Abstract
Different bacteria and fungi live as commensal organisms as part of the human microbiota, but shifts to a pathogenic state potentially leading to septic infections commonly occur in immunocompromised individuals. Several studies have reported synergistic or antagonistic interactions between individual bacteria and fungi which might be of clinical relevance. Here, we present first evidence for the interaction between Klebsiella pneumoniae and several Aspergillus species including A. fumigatus, A. terreus, A. niger and A. flavus which cohabit in the lungs and the intestines. Microbiological and molecular methods were employed to investigate the interaction in vitro, and the results indicate that Klebsiella pneumoniae is able to prevent Aspergillus spp. spore germination and hyphal development. The inhibitory effect is reversible, as demonstrated by growth recovery of Aspergillus spp. upon inhibition or elimination of the bacteria, and is apparently dependent on the physical interaction with metabolically active bacteria. Molecular analysis of Klebsiella-Aspergillus interaction has shown upregulation of Aspergillus cell wall-related genes and downregulation of hyphae-related genes, suggesting that Klebsiella induces cell wall stress response mechanisms and suppresses filamentous growth. Characterization of polymicrobial interactions may provide the basis for improved clinical management of mixed infections by setting the stage for appropriate diagnostics and ultimately for optimized treatment strategies.
Introduction
Microbial interactions are part of the highly complex human microbiome. Mapping of the human microbiome has shown a wide diversity of bacteria and fungi occupying specific niches1–4. The interplay between various microorganisms and their interactions with the host and the immune system may display beneficial or harmful effects. Importantly, changes induced by alterations including, for example, underlying diseases, the use of antibiotics, anti-cancer chemotherapy, or dietary changes have an enormous impact on microbial populations1,3,5–11. More recently, polymicrobial infections and bacterial-fungal interactions (BFI) have attracted greater attention. Several such interactions involving particularly Candida (C.) albicans have been reported12–20. The well-studied interaction between C. albicans and Pseudomonas (P.) aeruginosa has shown that synergistic and antagonistic effects can occur simultaneously, and the net results of the interactions can vary depending on external influences and the dominant intermicrobial dynamics12–14,21–24. A number of other bacteria were shown to interact with C. albicans, including Streptococcus spp., Lactobacillus spp., Staphylococcus (S.) aureus, Enterococus faecalis, and Escherichia coli12,15–17,19,20,25–27. P. aeruginosa has the capacity to inhibit the growth of various fungi, such as Aspergillus (A.) fumigatus and Cryptococcus spp28–34. These interactions occur through the production of quorum sensing molecules and virulence factors by P. aeruginosa including e.g. phenazines, decanol and 3-oxo-C12-homoserine lactone (3OC12HSL), which affect biofilm formation, inhibit yeast (Cryptococcus spp.) and hyphal development (C. albicans, A. fumigatus) through the generation of highly toxic reactive oxygen species (ROS)12–14,21–24,28–34. Adherence of P. aeruginosa and S. aureus to the hyphal form of C. albicans is thirty times higher compared to the yeast form16. The indicated interactions between bacteria and fungi occur when these pathogens share the same niches. Moreover, co-localization of A. fumigatus and P. aeruginosa in the lungs of patients with cystic fibrosis was associated with poorer outcomes when compared to single infections with these pathogens35,36. The same effect was observed in various studies reporting on the interaction of C. albicans with P. aeruginosa, which resulted in elevated mortality rates14,21,37, in line with other reports on polymicrobial infections16. Bacteria and fungi often live in highly organized structures termed biofilms rather than in planktonic state. Biofilms pose a higher risk for the development of serious infections because they often display greater resistance to antimicrobial treatment and to control by the immune system38–43. The biofilm structure protects against exogenous stresses including drug treatment, and provides a microenvironment facilitating nutrition and quorum sensing communication. Such features improve the fitness and resilience of biofilm structures providing a survival advantage in the host43–51. In our studies, we aimed to investigate the interactions between select opportunistic fungal pathogens including different Aspergillus species and the clinically important bacterium Klebsiella pneumoniae. Both species cohabit in various regions of the body including particularly the gut and the lungs1,2,52,53. Development of Aspergillus species encompasses the formation of spores or conidia and hyphal filaments. Under favorable environmental conditions, spores germinate into long hyphae which are responsible for tissue invasion and escape from the immune system54–57. Aspergillus species have the capacity to form biofilms in which hyphae play a leading role. The architecture of the lungs in conjunction with hyphal development of Aspergilli makes it difficult for the immune system to clear the fungal pathogens57–60. Next to C. albicans, A. fumigatus is the most prevalent cause of fungal infection in immunocompromised patients56,61,62, and is responsible for approximately 90% of invasive aspergilloses, based on clinical reports63,64. More recently, however, shifts from A. fumigatus to non-fumigatus Aspergillus species have been observed, involving particularly A. terreus, A. niger and A. flavus65–69. On the bacterial side, K. pneumoniae is an emerging pathogen displaying resistance to antibiotic treatment which has been associated with several nosocomial outbreaks53,70–83. The antimicrobial resistance and virulence of numerous clinical strains of K. pneumoniae have been associated with the presence of plasmids carrying resistance genes, the hypermucoviscosity phenotype, capsular polysaccharides and the capacity to form biofilms75,84–90. Bacteria and fungi are constantly exposed to stress conferred particularly by the host microenvironment and other pathogens. The cell wall is the first point of contact between microorganisms and the host or other pathogens. The cell wall plays an important role in mediating interactions with the external environment relevant for nutrient diffusion and molecule-based signalling. Simultaneously, it protects the cells from oxidative or osmotic stresses, and modulates the response to antimicrobial drugs91–95. Fungi are capable of adapting their cell walls in response to stress by activating multiple mechanisms directed towards repair or compensation for cell wall damage. In response to stress, C. albicans was shown to activate the MAPK and Ca2+/Calcineurin pathways, leading to upregulation of genes involved in the cell wall assembly, and Aspergillus species respond to stress in a similar manner96–104. It is important to point out that fungal spores and hyphae display different structures and compositions of the cell wall which can induce differential immune responses by the host105,106.
In the present study, we sought to investigate the interactions between four Aspergillus species including A. fumigatus, A. terreus, A. niger and A. flavus, and different strains of K. pneumoniae with low or high capacity of biofilm formation. Our studies provide new insights into the biological behavior of Aspergillus and K. pneumoniae in co-culture, by unravelling the type of interaction and response to stress. Our observations highlight the importance of identifying the presence of polymicrobial infections and potential interactions between the pathogens with regard to optimized diagnostic approaches and appropriate antimicrobial treatment.
Results
K. pneumoniae inhibits spore germination and hyphal development of Aspergillus species
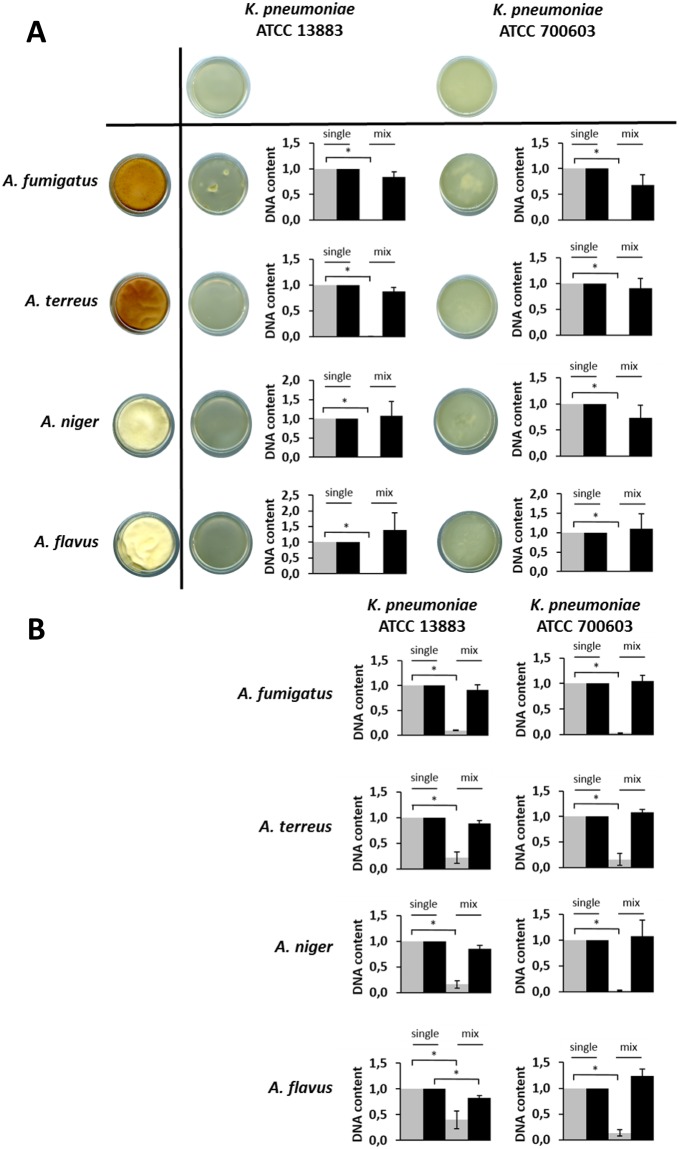
The in vitro interaction between K. pneumoniae and several Aspergillus species, including A. fumigatus, A. terreus, A. niger and A. flavus was characterized. K. pneumoniae and Aspergillus were grown alone and in co-culture for 24 and 48 h, as outlined in the Methods section. The level of inhibition of fungal spore germination was assessed by qPCR at 24 h (Fig. 1) and imaged by confocal microscopy (Fig. 2). The suppression of Aspergillus growth by K. pneumoniae strains is shown in Fig. 1. Our data have shown that, upon contact with K. pneumoniae, Aspergillus spores are not developing into hyphae (Fig. 1A). This is reflected by the unchanged DNA content of Aspergillus in the co-culture which corresponds to the initial fungal cell loads. In addition, this inhibition is independent of the biofilm forming capacity of K. pneumoniae strains, since the same effect was observed when using low-biofilm (ATCC 13883) and high-biofilm (ATCC 700603) forming strains. As shown in Fig. 1A, the inhibitory effect was observed for all Aspergillus species tested, indicating that this effect is Aspergillus species-independent.
Figure 1.
Inhibition of Aspergillus spore germination and hyphal development by K. pneumoniae strains. (A) Fungi and bacteria co-cultured since time zero. Scanner photos of the bottom of culture dishes taken after 48 h of cultivation are shown. (B) Fungal spores germinated for up to 12 h followed by addition of bacteria for a total of 24 h. The DNA content was quantified by qPCR at 24 h using species-specific primers for the conserved rRNA sequences 28S and 16S for fungi and bacteria, respectively. The grey bars in the plots correspond to fungi and the black bars to bacteria. The DNA content of single cultures was set to one, and the DNA content of each species in the co-culture was normalized to the corresponding single culture. Data are presented as mean + SE of three independent experiments.
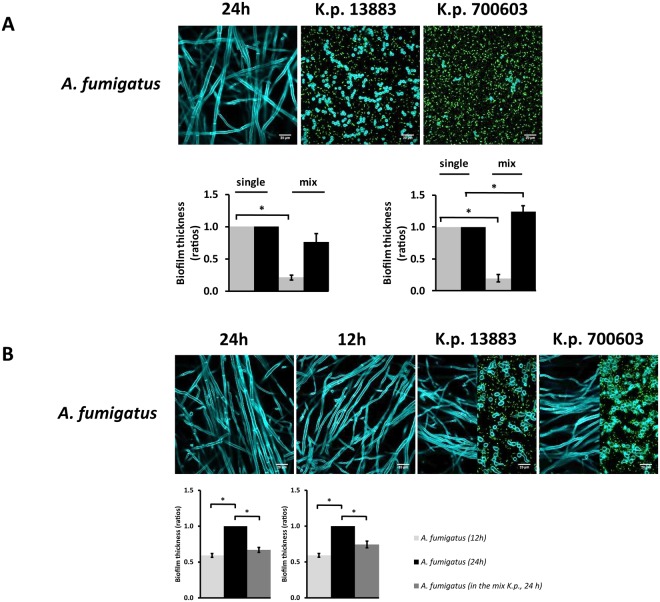
Figure 2.
Biofilm thickness of A. fumigatus vs K. pneumoniae analyzed by confocal microscopy. The fluorescent photos represent A. fumigatus growing alone or in co-culture with different strains of K. pneumoniae. Differential staining of the microorganisms upon growth in culture and measurement of biofilm thickness was performed as indicated in the Methods section. (A) Fungi and bacteria co-cultured since time zero. Grey bars correspond to fungi and black bars correspond to bacteria. (B) Fungal spores germinated for up to 12 h followed by addition of bacteria for a total of 24 h. The confocal microscopy images of A. fumigatus and K. pneumoniae co-cultures (K.p.13883, K.p.700603) show the levels of hyphae (left) and spores (right), respectively. The left plots indicate co-cultures of A. fumigatus with K. pneumoniae ATCC 13883 and the right plots indicate co-cultures of A. fumigatus with K. pneumoniae ATCC 700603. Biofilm thickness of the single cultures was set to one and the biofilm thickness of each species in the co-culture was normalized to the corresponding single culture. Data are presented as mean + SE of three independent experiments.
Due to the functional importance of hyphae for tissue invasion and escape from the immune system, we have addressed the effect of K. pneumoniae on hyphal development of the indicated Aspergillus species. We sought to understand whether K. pneumoniae also has an effect on hyphal development and if the inhibitory effect still occurs at the hyphal stage. The results have shown that, in the presence of K. pneumoniae, Aspergillus species were not able to progress with hyphal development by demonstrating significantly decreased DNA contents compared to Aspergillus in single cultures (Fig. 1B), revealing that the inhibitory effect of K. pneumoniae on Aspergillus species is independent of the fungal growth stage.
Biofilm formation of Aspergillus species is affected by the presence of K. pneumoniae
Both K. pneumoniae and Aspergillus species used in this study are capable of forming biofilms. Based on the observation that K. pneumoniae strains are able to suppress Aspergillus growth and hyphenation, we sought to investigate whether low- and high-biofilm forming strains of K. pneumoniae might also affect the biofilm formation of Aspergillus. A confocal microscopy approach was used to measure the biofilm thickness of bacteria and fungi growing alone and in co-culture. The results have shown that, upon contact with K. pneumoniae strains, the biofilm thickness of A. fumigatus was significantly decreased compared to A. fumigatus growing alone. These observations are supported by the confocal images showing that A. fumigatus in co-culture remained in the form of spores, independently of the K. pneumoniae strain tested (Fig. 2A). In addition, the high-biofilm forming strain of K. pneumoniae showed thicker biofilms when grown in the presence of A. fumigatus (Fig. 2A, right plot). We further investigated whether K. pneumoniae strains could also influence the biofilm formation of pre-formed A. fumigatus biofilms. To address this question, A. fumigatus spores were pre-germinated for 12 h, followed by addition of K. pneumoniae strains. Bacteria and fungi were grown alone for comparison with the co-cultures. A. fumigatus was grown alone for 12 and 24 h. The plots in Fig. 2B show that there was a significant increase in biofilm thickness between 12 and 24 h, indicative of normal fungal growth. Once bacteria were added to the culture at 12 h, we observed that the biofilm thickness of A. fumigatus was significantly decreased in the presence of either bacterial strain. The confocal images showed normal hyphal development at 12 and 24 h. Interestingly, once A. fumigatus was growing in co-culture with the K. pneumoniae strains, both morphologies of A. fumigatus were present, spores and hyphae (Fig. 2B). The images represent different slices of the Z-stacks indicating a level-dependent, predominant presence of hyphae (Fig. 2B, left-half image) or spores (Fig. 2B, right-half image). The density of bacterial cells was greater (green dots in the image) in areas revealing mainly fungal spores, in line with the observation that K. pneumoniae strains inhibit both spore germination and hyphal development.
Inhibition of Aspergillus growth by K. pneumoniae is dependent on direct contact
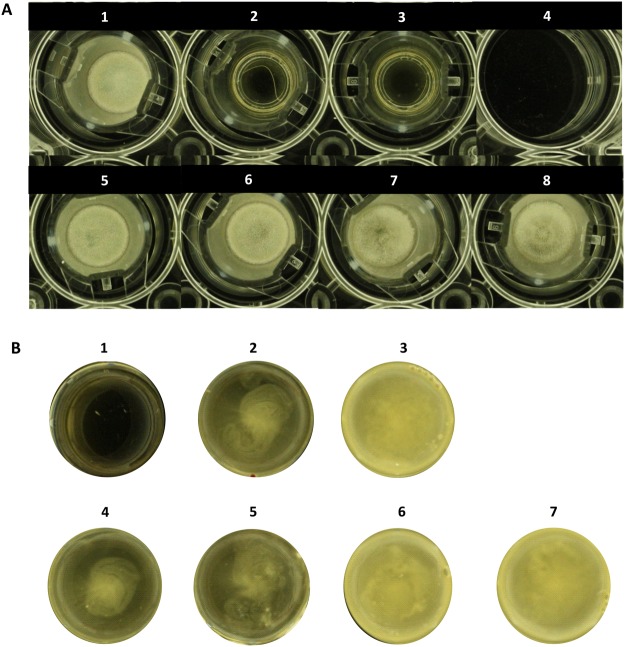
Based on the documented antagonistic effect of K. pneumoniae strains against the development and growth of Aspergillus species, we have addressed the contact-dependence of the interaction. Transwell plates permitting the growth of bacteria and fungi in physically separated compartments but allowing exchange of diffusible molecules through a porous membrane were employed for the analysis. In the wells (lower compartments), K. pneumoniae strains were grown alone or in co-culture with A. fumigatus, whereas in the inserts with the porous membrane (upper compartments), A. fumigatus was grown alone. This strategy allowed us to investigate whether the inhibitory effect on A. fumigatus was mediated via direct contact with K. pneumoniae or by the secretion of molecules conferring independence from physical contact. The results have shown that A. fumigatus was able to grow in the upper compartments physically separated from K. pneumoniae growing in the lower compartments (Fig. 3A), indicating dependence on direct contact for the inhibitory effect.
Figure 3.
Physical interaction between A. fumigatus and K. pneumoniae. The bacteria and fungi were grown in Transwell plates providing physical separation, but permitting interaction by exchange of secreted molecules. (A) Photos of the inserts (upper compartments). 1. Growth of A. fumigatus alone; 2./3. No growth of the K. pneumoniae strains K.p. 13883 and K.p. 700603 which had been seeded solely in the lower compartments (see B); 4. Negative control: YPD; 5. Aspergillus growing in the upper compartment in the presence of K.p. 13883 in the lower compartment; 6. Aspergillus growing in the upper compartment in the presence of a co-culture of A.f. vs K.p. 13883 in the lower compartment; 7. Aspergillus growing in the upper compartment in the presence of K.p. 700603 in the lower compartment; 8. Aspergillus growing in the upper compartment in the presence of a co-culture of A.f. vs K.p. 700603 in the lower compartment. (B) Photos of the wells (lower compartments) reflecting K. pneumoniae grown alone or in combination with A. fumigatus. The fungus seeded in the upper compartment cannot transgress to the lower compartment and vice versa. The wells show presence of bacterial growth only, without any evidence for A. fumigatus growth. 1. A. fumigatus without fungal growth as the fungus was seeded in the upper compartment (see A); 2. Growth of K.p. 13883 alone in the lower compartment; 3. Growth of K.p. 700603 alone in the lower compartment; 4. K.p. 13883 growing in co-culture with A.f. in the lower compartment, without any fungus in the upper compartment; 5. K.p. 13883 growing in co-culture with A.f. in the lower compartment with fungal growth in the upper compartment (see A); 6. K.p. 700603 growing in co-culture with A.f. in the lower compartment, without any fungus in the upper compartment; 7. K.p. 700603 growing in co-culture with A.f. in the lower compartment with fungal growth in the upper compartment (see A).
The lower compartments, where bacteria were grown alone or in co-culture with A. fumigatus, are shown in Fig. 3B. The presence of K. pneumoniae alone or in co-culture in the lower compartment did not result in any inhibitory effect on A. fumigatus in the upper compartment.
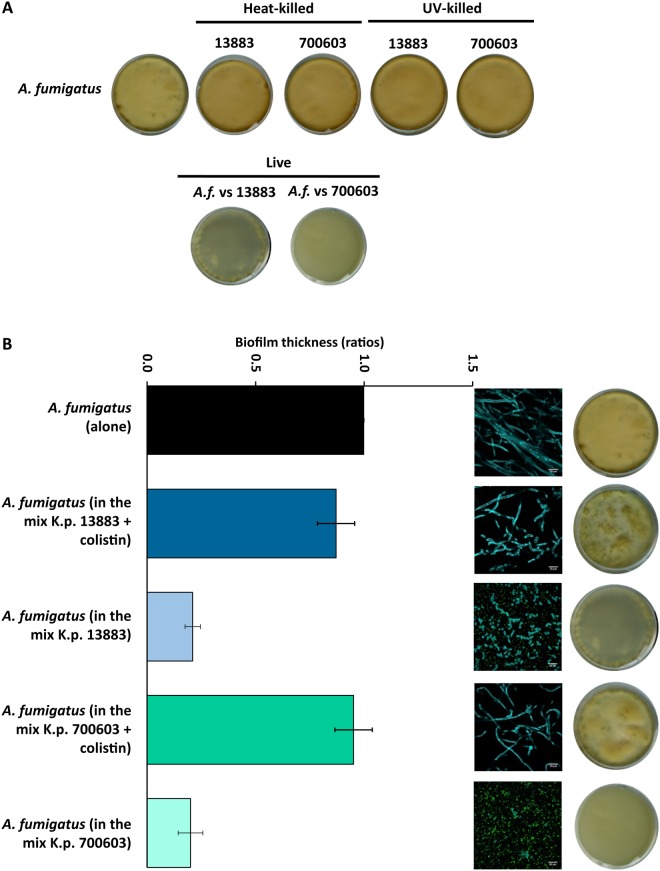
To further assess the putative secretion of inhibitory molecules, we investigated the effect of the bacterial supernatant (SN) on A. fumigatus growth. Supernatants of K. pneumoniae grown alone or in co-culture in biofilm mode were obtained at 12 h and 24 h. Collection of supernatants at different time points was performed to determine if the inhibitory effect is growth phase-dependent, because the secretion of molecules might vary during the bacterial growth cycle. The supernatants were added to cultures of A. fumigatus. After 48 h of incubation, the results have shown that, regardless of the culture conditions or growth phase of the bacteria, various preparations of bacterial SN were unable to inhibit growth of any of the fungal Aspergillus species studied (Fig. S1). By contrast, in the presence of bacteria, either in exponential or stationary growth phase, the inhibitory effect on Aspergillus growth was observed (Fig. S2). By contrast, co-cultures with heat- or UV-killed K. pneumoniae did not show any reduction of fungal growth, demonstrating the requirement of live bacteria for the inhibitory effect (Fig. 5A).
Figure 5.
Growth recovery of A. fumigatus after exposure to heat-killed, UV-killed and antibiotic-treated K. pneumoniae. (A) Scanner photos of the bottom of tissue culture dishes after 48 h of culture revealing fungal growth recovery in the presence of heat- and UV-killed K. pneumoniae strains. Controls are shown for live A. fumigatus vs K. pneumoniae 13883 and 700603, revealing inhibition of fungal growth. (B) The plot shows biofilm thickness of A. fumigatus alone and after exposure to the K. pneumoniae strains tested, upon treatment with colistin, indicating fungal growth recovery in the presence of antibiotic-treated bacteria. Decreased biofilm thickness of A. fumigatus in the presence of non-inhibited K. pneumoniae 13883 or 700603 is also shown as a control experiment. Confocal images and dishes are representative of single and co-cultures upon incubation for 48 h. Scale bars are 20 µm. Data are presented as mean + SE of three independent experiments.
Aspergillus species remain viable upon interaction with K. pneumoniae
In order to assess whether the observed inhibitory effect of K. pneumoniae on the growth and development of various Aspergillus species is sustainable or reversible, the fungi were grown alone or in co-culture with K. pneumoniae. After 24 h growth in biofilm mode, aliquots of the single cultures and the co-cultures were streaked onto YPD agar plates containing the antibiotic kanamycin to prevent bacterial growth. The cultures were evaluated after 48 h of incubation, and confirmed efficient elimination of the bacteria. The co-cultures of K. pneumoniae strains with Aspergillus species revealed recovery of fungal growth upon exposure to kanamycin (Fig. 4), demonstrating that the fungi maintain their viability and ability to re-initiate growth once the inhibitory effect of the bacteria is eliminated. The same effect was observed when using other antibiotics including colistin or tetracycline which target different structures of the bacteria (Fig. S3). Confocal microscopy was used to assess quantitatively the growth recovery of A. fumigatus upon interaction with K. pneumoniae and subsequent treatment with an antibiotic. Co-cultures of A. fumigatus and K. pneumoniae were initiated at time zero, followed by antibiotic treatment with colistin at 6 h for elimination of the bacteria. After 24 h growth in co-culture or alone, measurements of the biofilm thickness of A. fumigatus revealed that, in the antibiotic-treated cultures, A. fumigatus biofilms were similar to A. fumigatus in single culture. The respective confocal microscopy images also showed hyphal development, contributing to the biofilm thickness. In addition, the Petri dishes also showed growth recovery of A. fumigatus in co-culture after antibiotic treatment, similar to cultures of the fungus growing alone (Fig. 5B).
Figure 4.
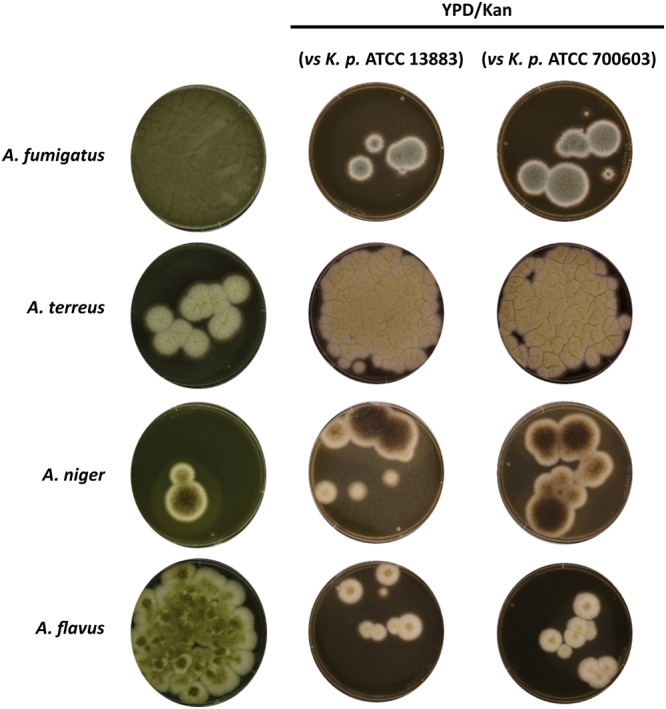
Viability of Aspergillus spp. upon interaction with K. pneumoniae. Aspergillus species grown alone and in co-culture with different K. pneumoniae strains for 24 h were aliquoted onto agar plates containing YPD (controls) and YPD plus the antibiotic kanamycin for selection of fungal growth. Images show fungal growth after incubation for 48 h.
Growth inhibition of Aspergillus species by K. pneumoniae is not affected by nutrient shortage
Nutrient competition for the carbon source is a potential limiting factor for fungal growth during the interaction of K. pneumoniae with Aspergillus spp. To address this notion, the interaction between A. fumigatus and K. pneumoniae (ATCC strain 700603), YPD containing a standard glucose concentration of 2% was compared with YPD containing 4% glucose. The microorganisms were grown alone and in co-culture for 48 h in YPD containing the indicated glucose concentrations. No major differences in fungal growth were observed during the interaction with bacteria (Fig. S4), suggesting that the availability or lack of carbon was not a relevant factor for the antagonistic effect exerted by the bacteria on fungal growth.
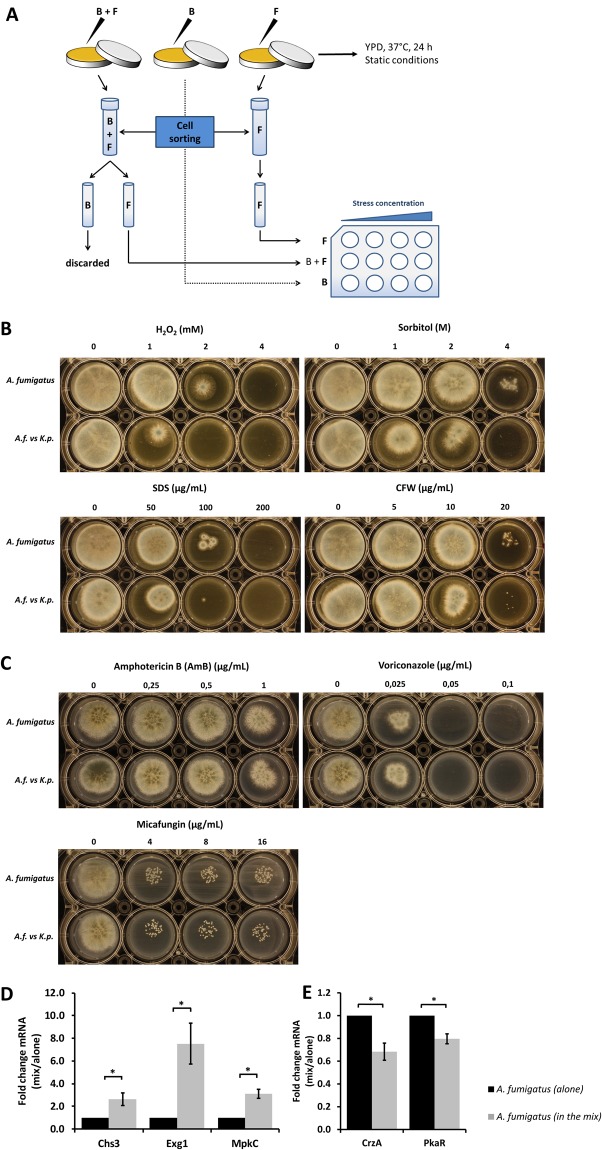
K. pneumoniae renders A. fumigatus sensitive to cell wall stress and induces upregulation of cell wall-related genes
We investigated whether K. pneumoniae could induce cell wall stress in A. fumigatus and render the fungus more or less susceptible to stressors including antifungal drug treatment. After 24 h of growth in single culture or co-culture, A. fumigatus and K. pneumoniae were isolated by flow sorting. Equal numbers of sorted cells were seeded into 12-well agar plates containing a combination of the antibiotics kanamycin and colistin to prevent bacterial growth. Moreover, the plates contained different concentrations of oxidative (H2O2), osmotic (sorbitol, sodium dodecyl sulfate (SDS)) and cell wall stressors (Calcofluor white (CFW)) as well as antifungal drugs (Fig. 6A). The sensitivity to these stresses was compared between A. fumigatus grown alone or upon exposure to K. pneumoniae, and the latter constellation revealed at least two-fold greater sensitivity (Fig. 6B). Additionally, three different classes of antifungal drugs including Amphotericin B, Voriconazole and Micafungin were tested to assess the sensitivity of A. fumigatus grown under the conditions indicated above. However, no differential sensitivity has been observed (Fig. 6C).
Figure 6.
Induction of cell wall stress and upregulation of cell wall-related genes in A. fumigatus mediated by K. pneumoniae. (A) Schematic view of flow sorting of A. fumigatus and K. pneumoniae (ATCC 700603). Sorted cells were plated onto 12-well agar plates containing kanamycin (1000 µg/mL) and colistin (100 µg/mL) for selection of fungal growth and prevention of any residual bacterial contamination. Plates also contained different concentration ranges of cell wall stressors and antifungal drugs to facilitate the observation of the sensitivity of fungi previously grown alone or exposed to K. pneumoniae. (B) Sensitivity of A. fumigatus to oxidative (H2O2), osmotic (sorbitol, SDS) and cell wall (CFW) stresses, with and without previous exposure to K. pneumoniae. (C) Sensitivity of A. fumigatus to antifungal drugs (amphothericin B, voriconazole, micafungin) with and without previous exposure to K. pneumoniae. (D) Upregulation of A. fumigatus cell wall-related genes in the presence of K. pneumoniae. (E) Downregulation of hyphae-related genes of A. fumigatus in the presence of K. pneumoniae determined by quantification of mRNA expression of hyphae-related genes of A. fumigatus upon exposure to K. pneumoniae (ATCC 700603). Fungal spores were germinated for up to 12 h followed by addition of bacteria for a total of 24 h. Transcript levels of the A. fumigatus single culture were set to one, and the transcript levels in the co-culture were normalized to the single culture. Additionally, normalization was also performed against the reference gene TUBA. Data are presented as mean + SE of four independent experiments.
To address the genetic basis of stress response of A. fumigatus, expression analysis of cell wall-related genes was performed at 24 hours after adding K. pneumoniae to A. fumigatus cultures pre-germinated for 12 h. Transcript levels of three genes were quantified by qPCR including CHS3, a regulator of chitin synthase expression, EXG1, a conveyer of glucan β-glucosidase activity, and MPKC, a putative mitogen-activated protein kinase (MAPK) involved in oxidative stress response. Normalization was performed against the reference gene TUBA107–109 and A. fumigatus grown alone. Significant upregulation of the A. fumigatus genes CHS3, EXG1 and MPKC was documented upon exposure to K. pneumoniae (Fig. 6D). Conversely, significant downregulation of genes associated with hyphal development such as CRZA and PKAR was identified (Fig. 6E). CRZA is a transcription factor in the calcineurin pathway, regulating conidial germination, hyphal growth and virulence110–113, and PKAR is a protein kinase in the cyclic adenosine monophosphate (cAMP) pathway also regulating hyphal formation114,115.
Discussion
We have demonstrated that the interaction between different strains of K. pneumoniae and various Aspergillus species in vitro mediates inhibition of fungal spore germination and hyphal growth. In addition, K. pneumoniae was also able to impair biofilm formation of Aspergillus species. This effect was shown to require the presence of live, actively growing bacteria and to be dependent on physical contact between the microorganisms. The inhibitory effect was reversible, as revealed by the resumption of fungal growth upon elimination of the bacteria by antibiotic treatment. During the interaction with K. pneumoniae, Aspergillus adopted a dormant or standby mode, during which the genes regulating hyphal development were downregulated, conferring low energy supply requirement for survival. This notion is in accordance with the observation that the effects of the interaction were apparently independent of more or less abundant carbon source, suggesting that competition for nutrients may not be a major factor in the inhibition of fungal growth.
Moreover, our data suggest that K. pneumoniae confers stress to the fungal cells resulting in the upregulation of protective mechanisms which involve remodelling and reinforcement of the fungal cell wall. This effect was demonstrated by increased sensitivity to stress induced by agents conferring osmotic, oxidative or cell wall-targeted challenges. In response to the stress effects, Aspergillus showed elevated expression of the CHS3, EXG1 and MPKC genes, which are involved in cell wall remodelling and oxidative stress response. This observation is in line with earlier studies indicating that, upon exposure to stress, fungi initiate a response based on activating cell wall salvage pathways that compensate for cell wall damage as a rescue mechanism96–100 including, for example, an increase in transcript abundance of MPKC in response to oxidative stress102–104,116–118. Since the formation of biofilms can per se lead to resistance against antimicrobial treatment owing to the decreased physical accessibility of the pathogens by drugs89,119–121, additional effects of microbial interactions within mixed biofilms affecting the protective mechanisms of individual pathogens may further enhance the resistance to therapy. The understanding of processes occurring during bacterial-fungal interactions (BFI) is therefore essential for the development of appropriate diagnostic and therapeutic approaches. The inhibitory or stimulating effect of BFI mediated by secreted molecules has been demonstrated for different constellations involving molecular cross-talk via production of quorum sensing molecules12–16,18–20,25,26,122. Earlier observations of BFI involving K. pneumoniae and C. albicans revealed a similar antagonistic effect mediated by the bacteria, but the mechanisms of the interaction have remained unclear123. By contrast, an example for synergistic effects is a report on the interaction between Klebsiella aerogenes and Cryptococcus neoformans, which revealed the induction of melanin production in the fungus mediated by bacterial secretion of dopamine, leading to enhanced protection of Cryptococcus from macrophages12. K. pneumoniae was also found to increase spore germination and hyphal growth of Glomus deserticola, a vesicular-arbuscular mycorrhizal fungus, and the authors suggested that K. pneumoniae may produce a diffusible compound resulting in hyphal extension124.
It is important to point out that the observations of BFI in vitro may not necessarily be reflected by identical implications in vivo, as demonstrated for the interaction between C. albicans and P. aeruginosa which were shown to display inhibitory effects in vitro13,32,125,126 but synergistic effects in mouse and zebrafish in vivo models127,128. Co-infections affecting the lungs of patients with cystic fibrosis resulted in worse clinical outcome in comparison to infections with each of the pathogens individually14,21,35–37.
Although this observation may not necessarily indicate discrepant in vitro and in vivo effects of the BFI, it highlights the need to verify observations made in vitro by exploiting models reflecting the complexity encountered in the human host organism. Such analyses may involve the use of organoids generated from primary biopsy materials or animal models. We are currently generating intestinal organoids derived from colon and jejunum/ileum biopsies from patients undergoing diagnostic endoscopy in order to facilitate studies of BFI in a system mimicking the human bowel. Moreover, we are planning to study the interactions in a zebrafish model which permits analyses in the presence of an immune system closely resembling the situation in the human host129,130. Once confirmed in additional model systems, the observations presented may have important clinical implications. If, in the presence of co-localized infections by K. pneumoniae and various Aspergillus species, treatment with antibiotics could indeed unleash fungal growth, it may lead to rapid expansion of Aspergillus requiring timely therapeutic intervention to prevent severe and potentially life-threatening disease in immunocompromised hosts. The identification of metabolites, such as specific quorum sensing molecules released during BFI, could be exploited for clinical diagnosis of these co-infections, and provide a basis for appropriate treatment measures. Proteomic, metabolomic and genetic analyses of the interaction between K. pneumoniae and Aspergilli are currently ongoing, and will expectedly reveal candidate molecules for future diagnostic exploitation, with the ultimate aim to improve the control and outcome of polymicrobial infections, particularly in the immunocompromised patient setting.
Materials and Methods
Strains and growth conditions
Aspergillus and Klebsiella pneumoniae strains used in this study are listed in Table 1. Two different strains of K. pneumoniae were used, a low-biofilm (ATCC 13883) and a high-biofilm (ATCC 700603) forming strain75. Aspergillus strains were maintained in Malt Extract agar (MEA, Sigma) plates for 3–4 days at 37 °C. Spores were collected into 1x PBS + 0.1% Tween 20, following filtration by using a 40 μm cell strainer. Spores were stored at 4 °C for up to one week for subsequent use in the experiments. K. pneumoniae strains were maintained in Luria-Bertani (LB) agar plates and grown overnight in liquid LB medium at 37 °C with agitation at 180 rpm. Equal cell numbers of Aspergillus spp. and K. pneumoniae (106 cells/mL) were used in co-culture experiments. For qualitative assessment of bacterial-fungal interaction, Aspergillus spp. and K. pneumoniae were grown alone and in co-culture in sterile-filtered yeast extract peptone dextrose (YPD) medium (Formedium, Norfolk, UK) at 37 °C. The microorganisms were grown as biofilms, with static incubation in 35 × 10 mm tissue culture dishes (CytoOne, Starlab GmbH, Ahrensburg, Germany). For co-culture experiments, fungi and bacteria were either mixed at time zero or K. pneumoniae strains were added after pre-germination of Aspergillus spores, permitting the formation of hyphae for up to 12 hours (h). For analysis by confocal microscopy, the same growth procedure was performed using IBIDI plates (µ-dish, 35 mm high, ibiTreat, 35 mm). Growth phase tests of K. pneumoniae included overnight growth for 16 h (stationary phase). For exponential phase, K. pneumoniae was grown overnight, followed by growth in fresh medium for 3 additional h.
Table 1.
Microbial strains and primers used for PCR analysis.
| Designation | ||
|---|---|---|
| Strains | Species | Code |
| A. fumigatus | ATCC 204305 | |
| A. terreus | ATCC 1012 | |
| A. niger | DSM 1959 | |
| A. flavus | CM 5095 | |
| K. pneumoniae | ATCC 13883 | |
| ATCC 700603 | ||
| Primers | Aspergillus | Sequence (5′ to 3′) |
| 28S_Fw | GTTGTTTGGGAATGCAGCTCTA | |
| 28S_Rv | TCTCCGGCCAGTATTTAGCTTT | |
| Chs3_Fw | TAGCCAGAACAACTCCTCCC | |
| Chs3_Rv | TGAGTGCGACCTTAGAATTACGA | |
| Exg1_Fw | AGATTACTACAACCAGATTGCGG | |
| Exg1_Rv | GTATCCATGACCACATCCTCAC | |
| MpkC_Fw | CCACCTCATCACAAACATCCT | |
| MpkC_Rv | GGCATCGAAATCAGTATCTTTGG | |
| CrzA_Fw | GAGAACTTCACCTTGTCCGAG | |
| CrzA_Rv | GGCATCATTTCCTGTCCCTG | |
| PkaR_Fw | CATCCGAAGACCGAAGAACAG | |
| PkaR_Rv | CCAAAGCGTCAAGTACAGTCC | |
| TubA_Fw | GGTAACCAAATCGGTGCTGCTTTC | |
| TubA_Rv133 | ACCCTCAGTGTAGTGACCCTTGGC | |
| Klebsiella | Sequence (5′ to 3′) | |
| 16S_Fw | CCAGCAGCCGCGGTAA | |
| 16S_Rv | TTACGCCCAGTAATTCCGATTAA |
ATCC (American type culture collection), DSM (Deutsche sammlung von mikroorganismen und zellkulturen), CM (Centro nacional de microbiologia), Fw (forward), Rv (reverse). With the exception of primers for TubA133, all primers employed were specifically designed for the present study.
Primer design and qPCR
Genomic sequences of Aspergillus spp. and K. pneumoniae were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/), and species-specific primers were designed for amplification of the conserved 28 S and 16 S rRNA sequences for fungi and bacteria, respectively. For gene expression analysis by qPCR, target sequences of Aspergillus spp. and K. pneumoniae were retrieved from the Aspergillus genome database (http://www.aspgd.org/) and the Kyoto encyclopedia of genes and genomes (KEGG) (http://www.genome.jp/kegg/), respectively. Primer design was performed by using the software PerlPrimer (version 1.1.21) (Open-source PCR primer design, Parkville, Australia)131. Fasta files were uploaded and the features selected included annealing temperatures of 60–62 °C and amplicon sizes of 100–150 bases. Primers used in this study are listed in Table 1. Primers were pre-tested for correct amplification prior to use in qPCR analysis. The thermocycler used for qPCR reactions was the Realplex Mastercycler epgradient S, Eppendorf (Hamburg, Germany).
Transformation of K. pneumoniae with GFP
Strains of K. pneumoniae ATCC 13883 and ATCC 700603 were transformed with the plasmid pUA-PrpsM-gfp, ORI: SC101, with GFP and kanamycin markers132. Briefly, competent cells were prepared with CaCl2, following transformation by electroporation (2,5 KV for 0,2 cm gap cuvette or 1,75 KV for 0,1 cm gap cuvette, 200 Ω, 25 μF, time constant ≥4 msec), using 1 µL plasmidic DNA. Cells were then recovered in pre-warmed LB medium and positive transformants selected on kanamycin agar plates. Uptake of the plasmid was confirmed by flow cytometry and fluorescence microscopy.
Imaging by confocal microscopy and biofilm thickness measurements
Aspergillus and K. pneumoniae cells were visualized under a confocal laser scanning microscope (CLSM) using IBIDI plates (µ-dish, 35 mm high, ibiTreat, 35 mm). Prior to imaging, cells were fixed with 4% paraformaldehyde for 30 min, followed by calcofluor white (CFW) staining (10 µg/mL) of single and co-cultures of Aspergillus for 20 min in the dark, at room temperature. K. pneumoniae strains were GFP-tagged as described above. The fluorescence channels for DAPI and EGFP were applied to image Aspergillus and K. pneumoniae cells, respectively. For biofilm thickness (µm) measurements of single and co-cultures, the Z-stack function was selected. Images were analyzed using the Fiji software (Open source Java image processing, NIH image).
Transwell plates assay
The effect of physical contact on the interaction between Aspergillus and K. pneumoniae was assessed in Transwell® plates (6.5 mm diameter inserts with 0.4 µm Pore Polyester Membrane, tissue culture-treated, Corning, Costar). These plates contain an upper compartment or insert which is separated from the lower compartments or wells. Briefly, YPD medium was pre-warmed and 600 µL were placed in the wells, while 100 µL were placed into the inserts. A. fumigatus was seeded into the inserts and the wells contained either K. pneumoniae alone or in co-culture with A. fumigatus. Imaging of the plates was performed after 24 h incubation at 37 °C with a camera (Canon Macro Lens EF-S 60 mm). Potential cross-over of microorganisms between the upper and lower compartments was excluded by microscopy-based control.
Supernatant assay
Supernatant (SN) of K. pneumoniae growing alone and in co-culture with Aspergillus spp. was obtained after 12 and 24 h growth in biofilm mode in 35 × 10 mm tissue culture dishes (CytoOne, Starlab GmbH, Ahrensburg, Germany). After growth, cells and supernatants were collected into Eppendorf tubes and spun down 2x at maximum speed (25 000 g) at 4 °C (centrifuge 5417 R, Eppendorf, Hamburg, Germany). The SNs corresponding to each growth condition were pooled into 50 mL Falcon tubes, followed by filtration with vacuum filter units (Millipore Express PLUS (PES) 0.22 µm membrane). The freshly collected SN was immediatelly used for growth of Aspergillus spp. Different concentrations of YPD medium were used, including 0.5 volumes SN and 0.5 volumes of 2x YPD or 1x YPD or H2O. These conditions were intended to mimic exhaustion of nutrients, availability of nutrients and non-exhausted SN, respectively. Imaging of the plates was performed after 48 h incubation. As a control, to check for cell-free supernatants, an aliquot (100 µL) of each SN was distributed on LB agar plates and incubated at 37 °C.
Testing for recovery of fungal viability
Aspergillus spp. and K. pneumoniae were grown alone and in co-culture in sterile-filtered YPD medium (Formedium, Norfolk, UK) at 37 °C. These were grown as biofilms with static incubation in 35 × 10 mm tissue culture dishes (CytoOne, Starlab GmbH, Ahrensburg, Germany). Co-cultures of fungi and bacteria were mixed together at time zero and incubated for 24 h. An aliquot of these cultures (100 µL) was then streaked on YPD agar plates containing the antibiotic kanamycin 1000 µg/mL to select for fungal growth. Imaging was performed (Canon Macro Lens EF-S 60 mm) after 48 h incubation at 37 °C. As a negative control, to check for absence of bacterial growth, K. pneumoniae strains were streaked alone onto YPD/Kanamycin agar plates. Positive controls were implemented by growing Aspergillus spp. alone in YPD/Kanamycin agar plates to check for unaffected fungal growth in the presence of this antibiotic.
Heat-, UV-killed and antibiotic-treated bacteria
Heat-killing (HK) of K. pneumoniae cells was performed in a Thermomixer comfort (Eppendorf) at 95 °C for 30 min in PBS, whereas UV-killing (UVK) was performed in a Stratalinker for 3 cycles at 9999 × 100 µjoules with plate shaking between the cycles. Following HK or UVK, K. pneumoniae cells were added to Aspergillus spp in co-culture in 35 × 10 mm tissue culture dishes (CytoOne, Starlab GmbH, Ahrensburg, Germany) as described above (Strains and growth conditions). Imaging was performed after 48 h incubation. As a control, to check for efficiency of killing, an aliquot (100 µL) of HK and UVK cells was streaked onto LB agar plates and incubated at 37 °C.
Treatment of K. pneumoniae with the antibiotics colistin, kanamycin or tetracycline was performed at the concentrations of 100, 2000 and 10 µg/mL, respectively. Aspergillus spp. and K. pneumoniae were grown alone and in co-culture in YPD medium at 37 °C in 35 × 10 mm tissue culture dishes (CytoOne, Starlab GmbH, Ahrensburg, Germany). Co-cultures of fungi and bacteria were mixed together at time zero, and antibiotics were added to the culture at 6 h. Imaging was performed after 48 h incubation.
Cell sorting of A. fumigatus and cell wall stress assay
Sorting of A. fumigatus cells growing alone or in co-culture with K. pneumoniae was performed after 24 h growth in 35 × 10 mm tissue culture dishes (CytoOne, Starlab GmbH, Ahrensburg, Germany). Following growth, cultures were collected into 15 mL Falcon tubes, spun down for 5 min at 4.400 g and SN was discarded. A volume of 2 mL 1x PBS was added and samples were vortexed vigorously. An aliquot (1 mL) was added into a FACS tube through a membrane on the lid. These lids were removed and CFW was added to the cells at a final concentration of 10 µg/mL. Samples were then processed using a FACSaria cell sorter (BD Biosciences) by sorting the cells based on CFW fluorescence (BV421 detector). A total of 20 000 cells were sorted into 1 mL 1x PBS. From these, a volume of 10 µL was spotted onto 12-well agar plates.
To perform the cell wall stress assay, 12-well agar plates were prepared using YPD agar containing the antibiotics colistin at 100 µg/mL and kanamycin at 1000 µg/mL to preclude any possibility of bacterial growth. In addition, each plate was complemented with different concentrations of chemical cell wall stressors and antifungals. The cell wall stresses included: H2O2 1 mM, 2 mM and 4 mM; sorbitol 1 M, 2 M and 4 M; sodium dodecyl sulfate (SDS) 50 µg/mL, 100 µg/mL and 200 µg/mL; CFW 5 µg/mL, 10 µg/mL and 20 µg/mL. The antifungal drugs and their corresponding concentrations included amphotericin B (AmB) 0.25 µg/mL, 0.5 µg/mL and 1 µg/mL; voriconazole 0.025 µg/mL, 0.05 µg/mL and 0.1 µg/mL; micafungin 4 µg/mL, 8 µg/mL and 16 µg/mL. Plates were incubated at 37 °C and imaged (Canon Macro Lens EF-S 60 mm) after 2 days. K. pneumoniae growing alone was also spotted onto these plates as a control for complete growth inhibition of bacterial cells.
Statistics
The significance of differences between Aspergillus-K. pneumoniae co-cultures and single cultures was determined by using the T-test with one-tailed distribution for paired samples (Excel software). P values < 0.05 were considered significant. The calculations were based on at least three independent biological replicates.
Electronic supplementary material
Acknowledgements
This work was supported by the European Commission within the FP7 Framework Programme [Fungitect-Grant No. 602125]. We also thank Thomas Sauer, Vienna Biocenter Campus (VBC), Austria, for technical support at the FACS facility of the MFPL. The authors thank Nela Nikolic (Isabella Moll group, MFPL, Vienna Biocenter Campus, Austria) for technical support and gift of the bacterial plasmid pUA-PrpsM, and Steffen Rupp, Fraunhofer IGB, Stuttgart, Germany, for providing A. niger and A. flavus strains.
Author Contributions
Wrote the manuscript: F.N. and T.L. Conceived and designed experiments: F.N., L.P., S.J. and T.L. Performed experiments: F.N. Analyzed data: F.N., S.J. and T.L. Contributed materials: K.K.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36524-8.
References
- 1.Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. 2014;14:827–835. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- 2.Nash AK, et al. The gut mycobiome of the Human. Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witherden, E. A., Shoaie, S., Hall, R. A. & Moyes, D. L. The Human Mucosal Mycobiome and Fungal Community Interactions. J Fungi (Basel)3 (2017). [DOI] [PMC free article] [PubMed]
- 4.Jo JH, Kennedy EA, Kong HH. Topographical and physiological differences of the skin mycobiome in health and disease. Virulence. 2017;8:324–333. doi: 10.1080/21505594.2016.1249093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galloway-Pena J, Brumlow C, Shelburne S. Impact of the Microbiota on Bacterial Infections during Cancer Treatment. Trends Microbiol. 2017;25:992–1004. doi: 10.1016/j.tim.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Urbaniak C, et al. Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome. 2014;2:2049–2618. doi: 10.1186/2049-2618-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heisel, T. et al. High-Fat Diet Changes Fungal Microbiomes and Interkingdom Relationships in the Murine Gut. mSphere2 (2017). [DOI] [PMC free article] [PubMed]
- 8.Lof, M., Janus, M. M. & Krom, B. P. Metabolic Interactions between Bacteria and Fungi in Commensal Oral Biofilms. J Fungi (Basel)3 (2017). [DOI] [PMC free article] [PubMed]
- 9.Raskov H, Burcharth J, Pommergaard HC. Linking Gut Microbiota to Colorectal Cancer. J Cancer. 2017;8:3378–3395. doi: 10.7150/jca.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsilimigras MC, Fodor A, Jobin C. Carcinogenesis and therapeutics: the microbiota perspective. Nat Microbiol. 2017;2:17008. doi: 10.1038/nmicrobiol.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zitvogel L, Daillere R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15:465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 12.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 13.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 14.Mear JB, et al. Candida albicans and Pseudomonas aeruginosa interactions: more than an opportunistic criminal association? Med Mal Infect. 2013;43:146–151. doi: 10.1016/j.medmal.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Shirtliff ME, Peters BM, Jabra-Rizk MA. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009;299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pammi M, Liang R, Hicks J, Mistretta TA, Versalovic J. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol. 2013;13:1471–2180. doi: 10.1186/1471-2180-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014;16:214–231. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham CE, Cruz MR, Garsin DA, Lorenz MC. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc Natl Acad Sci USA. 2017;114:4507–4512. doi: 10.1073/pnas.1620432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz MR, Graham CE, Gagliano BC, Lorenz MC, Garsin DA. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect Immun. 2013;81:189–200. doi: 10.1128/IAI.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandara HM, Lam OL, Watt RM, Jin LJ, Samaranayake LP. Bacterial lipopolysaccharides variably modulate in vitro biofilm formation of Candida species. J Med Microbiol. 2010;59:1225–1234. doi: 10.1099/jmm.0.021832-0. [DOI] [PubMed] [Google Scholar]
- 21.Bergeron, A. C. et al. Candida and Pseudomonas interact to enhance virulence of mucosal infection in transparent zebrafish. Infect Immun (2017). [DOI] [PMC free article] [PubMed]
- 22.Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 2009;75:504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan DA. Talking to themselves: autoregulation and quorum sensing in fungi. Eukaryot Cell. 2006;5:613–619. doi: 10.1128/EC.5.4.613-619.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mear JB, et al. Candida albicans airway exposure primes the lung innate immune response against Pseudomonas aeruginosa infection through innate lymphoid cell recruitment and interleukin-22-associated mucosal response. Infect Immun. 2014;82:306–315. doi: 10.1128/IAI.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8:1658–1664. doi: 10.1128/EC.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. 2017;7:41332. doi: 10.1038/srep41332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levison ME, Pitsakis PG. Susceptibility to experimental Candida albicans urinary tract infection in the rat. J Infect Dis. 1987;155:841–846. doi: 10.1093/infdis/155.5.841. [DOI] [PubMed] [Google Scholar]
- 28.Mowat E, et al. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol Lett. 2010;313:96–102. doi: 10.1111/j.1574-6968.2010.02130.x. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira, J. A. et al. Inhibition of Aspergillus fumigatus and Its Biofilm by Pseudomonas aeruginosa Is Dependent on the Source, Phenotype and Growth Conditions of the Bacterium. PLoS One10 (2015). [DOI] [PMC free article] [PubMed]
- 30.Sass G, et al. Studies of Pseudomonas aeruginosa Mutants Indicate Pyoverdine as the Central Factor in Inhibition of Aspergillus fumigatus Biofilm. J Bacteriol. 2017;200:00345–00317. doi: 10.1128/JB.00345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerr JR. Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J Clin Microbiol. 1994;32:525–527. doi: 10.1128/jcm.32.2.525-527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr JR, et al. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol. 1999;52:385–387. doi: 10.1136/jcp.52.5.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rella A, et al. Pseudomonas aeruginosa inhibits the growth of Cryptococcus species. Mycopathologia. 2012;173:451–461. doi: 10.1007/s11046-011-9494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teoh-Chan H, Chau PY, Ng MH, Wong PC. Inhibition of Cryptococcus neoformans by Pseudomonas aeruginosa. J Med Microbiol. 1975;8:77–81. doi: 10.1099/00222615-8-1-77. [DOI] [PubMed] [Google Scholar]
- 35.Reece E, et al. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: an Irish registry analysis. BMC Pulm Med. 2017;17:70. doi: 10.1186/s12890-017-0416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith K, et al. Aspergillus fumigatus enhances elastase production in Pseudomonas aeruginosa co-cultures. Med Mycol. 2015;53:645–655. doi: 10.1093/mmy/myv048. [DOI] [PubMed] [Google Scholar]
- 37.Neely AN, Law EJ, Holder IA. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect Immun. 1986;52:200–204. doi: 10.1128/iai.52.1.200-204.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser C, et al. Biofilms and host response - helpful or harmful. APMIS. 2017;125:320–338. doi: 10.1111/apm.12674. [DOI] [PubMed] [Google Scholar]
- 39.Schroeder, M., Brooks, B. D. & Brooks, A. E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes (Basel)8 (2017). [DOI] [PMC free article] [PubMed]
- 40.Ramage G, Mowat E, Jones B, Williams C, Lopez-Ribot J. Our current understanding of fungal biofilms. Crit Rev Microbiol. 2009;35:340–355. doi: 10.3109/10408410903241436. [DOI] [PubMed] [Google Scholar]
- 41.Arciola CR, Campoccia D, Ravaioli S, Montanaro L. Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Microbiol. 2015;5:7. doi: 10.3389/fcimb.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H, et al. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J Hosp Infect. 2015;91:35–44. doi: 10.1016/j.jhin.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 43.Nobile CJ, Johnson AD. Candida albicans Biofilms and Human Disease. Annu Rev Microbiol. 2015;69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong, E. F. et al. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. MBio7 (2016). [DOI] [PMC free article] [PubMed]
- 45.Kragh KN, et al. Role of Multicellular Aggregates in Biofilm Formation. MBio. 2016;7:e00237. doi: 10.1128/mBio.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 47.Hoiby N. A short history of microbial biofilms and biofilm infections. APMIS. 2017;125:272–275. doi: 10.1111/apm.12686. [DOI] [PubMed] [Google Scholar]
- 48.Jensen PO, Kolpen M, Kragh KN, Kuhl M. Microenvironmental characteristics and physiology of biofilms in chronic infections of CF patients are strongly affected by the host immune response. APMIS. 2017;125:276–288. doi: 10.1111/apm.12668. [DOI] [PubMed] [Google Scholar]
- 49.Vipulanandan G, et al. Dynamics of Mixed- Candida Species Biofilms in Response to Antifungals. J Dent Res. 2018;97:91–98. doi: 10.1177/0022034517729351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa-Orlandi, C. B. et al. Fungal Biofilms and Polymicrobial Diseases. J Fungi (Basel)3 (2017). [DOI] [PMC free article] [PubMed]
- 52.Rosen, D. A., Twentyman, J., Hunstad, D. A. High Levels of Cyclic Di-GMP in Klebsiella pneumoniae Attenuate Virulence in the Lung. Infect Immun86 (2018). [DOI] [PMC free article] [PubMed]
- 53.Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/CMR.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tekaia F, Latge JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol. 2005;8:385–392. doi: 10.1016/j.mib.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 56.Mowat E, Butcher J, Lang S, Williams C, Ramage G. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J Med Microbiol. 2007;56:1205–1212. doi: 10.1099/jmm.0.47247-0. [DOI] [PubMed] [Google Scholar]
- 57.Dagenais TR, Keller NP. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin Microbiol Rev. 2009;22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hohl TM, et al. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 2005;1:e30. doi: 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kernien JF, Snarr BD, Sheppard DC, Nett JE. The Interface between Fungal Biofilms and Innate Immunity. Front Immunol. 2017;8:1968. doi: 10.3389/fimmu.2017.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taghavi M, Khosravi A, Mortaz E, Nikaein D, Athari SS. Role of pathogen-associated molecular patterns (PAMPS) in immune responses to fungal infections. Eur J Pharmacol. 2017;808:8–13. doi: 10.1016/j.ejphar.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 61.Ellis M, Richardson M, de Pauw B. Epidemiology. Hosp Med. 2000;61:605–609. doi: 10.12968/hosp.2000.61.9.1415. [DOI] [PubMed] [Google Scholar]
- 62.Schmiedel Y, Zimmerli S. Common invasive fungal diseases: an overview of invasive candidiasis, aspergillosis, cryptococcosis, and Pneumocystis pneumonia. Swiss Med Wkly. 2016;146:w14281. doi: 10.4414/smw.2016.14281. [DOI] [PubMed] [Google Scholar]
- 63.Kohler, J. R., Hube, B., Puccia, R., Casadevall, A. & Perfect, J. R. Fungi that Infect Humans. Microbiol Spectr5 (2017). [DOI] [PubMed]
- 64.Schmitt HJ, Blevins A, Sobeck K, Armstrong D. Aspergillus species from hospital air and from patients. Mycoses. 1990;33:539–541. doi: 10.1111/myc.1990.33.11-12.539. [DOI] [PubMed] [Google Scholar]
- 65.Denning DW. Invasive aspergillosis. Clin Infect Dis. 1998;26:781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 66.Geltner C, Lass-Florl C. Invasive pulmonary Aspergillosis in organ transplants–Focus on lung transplants. Respir Investig. 2016;54:76–84. doi: 10.1016/j.resinv.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Lass-Florl C. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses. 2009;52:197–205. doi: 10.1111/j.1439-0507.2009.01691.x. [DOI] [PubMed] [Google Scholar]
- 68.Lass-Florl C, Cuenca-Estrella M. Changes in the epidemiological landscape of invasive mould infections and disease. J Antimicrob Chemother. 2017;72:i5–i11. doi: 10.1093/jac/dkx028. [DOI] [PubMed] [Google Scholar]
- 69.Binder U, Lass-Florl C. New insights into invasive aspergillosis–from the pathogen to the disease. Curr Pharm Des. 2013;19:3679–3688. doi: 10.2174/13816128113199990366. [DOI] [PubMed] [Google Scholar]
- 70.Campos AC, et al. Outbreak of Klebsiella pneumoniae carbapenemase-producing K pneumoniae: A systematic review. Am J Infect Control. 2016;44:1374–1380. doi: 10.1016/j.ajic.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 71.Giani T, et al. Large Nosocomial Outbreak of Colistin-Resistant, Carbapenemase-Producing Klebsiella pneumoniae Traced to Clonal Expansion of an mgrB Deletion Mutant. J Clin Microbiol. 2015;53:3341–3344. doi: 10.1128/JCM.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Duin D, Doi Y. Outbreak of Colistin-Resistant, Carbapenemase-Producing Klebsiella pneumoniae: Are We at the End of the Road? J Clin Microbiol. 2015;53:3116–3117. doi: 10.1128/JCM.01399-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62:867–874. doi: 10.18388/abp.2015_1148. [DOI] [PubMed] [Google Scholar]
- 74.Hennequin C, Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2016;35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 75.Naparstek L, Carmeli Y, Navon-Venezia S, Banin E. Biofilm formation and susceptibility to gentamicin and colistin of extremely drug-resistant KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2014;69:1027–1034. doi: 10.1093/jac/dkt487. [DOI] [PubMed] [Google Scholar]
- 76.Arnold RS, et al. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. 2011;104:40–45. doi: 10.1097/SMJ.0b013e3181fd7d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barwa R, Shaaban M. Molecular Characterization of Klebsiella pneumoniae Clinical Isolates with Elevated Resistance to Carbapenems. Open Microbiol J. 2017;11:152–159. doi: 10.2174/1874285801711010152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cienfuegos-Gallet, AV, Chen, L., Kreiswirth, B. N. & Jimenez, J. N. Colistin Resistance in Carbapenem-Resistant Klebsiella pneumoniae Mediated by Chromosomal Integration of Plasmid DNA. Antimicrob Agents Chemother61 (2017). [DOI] [PMC free article] [PubMed]
- 79.Kidd TJ, et al. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med. 2017;9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kittinger C, et al. Enterobacteriaceae Isolated from the River Danube: Antibiotic Resistances, with a Focus on the Presence of ESBL and Carbapenemases. PLoS One. 2016;11:e0165820. doi: 10.1371/journal.pone.0165820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanchez GV, et al. Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998–2010. Emerg Infect Dis. 2013;19:133–136. doi: 10.3201/eid1901.120310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shields RK, Clancy CJ, Press EG, Nguyen MH. Aminoglycosides for Treatment of Bacteremia Due to Carbapenem-Resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2016;60:3187–3192. doi: 10.1128/AAC.02638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trecarichi EM, et al. Bloodstream infections caused by Klebsiella pneumoniae in onco-hematological patients: clinical impact of carbapenem resistance in a multicentre prospective survey. Am J Hematol. 2016;91:1076–1081. doi: 10.1002/ajh.24489. [DOI] [PubMed] [Google Scholar]
- 84.Kim YJ, et al. Virulence factors and clinical patterns of hypermucoviscous Klebsiella pneumoniae isolated from urine. Infect Dis (Lond) 2017;49:178–184. doi: 10.1080/23744235.2016.1244611. [DOI] [PubMed] [Google Scholar]
- 85.Pan YJ, et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci Rep. 2015;5:15573. doi: 10.1038/srep15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rivero A, Gomez E, Alland D, Huang DB, Chiang T. K2 serotype Klebsiella pneumoniae causing a liver abscess associated with infective endocarditis. J Clin Microbiol. 2010;48:639–641. doi: 10.1128/JCM.01779-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bengoechea JA. Klebsiella sweet deadly kiss. Virulence. 2016;7:742–744. doi: 10.1080/21505594.2016.1204509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seifi K, et al. Evaluation of Biofilm Formation Among Klebsiella pneumoniae Isolates and Molecular Characterization by ERIC-PCR. Jundishapur J Microbiol. 2016;9:e30682. doi: 10.5812/jjm.30682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vuotto C, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol. 2017;123:1003–1018. doi: 10.1111/jam.13533. [DOI] [PubMed] [Google Scholar]
- 90.Zowawi, H. M. et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep5 (2015). [DOI] [PMC free article] [PubMed]
- 91.Bucher T, Oppenheimer-Shaanan Y, Savidor A, Bloom-Ackermann Z, Kolodkin-Gal I. Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ Microbiol Rep. 2015;7:990–1004. doi: 10.1111/1758-2229.12346. [DOI] [PubMed] [Google Scholar]
- 92.Latge JP. 30 years of battling the cell wall. Med Mycol. 2017;55:4–9. doi: 10.1093/mmy/myw076. [DOI] [PubMed] [Google Scholar]
- 93.Lee MJ, Sheppard DC. Recent advances in the understanding of the Aspergillus fumigatus cell wall. J Microbiol. 2016;54:232–242. doi: 10.1007/s12275-016-6045-4. [DOI] [PubMed] [Google Scholar]
- 94.Yoshimi A, Miyazawa K, Abe K. Cell wall structure and biogenesis in Aspergillus species. Biosci Biotechnol Biochem. 2016;80:1700–1711. doi: 10.1080/09168451.2016.1177446. [DOI] [PubMed] [Google Scholar]
- 95.Hopke, A. et al. Neutrophil Attack Triggers Extracellular Trap-Dependent Candida Cell Wall Remodeling and Altered Immune Recognition. PLoS Pathog12 (2016). [DOI] [PMC free article] [PubMed]
- 96.Beyda ND, Lewis RE, Garey KW. Echinocandin resistance in Candida species: mechanisms of reduced susceptibility and therapeutic approaches. Ann Pharmacother. 2012;46:1086–1096. doi: 10.1345/aph.1R020. [DOI] [PubMed] [Google Scholar]
- 97.Munro CA, et al. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walker LA, Gow NA, Munro CA. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob Agents Chemother. 2013;57:146–154. doi: 10.1128/AAC.01486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walker LA, et al. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4:1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang J, et al. Calcineurin is required for pseudohyphal growth, virulence, and drug resistance in Candida lusitaniae. PLoS One. 2012;7:31. doi: 10.1371/journal.pone.0044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Walker LA, Lee KK, Munro CA, Gow NA. Caspofungin Treatment of Aspergillus fumigatus Results in ChsG-Dependent Upregulation of Chitin Synthesis and the Formation of Chitin-Rich Microcolonies. Antimicrob Agents Chemother. 2015;59:5932–5941. doi: 10.1128/AAC.00862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Altwasser R, et al. Network Modeling Reveals Cross Talk of MAP Kinases during Adaptation to Caspofungin Stress in Aspergillus fumigatus. PLoS One. 2015;10:e0136932. doi: 10.1371/journal.pone.0136932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fujioka T, et al. MpkA-Dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot Cell. 2007;6:1497–1510. doi: 10.1128/EC.00281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Valiante V, Macheleidt J, Foge M, Brakhage AA. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front Microbiol. 2015;6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beauvais A, Fontaine T, Aimanianda V, Latge JP. Aspergillus cell wall and biofilm. Mycopathologia. 2014;178:371–377. doi: 10.1007/s11046-014-9766-0. [DOI] [PubMed] [Google Scholar]
- 106.Gow, N. A. R., Latge, J. P. & Munro, C. A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol Spectr5 (2017). [DOI] [PubMed]
- 107.Pfaller MA, Woosley LN, Messer SA, Jones RN, Castanheira M. Significance of molecular identification and antifungal susceptibility of clinically significant yeasts and moulds in a global antifungal surveillance programme. Mycopathologia. 2012;174:259–271. doi: 10.1007/s11046-012-9551-x. [DOI] [PubMed] [Google Scholar]
- 108.Bain JM, et al. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. J Clin Microbiol. 2007;45:1469–1477. doi: 10.1128/JCM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bohle K, et al. Selection of reference genes for normalisation of specific gene quantification data of Aspergillus niger. J Biotechnol. 2007;132:353–358. doi: 10.1016/j.jbiotec.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 110.Cramer RA, Jr., et al. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot Cell. 2008;7:1085–1097. doi: 10.1128/EC.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Soriani FM, et al. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol Microbiol. 2008;67:1274–1291. doi: 10.1111/j.1365-2958.2008.06122.x. [DOI] [PubMed] [Google Scholar]
- 112.Fortwendel JR, et al. Differential effects of inhibiting chitin and 1,3-{beta}-D-glucan synthesis in ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob Agents Chemother. 2009;53:476–482. doi: 10.1128/AAC.01154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Soriani FM, et al. Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiol. 2010;10:1471–2180. doi: 10.1186/1471-2180-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liebmann B, Muller M, Braun A, Brakhage AA. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect Immun. 2004;72:5193–5203. doi: 10.1128/IAI.72.9.5193-5203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao W, et al. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect Immun. 2006;74:4865–4874. doi: 10.1128/IAI.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev. 2011;75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hohl TM, Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell. 2007;6:1953–1963. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wiedemann A, Spadinger A, Lowe A, Seeger A, Ebel F. Agents that activate the High Osmolarity Glycerol pathway as a means to combat pathogenic molds. Int J Med Microbiol. 2016;306:642–651. doi: 10.1016/j.ijmm.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 119.Schroeder, M., Brooks, B. D. & Brooks, A. E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes8 (2017). [DOI] [PMC free article] [PubMed]
- 120.Zhao J, Shen Y, Haapasalo M, Wang Z, Wang Q. A 3D numerical study of antimicrobial persistence in heterogeneous multi-species biofilms. J Theor Biol. 2016;392:83–98. doi: 10.1016/j.jtbi.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 121.Azevedo AS, Almeida C, Melo LF, Azevedo NF. Impact of polymicrobial biofilms in catheter-associated urinary tract infections. Crit Rev Microbiol. 2017;43:423–439. doi: 10.1080/1040841X.2016.1240656. [DOI] [PubMed] [Google Scholar]
- 122.Xu L, et al. Pseudomonas aeruginosa inhibits the growth of pathogenic fungi: In vitro and in vivo studies. Exp Ther Med. 2014;7:1516–1520. doi: 10.3892/etm.2014.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fox EP, et al. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 2014;24:2411–2416. doi: 10.1016/j.cub.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Will ME, Sylvia DM. Interaction of rhizosphere bacteria, fertilizer, and vesicular-arbuscular mycorrhizal fungi with sea oats. Appl Environ Microbiol. 1990;56:2073–2079. doi: 10.1128/aem.56.7.2073-2079.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morales DK, et al. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol. 2010;78:1379–1392. doi: 10.1111/j.1365-2958.2010.07414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Morales DK, et al. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio. 2013;4:00526–00512. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Roux D, et al. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit Care Med. 2009;37:1062–1067. doi: 10.1097/CCM.0b013e31819629d2. [DOI] [PubMed] [Google Scholar]
- 128.Trejo-Hernandez A, Andrade-Dominguez A, Hernandez M, Encarnacion S. Interspecies competition triggers virulence and mutability in Candida albicans-Pseudomonas aeruginosa mixed biofilms. ISME J. 2014;8:1974–1988. doi: 10.1038/ismej.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meijer AH, Spaink HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets. 2011;12:1000–1017. doi: 10.2174/138945011795677809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Torraca V, Mostowy S. Zebrafish Infection: From Pathogenesis to Cell Biology. Trends Cell Biol. 2018;28:143–156. doi: 10.1016/j.tcb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- 132.Zaslaver A, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 133.Nasri T, et al. PCR-RFLP on beta-tubulin gene for rapid identification of the most clinically important species of Aspergillus. J Microbiol Methods. 2015;117:144–147. doi: 10.1016/j.mimet.2015.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.