Abstract
Introduction
To identify the association between the TMPRSS2:ERG fusion gene, their variants and the onset of localized prostate cancer.
Material and methods
A systematic search strategy was carried out through MEDLINE, EMBASE, LILACS, CENTRAL and unpublished literature. We included randomized control trials, cohort, case-control and cross-sectional studies that involved patients >18 years-old assessing the association between TMPRSS2 fusion gene, its single nucleotide polymorphisms and prostate cancer. The primary outcome was prostate cancer defined by histology of the tumor coming from transrectal ultrasound guided biopsy, transurethral resection of the prostate or radical prostatectomy. We assessed the risk of bias with QUADAS2 and performed a meta-analysis with Stata 14.
Results
We found 241 records with the search strategies. After duplicates were removed, 18 studies were included in qualitative analysis and 15 studies in meta-analysis. All included studies that had no applicability concerns and low risk of bias for flow and timing. Nine studies had an unclear risk of bias for index and reference tests, since they did not describe the blinding assessment appropriately. Regarding the association between TMPRSS2:ERG and prostate cancer, we found an odds ratio (OR) 2.24 and a 95% confidence interval (CI) (1.29 to 3.91). Regarding the kind of sample, urine showed an OR 2.79 and a 95% CI (1.12 to 6.98) and when using a DNA molecular template, the OR was 3.55 with a 95% CI (1.08 to 11.65).
Conclusions
There was an association between TMPRSS2:ERG fusion gene with the diagnosis of prostate cancer, mainly in urine samples and DNA-based molecular templates. TMPRSS2:ERG might be used as the gold standard biomarker for diagnosis and stratification of PCa.
Keywords: gene, meta-analysis, prostate neoplasia, TMPRSS2, ERG
INTRODUCTION
Prostate cancer (PCa) is a heterogeneous disease with a variable natural history and slow growth pattern. It may display latency periods of up to 20 years in which it remains organ-confined. Although PCa lesions can remain localized for long periods, more aggressive forms might occur and when metastasis occurs, lymph nodes and bones are affected predominantly with detrimental results [1]. PCa is the second most commonly diagnosed worldwide cancer among men (mainly >65 years). It is a public health concern in developed countries, in which elderly men correspond to the greatest affected proportion of the general population [2].
Patients at high risk and susceptibility to develop PCa, require screening over time. Age, race and family history are the most important risk factors [3]. A 50% higher risk in monozygotic twins than in dizygotic twins and the higher incidence in African Americans (and the lower rate in Americans of Asian ancestry) supports genetic factors as an important determinant of the variation risk at the population level [4]. There have been concerns about the diagnosis and early treatment of this disease due to the absence of specific markers [5]. Until now, the gold standard for the diagnosis of PCa has been an invasive procedure consisting in the histopathological evaluation of the prostate, a procedure with significant morbidity [6].
Currently, the use of prostate-specific antigen (PSA) as a screening and monitoring marker for prostate cancer is widespread and is currently the only widely used serum biomarker for PCa [7], although there is still debate about the screening for PCa among men in the general population. This biomarker is prostate-specific, but it has a low specificity and could also increase unnecessary biopsies, without lowering mortality [8].
The transmembrane protease serine 2:vets erythroblastosis virus E26 oncogene homolog (TMPRSS2:ERG) gene fusion has been assessed as a specific biomarker for PCa, since 2005 [9]. This transmembrane protease serine 2 (TMPRSS2) is a promising biomarker located at 21q22.2 and expressed in normal and malignant prostatic epithelium. Additionally, ERG is a member of the E-twenty six family members (ETS), which are key regulators of differentiation, apoptosis, embryonic development, cell proliferation and inflammation. Currently, there are different studies trying to look for the association between this gene, the fusion and the advanced prostate cancer, but less research for this gene as a diagnostic tool [10].
The primary aim of this study was to identify the association between the TMPRSS2:ERG fusion gene, their variants and the onset of localized prostate cancer.
MATERIAL AND METHODS
We performed this review according to the recommendations of the Cochrane Collaboration [11] and following the PRISMA Statement [12]. The PROSPERO registration number is CRD42018087071.
Eligibility criteria
We included randomized control trials (RCTs), cohort, case-control and cross-sectional studies that involved patients >18 years-old assessing the association between TMPRSS2 fusion gene, its single nucleotide polymorphisms (SNPs) and PCa. Studies from molecular biology, translational and clinical research and those that compare men with prostate cancer and those without prostate cancer, were also included. We excluded observational descriptive studies, studies with no human subjects and advanced prostate cancer. There was no setting or language restrictions.
TMPRSS2 and associated SNPs
We previously performed a search in Emsemble.org with the ‘prostate adenocarcinoma’ as keyword, and then we identified associated GENES and chose TMPRSS2 variants in humans. We applied the chosen gene in Genecards.org and identified the associated phenotype, additionally identified superpathways for PCa in Kyoto Encyclopedia of Genes and Genomes (KEGG) and performed a final search in University of California Santa Cruz (UCSC) Genome Browser on Human Dec. 2013 (GRCh38/hg38) assembly. After all, we found the following SNPs that could be a missense variant when transcribed into a protein: rs572530227, rs537370123, rs570454392, rs545726689, rs185312677, rs540987630, rs546335233, rs561063944, rs147233451, rs544474510, rs565237319, rs181414852, rs577684898, rs547544037, rs530689404, rs12329760, rs75603675.
Primary outcome: Prostate cancer defined by histology of the tumor coming from transrectal ultrasound guided (TRUS) biopsy, transurethral resection of the prostate (TURP) or radical prostatectomy
Information sources
Literature search was conducted in accordance to recommendations by Cochrane. We used medical subject headings (MeSh), Emtree language, health science descriptors (Decs) and related text words.
We searched Medline (OVID), EMBASE, LILACS and the Cochrane Central Register of Controlled Trials (CENTRAL) from inception to February 2018 (Appendix 1). To ensure literature saturation, we scanned references from relevant articles identified through the search, conferences, thesis databases, Open Grey, Google scholar and clinicaltrials.gov, among others. We tried to contact authors by e-mail in case of missing information.
Additionally, we looked for information in the following specific databases: single nucleotide polymorphism database (dbSNP), GeneSNP, Polyphen, Human Genome Database, Ensemble, among others.
Data collection
Two researchers reviewed each reference by title and abstract. Then scanned full-texts of relevant studies, applied pre-specified inclusion and exclusion criteria and extracted the data. Disagreements were resolved by consensus and where disagreement could not be solved, a third reviewer dissolved the conflict.
Two trained reviewers using a standardized form independently extracted the following information from each article: study design, geographic location, authors names, title, objectives, inclusion and exclusion criteria, number of patients included, losses to follow-up, timing, definitions of outcomes, outcomes and association measures and funding sources.
Risk of bias
The assessment of the risk of bias for each study was using QUADAS2 tool as recommended by Cochrane [13].
Data analysis / Synthesis of results
The statistical analysis was performed by using Stata 14® and Review Manager 5.3 (RevMan® 5.3). For categorical outcomes we reported information about the odds ratio (OR), with 95% confidence intervals according to the type of variables and we pooled the information with a random effect meta-analysis according to the heterogeneity expected. The results reported in forest plots of the estimated effects of the included studies with a 95% confidence interval (95% CI). Heterogeneity was evaluated by using the I2 test. For the interpretation, it was determined that the values of 25%, 50%, and 75% in the I2 test correspond to low, medium, and high levels of heterogeneity, respectively.
Publication bias
An evaluation was conducted to identify reporting or publication bias using the funnel plot.
Sensitivity analysis
We performed sensitivity analysis extracting weighted studies and running the estimated effect to find differences.
Subgroup analysis
We performed the subgroup analysis by: geographical setting, sample and technique.
RESULTS
Study selection
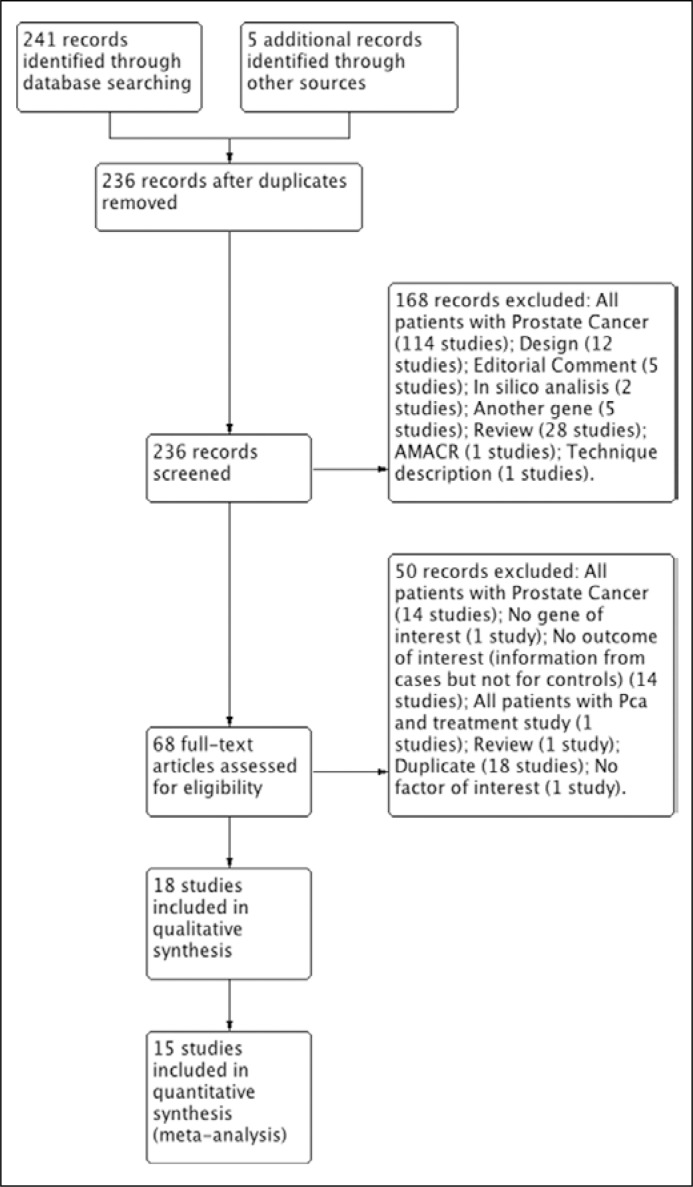
We found 241 records with the search strategies. After duplicates were removed, there were 223 records. Finally, 18 studies were included in qualitative analysis and 15 studies in meta-analysis (Albadine 2009; Chan 2013; Dimitriadis 2013; Huang 2011; Laxman 2008; Leyten 2012; Lin 2013; Maekawa 2014; Mosquera 2009; Nguyen 2011; Park 2014; Penney 2016; Robert 2013; Salami 2013; Sanda 2015; Cornu 2013; Tomlins 2011; Tavukcu 2013) [5, 6, 7, 9, 14–27] (Figure 1).
Figure 1.
Flowchart of included studies.
Included studies
A total of 1057 patients were included, with a median of 25.5 patients per study. All fifteen studies evaluated new biomarkers in different samples for early diagnostic of PCa (Table 1).
Table 1.
Characteristics of included studies
| Author | Gene | Technique | Molecular Template | Sample | Setting | Age | Study Design | N |
|---|---|---|---|---|---|---|---|---|
| Penney 2016 | TMPRSS2:ERG | Immunohistochemical x Microarrays | Protein | Tissue | USA | 40–84 | Cohort | 487 |
| Sanda 2015 | TMPRSS2:ERG | mRNA | RNA | Urine | USA | 55–60 | Prospective multi-center study | 1077 (516) |
| Park 2014 | TMPRSS2:ERG | Inmunohistochemical x Monoclonal ERG antibody | Protein | Tissue | USA | Median 65 | Randomized phase III, double-blinded, placebo-controlled clinical | 1590 (461) |
| Tavukcu 2013 | TMPRSS2:ERG | RT-PCR | RNA | Tissue, Peripheral blood, Urine, Pubic hair | Turkey | 43–78 | Cohort | 50 |
| Lin 2013 | TMPRSS2:ERG | RT-PCR | RNA | Urine | USA | NA | Prospective, observational, active surveillance study | 387 |
| Leyten 2012 | TMPRSS2:ERG | mRNA | RNA | Urine | Netherlands | 44–86 | Prospective multicenter cohort | 443 |
| Tomlins 2011 | TMPRSS2:ERG | Transcription-mediated amplification (Malign)/FISH assay (Benign) | RNA/DNA | Urine (Malign) /Tissue (Benign) | USA/Canada | 56–65 | Cohort | 1312 |
| Mosquera 2009 | TMPRSS2:ERG | FISH | DNA | Tissue | USA | 54–70 | Cohort | 140 (134) |
| Laxman 2008 | TMPRSS2:ERG | RT-PCR | RNA | Urine | USA | NA | Cohort | 234 |
| Maekawa 2014 | TMPRSS2 | Real Time PCR SNP SondaTagMan | DNA | Blood | Japan | 48–100 | Cases and controls | 518 |
| Chan 2013 | TMPRSS2:ERG | RT-PCR | RNA | Urine | Canada | Median 68 | Cohort | 92 |
| Cornu 2013 | TMPRSS2:ERG | TaqManTM assays | DNA | Urine | France | 59–68 | Cohort | 291 |
| Robert 2013 | TMPRSS2:ERG | RT-PCR | DNA | Tissue | Netherlands | NA | Cohort? | 128 (96) |
| Huang 2011 | TMPRSS2:ERG | Immunohistochemical microarray | Protein | Tissue | USA | NA | Retrospective cohort | 80 |
| Nguyen 2011 | TMPRSS2:ERG | RT-PCR | RNA | Urine | Canada | 19–88 | Cases and controls | 101 |
| Albadine 2009 | TMPRSS2:ERG | FISH | DNA | Tissue | USA | NA | Cohort | 92 |
| Dimitriadis 2013 | TMPRSS2:ERG | RT-PCR TagMan | DNA | Urine | Greece | 45–83 | Cohort | 66 |
| Salami 2013 | TMPRSS2:ERG | RT-PCR | RNA | Urine | USA | 56–71 | Cohort | 48 (45) |
Albadine 2009; Dimitriadis 2013; Huang 2011; Laxman 2008; Lin 2013; Mosquera 2009; Park 2014; Penney 2016; Salami 2013; Sanda 2015 performed their analysis based on data from the United States (USA) [5, 7, 9, 16, 18, 22, 23, 24, 26, 27]. On the other side, Chan 2013 and Nguyen 2011 brought data from Canada [15, 25]. Netherlands was the based for the analysis of Leyten 2012 and Robert 2013 [6, 17] and Maekawa 2014 was the only one coming from Japan [14].
Regarding the samples: Albadine 2009; Huang 2011; Mosquera 2009; Penney 2016; Robert 2013 and Park 2014 performed their analysis based on tissue samples [9, 17, 18, 22, 23, 24]. On the other side, Chan 2013; Dimitriadis 2013; Laxman 2008; Leyten 2012; Lin 2013; Nguyen 2011; Salami 2013; Sanda 2015 were based on urine [5, 6, 7, 15, 16, 25, 26, 27]. Only Maekawa 2014 used blood samples for their analysis [14].
Regarding the molecular templates: Huang 2011; Penney 2016; and Park 2014 performed their analysis based on protein expression [9, 18, 23]. On the other side, Chan 2013; Laxman 2008; Leyten 2012; Lin 2013; Nguyen 2011; Salami 2013 and Sanda 2015 were based on techniques related to RNA [6, 7, 15, 16, 25, 26, 27] and Albadine 2009; Dimitriadis 2013; Maekawa 2014; Mosquera 2009; Robert 2013 based their analysis on techniques related to DNA [5, 14, 17, 22, 24].
Regarding genetic polymorphisms: Cornu 2013 evaluated two SNPs at 8q24 locus (rs1447295 and rs6983267) in all patients. These two SNPs correlated to the biopsy outcome in clinical practice [19]. Additionally, Penney 2016 evaluated six SNPs, comparing ERG+ to ERG- cancers. They found four significantly associated with ERG+ compared to controls (rs7679673, rs902774, rs11672691, rs1859962). Three were significantly associated with ERG- compared to controls (rs2660753, rs7629490, rs1016343), and the associations trending in opposite directions for ERG+ and ERG- for three of them (rs12653946, rs1512268, rs11704416) [18].
Furthermore, Maekawa et al. showed that the rs12329760 polymorphism was significantly associated with the risk of sporadic prostate cancer in Japanese men [14].
Risk of bias within and across the studies
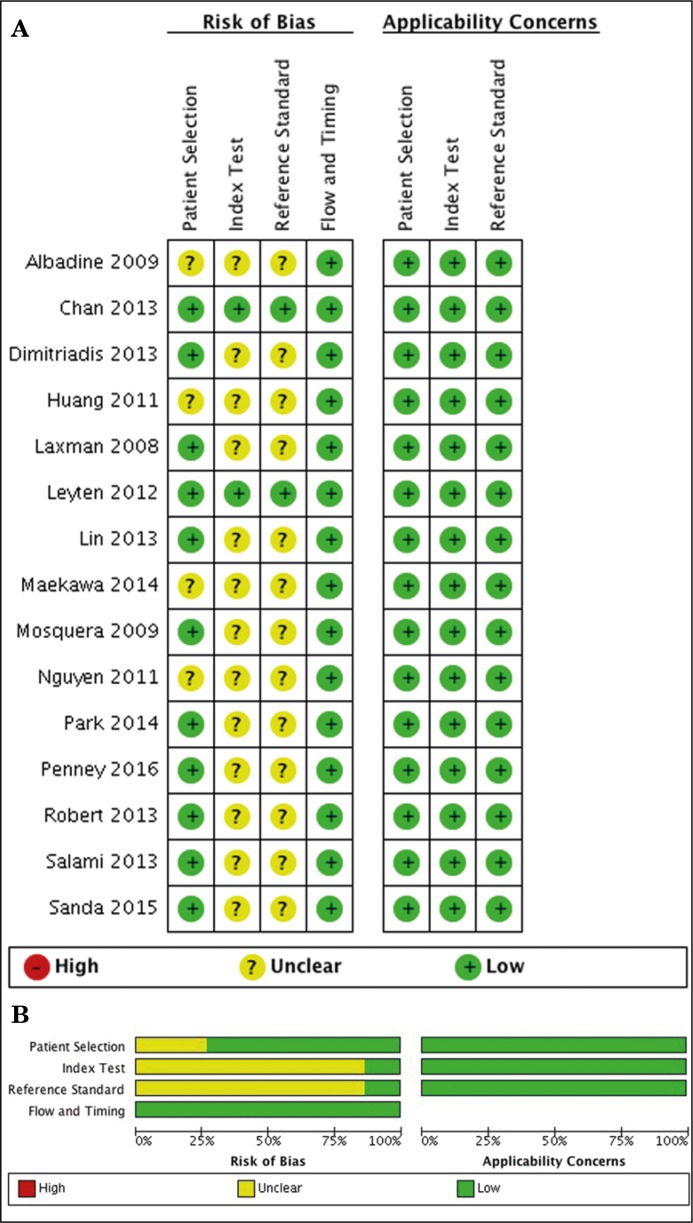
All included studies had no applicability concerns and low risk of bias for flow and timing [5, 6, 7, 9, 14–18, 22–27]. Regarding patient selection, index test and reference standard risk of bias, we found that Chan 2013 and Leyten 2012 had low risk; however, Albadine 2009, Huang 2011, Maekawa 2014 and Nguyen 2011 had an unclear risk of bias since they used case-controlled studies to perform their analysis and did not describe the blinding assessment for index and reference tests (Figure 2).
Figure 2.
Risk of bias within and across the studies. A. Within studies. B. Across the studies.
Dimitriadis 2013, Laxman 2008, Lin 2013, Mosquera 2009, Park 2014, Penney 2016; Robert 2013, Salami 2013 and Sanda 2015 had an unclear risk of bias for index and reference tests since they did not describe the blinding assessment appropriately (Figure 2).
TMPRSS2:ERG and prostate cancer
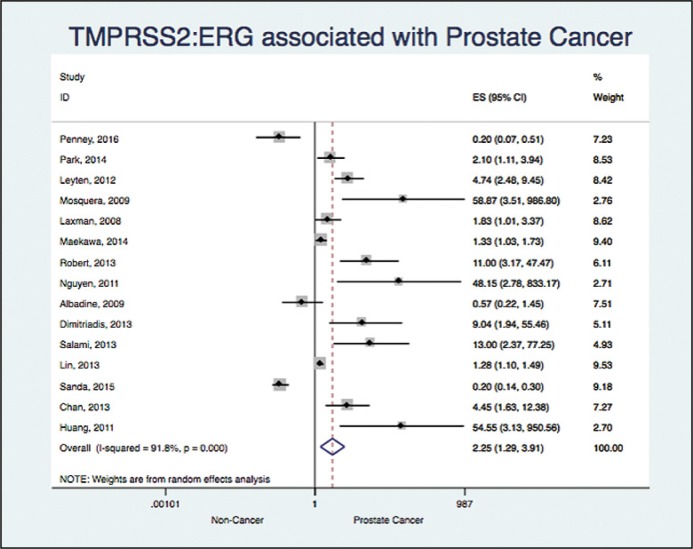
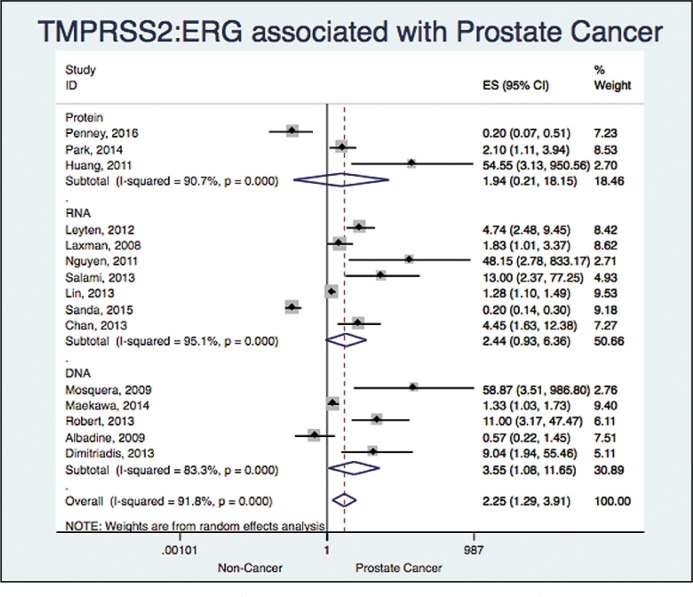
15 studies assessed the association between TMPRSS2:ERG and prostate cancer (Albadine 2009; Chan 2013; Dimitriadis 2013; Huang 2011; Laxman 2008; Leyten 2012; Lin 2013; Maekawa 2014; Mosquera 2009; Nguyen 2011; Park 2014; Penney 2016; Robert 2013; Salami 2013; Sanda 2015) [5, 6, 7, 9, 14–18, 22–27]. We found an OR 2.24 with a 95% CI (1.29 to 3.91) and an I2 = 91%, showing a significant association, but a high heterogeneity (Figure 3).
Figure 3.
Association between TMPRSS2:ERG and prostate cancer.
Publication bias
We did not identify reporting or publication bias using the Begg's and Egger's tests (p = 0.631 and p = 0.716, respectively).
Sensitivity analysis
We did not find any differences in OR when we performed sensitivity analysis.
Subgroup analysis
Based on sample
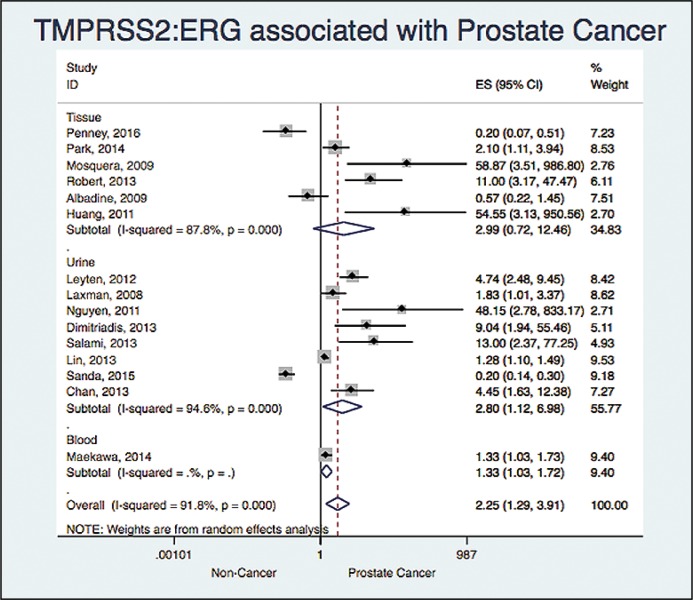
When analyzing based on the sample we found: OR 2.98 and a 95% CI (0.71 to 12.46) with a I2 = 87% (six studies), OR 2.79 and a 95% CI (1.12 to 6.98) with a I2 = 94% (eight studies) and OR 1.33 and a 95% CI (1.02 to 1.72) (one study) for tissue, urine and blood respectively (Figure 4).
Figure 4.
Association between TMPRSS2:ERG and prostate cancer based on the sample.
Based on the molecular template
When analyzing based on the Molecular template, we found: OR 1.93 and a 95% CI (0.20 to 18.14) with a I2 = 90% (Three studies), OR 2.43 and a 95% CI (0.93 to 6.36) with a I2 = 95% (Seven studies) and OR 3.55 and a 95% CI (1.08 to 11.65) with a I2 = 83% (Five studies) for protein, RNA and DNA, respectively (Figure 5).
Figure 5.
Association between TMPRSS2:ERG and prostate cancer based on the technique.
Based on geographical setting
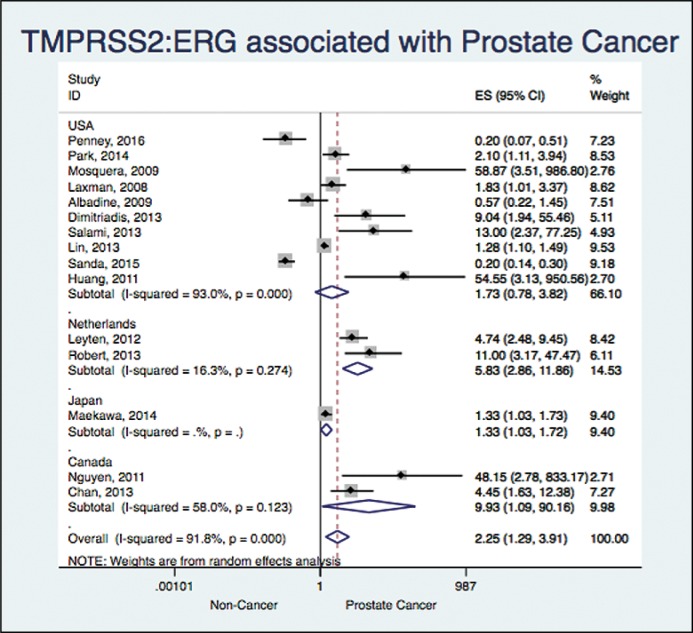
When analyzing based on the geographical setting, we found: OR 1.73 and a 95% CI (0.77 to 3.82) with a I2 = 93% (10 studies), OR 5.83 and a 95% CI (2.86 to 11.86) with a I2 = 16% (two studies), OR 1.33 and a 95% CI (1.02 to 1.72) (one study) and OR 9.92 and a 95% CI (1.09 to 90.16) with a I2 = 58% (two studies) for USA, Netherlands, Japan and Canada, respectively (Figure 6).
Figure 6.
Association between TMPRSS2:ERG and prostate cancer based on the geographical setting.
DISCUSSION
Summary of the main findings
We found that TMPRSS2:ERG fusion was significantly associated with diagnosing PCa, mainly in urine samples and DNA-based molecular templates, however, there was an important heterogeneity that could not be explained with the planned subgroups.
Contrast with literature
Penney et al., showed that tumors that develop TMPRSS2-ERG fusion have different genetic etiology, suggesting also that SNPs are differently associated with the risk of developing prostate tumors either with or without fusion [18]. Park et al., described that 53% of patients with ERG-positive High-Grade Prostatic Intraepithelial Neoplasia (HGPIN) revealed progression to PCa, which needs to be addressed in other studies and systematic reviews [9]. Additionally, Huang et al., determined that the fusion between the androgen regulated TMPRSS2 gene and the members of the ETS transcription factor family has been identified as a genomic aberration in prostate cancer [23].
Tavukcu et al., compared different samples (peripheral blood, pubic hair and urine). They identified a higher number of copies of the fusion gene in post- three-colour assay urine and tissue samples compared with blood and pubic hair samples. They concluded that peripheral blood and pubic hair polymerase chain reaction (PCR) analysis of TMPRSS2:ERG gene fusion seemed to be suboptimal and emphasizing that the samples obtained from urine after prostatic massage seemed to be as effective as direct tissue sampling [21].
High prevalence of TMPRSS2-ERG positive prostate cancer has been found clinically relevant. Studies have shown that TMPRSS2-ERG fusion prostate cancer is associated with higher tumor stage and cancer-specific survival or metastasis [22]. Nonetheless, we evaluated the association with TMPRSS2-ERG gene fusion in the early detection of PCa. The PSA level of subjects with the TMPRSS2-ERG fusion was also significantly higher than those one who do not have it.
The main mechanisms by which the TMPRSS2-ERG fusion genes are produced are interstitial deletion and balanced translocation [10, 28]. Because of their specificity, detection of these fusion genes could be a valuable ancillary diagnostic tool in the early detection of PCa. In fact, these rearranged genes can be detected either by fluorescence in situ hybridization (FISH), reverse transcription polymerase chain reaction (RT-PCR) techniques [29], or branched DNA (bDNA) analysis that is a very sensitive approach [28].
FISH has been considered a standard for the detection of fusion rearrangements; the break-apart strategy is the main approach used for this purpose [30]. Yoshimoto et al. developed a three-colour assay, which is able to distinguish between the two main mechanisms of gene rearrangement for TMPRSS2-ERG, the interstitial deletion, or the reciprocal translocation [31].
Different questions arise regarding FISH and the discussion of the results, for instance, the presence of multiple signals showing multiple copies of the fusion gene are difficult to interpret [32], the number of nuclei and the score of rearranged nuclei to be assessed as positive [32, 33].
On the other side, RT-PCR provides some advantages such as the lower cost and its capacity of discriminating different variants of the TMPRSS2-ERG fusion gene. In this regard, some authors have shown an association between some of these fusion subtypes with good [34] and poor prognoses [35]. However, because of its high sensitivity and cross-contamination, RT-PCR may show false positive results. Hence, RT-PCR is an interesting and useful technique in the diagnostic setting and should be considered as potential complement to FISH [28].
Labor-intensive and cost prohibitive methods such as long-range PCR followed by Sanger sequencing or whole genome sequencing have thus far yielded only a handful of TMPRSS2-ERG genomic breakpoints [36].
Assays for detecting TMPRSS2-ERG fusion have been limited to those based on RT-PCR or FISH. RT-PCR requires the presence of a stable, full-length transcript that can be difficult to retain in routine clinical processing, whereas FISH requires subspecialty molecular pathology expertise that is not uniformly available [37].
Detection of TMPRSS2-ERG fusions is most commonly carried out using either fluorescence in situ hybridization (FISH) or reverse transcription polymerase chain reaction (RT-PCR), but these methods are costly and require considerable infrastructure and expertise [38].
Strengths and limitations
This is the first systematic review related to the association of this important gene and the prostate cancer in early stages, following the international recommendations for systematic reviews and meta-analysis. There was a very sensitive search strategy, enhanced with specific findings for SNPs searched in Genecards.org, KEGG and UCSC Genome Browser.
The most important limitation of this review is the high heterogeneity, which might be explained by geographical setting, molecular template and type of sample. Even though we analyzed and identified some important findings we could not explain the heterogeneity, therefore there might be other important variables to have in mind for future studies, making data more homogeneous.
Clinical and population importance
Prostate cancer (PCa) is characterized by its extensive clinical heterogeneity and so early stratification of this aggressive disease from a majority of indolent cancers at diagnosis is a critical clinical task in cancer management and treatment.
Based on these findings, we state that the TMPRSS2:ERG fusion gene might be the new gold standard biomarker for the diagnosis and stratification of PCa in the very early stages, allowing clinicians to identify and treat locally the cancer with the advent of new technology and to establish which one will become a very aggressive tumor.
As a conclusion, there is an association between TMPRSS2:ERG fusion gene with the diagnosis of prostate cancer, mainly based on urine samples and DNA-based molecular templates. TMPRSS2: ERG might be used as the gold standard biomarker for diagnosis and stratification of PCa.
There is still too much work for standardizing the molecular template, the specific technique and the sample to be used. We recommend that there be an increase in our efforts to elucidate these issues.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
APPENDIX 1. SEARCH STRATEGY
Medline through OVID
TMPRSS2.mp
(Transmembrane Protease, Serine 2).mp
(TMPRSS2 protein, human).mp
(rs572530227.mp or rs537370123.mp or rs570454392.mp or rs545726689.mp or rs185312677.mp or rs540987630.mp or rs546335233.mp or rs561063944.mp or rs147233451.mp or rs544474510.mp or rs181414852.mp or rs565237319.mp or rs577684898.mp or rs547544037.mp or rs530689404.mp or rs12329760.mp or rs75603675.mp)
or/
exp Prostatic Neoplasms/
exp prostatic intraepithelial neoplasia/
(prostatic adj2 malignanc$).mp
(prostatic adj2 cancer).mp
or/
exp randomized controlled trial/
(randomi*ed adj2 controlled adj2 trial).mp.
exp clinical trial/
(clinical adj2 trial).mp
exp double-blind method/
Exp cohort studies/
(cohort adj2 stud$).mp
exp case-control studies
exp Cross-sectional studies
Embase:
'tmprss2 gene'/exp
'transmembrane protease serine 2'/exp
TMPRSS2:ti,ab
(TMPRSS2 next/3 protein):ti,ab
(Transmembrane next/3 Protease next/3 Serine):ti,ab
(rs572530227 or rs537370123 or rs570454392 or rs545726689 or rs185312677 or rs540987630 or rs546335233 or rs561063944 or rs147233451 or rs544474510 or rs181414852 or rs565237319 or rs577684898 or rs547544037 or rs530689404 or rs12329760 or rs75603675):ti,ab
or/
'prostate cancer'/exp
'prostatic intraepithelial neoplasia'/exp
(prostatic next/3 malignanc*):ti,ab
(prostatic next/3 cancer):ti,ab
Or/
'randomized controlled trial'/exp
(randomi*ed NEXT/2 controlled NEXT/2 trial):ti,ab
'clinical trial'/exp
(clinical NEXT/2 trial):ti,ab
'double blind procedure'/exp
'cross-sectional study'/exp
'case control study'/exp
'cohort analysis/exp
or/
Central (Ovid)
TMPRSS2.mp
(Transmembrane Protease, Serine 2).mp
(TMPRSS2 protein, human).mp
(rs572530227.mp or rs537370123.mp or rs570454392.mp or rs545726689.mp or rs185312677.mp or rs540987630.mp or rs546335233.mp or rs561063944.mp or rs147233451.mp or rs544474510.mp or rs181414852.mp or rs565237319.mp or rs577684898.mp or rs547544037.mp or rs530689404.mp or rs12329760.mp or rs75603675.mp)
or/
exp Prostatic Neoplasms/
exp prostatic intraepithelial neoplasia/
(prostatic adj2 malignanc$).mp
(prostatic adj2 cancer).mp
References
- 1.Wang Z-Y, Li H-Y, Jiang Z, Zhou T-B, Drummen GPC. GSTM1 Gene Polymorphism is Implicated in Increased Susceptibility to Prostate Cancer in Caucasians and Asians. Technol Cancer Res Treat. 2016;15:NP69–NP78. doi: 10.1177/1533034615617650. [DOI] [PubMed] [Google Scholar]
- 2.Mottet N, Bellmunt J, Briers E, et al. Guidelines on Prostate Cancer. European Association of Urology; 2017. [Google Scholar]
- 3.Helfand B, Kearns J, Conran C, Xu J. Clinical validity and utility of genetic risk scores in prostate cancer. Asian J Androl. 2016;18:509–514. doi: 10.4103/1008-682X.182981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 5.Dimitriadis E, Kalogeropoulos T, Velaeti S, et al. Study of genetic and epigenetic alterations in urine samples as diagnostic markers for prostate cancer. Anticancer Res. 2013;33:191–197. [PubMed] [Google Scholar]
- 6.Leyten G, Hessels D, Jannink SA, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2014;65:534–542. doi: 10.1016/j.eururo.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Salami S, Schmidt F, Laxman B, et al. Combining urinary detection of TMPRSS2: ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol. 2013;31:566–571. doi: 10.1016/j.urolonc.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilic D, Neuberger MM, Djulbegovic M, Dahm P. Screening for prostate cancer. Cochrane Database Syst Rev. 2013;1:CD004720. doi: 10.1002/14651858.CD004720.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park K, Dalton JT, Narayanan R, et al. TMPRSS2: ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J Clin Oncol. 2014;32:206–211. doi: 10.1200/JCO.2013.49.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández-Serra A, Rubio-Briones J, García-Casado Z, Solsona E, López-Guerrero JA. Cáncer de próstata: la revolución de los genes de fusión. Actas Urol Esp. 2011;35:420–428. doi: 10.1016/j.acuro.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Higgins J, Green S. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0. The Cochrane Collaboration; 2011. [Google Scholar]
- 12.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:1–6. [PMC free article] [PubMed] [Google Scholar]
- 13.Whiting PF, Rutjes AWS, Westwood ME, et al. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 14.Maekawa S, Suzuki M, Arai T, et al. TMPRSS2 Met160Val polymorphism: Significant association with sporadic prostate cancer, but not with latent prostate cancer in Japanese men. Int J Urol. 2014;21:1234–1238. doi: 10.1111/iju.12578. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen PN, Violette P, Chan S, et al. A panel of TMPRSS2:ERG fusion transcript markers for urine-based prostate cancer detection with high specificity and sensitivity. Eur Urol. 2011;59:407–414. doi: 10.1016/j.eururo.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Sanda M, Feng Z, Wei J, et al. Pi-08 Combining Urine Pca3 and Tmprss2: Erg Tests To Refine Prostate Cancer Detection - Validation Study and Health Economic Analysis. J Urol. 2015;193:e982–983. [Google Scholar]
- 17.Robert G, Jannink S, Smit F, et al. Rational basis for the combination of PCA3 and TMPRSS2:ERG gene fusion for prostate cancer diagnosis. Prostate. 2013;73:113–120. doi: 10.1002/pros.22546. [DOI] [PubMed] [Google Scholar]
- 18.Penney KL, Pettersson A, Shui IM, et al. Association of Prostate Cancer Risk Variants with TMPRSS2:ERG Status: Evidence for Distinct Molecular Subtypes. Cancer Epidemiol Biomarkers Prev. 2016;25:745–749. doi: 10.1158/1055-9965.EPI-15-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornu JN, Cancel-Tassin G, Egrot C, Gaffory C, Haab F, Cussenot O. Urine TMPRSS2: ERG fusion transcript integrated with PCA3 score, genotyping, and Biological features are correlated to the Results of prostatic biopsies in men at risk of prostate cancer. Prostate. 2013;73:242–249. doi: 10.1002/pros.22563. [DOI] [PubMed] [Google Scholar]
- 20.Tomlins SA, Aubin S, Siddiqui J, et al. Urine TMPRSS2:ERG for prostate cancer risk stratification in men with elevated serum PSA. Cancer Res. 2011;71(8 Suppl 1) abs. 2815. [Google Scholar]
- 21.Tavukcu HH, Mangir N, Ozyurek M, Turkeri L. Preliminary Results of Noninvasive Detection of TMPRSS2 : ERG Gene Fusion in a Cohort of Patients With Localized Prostate Cancer. Korean J Urol. 2013;54:359–363. doi: 10.4111/kju.2013.54.6.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosquera J, Mehra R, Regan MM, et al. Prevalence of TMPRSS2-ERG Fusion Prostate Cancer among Men Undergoing Prostate Biopsy in the United States. Clin Cancer Res. 2009;15:4706–4712. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Schauer IG, Zhang J, Mercado-uribe I, Deavers MT. The oncogenic gene fusion TMPRSS2 : ERG is not a diagnostic or prognostic marker for ovarian cancer. Int J Clin Exp Pathol. 2011;4:644–650. [PMC free article] [PubMed] [Google Scholar]
- 24.Albadine R, Latour M, Platz E, Meeker A, Demazo A, Netto G. Tmprss2-Erg Gene Fusions in Minimal Prostatic Carcinoma. J Urol. 2009;181:814. [Google Scholar]
- 25.Chan SW, Nguyen P-N, Violette P, et al. Early detection of clinically significant prostate cancer at diagnosis: a prospective study using a novel panel of TMPRSS2:ETS fusion gene markers. Cancer Med. 2013;2:63–75. doi: 10.1002/cam4.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin DW, Newcomb LF, Brown EC, et al. Urinary TMPRSS2:ERG and PCA3 in an active surveillance cohort: results from a baseline analysis in the canary prostate active surveillance study. Clin Cancer Res. 2013;19:2442–2450. doi: 10.1158/1078-0432.CCR-12-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Serra A, Rubio L, Calatrava A, et al. Molecular characterization and clinical impact of TMPRSS2-ERG rearrangement on prostate cancer: comparison between FISH and RT-PCR. Biomed Res Int. 2013;2013:1–10. doi: 10.1155/2013/465179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 30.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimoto M, Joshua AM, Chilton-Macneill S, et al. Three-Color FISH Analysis of TMPRSS2/ERG Fusions in Prostate Cancer Indicates That Genomic Microdeletion of Chromosome 21 Is Associated with Rearrangement 1. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machado I, Noguera R, Pellin A, et al. Molecular diagnosis of Ewing sarcoma family of tumors: a comparative analysis of 560 cases with FISH and RT-PCR. Diagnostic Mol Pathol. 2009;18:189–199. doi: 10.1097/PDM.0b013e3181a06f66. [DOI] [PubMed] [Google Scholar]
- 33.Salgado R, Llombart B, M, Pujol R, et al. Molecular diagnosis of dermatofibrosarcoma protuberans: A comparison between reverse transcriptase-polymerase chain reaction and fluorescence in situ hybridization methodologies. Genes Chromosomes Cancer. 2011;50:510–517. doi: 10.1002/gcc.20874. [DOI] [PubMed] [Google Scholar]
- 34.Hermans KG, Boormans JL, Gasi D, et al. Overexpression of prostate-specific TMPRSS2(exon 0)-ERG fusion transcripts corresponds with favorable prognosis of prostate cancer. Clin Cancer Res. 2009;15:6398–6403. doi: 10.1158/1078-0432.CCR-09-1176. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 36.Weier C, Haffner MC, et al. Nucleotide resolution analysis of TMPRSS2 and ERG rearrangements in prostate cancer. J Pathol. 2013;230:174–183. doi: 10.1002/path.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu B, Maqsodi B, Yang W, et al. Detection of TMPRSS2-ERG fusion gene expression in prostate cancer specimens by a novel assay using branched DNA. Urology. 2009;74:1156–1161. doi: 10.1016/j.urology.2009.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaux A, Albadine R, Toubaji A, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35:1014–1020. doi: 10.1097/PAS.0b013e31821e8761. [DOI] [PMC free article] [PubMed] [Google Scholar]