function as a scaffolding protein to maintain the AMPK–CaMKK2 interaction and to regulate calcium-mediated, but not energy stress–mediated, AMPK activation. Our study thus uncovered a hitherto unrecognized regulatory mechanism of calcium-regulated AMPK signaling.
MATERIALS AND METHODS
Cell culture studies
Human embryonic kidney (HEK) 293T, 786-O, and HeLa cells were obtained from American Type Culture Collection (Manassas, VA, USA). All cell lines were free of mycoplasma contamination (tested by the vendor). No cell lines used in this study are found in the International Cell Line Authentication Committee (ICLAC) database (http://iclac.org) of commonly misidentified cell lines, based on short tandem repeat profiling performed by the vendor. All cell lines were cultured and maintained in DMEM (D6429; MilliporeSigma, Burlington, MA, USA) supplemented with 10% (v/v) fetal bovine serum (F4135; MilliporeSigma). Plasmid transfection in cells was carried out using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA). For experiments to induce energy stress, 786-O and HeLa cells were cultured in either glucose-containing or glucose-free DMEM supplemented with 10% dialyzed fetal bovine serum (F0392; MilliporeSigma) or with DMEM containing 2 deoxy-glucose (5 mM) (D6134; MilliporeSigma) as previously described (10, 33, 34). For experiments to induce intracellular calcium levels, cells were treated with ionomycin (1 μM) (I9657; MilliporeSigma) or thapsigargin (1 μM) (T9033; MilliporeSigma) for 15 or 30 min.
Constructs and viruses
Full-length STIM2 and its truncated mutants were amplified using PCR and subcloned in entry vector using a pENTR/D-TOPO Cloning Kit (K240020; Thermo Fisher Scientific) following the manufacturer’s instructions. Subsequently, STIM2-containing entry vector was used to generate gateway vectors with C-terminal streptavidin binding peptide (SFB) tag lentiviral expression vector (kindly gifted by Dr. Junjie Chen, M. D. Anderson Cancer Center) (35). The STIM2–yellow fluorescent protein (YFP) construct was generated by inserting amplified STIM2 into the pEYFP-N1 vector (ClonTech Laboratories, Mountain View, CA, USA). The STIM1 Myc-His construct was generated by inserting mouse STIM1 cDNA between the BamHI and XbaI sites in pEF4 vector. AMPK-α, AMPK-β, AMPK-γ, and CaMKK2 entry clones were obtained from the Human ORFeome V5.1 library. These entry clones were used to generate gateway vectors with N-terminal myc-tag using the gateway-based LR reaction (12538; Thermo Fisher Scientific). To construct AMPK-α–mCherry vector, AMPK-α was amplified using PCR and cloned into the mCherry-N1 vector (54517; Addgene, Cambridge, MA, USA) using Xho1 and EcoR1 restriction sites. To generate knockout (KO) cells using the clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated protein 9 (Cas9) approach, single guide RNAs (sgRNAs) targeting STIM1 and STIM2 were cloned in the lentiviral pLenti-CRISPR-V2 plasmid (52961; Addgene) following the manufacturer’s instructions. HEK 293T cells were transfected with sgRNA vector along with the ViraPower Lentiviral Expression System (K496000; Thermo Fisher Scientific) to generate lentiviral supernatant. Forty-eight hours after transfection, the cell supernatant was collected, filtered using 0.45-μm filters (SLHV033RS; Thermo Fisher Scientific), and used to infect target cells. Stably transfected cells were selected using 2 μg/ml puromycin (ant-pr-1; Invivogen, San Diego, CA, USA).
Immunoprecipitation
Cultured cells were washed with PBS and lysed using NP40 lysis buffer [50 mM Tris (pH 7.4), 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1% Nonidet P40 (NP40) (pH 8.0)] containing complete mini protease inhibitors (11836153001; Roche, Basel, Switzerland) and phosphatase inhibitor cocktail (524625; Calbiochem, San Diego, CA, USA) as previously described (36). Cell lysates were then centrifuged at 12,000 g for 10 min at 4°C to clear the cellular debris. For immunoprecipitation of SFB-tagged proteins, 30 μl S-protein agarose beads (69704; MilliporeSigma) were washed twice with NP40 lysis buffer and incubated with the cell lysate overnight at 4°C. Immunoprecipitates were washed 3 times with NP40 lysis buffer and subjected to denaturation in 30 μl LDS sample buffer (1610747; Bio-Rad, Hercules, CA, USA). Coimmunoprecipitated proteins were then resolved on 10% SDS-PAGE gel and analyzed by immunoblotting after Western blotting. For immunoprecipitation of endogenous AMPK proteins or Myc tag STIM1 proteins, 30 μl Protein G magnetic beads (9006S; Cell Signaling Technology, Danvers, MA, USA) were washed twice with NP40 lysis buffer and incubated with the cell lysate with anti-AMPKα1+ α2 antibody (ab80039; Abcam, Cambridge, MA, USA) or anti-Myc antibody (2276S; Cell Signaling Technology, Danvers, MA, USA) overnight at 4°C. Immunoprecipitates were washed 3 times with NP40 lysis buffer and subjected to denaturation in 30 μl LDS sample buffer. Coimmunoprecipitated proteins were resolved on 10% SDS-PAGE gel and analyzed by immunoblotting.
Western blotting analysis
For Western blotting analysis, 20–40 μg cell lysate or immunoprecipitated samples were denatured in 3× LDS sample buffer and separated on either 10 or 12% SDS-PAGE gel. Separated proteins were transferred onto PVDF membrane (1620177; Bio-Rad). The following antibodies were used in our study: phospho–AMPK-α (2535S, 1:1000 dilution; Cell Signaling Technology) rabbit mAb, AMPK-α (5832S, 1:1000 dilution; Cell Signaling Technology) rabbit mAb, AMPK-β (4150S, 1:1000 dilution; Cell Signaling Technology) rabbit mAb, AMPK-γ (4187S, 1:1000 dilution; Cell Signaling Technology) rabbit polyclonal Ab, phospho-ACC (3661S, 1:1000 dilution; Cell Signaling Technology) rabbit pAb, acetylCoA carboxylase (ACC) (3662S, 1:1000 dilution; Cell Signaling Technology) rabbit pAb, phospho-S6K (9205S, 1:1000 dilution; Cell Signaling Technology) rabbit pAb, S6K (9202S, 1:1000 dilution; Cell Signaling Technology) rabbit pAb, phospho-S6 (2215S, 1:3000 dilution; Cell Signaling Technology) rabbit pAb, S6 (2217S, 1:5000 dilution; Cell Signaling Technology) rabbit mAb, STIM1 (5668S, 1:1000 dilution; Cell Signaling Technology) rabbit mAb, STIM2 (ACC-064, 1:1000 dilution; Alomone Labs, Jerusalem, Israel) rabbit polyclonal Ab, vinculin (V4505, 1:10,000 dilution; MilliporeSigma) mouse mAb, c-Myc (5605P, 1:1000 dilution; Cell Signaling Technology) rabbit mAb, and Flag-tag (14793S, 1:5000 dilution; Cell Signaling Technology) rabbit mAb.
Cell death assay
Cell death was measured by propidium iodide (PI) staining as previously described (34, 37, 38). Briefly, 1 × 106 cells were counted and washed twice with PBS. Cells were resuspended in 100 μl PBS containing 2 μg/ml PI and incubated for 30 min at room temperature. Percentages of PI-stained cells were analyzed using a flow cytometer (Accuri C6; BD Biosciences, San Jose, CA, USA) and compared with unstained cells.
Light microscopy and immunofluorescence
For light microscopy, cells were cultured in 6-well plates and incubated in glucose-containing and glucose-free medium for the indicated time points. After treatment, phase contrast images were acquired using an EVOSf1 microscope (Advanced Microscopy Group, Bothell, WA, USA) with ×10 phase contrast objective. For each condition, images from at least 3 different fields were taken, and representative images from the single field were shown. For immunofluorescence microscopy, HeLa cells (5 × 105) were plated on glass coverslips in 6-well plates and transfected with AMPK-α–mCherry (2 μg) and STIM2-YFP (2 μg). After 36 h, cells were incubated with ionomycin (1 μM) or thapsigargin (1 μM) containing medium for 30 min before proceeding to fixation. Cells were washed 3 times with PBS and fixed using 4% paraformaldehyde (sc-281692; Santa Cruz Biotechnology, Dallas, TX, USA) for 15 min at room temperature. Cells were washed 3 times after fixation, and coverslips were mounted on glass slides using Vectashield (H-1200; Vector Laboratories, Burlingame, CA, USA) containing DAPI. In other experiments, HeLa cells were transfected with CaMKK2-myc (2 μg). Thirty-six hours after transfection, cells were treated with thapsigargin (1 μM) for 30 min before washing and fixation. After fixation, cells were permeabilized with 0.5% Triton X-100 (T9284; MilliporeSigma) in PBS for 5 min at room temperature. Cells were then washed and blocked with Odyssey locking buffer (927-40100; Li-Cor Biosciences, Lincoln, NE, USA) for 60 min and incubated with the following primary antibodies: rabbit anti-myc (ab9106, 1:500 dilution; Abcam) mAb and anti-AMPKα (NBP2-22127, 1:200 dilution; Novus Biologic, Littleton, CO, USA) mouse mAb prepared in Odyssey blocking buffer overnight at 4°C. After incubation with primary antibodies, cells were washed 3 times and incubated with the following secondary antibodies: Alexa Fluor 488 goat anti-mouse IgG antibody (A-11001, 1:500 dilution; Thermo Fisher Scientific) and Alexa Fluor 633 goat anti-rabbit IgG antibody (A21070; Thermo Fisher Scientific). Images were acquired using a confocal microscope (LSM 880; Carl Zeiss, Oberkochen, Germany) with ×63 oil immersion objective and captured using 488, 561, and 640 nm excitation lasers and a pinhole aperture of 1 AU. The overlap coefficient was calculated using Zen software (Carl Zeiss).
Real-time intracellular Ca2+ measurements
Real-time intracellular [Ca2+] was measured with Fura-2 AM as previously described (39, 40). In brief, 786-O or HeLa cells were seeded and cultured on a glass-bottom plate for 24 h before imaging. The cells were loaded with 2 µM Fura-2 AM in the dark for 30 min at room temperature. Intracellular [Ca2+] levels are shown as the F340/F380 ratio, which was collected every 5 s by the Nikon A1 confocal microscope.
Oligonucleotide and primer sequences
Primers used for protein expression:
STIM2 forward: 5′-CACCTTGCTGGTGCTCGGGCTG-3′
STIM2 reverse: 5′-CTTAGATTTCTTCTTAAAAAGGC-3′
STIM2-242 forward: 5′-CACCAAAGAACATGTTGCAAAAATGATG-3′
STIM2-349 forward: 5′-CACCGATGCACTTCAGAAATGGCTTC-3′
STIM2-442 forward: 5′-CACCGGCTTTCAGATAGCCCATAAC-3′
STIM2-341 reverse: 5′-CAGTTCAAATTCTTTTTCGGCC-3′
STIM2-441 reverse: 5′-ACAGATCTTCTCAATTTGTTGC-3′
STIM2-P/H forward: 5′-TTCACGCCGCAGCATTGTGGATATCCTCTCAGTGTCA-3′
STIM2-P/H reverse: 5′-TGACACTGAGAGGATATCCACAATGCTGCGGCGTGAA-3′
STIM2-730 reverse: 5′-TTTCTCTCCATTATGACAAAGGTC-3′
AMPKa1 forward: 5′-CGGCTCGAGATGCGCAGACTCAGTTCCTG -3′
AMPKa1 reverse: 5′-CGGGAATTCGTTGTGCAAGAATTTTAATTAG -3′
Guide RNA sequences used for generation of CRISPER knockout cells:
STIM1 sg: 5′-GTATGCGTCCGTCTTGCCCTG-3′
STIM2 sg1: 5′-GCCCTTGAAGACACTCTTCAG-3′
STIM2 sg2: 5′-GACCACTGAAGAGTGTCTTCA-3′.
Statistical analysis and reproducibility
All experiments were performed at least 3 times. A Student’s t test was used to calculate P values. A value of P < 0.05 was considered statistically significant.
RESULTS
STIM2 interacts with AMPK
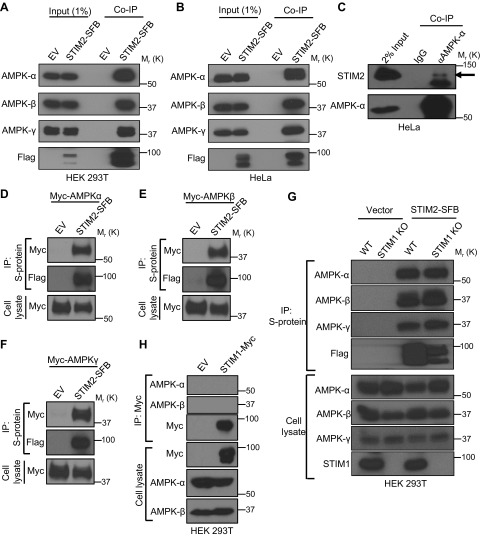
To identify potential novel regulators of AMPK signaling, we mined the BioPlex protein–protein interaction database (41) to search for potential AMPK-interacting proteins. Analysis of AMPK interactome identified STIM2 as a putative AMPK binding protein. AMPK consists of 1 catalytic subunit (α subunit) and 2 regulatory subunits (β and γ subunits), which together form a stable heterotrimeric complex (4). Further analysis of the BioPlex database revealed that immunoprecipitation of overexpressed STIM2 followed by mass spectrometry identified all 3 subunits of the AMPK complex as potential STIM2-binding proteins (Table 1). To confirm STIM2 interaction with AMPK, we generated stable cells expressing STIM2-SFB (S protein–Flag-SFB tagged at the C-terminal cytoplasmic region of STIM2). Pulldown of overexpressed STIM2 coprecipitated all 3 subunits of endogenous AMPK complex in both HEK 293T and HeLa cells (Fig. 1A, B). To further validate our findings, we immunoprecipitated endogenous AMPK using anti–AMPK-α antibody and detected coprecipitated endogenous STIM2 in HeLa cells, confirming the AMPK–STIM2 interaction at the endogenous level (Fig. 1C). Because HeLa is an LKB1-deficient cell line (10), our data also indicated that the STIM2–AMPK interaction is independent of LKB1.
TABLE 1.
STIM2-interacting proteins identified by mass spectrometry and ranked based on the priority score
| Gene | Priority score |
|---|---|
| STIM1 | 1 |
| PRKAA1 (AMPK-α1) | 1 |
| PRKAA2 (AMPK-α2) | 0.9957 |
| PRKAB1 (AMPK-β1) | 0.9988 |
| PRKAB2 (AMPK-β2) | 0.9986 |
| PRKAG1 (AMPK-γ1) | 0.9988 |
| PRKAG2 (AMPK-γ2) | 0.9993 |
| FAM5C | 1 |
Priority scores from the BioPlex database (Harvard Medical School, Boston, MA, USA; http://bioplex.hms.harvard.edu/).
Figure 1 .
STIM2 interacts with AMPK. A, B) Coimmunoprecipitation (Co-IP) of STIM2 and AMPK subunits in HEK 293T (A) and HeLa (B) cells. Protein extracts from cells stably expressing STIM2-SFB were immunoprecipitated using S-protein beads after immunoblotting with the indicated antibodies. Cells expressing empty vector (EV) were used as the control. C) Endogenous interaction of STIM2 and AMPK. HeLa cell lysates were subjected for immunoprecipitation using mouse anti–AMPK-α antibody and IgG control. Coprecipitated STIM2 was detected after immunoblotting. The arrow denotes STIM2 in AMPK immunoprecipitation. D–F) STIM2 interacts with all 3 subunits of AMPK. HEK 293T cells were cotransfected with STIM2-SFB or EV along with myc–AMPK-α (D), myc–AMPK-β (E), or myc–AMPK-γ (F), and then subjected for STIM2-SFB immunoprecipitation. Coprecipitated proteins were detected by immunoblotting with anti-myc antibody. G) STIM2 interacts with AMPK independent of STIM1. STIM1 wild-type (WT) or knock-out (KO) cells expressing STIM2-SFB or control vector were immunoprecipitated with S-protein beads. Coprecipitated proteins were probed by indicated antibodies. H) STIM1 does not interact with AMPK. Protein extracts from transiently transfected HEK293T cells with STIM1-myc were immunoprecipitated using S-protein beads after immunoblotting with the indicated antibodies. Cells expressing EV were used as control.
To study which subunit of the AMPK complex mediates its interaction with STIM2, we coexpressed STIM2-SFB with each subunit of AMPK in HEK 293T cells and examined which subunit of AMPK interacts with STIM2. Coimmunoprecipitation analysis revealed that STIM2 interacted with all 3 subunits of AMPK (Fig. 1D–F). STIM2 has been shown to form a complex with STIM1 (42). Consistent with this, analysis of the BioPlex database also identified STIM1 as an STIM2-interacting protein (Table 1). To study whether the STIM2–AMPK interaction is dependent on STIM1, we generated STIM1 KO HEK 293T cells by CRISPR-Cas9 technology. Coimmunoprecipitation experiments carried out in STIM1 wild-type and KO cells revealed that STIM1 deletion did not affect STIM2 interaction with AMPK, suggesting that the STIM2–AMPK interaction is independent of STIM1 (Fig. 1G). Because STIM2 is an STIM1 homolog, we also checked for interactions between STIM1 and AMPK. Despite the high homology between STIM1 and STIM2, coimmunoprecipitation experiments revealed that STIM1 did not interact with AMPK (Fig. 1H). Together, these results identified STIM2 as an interacting protein of the AMPK complex.
STIM2 interaction with AMPK modulates AMPK activation
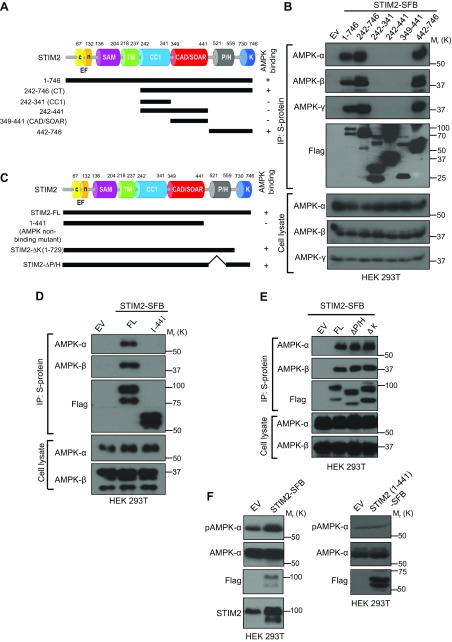
We carried out experiments to identify the regions in STIM2 that mediate its interaction with AMPK. STIM2 is a single transmembrane protein and consists of a luminal N-terminal region, a transmembrane region, and a cytoplasmic C-terminal region that contains a coiled-coil domain, a calcium release-activated channel–activating domain/STIM1 Orai1–activating region, a Pro/His-rich (P/H) domain, and a Lys-rich (K) domain (Fig. 2A) (26, 43, 44). Because AMPK mainly localizes in cytoplasm, we generated a series of truncation mutants of the cytoplasmic region in STIM2 (Fig. 2A) and examined their interactions with endogenous AMPK. Such analyses revealed that the C-terminal portion of the STIM2 cytoplasmic region (442–776 aa, which contains P/H and K domains) was both required and sufficient for mediating STIM2 interaction with AMPK (Fig. 2A, B). We then truncated 442–776 aa in the context of full-length STIM2 (1–441 aa; Fig. 2C) and confirmed that full-length STIM2, but not the 1–441 mutant, interacted with AMPK (Fig. 2D). To further characterize the STIM2 interaction with AMPK, we generated STIM2 truncation mutants lacking either the P/H domain (∆P/H) or the K domain (∆K). Coimmunoprecipitation experiment revealed that ∆P/H and ∆K mutants of STIM2 could still interact with AMPK (Fig. 2E). These data suggest that the P/H and K domains are not required for STIM2 interaction with AMPK. Because the SOAR, P/H, and K domains are conserved between STIM1 and STIM2, our data showing that none of these domains is critical for STIM2 interaction with AMPK explains why AMPK selectively interacts with STIM2 but not STIM1 and suggests that a divergent region in the C-terminal portion of STIM2, such as the linker region between P/H and K domains, might be involved in STIM2 interaction with AMPK.
Figure 2 .
STIM2 interaction with AMPK promotes AMPK activation. A) Schematic representation of STIM2 full-length (FL) and deletion mutants used in this study. B) STIM2 interacts with AMPK through its C-terminal region. Protein extracts of HEK 293T cells expressing FL and truncated mutants of STIM2 were immunoprecipitated with S-protein beads and analyzed for coprecipitated AMPK subunits after immunoblotting with the indicated antibodies. C) Schematic representation of STIM2 FL and other deletion mutants in this study. D) HEK 293T cells expressing STIM2 FL (1–746) and STIM2 1–441 were immunoprecipitated using S-protein beads and analyzed for coprecipitated proteins using the indicated antibodies. E) HEK 293T cells expressing FL STIM2, STIM2 deletion mutants for the P/H domain (∆P/H) or the K domain (∆K) were immunoprecipitated using S-protein beads and analyzed for coprecipitated proteins using indicated antibodies. F) Transient overexpression of STIM2 FL, but not its AMPK-nonbinding mutant, promotes AMPK phosphorylation. Cell lysates from HEK 293T cells expressing either empty vector, STIM2 FL, or its AMPK-nonbinding mutant (STIM2 1–441) were analyzed by immunoblotting for the phosphorylated and total levels of AMPK.
To determine any possible role of STIM2 in regulating AMPK signaling, we overexpressed full-length STIM2 and its AMPK nonbinding mutant (1–441) in HEK 293T cells. Overexpression of full-length STIM2, but not its AMPK nonbinding mutant, increased AMPK phosphorylation (Fig. 2F), suggesting that STIM2 promotes AMPK activation likely through its interaction with AMPK. Together, our results revealed that the C-terminus of the STIM2 cytoplasmic region mediates its interaction with AMPK and suggested that STIM2 interaction with AMPK promotes AMPK activation.
STIM2 does not regulate AMPK-mTORC1 signaling upon energy stress
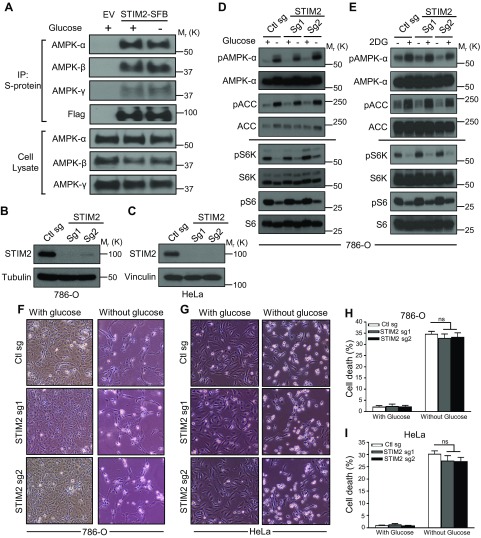
Given the well-established role of AMPK in mediating energy stress response (2), we studied the potential role of STIM2 in regulating AMPK signaling in response to energy stress. Previous studies have shown that glucose starvation–induced energy stress modulates AMPK interaction with its regulators, such as LKB1 and NBR2 (10, 14). Thus, we examined whether glucose starvation also affected STIM2–AMPK complex formation. Our analysis showed that glucose starvation did not obviously affect STIM2 interaction with any of the AMPK subunits (Fig. 3A). We then studied the potential role of STIM2 in regulating energy stress–induced AMPK activation. To this end, we used CRISPR-Cas9 technology to generate STIM2 KO cells in both 786-O and HeLa cells by 2 independent guide RNAs (Fig. 3B, C). Consistent with our previous report (10), glucose starvation in 786-O cells potently induced phosphorylation of AMPK and the AMPK substrate ACC and inhibited phosphorylation of S6K and S6, the biochemical markers of mTORC1 activation; however, STIM2 deficiency did not affect glucose starvation–induced AMPK activation or mTORC1 inhibition (Fig. 3D). Similarly, STIM2 deletion did not affect AMPK or mTORC1 signaling upon treatment of 2 deoxy-glucose, another energy stress inducer that inhibits hexokinase and blocks glycolysis (10) (Fig. 3E). Because energy stress–induced AMPK activation is known to be defective in the HeLa cell line (which is LKB1 deficient) (10), we did not further study whether STIM2 deficiency affects energy stress–induced AMPK activation in HeLa cells. In response to energy stress, AMPK promotes catabolic processes and inhibits anabolic processes to conserve ATP expenditure and to maintain cell survival during energy stress; deficiency of AMPK or its activators, such as LKB1 or NBR2, results in defective energy stress response and impaired survival under energy deprivation (2, 10). Consistent with these data, we did not observe any significant difference in glucose starvation–induced cell death in both control and STIM2 KO 786-O or HeLa cells (Fig. 3F–I). Together, these results suggested that STIM2 is not required for energy stress–induced AMPK activation.
Figure 3 .
STIM2 does not regulate AMPK-mTORC1 signaling upon energy stress. A) Glucose starvation does not alter AMPK binding to STIM2. Cells stably expressing STIM2-SFB were cultured in glucose-containing and glucose-free medium for 6 h and subjected for immunoprecipitation by S-protein beads. Coprecipitated proteins were analyzed after Western blotting. B, C) STIM2 levels in 786-O (B) and HeLa (C) cells infected with control sgRNA (Ctl sg), STIM2 sgRNA-1 (STIM2 sg1), and STIM2 sgRNA-2 (STIM2 sg2) were measured by Western blotting. D, E) Energy stress does not alter AMPK and mTORC1 activation in STIM2-depleted cells. STIM2-deficient 786-O cells were cultured in glucose-containing or glucose-free medium for 24 h (D) or treated with 2 deoxy-glucose (5 mM) for 18 h (E). Cell lysates were analyzed by immunoblotting for the phosphorylated and total levels of indicated proteins. F–I) STIM2 depletion does not affect glucose starvation–induced cell death. STIM2-deficient 786-O (F, H) and HeLa (G, I) cells were cultured in glucose-containing or glucose-free medium for 48 or 6 h, respectively. Representative images (F, G) and quantification of the cell death (H, I) were analyzed after glucose starvation. Ns, not significant. P > 0.05.
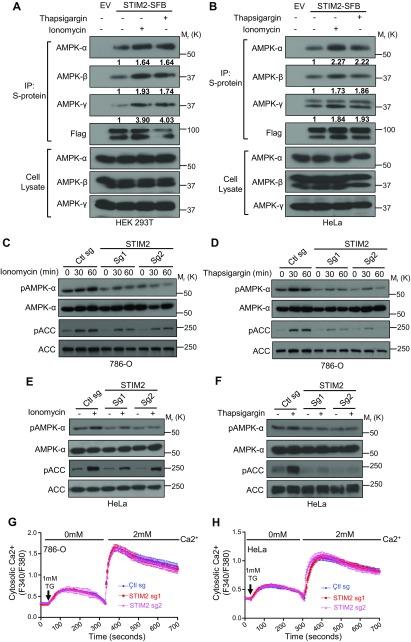
STIM2 regulates calcium-induced AMPK signaling
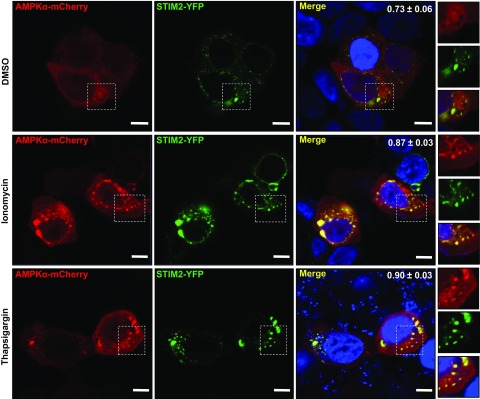
Similar to energy stress–mediated AMPK activation, alteration in intracellular calcium levels can also modulate AMPK signaling (45). Given the established role of STIM2 as a calcium sensor (26), we studied the potential involvement of STIM2 in calcium-induced AMPK activation. We first analyzed the subcellular localization of STIM2 and AMPK by immunofluorescence and observed an increase in colocalization between YFP-tagged STIM2 and mCherry-tagged AMPK-α after treatment of ionomycin or thapsigargin (Fig. 4). Further supporting our observation, coimmunoprecipitation analysis revealed an increase in the interaction between STIM2 and AMPK upon treatment of calcium ionophores (Fig. 5A, B). Next, we examined the effect of STIM2 deficiency on calcium-induced AMPK activation. As expected, ionomycin or thapsigargin treatment increased phosphorylation levels of both AMPK and AMPK substrate ACC. STIM2 deletion significantly attenuated ionomycin- or thapsigargin-induced AMPK activation (Fig. 5C, D). We made a similar observation in HeLa cells, an LKB1-deficient cell line (Fig. 5E, F), thus suggesting that STIM2 regulation of calcium-induced AMPK activation is independent of LKB1. We also examined the effect of STIM2 deficiency on SOCE in the cell lines used in our study. Consistent with previously reported results (32), our data revealed that STIM2 deletion did not affect SOCE in either 786-O or HeLa cells (Fig. 5G, H), suggesting that it is less likely that STIM2 regulates AMPK indirectly through SOCE. Collectively, our results indicated that an increase in the cytosolic calcium levels promotes complex formation between STIM2 and AMPK and that STIM2 promotes calcium-induced AMPK activation independent of SOCE.
Figure 4 .
Cytosolic calcium influx regulates AMPK-STIM2 colocalization. HeLa cells transiently expressing AMPK-α–mCherry and STIM2-YFP were incubated with medium containing ionomycin (1 μM) or thapsigargin (1 μM) for 30 min and processed for immunofluorescence imaging. Represented images from each condition are presented. Overlap coefficients for the STIM2 and AMPK signal are presented in the upper right corner of the merge panels (n = 12 for DMSO- or ionomycin-treated cells; n = 15 for thapsigargin-treated cells). In all images, insets display the selected area along with the overlays. Scale bars, 5 μm. P < 0.0001.
Figure 5 .
STIM2 regulates calcium-induced AMPK activation. A, B) Cytosolic calcium influx promotes STIM2–AMPK interaction. HEK 293T (A) and HeLa (B) cells stably expressing STIM2-SFB were incubated with medium containing ionomycin (1 μM) or thapsigargin (1 μM) for 30 min. Cell lysates were subjected to immunoprecipitation using S-protein beads. Coprecipitates were analyzed by Western blotting. C–F) STIM2 modulates calcium-induced AMPK activation. STIM2-deficient 786-O (C, D) or HeLa (E, F) cells were incubated with medium containing ionomycin (1 μM) or thapsigargin (1 μM) for the indicated time points for 786-O cells or for 30 min for HeLa cells. Cell lysates were then analyzed by immunoblotting for the phosphorylated and total levels of indicated proteins. G, H) Thapsigargin-induced Ca2+ influx in control and STIM2-depleted 786-O (G) or HeLa cells (H) was measured with Fura-2 AM. Error bars denote sem [n = 20 (control), n = 20 (STIM2 sg1), n = 20 (STIM2 sg2) (G); n = 50 (control), n = 83 (STIM2 sg1), n = 67 (STIM2 sg2)] (H).
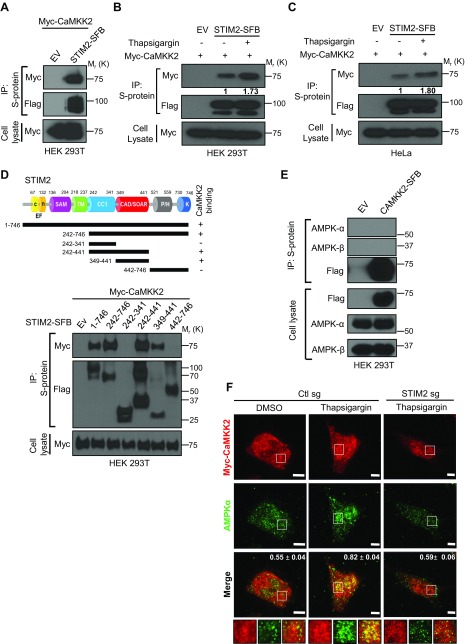
STIM2 interacts with CaMKK2 and regulates CaMKK2 colocalization with AMPK
The aforementioned data prompted us to further study the potential mechanism by which STIM2 regulates calcium-mediated AMPK activation. Because CaMKK2 functions as the main upstream kinase for calcium-induced AMPK phosphorylation (19, 21), we analyzed the potential interaction of STIM2 with CaMKK2. Our results showed that ectopically expressed STIM2 interacted with CaMKK2 (Fig. 6A). Similar to our observation with calcium-stimulated STIM2 interaction with AMPK (Fig. 5A, B), treatment of cells with thapsigargin also increased STIM2 interaction with CaMKK2 in both HEK 293T and HeLa cells (Fig. 6B, C). To map the regions of STIM2 responsible for its interaction with CaMKK2, we cotransfected various truncation mutants of STIM2 with CaMKK2 into cells and examined their binding with CaMKK2. Coimmunoprecipitation analysis revealed that STIM2 interacted with CaMKK2 through its CAD/SOAR domain spanning residues 349–441 (Fig. 6D). The CaMKK2-binding region in STIM2 is different from and adjacent to its AMPK-interacting region (residues 442–746; Fig. 2A, B), indicating that STIM2 might act as a scaffolding protein for AMPK and CaMKK2. To test this hypothesis, we examined whether STIM2 deficiency affected AMPK interaction with CaMKK2. Consistent with a previous report (46), we failed to detect an interaction between CaMKK2 and AMPK by coimmunoprecipitation analysis (Fig. 6E), likely due to a weak and/or transient interaction between CaMKK2 and AMPK, which is common for interactions between kinases and substrates. As more indirect evidence, our analysis by immunofluorescence microscopy showed that CaMKK2 partially colocalized with AMPK upon thapsigargin treatment, and the CaMKK2–AMPK colocalization was significantly attenuated in STIM2 KO cells (Fig. 6F). In summary, our results showed that STIM2 also interacts with CaMKK2 and suggest that STIM2 might serve as a scaffolding protein to promote CaMKK2 interaction with and phosphorylation of AMPK in response to calcium stimulation.
Figure 6 .
STIM2 recruits CaMKK2 to regulate calcium-induced AMPK activation. A) STIM2 interacts with CaMKK2. HEK 293T cells were cotransfected with STIM2-SFB [or empty vector (EV)] and myc-CaMKK2. STIM2-SFB was immunoprecipitated using S-protein beads, and coprecipitated myc-CaMKK2 was detected after immunoblotting using anti-myc antibody. B, C) Cytosolic calcium influx promotes STIM2 interaction with CaMKK2. HEK 293T (B) and HeLa (C) cells stably expressing STIM2-SFB alone were transiently transfected with myc-CaMKK2 and then incubated with medium containing thapsigargin (1 μM) for 30 min. Cell lysates were subjected to immunoprecipitation using S-protein beads. Coprecipitates were analyzed after Western blotting with anti-myc antibody. D) STIM2 interacts with CaMKK2 through its CAD/SOAR region. Upper panel: Schematic representation of STIM2 full-length and deletion mutants used in this study. Lower panel: Protein extracts of HEK 293T cells expressing full-length and truncated mutants of STIM2-SFB along with myc-CaMKK2 were immunoprecipitated with S-protein beads and analyzed for coprecipitated myc-CaMKK2 using immunoblotting with anti-myc antibody. E) AMPK does not form a stable complex with CaMKK2. Protein extracts of HEK 293T cells expressing full-length CaMKK2-SFB were immunoprecipitated with S-protein beads and analyzed for coprecipitated proteins using indicated antibodies. F) Calcium-induced colocalization between AMPK and CaMKK2 depends on STIM2. Control and STIM2-depleted HeLa cells were transiently transfected with myc-CaMKK2, incubated with medium containing thapsigargin (1 μM) for 30 min, and processed for immunofluorescence imaging. Rabbit anti-myc antibody, goat anti-rabbit IgG Alexa Fluor 633 secondary antibody for myc (red), and mouse anti–AMPK-α goat anti-mouse IgG Alexa Fluor 488 secondary for AMPK-α (green) were used. Representative images from each condition are presented. Overlap coefficients for the AMPK and CaMKK2 signal are presented in the upper right corner of the merge panels (n = 19 for Ctl sg + DMSO, n = 22 for Ctl sg + thapsigargin, and n = 18 for STIM2 sg + thapsigargin). In all images, insets display the selected area along with the overlays. Scale bars, 5 μm. P < 0.0001.
DISCUSSION
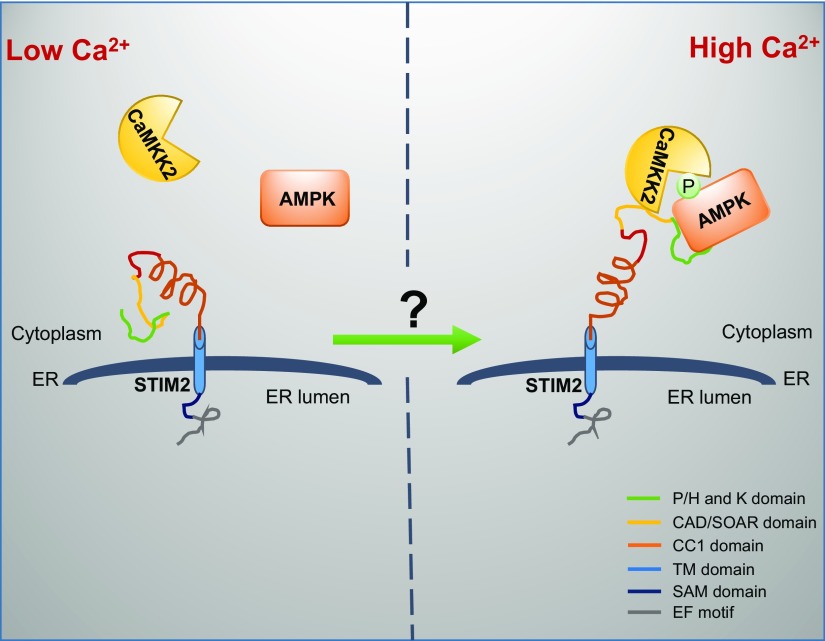
Regulation of AMPK activation is central to the maintenance of cellular homeostasis. AMPK activation by alterations of intracellular energy or calcium levels is mediated through distinctive mechanisms: LKB1 mediates energy stress–induced AMPK activation, whereas CaMKK2 mainly mediates calcium-induced AMPK activation. Our study identifies STIM2 as a regulator of calcium-induced, but not energy stress–induced, AMPK activation and suggests that the increase in intracellular calcium levels promotes STIM2 interaction with both AMPK and CaMKK2 and that STIM2 functions as a scaffolding protein to tether CaMKK2 to phosphorylate AMPK and thus promotes calcium-induced AMPK activation (Fig. 7). This model is supported by several lines of evidence. The rise in intracellular calcium level, either through depletion of ER calcium levels (via thapsigargin treatment) or influx from extracellular space (via ionomycin treatment), promotes the interaction between STIM2 and AMPK (as well as CaMKK2) (Figs.5A, B and 6B, C); AMPK- and CaMKK2-interacting regions in STIM2 are adjacent to each other, raising the possibility that STIM2 can tether CaMKK2 to interact with AMPK (Figs. 2A, B and 6D); and CaMKK2 colocalization with AMPK and calcium-induced AMPK activation is significantly compromised in STIM2-deficient cells (Figs. 5C–F and 6F). Our model is thus analogous to another model wherein energy stress promotes AXIN–LKB1–AMPK complex formation and the scaffolding protein AXIN tethers LKB1 to phosphorylate AMPK in response to energy stress (14).
Figure 7 .
The working model of STIM2 regulation of calcium-stimulated AMPK activation.
The underlying mechanisms by which intracellular calcium regulates STIM2 interaction with AMPK (or CaMKK2) remain elusive. STIM proteins contain a luminal EF-hand motif that can associate with calcium in ER. It has been proposed that, after depletion of ER calcium stores, the dissociation of calcium from the luminal EF-hand motif in STIM triggers STIM conformational changes to a more extended state, promoting its oligomerization and interaction with plasma membrane–localized ORAI1, thereby opening ORAI1 for calcium entry (26). STIM2 is a more sensitive calcium sensor but is a much weaker activator of SOCE than STIM1. It has been shown that phenylalanine residue (Phe-394) of STIM1 is crucial for its interaction with ORAI; however, this residue is replaced by leucine in STIM2, resulting in much reduced affinity for STIM2 to interact with ORAI1 (47). Our study raises an intriguing hypothesis that the release of calcium from the luminal EF-hand motif in STIM2 might cause a conformational change of STIM2 and expose the AMPK- and CaMKK2-interacting sites located in the cytosolic region of STIM2, resulting in STIM2 interaction with AMPK and CaMKK2 and subsequent CaMKK2-mediated AMPK phosphorylation (Fig. 7). This hypothesis awaits further investigation in future studies.
Our study has convincingly showed the interaction between STIM2 and AMPK; however, which subunit of the AMPK complex mediates STIM2 interaction with AMPK remains unclear. Our coimmunoprecipitation analysis showed that STIM2 interacted with all 3 ectopically expressed AMPK subunits (Fig. 1D–F). Thus, it is likely that STIM2 can directly interact with all 3 subunits of AMPK. However, because the cell lines used in our experiments (HEK 293T or HeLa cells) express endogenous AMPK-α, -β, and -γ subunits, it is possible that STIM2 interaction with some AMPK subunits might be indirect and mediated through another AMPK subunit or a third partner. Future studies will be directed to further characterize the interaction between STIM2 and AMPK.
ACKNOWLEDGMENTS
The authors thank all members of the B.G. laboratory for their advice and technical assistance. This research was supported by the Andrew Sabin Family Fellow Award, the Sister Institution Network Fund, the Anna Fuller Fund, U.S. National Institutes of Health (NIH) National Cancer Institute Grant CA181196 (to B.G.), NIH National Institute of General Medical Sciences Grant R01GM112003, Welch Foundation Grant BE-1913, American Cancer Society Grant RSG-16-215-01 TBE (to Y.Z.), and NIH Cancer Center Support Grant P30CA016672 to The University of Texas M. D. Anderson Cancer Center. The authors declare no conflicts of interest.
Glossary
- ACC
acetylCoA carboxylase
- CaMKK2
calcium/calmodulin-dependent protein kinase 2
- Cas9
clustered regularly interspaced short palindromic repeat–associated protein 9
- CRISPR
clustered regularly interspaced short palindromic repeat
- ER
endoplasmic reticulum
- HEK
human embryonic kidney
- KO
knockout
- LKB1
liver kinase B1
- mTORC1
mechanistic target of rapamycin complex 1
- NBR2
neighbor of BRCA1 gene 2
- P/H
Pro/His-rich
- PI
propidium iodide
- SFB
streptavidin binding peptide
- sgRNA
single guide RNA
- SOCE
store-operated calcium entry
- STIM
stromal interaction molecule
- TSC
tuberous sclerosis complex
- YFP
yellow fluorescent protein
AUTHOR CONTRIBUTIONS
X. Liu performed the experiments shown in Figs. 1C, H, 2C–F, 5E, and 6E; J. Jing performed experiments shown in Fig. 5G, H and generated AMPK-α–mCherry construct; A. S. Chauhan performed most of the other experiments with assistance from H. Lee and R. K. Yadav; J. Liu generated the STIM1 KO cells used in Fig. 1G; B. Gan and Y. Zhou supervised the study; A. S. Chauhan, Y. Zhou, and B. Gan designed the experiments and wrote the manuscript; and all authors commented on the manuscript.
REFERENCES
- 1.Hardie, D. G., Schaffer, B. E., Brunet, A. (2016) AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 26, 190–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mihaylova, M. M., Shaw, R. J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross, F. A., MacKintosh, C., Hardie, D. G. (2016) AMP-activated protein kinase: a cellular energy sensor that comes in 12 flavours. FEBS J. 283, 2987–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin, S. C., Hardie, D. G. (2018) AMPK: sensing glucose as well as cellular energy status. Cell Metab. 27, 299–313 [DOI] [PubMed] [Google Scholar]
- 5.Ross, F. A., Jensen, T. E., Hardie, D. G. (2016) Differential regulation by AMP and ADP of AMPK complexes containing different γ subunit isoforms. Biochem. J. 473, 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao, B., Sanders, M. J., Underwood, E., Heath, R., Mayer, F. V., Carmena, D., Jing, C., Walker, P. A., Eccleston, J. F., Haire, L. F., Saiu, P., Howell, S. A., Aasland, R., Martin, S. R., Carling, D., Gamblin, S. J. (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472, 230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw, R. J., Kosmatka, M., Bardeesy, N., Hurley, R. L., Witters, L. A., DePinho, R. A., Cantley, L. C. (2004) The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101, 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods, A., Johnstone, S. R., Dickerson, K., Leiper, F. C., Fryer, L. G., Neumann, D., Schlattner, U., Wallimann, T., Carlson, M., Carling, D. (2003) LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13, 2004–2008 [DOI] [PubMed] [Google Scholar]
- 9.Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Mäkelä, T. P., Alessi, D. R., Hardie, D. G. (2003) Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2, 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, X., Xiao, Z. D., Han, L., Zhang, J., Lee, S. W., Wang, W., Lee, H., Zhuang, L., Chen, J., Lin, H. K., Wang, J., Liang, H., Gan, B. (2016) LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat. Cell Biol. 18, 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, X., Xiao, Z. D., Gan, B. (2016) An lncRNA switch for AMPK activation. Cell Cycle 15, 1948–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao, Z. D., Liu, X., Zhuang, L., Gan, B. (2016) NBR2: a former junk gene emerges as a key player in tumor suppression. Mol. Cell. Oncol. 3, e1187322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, C. S., Jiang, B., Li, M., Zhu, M., Peng, Y., Zhang, Y. L., Wu, Y. Q., Li, T. Y., Liang, Y., Lu, Z., Lian, G., Liu, Q., Guo, H., Yin, Z., Ye, Z., Han, J., Wu, J. W., Yin, H., Lin, S. Y., Lin, S. C. (2014) The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 20, 526–540 [DOI] [PubMed] [Google Scholar]
- 14.Zhang, Y. L., Guo, H., Zhang, C. S., Lin, S. Y., Yin, Z., Peng, Y., Luo, H., Shi, Y., Lian, G., Zhang, C., Li, M., Ye, Z., Ye, J., Han, J., Li, P., Wu, J. W., Lin, S. C. (2013) AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab. 18, 546–555 [DOI] [PubMed] [Google Scholar]
- 15.Gwinn, D. M., Shackelford, D. B., Egan, D. F., Mihaylova, M. M., Mery, A., Vasquez, D. S., Turk, B. E., Shaw, R. J. (2008) AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoki, K., Zhu, T., Guan, K. L. (2003) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 17.Carling, D. (2017) AMPK signalling in health and disease. Curr. Opin. Cell Biol. 45, 31–37 [DOI] [PubMed] [Google Scholar]
- 18.Huang, J., Manning, B. D. (2008) The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 412, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawley, S. A., Pan, D. A., Mustard, K. J., Ross, L., Bain, J., Edelman, A. M., Frenguelli, B. G., Hardie, D. G. (2005) Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 20.Hurley, R. L., Anderson, K. A., Franzone, J. M., Kemp, B. E., Means, A. R., Witters, L. A. (2005) The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J. Biol. Chem. 280, 29060–29066 [DOI] [PubMed] [Google Scholar]
- 21.Woods, A., Dickerson, K., Heath, R., Hong, S. P., Momcilovic, M., Johnstone, S. R., Carlson, M., Carling, D. (2005) Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2, 21–33 [DOI] [PubMed] [Google Scholar]
- 22.Racioppi, L., Means, A. R. (2012) Calcium/calmodulin-dependent protein kinase kinase 2: roles in signaling and pathophysiology. J. Biol. Chem. 287, 31658–31665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clapham, D. E. (2007) Calcium signaling. Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 24.Gudlur, A., Zhou, Y., Hogan, P. G. (2013) STIM-ORAI interactions that control the CRAC channel. Curr. Top. Membr. 71, 33–58 [DOI] [PubMed] [Google Scholar]
- 25.Hogan, P. G., Rao, A. (2015) Store-operated calcium entry: mechanisms and modulation. Biochem. Biophys. Res. Commun. 460, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soboloff, J., Rothberg, B. S., Madesh, M., Gill, D. L. (2012) STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 13, 549–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wedel, B., Boyles, R. R., Putney, J. W., Jr., Bird, G. S. (2007) Role of the store-operated calcium entry proteins Stim1 and Orai1 in muscarinic cholinergic receptor-stimulated calcium oscillations in human embryonic kidney cells. J. Physiol. 579, 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandman, O., Liou, J., Park, W. S., Meyer, T. (2007) STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell 131, 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soboloff, J., Spassova, M. A., Hewavitharana, T., He, L. P., Xu, W., Johnstone, L. S., Dziadek, M. A., Gill, D. L. (2006) STIM2 is an inhibitor of STIM1-mediated store-operated Ca2+ Entry. Curr. Biol. 16, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 30.Roos, J., DiGregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., Safrina, O., Kozak, J. A., Wagner, S. L., Cahalan, M. D., Veliçelebi, G., Stauderman, K. A. (2005) STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liou, J., Kim, M. L., Heo, W. D., Jones, J. T., Myers, J. W., Ferrell, J. E., Jr., Meyer, T. (2005) STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird, G. S., Hwang, S. Y., Smyth, J. T., Fukushima, M., Boyles, R. R., Putney, J. W., Jr. (2009) STIM1 is a calcium sensor specialized for digital signaling. Curr. Biol. 19, 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, Z. D., Han, L., Lee, H., Zhuang, L., Zhang, Y., Baddour, J., Nagrath, D., Wood, C. G., Gu, J., Wu, X., Liang, H., Gan, B. (2017) Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 8, 783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koppula, P., Zhang, Y., Shi, J., Li, W., Gan, B. (2017) The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 292, 14240–14249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W., Li, N., Li, X., Tran, M. K., Han, X., Chen, J. (2015) Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Rep. 13, 524–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin, A., Piao, H. L., Zhuang, L., Sarbassov, D. D., Ma, L., Gan, B. (2014) FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacological inhibition of the PI3K-AKT pathway. Cancer Res. 74, 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai, F., Lee, H., Zhang, Y., Zhuang, L., Yao, H., Xi, Y., Xiao, Z. D., You, M. J., Li, W., Su, X., Gan, B. (2017) BAP1 inhibits the ER stress gene regulatory network and modulates metabolic stress response. Proc. Natl. Acad. Sci. USA 114, 3192–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, A., Yao, J., Zhuang, L., Wang, D., Han, J., Lam, E. W., Gan, B. (2014) The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene 33, 3183–3194; erratum: 5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing, J., He, L., Sun, A., Quintana, A., Ding, Y., Ma, G., Tan, P., Liang, X., Zheng, X., Chen, L., Shi, X., Zhang, S. L., Zhong, L., Huang, Y., Dong, M. Q., Walker, C. L., Hogan, P. G., Wang, Y., Zhou, Y. (2015) Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol. 17, 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, Y., Ramachandran, S., Oh-Hora, M., Rao, A., Hogan, P. G. (2010) Pore architecture of the ORAI1 store-operated calcium channel. Proc. Natl. Acad. Sci. USA 107, 4896–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttlin, E. L., Bruckner, R. J., Paulo, J. A., Cannon, J. R., Ting, L., Baltier, K., Colby, G., Gebreab, F., Gygi, M. P., Parzen, H., Szpyt, J., Tam, S., Zarraga, G., Pontano-Vaites, L., Swarup, S., White, A. E., Schweppe, D. K., Rad, R., Erickson, B. K., Obar, R. A., Guruharsha, K. G., Li, K., Artavanis-Tsakonas, S., Gygi, S. P., Harper, J. W. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, R. T., Manji, S. S., Parker, N. J., Hancock, M. S., Van Stekelenburg, L., Eid, J. P., Senior, P. V., Kazenwadel, J. S., Shandala, T., Saint, R., Smith, P. J., Dziadek, M. A. (2001) Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem. J. 357, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prakriya, M., Lewis, R. S. (2015) Store-operated calcium channels. Physiol. Rev. 95, 1383–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosado, J. A., Diez, R., Smani, T., Jardín, I. (2016) STIM and Orai1 variants in store-operated calcium entry. Front. Pharmacol. 6, 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fogarty, S., Ross, F. A., Vara Ciruelos, D., Gray, A., Gowans, G. J., Hardie, D. G. (2016) AMPK causes cell cycle arrest in LKB1-deficient cells via activation of CAMKK2. Mol. Cancer Res. 14, 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fogarty, S., Hawley, S. A., Green, K. A., Saner, N., Mustard, K. J., Hardie, D. G. (2010) Calmodulin-dependent protein kinase kinase-beta activates AMPK without forming a stable complex: synergistic effects of Ca2+ and AMP. Biochem. J. 426, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X., Wang, Y., Zhou, Y., Hendron, E., Mancarella, S., Andrake, M. D., Rothberg, B. S., Soboloff, J., Gill, D. L. (2014) Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nat. Commun. 5, 3183 [DOI] [PMC free article] [PubMed] [Google Scholar]