Significance
In bacteria, small RNAs pair with target mRNAs to rapidly adjust gene expression in response to environmental and physiological changes. In this study, we identified a small RNA, SdhX, in Escherichia coli, whose expression is dependent on expression of tricarboxylic acid cycle genes. SdhX adjusts carbon flux by negatively regulating acetate kinase levels, improving growth on acetate. SdhX also leads to increased resistance to hydroxyurea, as do deletions of acetate kinase. Balancing metabolic flux is important for all bacterial cells, and SdhX plays a major role in this process by linking expression of tricarboxylic acid cycle genes to regulation of acetate metabolism, thus playing a role in gating overflow metabolism.
Keywords: RybD, acetate kinase, hydroxyurea, acetyl-phosphate, Hfq
Abstract
Bacterial regulatory small RNAs act as crucial regulators in central carbon metabolism by modulating translation initiation and degradation of target mRNAs in metabolic pathways. Here, we demonstrate that a noncoding small RNA, SdhX, is produced by RNase E-dependent processing from the 3′UTR of the sdhCDAB-sucABCD operon, encoding enzymes of the tricarboxylic acid (TCA) cycle. In Escherichia coli, SdhX negatively regulates ackA, which encodes an enzyme critical for degradation of the signaling molecule acetyl phosphate, while the downstream pta gene, encoding the enzyme critical for acetyl phosphate synthesis, is not significantly affected. This discoordinate regulation of pta and ackA increases the accumulation of acetyl phosphate when SdhX is expressed. Mutations in sdhX that abolish regulation of ackA lead to more acetate in the medium (more overflow metabolism), as well as a strong growth defect in the presence of acetate as sole carbon source, when the AckA-Pta pathway runs in reverse. SdhX overproduction confers resistance to hydroxyurea, via regulation of ackA. SdhX abundance is tightly coupled to the transcription signals of TCA cycle genes but escapes all known posttranscriptional regulation. Therefore, SdhX expression directly correlates with transcriptional input to the TCA cycle, providing an effective mechanism for the cell to link the TCA cycle with acetate metabolism pathways.
Bacterial survival and successful competition rely on efficiently extracting and consuming available carbon and energy sources from the environment. Bacteria have evolved complex regulatory networks enabling rapid adaption to environmental changes. Posttranscriptional regulation by small RNAs (sRNAs) can have significant effects on gene expression by reinforcing transcriptional regulation and providing links between different regulatory modules (1). Many sRNAs base pair with specific mRNA targets to regulate their stability and translation efficiency (reviewed in ref. 2). In Escherichia coli and many other bacteria, base-pairing is facilitated by the RNA chaperone Hfq, which also protects sRNAs from degradation by cellular nucleases (3–5). Here, we have investigated the role of sRNAs in regulation of acetate metabolism.
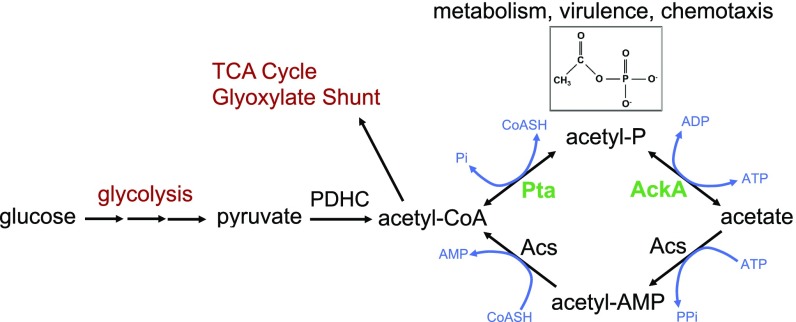
Acetate is ubiquitously found in natural environments, and is one of the major short-chain fatty acids in the gut, presumably providing a relevant nutrient for Enterobacteria as well as other bacteria (6, 7). Acetate is also excreted by cells, even during optimal growth under aerobic conditions, in a process called acetate overflow (8, 9). In E. coli, the primary pathway of acetate production involves two enzymes that are intimately connected to central metabolism, phosphotransacetylase (Pta) and acetate kinase (AckA). During exponential growth, acetyl-CoA, the product of glycolysis and the consumable substrate for the tricarboxylic acid (TCA) cycle, can be converted into acetyl-phosphate (AcP) by Pta and then into acetate by AckA (Fig. 1) (7). The relative activities of Pta and AckA serve to modulate intracellular levels of AcP, a high-energy molecule that regulates many cellular processes in Enterobacteria by phosphorylating or acetylating proteins and other molecules (10, 11). E. coli also takes up acetate, using the Pta-AckA pathway in reverse, resulting in synthesis of acetyl-CoA. This pathway typically operates at high extracellular acetate concentrations (≥8 mM) (9). While the Pta-AckA pathway is the primary one for consumption of acetate when glycolytic carbon sources are available, an alternative pathway for acetate conversion to acetyl-CoA is catalyzed by acetyl-CoA synthetase (Acs) (10, 12). Cells induce this second high-affinity irreversible pathway when carbon is limiting, during stationary phase and under low oxygen pressure, but AckA and Pta are critical for growth on higher levels of acetate as a sole carbon source (12–14).
Fig. 1.
Acetate activation pathways in E. coli. AckA, acetate kinase; Acs, AMP-forming acetyl-CoA synthetase; PDHC, pyruvate dehydrogenase complex, Pta, phosphotransacetylase; TCA, tricarboxylic acid. Acetyl-phosphate (AcP) is a high-energy form of phosphate and the most unstable phosphorylated compound in the cell. The size of its pool can vary considerably depending on the cell metabolic state and environmental factors (40, 41). AcP can function as a global signal by using either the phosphoryl or acetyl group to regulate many cellular processes such as carbon metabolism, flagellar biogenesis, biofilm development, capsule biosynthesis, and pathogenicity (7).
This study identifies an sRNA, SdhX [previously called RybD (15)], derived from the 3′UTR of sdhCDAB-sucABCD transcripts, as a major regulator of ackA gene expression, thereby affecting cellular levels of the signaling molecule AcP and the flux from acetyl-CoA to acetate and back. Unexpectedly, we also found that SdhX, via its effect on ackA, can reduce sensitivity to damage caused by the DNA replication inhibitor hydroxyurea. Our results suggest an efficient and effective mechanism for cells to coordinate the expression and metabolic state of the TCA cycle with acetate metabolism pathways and uncover cellular consequences of disrupting acetate metabolism (9).
Results
Acetate Metabolism Is Subject to sRNA Regulation.
Our previous RNA coimmunoprecipitation experiments with the Hfq chaperone in E. coli identified a number of potential new targets (15). Although posttranscriptional regulation of the genes of acetate metabolism has not been reported, the mRNA of ackA, encoding acetate kinase, was highly enriched in one of two experiments (39-fold compared with total mRNA), as well as in a number of other studies in enterobacteria (16, 17), suggesting it could be a target of Hfq-dependent sRNAs. Given the importance of the Pta-AckA pathway in the acetate switch (9) and AcP production (7), we further investigated this possibility. Posttranscriptional regulation of pta, although not enriched in Hfq immunoprecipitation experiments, was also examined.
To investigate potential regulation by sRNAs, translational fusions were created for ackA and pta, carrying the leader sequence and the first few codons of each gene, fused in-frame to lacZ, downstream from an arabinose-inducible PBAD promoter. Transcription start sites (TSS) of these genes were selected based on differential RNA sequencing data (SI Appendix, Fig. S1A) (18). The basal activity without arabinose was low for both fusions and showed significantly enhanced expression in the presence of arabinose (SI Appendix, Fig. S1B).
An sRNA plasmid library containing 30 individually cloned Hfq-binding sRNAs expressed from a PLAC promoter was transformed into each of the reporter fusion strains (19), and the β-galactosidase activities for strains with each plasmid were compared with the vector control. A change of twofold or higher was considered significant.
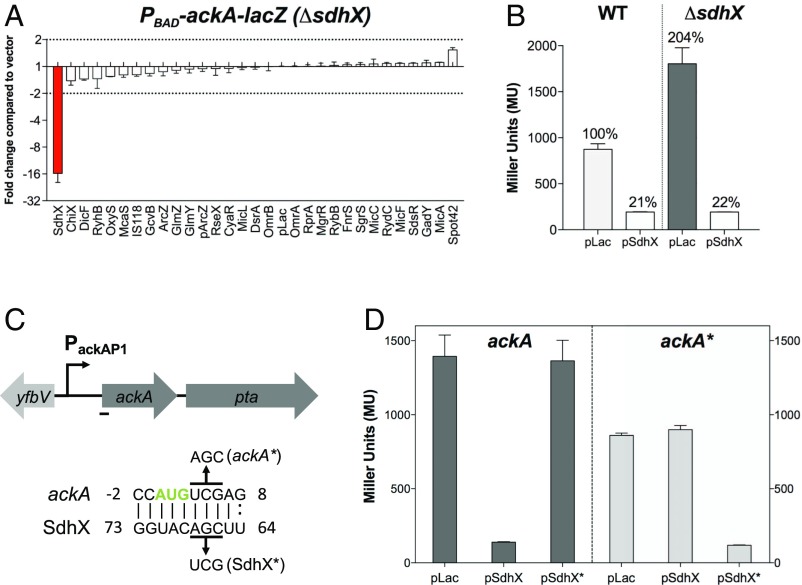
The pta-lacZ fusion was not significantly regulated by any of the tested sRNAs (SI Appendix, Fig. S2A). Interestingly, the activity of the ackA-lacZ translational fusion was strongly repressed by the overexpression of SdhX compared with the vector control (SI Appendix, Fig. S2B); the activity of the fusion was higher than the vector control (in the range of twofold) in cells overexpressing two other sRNAs (SdsR and Spot42). However, when the same fusion was screened in a strain deleted for the endogenous sdhX, SdhX down-regulation was stronger but the positive regulation by SdsR and Spot42 was not seen (Fig. 2A; compare with SI Appendix, Fig. S2B). These results suggest that SdsR and Spot42 primarily act indirectly, by interfering with regulation by the endogenous SdhX in wild-type cells. One explanation might be titration of the RNA chaperone Hfq (20) from either the sRNA or the target mRNA, although we cannot rule out a modest positive (direct or indirect) regulation by Spot 42. Consistent with regulation by the endogenous SdhX, the basal level of expression of the fusion was twofold higher in strains deleted for sdhX (Fig. 2B). Therefore, the native copy of SdhX has a strong effect on ackA gene expression when cells are growing in rich medium. Together, these results suggest that SdhX is the only sRNA in the library that significantly regulates ackA expression. This conclusion is consistent with a recent report using a system-wide approach for isolating chimeras of sRNAs and their target mRNAs, termed RIL-seq (RNA interaction by ligation and sequencing), published during this study, that implicated ackA as an SdhX target but found no other sRNAs associated with ackA (21).
Fig. 2.
SdhX regulates ackA gene expression at the posttranscriptional level. (A) Screening of the sRNA library with the PBAD-ackA-lacZ fusion in a ΔsdhX background (FDM1701). Cells were grown and assayed as described in Materials and Methods, using 0.02% arabinose. Vector control expressed 156 machine units. Bar showing significant change (>2×) is highlighted in red. (B) β-Galactosidase assays of wild-type (FDM1700) and ΔsdhX (FDM1701) strains carrying a PBAD-ackA-lacZ translational fusion and transformed either with the vector control PLAC or pSdhX. Cells were grown in flasks at 37 °C in LB containing ampicillin (100 μg/mL), IPTG (100 μM), and 0.02% arabinose to stationary phase (OD600 of between 2.0 and 2.5) and assayed for β-galactosidase. (C) Schematic of ackA-pta genes and in silico base-pairing prediction between the single-stranded region of SdhX and ackA mRNA showing base pairing overlapping the ackA start codon (green); mutations created to test this pairing are shown above (ackA*) and below (SdhX*) the sequences. The ackA promoter is indicated as a black arrow (SI Appendix, Fig. S1C). (D) β-Galactosidase assays of ΔsdhX cells carrying PBAD-ackA–lacZ (FDM1701) or PBAD-ackA*–lacZ (FDM1707) translational fusions and transformed either with PLAC, pSdhX, or pSdhX*. Cells were grown as in B for 4 h before being assayed for β-galactosidase. Three independent cultures were assayed for each strain and experiment; data are plotted as mean ± SD.
SdhX Directly Represses ackA Gene Expression.
Both the RNA folding algorithm Mfold (22) and the comparative prediction algorithm for sRNA targets (CopraRNA) (23) predict pairing of SdhX with a region overlapping the start codon of ackA mRNA (Fig. 2C). The pairing region in SdhX is within the most conserved sequence (discussed further below). The predicted pairing was confirmed using mutations to disrupt pairing and compensating mutations to restore pairing within SdhX and the ackA–lacZ translational fusion (Fig. 2D). Overexpression of a mutant variant of SdhX, SdhX*, carrying a 3-nt substitution in the predicted base-pairing region (Fig. 2C), was unable to repress the ackA–lacZ fusion. Introducing a compensatory mutation in the ackA–lacZ translational fusion (ackA*), by creating a silent/synonymous mutation in the second codon of ackA, resulted in modestly decreased expression of the fusion and fully prevented repression by the wild-type SdhX. The compensatory mutation was efficiently suppressed by overexpression of SdhX*. Therefore, translational repression of ackA by SdhX is mediated by direct base pairing in a region overlapping the translation start codon.
SdhX Contributes to the Discoordinate Expression of the ackA-pta Operon.
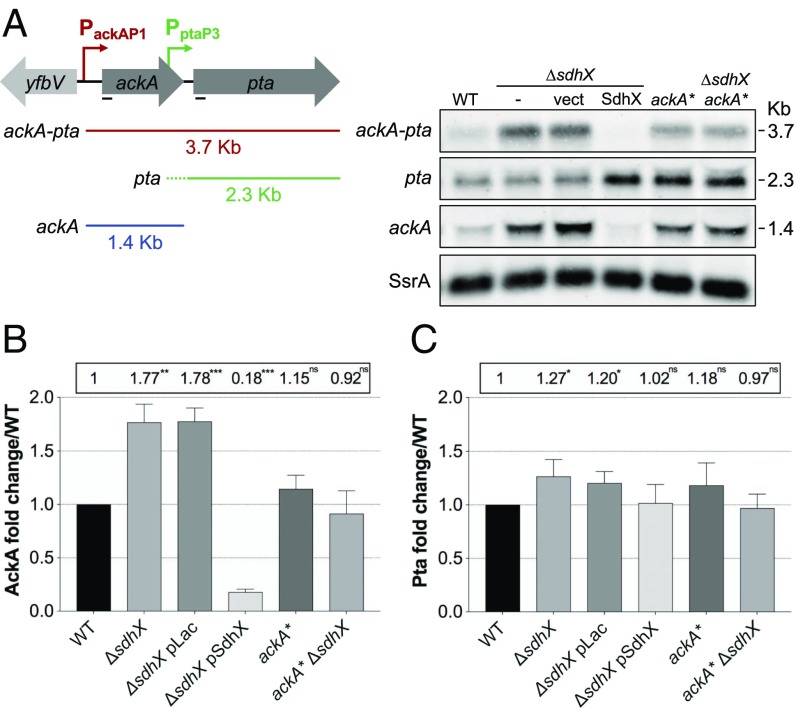
We investigated the effect of SdhX on the native ackA and downstream pta genes at the posttranscriptional and translational levels. In a wild-type strain, the full-length ackA-pta transcript (∼3.7 kb) accounts for around 25–30% of the total signal for ackA mRNA; the remaining 70–75% is seen as an ∼1.45-kb RNA, the size expected for the ackA gene and its leader (Fig. 3A and SI Appendix, Fig. S3 A and B). The size of this transcript suggests that it derives from termination and processing within the ackA-pta intergenic region (Fig. 3A and SI Appendix, Fig. S3A). A transcript of ∼2.3 kb was detected for the pta gene, consistent with transcription initiation and processing within the ackA-pta intergenic region; this transcript represents ∼75% of total pta transcripts in wild-type cells.
Fig. 3.
SdhX enhances discoordinate expression within the ackA-pta operon. (A, Left) A scheme of ackA-pta (red), pta (green), and ackA (blue) transcripts derived from the ackA promoter (PackA) and partially from the pta promoter (Ppta, 263-nt upstream of pta CDS). The ackA-pta intergenic region is 74 nt. (Right) A Northern blot analysis of ackA and pta expression in the wild-type strain (NM525) and its derivatives ΔsdhX (FDM1708) carrying either the vector control or a plasmid expressing SdhX sRNA, ackA* (FDM1712), and ackA* ΔsdhX (FDM1713). Cells were grown at 37 °C in LB containing 100 μg/mL ampicillin and 100 μM IPTG to induce sRNA expression. Total RNA was extracted at OD600 of 1 and transcripts were probed for both ackA and pta (black lines, Left). Transcripts were quantified with the same samples on an independently probed gel and are plotted in SI Appendix, Fig. S3 A and B. Band intensities were normalized with SsrA as a loading control. (B and C) Fold-changes of AckA (B) and Pta (C) proteins relative to the wild-type strain, set to 1, in the same growth conditions as in A and normalized with EF-Tu loading control. Strains are as listed in legend for SI Appendix, Fig. S3C. Error bars indicate SD of three biological replicates. Unpaired Student’s t test was used to calculate statistical significance (ns, not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001). A representative Western blot is shown in SI Appendix, Fig. S3C.
SdhX had differential effects on expression of ackA and pta mRNAs. Deletion of the sdhX allele increased expression of both the full-length transcript and the ackA-specific transcript, producing a net increase in ackA-encoding transcripts of three- to fourfold (SI Appendix, Fig. S3B, Left graph). The pta-specific mRNA levels did not increase (Fig. 3A and SI Appendix, Fig. S3B, Right graph, green bars), although the increase in the full-length transcript doubled the total transcripts containing pta (SI Appendix, Fig. S3B, red bars). Overexpressing SdhX in a ΔsdhX strain eliminated the full-length ackA-pta transcript and drastically reduced overall ackA transcript levels, about 25-fold compared with the ΔsdhX strain containing the vector and more than 6-fold compared with the wild-type (sdhX+) strain (SI Appendix, Fig. S3B). In striking contrast, the 2.3-kb pta band was not decreased and in fact was slightly increased, for unknown reasons, upon SdhX overexpression. The effect of SdhX was dependent upon pairing to ackA; strains carrying the ackA* mutation in the native ackA gene, which renders ackA resistant to repression by chromosomally encoded SdhX, had full-length ackA-pta mRNA levels slightly higher than the wild-type strain. Neither ackA mRNA nor pta mRNA levels were affected by deleting sdhX in the ackA* strain (Fig. 3A and SI Appendix, Fig. S3B).
Levels of the AckA protein and a FLAG-tagged version of the Pta protein were also measured as a function of changes in SdhX and its ability to pair with ackA (Fig. 3 B and C and SI Appendix, Fig. S3C). In the absence of SdhX, the amount of AckA relative to the wild-type strain increased (Fig. 3B) but Pta levels increased only slightly (Fig. 3C). In the presence of overproduced SdhX, AckA protein was reduced by fivefold (Fig. 3B, fourth vs. first bar), while Pta was not affected (Fig. 3C, fourth vs. first bar). The ackA* mutation did not significantly affect AckA and Pta levels, either in the presence or absence of SdhX (Fig. 3 B and C), consistent with the inability of SdhX to pair with this mutant. Note that the ackA* mutation has two effects, decreasing translation in the absence of SdhX (see Fig. 2D, compare first bar in each panel, or compare bar 2 to bar 6 in Fig. 3B), but also relieving SdhX repression. These effects balance out, so that the AckA levels were not significantly different in the wild-type and ackA* strains, in the presence or absence of SdhX.
Overall, our results demonstrate that SdhX specifically reduces ackA-pta and ackA transcripts; pta transcripts are not affected. At both the level of mRNA and protein, SdhX has discoordinate effects on AckA and Pta.
ackA and pta are generally considered to constitute an operon in E. coli, consistent with our detection of a full-length ackA-pta transcript, particularly in the absence of SdhX. However, the existence of a pta specific promoter has been suggested (24), and would be consistent with our finding that a functional pta transcript is present even in the absence of detectable ackA transcripts (Fig. 3A and SI Appendix, Fig. S3 A and B). Transcriptome analysis of E. coli mapped a putative transcription initiation site for pta, at the 3′ end of the ackA coding sequence (18); we will refer to this promoter, confirmed here, as P3.
A series of pta–lacZ transcriptional reporter fusions containing various regions upstream of the predicted pta P3 promoter were tested for activity (SI Appendix, Fig. S4A). The results suggest the presence of a promoter within ackA, accounting for about a third of the total pta expression in minimal medium supplemented with glucose. A 5′ RACE of this ackA 3′ region identified a TSS at −261 from the pta start codon (upstream from the ackA stop codon) (SI Appendix, Fig. S4B, *), as well as a processing site in the ackA-pta intergenic region (SI Appendix, Fig. S4B, **). Therefore, a promoter capable of expressing pta is present in the 3′ end of the ackA ORF (SI Appendix, Fig. S4B), consistent with previous reports (18, 24). A transcript initiating at this promoter will not carry the SdhX pairing region, and thus should be resistant to SdhX repression.
Processing within the ackA-pta intergenic region may also contribute to insulating the pta transcript from SdhX repression. This processing is at a conserved GTTTT sequence, a consensus motif for RNase E, at the position shown as double asterisks (**) in SI Appendix, Fig. S4B (25). The cleavage occurs downstream of the first highly structured hairpin, leaving a second hairpin at the 5′ end of processed pta transcripts (SI Appendix, Fig. S4C) and presumably releasing the 1.45-kb ackA band from the full-length transcript. Consistent with a role for RNase E, levels of the full-length transcript and of a slightly larger pta transcript, presumably initiating at the newly defined pta P3 promoter, were increased under conditions in which RNase E is inactive (SI Appendix, Fig. S4C).
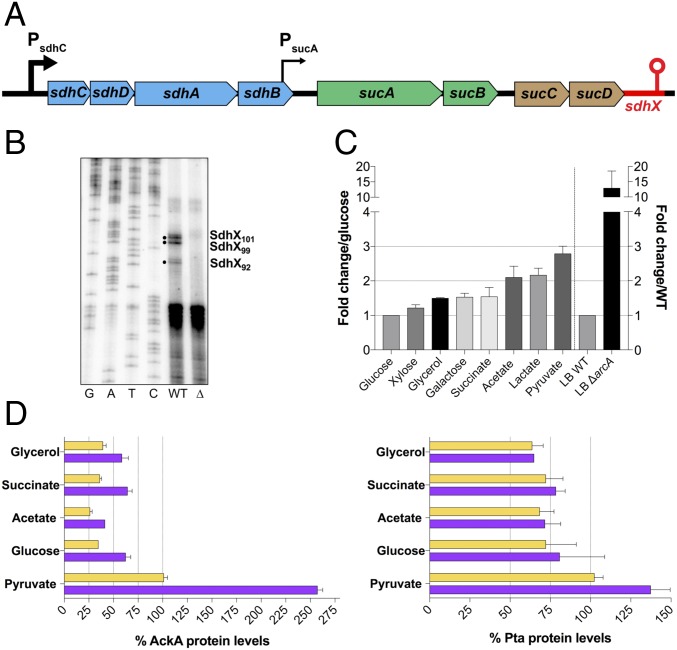
SdhX Is a Processed Hfq-Dependent sRNA Expressed from the sdhC Promoter.
SdhX, originally named RybD, was detected among the RNA species coimmunoprecipitated with Hfq (15, 26), suggesting it is an Hfq-dependent sRNA. It is encoded immediately downstream of the sdhCDAB-sucABCD gene cluster (Fig. 4A), adding to the number of sRNAs that have been reported to be encoded in mRNA 3′UTRs (21, 27). Some of these sRNAs are synthesized from specific promoters within an upstream ORF (28), but others are transcriptionally dependent on their upstream genes (29). We investigated conditions under which functional SdhX is produced. The 5′ end was mapped by primer extension; three prominent bands were detected that correspond to lengths of SdhX of 101, 99, and 92 nt (Fig. 4B). All SdhX species were sensitive to treatment with the terminator 5′-P-dependent exonuclease, indicating that all species are processed from longer transcripts (SI Appendix, Fig. S5A). Based on the first identified 5′ end, the longest form of E. coli SdhX is a 101-nt sRNA, with the processing site immediately downstream of the sucD stop codon (SI Appendix, Fig. S5B). The shortest form (SdhX-S) corresponds to an ∼38-nt sRNA that includes only a short sequence upstream of the predicted Rho-independent terminator (SI Appendix, Fig. S5B) and was not abundant in our experiments (SI Appendix, Fig. S5A). This region, conserved in other Enterobacterial species, includes the ackA pairing site (Fig. 2C and marked with asterisks in SI Appendix, Fig. S5B), and is thus likely to provide the seed domain, defined as the primary site for SdhX pairing and regulation of most of its mRNA targets. In silico predictions and in vitro structure probing of SdhX suggests the sRNA is highly structured, with only the 5′ end and the seed domain (nucleotides 64–75) likely to be single-stranded (SI Appendix, Fig. S5 C and D).
Fig. 4.
Characterization of SdhX as a processed 3′UTR-derived sRNA. (A) Schematic shows the sdh and suc genes, PsdhC and weak constitutive PsucA promoters (35) (black arrows) and sRNA gene (red). Genes coding for succinate dehydrogenase are colored in blue, 2-oxoglutarate dehydrogenase in green, and succinyl-CoA synthetase in brown. (B) Mapping of SdhX 5′ ends by primer extension. Analysis was carried out using primer FDM17 (SI Appendix, Table S3) on total RNA isolated from NM525 (wild type) and isogenic ΔsdhX (Δ) (FDM1708) cells grown to OD600 of 4 in LB medium. Solid bullets indicate prominent extension products present in the wild-type strain but absent in the ΔsdhX strain. The first four lanes are sequencing ladders. (C) Quantification of endogenous SdhX expression for cells (NM525) grown in MOPS minimal medium supplemented with different carbon sources plotted relative to growth in glucose (left side of graph) or in LB rich medium with ΔarcA derivative (FDM1733) relative to wild type (right side of graph). Cells were grown overnight with the indicated carbon source in minimal medium and diluted to an OD600 of 0.05 into the same type of media. Cells were then grown until midexponentional phase (OD600 ∼ 0.4) before isolation of total RNA. Wild-type cells in LB medium express about 10 times less SdhX than cells grown in MOPS minimal medium supplemented with glucose during midexponential growth. The concentration of specific carbon sources was 0.5% glycerol, 0.2% d-glucose, 0.2% d-xylose, 0.2% d-galactose, 60 mM sodium acetate, 0.6% sodium DL-lactate, 0.6% sodium pyruvate, and 0.8% sodium succinate hexahydrate. Results are representative of two independent experiments; data are plotted as mean ± SD. SI Appendix, Fig. S6C shows one of these Northern blots. (D) Western blot quantification of AckA and Pta relative protein levels from a wild-type strain (FDM1714) (yellow bars) and isogenic ΔsdhX (FDM1715) (purple bars) grown with the indicated carbon sources during midexponential growth (OD600 ∼ 0.4). Experiments were repeated two times; a representative blot is shown in SI Appendix, Fig. S7.
In cells lacking Hfq, very little SdhX was detected (SI Appendix, Fig. S5E) and repression of ackA was lost (SI Appendix, Fig. S5F), consistent with its identification as an Hfq-dependent regulatory sRNA. Aberrant cleavage products were detected from the endogenous SdhX copy in the absence of Hfq (SI Appendix, Fig. S5E), suggesting a role for Hfq in the accurate processing of SdhX.
The data above indicate that SdhX is processed from the 3′UTR of sucD mRNA. sucD is part of the sdhCDAB-sucABCD operon and has been reported to be transcribed primarily from the promoter upstream of sdhC (30, 31). Previous studies suggest that RNase III, which cuts double-stranded RNAs, separates the sdhCDAB and sucABCD RNAs by cleavage within a hairpin in the sdhB-sucA intergenic region (32). RNase E has also been found to play a role in sdhCDAB-sucABCD processing and degradation (33).
We examined the role of these ribonucleases in SdhX biogenesis using isogenic strains carrying wild-type or mutant alleles of RNase III (Δrnc; rnc::catR) or RNase E (rne-3071, expressing a thermosensitive protein), or both. After a short incubation at the nonpermissive temperature (43.5 °C), the rne-3071 mutant and the rne-3071 rnc double mutant accumulate higher molecular weight precursors but lose much of the mature SdhX band (SI Appendix, Fig. S6A, SdhX probe, compare lane 6 to lane 2), suggesting that the RNase E endoribonuclease is critical for formation of SdhX, and the preexisting SdhX was degraded during this high-temperature incubation. RNase III may contribute modestly to production of SdhX as well, either directly or possibly indirectly (SI Appendix, Fig. S6A, lower levels of SdhX in lanes 3 and 4 compared with 1 and 2, not a consistent finding). High molecular weight bands consistent with sucABCD-SdhX transcripts accumulate in the rne-3071 strain at the nonpermissive temperature (light green and purple arrows, SI Appendix, Fig. S6A, SdhX probe, lane 6), and a full-length sdhCDAB-sucABCD-SdhX transcript was seen in the absence of both RNase E and RNase III activities (10 kb) (cyan arrow, SI Appendix, Fig. S6A, SdhX probe, lane 8). The origin of this transcript was confirmed by probing the blot for sdhC (SI Appendix, Fig. S6A, sdhC probe). These results are consistent with previously reported RNase III processing in the sdhB-sucA intergenic region (32) and with SdhX being processed from this sdhCDAB-sucABCD mRNA.
The critical importance of RNase E for production of SdhX was confirmed by monitoring the appearance of SdhX after reactivation of RNase E in the rne-3071 thermosensitive strain (SI Appendix, Fig. S6B). After inactivation of RNase E, processed SdhX levels fell to very low levels (SI Appendix, Fig. S6B, 0 min lane), whereas substantial amounts of the longer suc-sdhX mRNA remained. The culture was then shifted back to 32 °C in the presence of rifampicin to prevent new rounds of transcription, and the rate at which mature SdhX appeared was monitored. Mature SdhX increased over time, reaching levels about 35-fold higher after 20 min (SI Appendix, Fig. S6B, red line), while the sucABCD-sdhX transcripts (SI Appendix, Fig. S6B, light green line) disappeared, consistent with processing of this longer transcript by RNase E to produce SdhX.
Our data support a model in which SdhX is transcribed as part of the sdh-suc operon and functional SdhX sRNA is the product of processing of the precursor sdhCDAB-sucABCD and sucABCD transcripts. Furthermore, RNase E is required for this processing. Based on this model, we would expect expression of this sRNA to depend upon expression of the sdh-suc operon.
If SdhX is processed from the sdh-suc transcript, its transcription should reflect transcription of the operon. The sdhC promoter is known to be positively regulated by CRP/cAMP, and is thus poorly expressed in the presence of glucose (30). Accordingly, we found that SdhX expression was lower in medium with glucose, compared with its expression in other carbon sources (Fig. 4C, left side of graph, and SI Appendix, Fig. S6C). Pyruvate is the terminal product of glycolysis and enters immediately into the TCA cycle under aerobic growth conditions. Consistent with expected induction of TCA cycle genes in cells grown with pyruvate as the carbon source, SdhX expression was about threefold higher in pyruvate-grown cells compared with cells grown in glucose. In addition, deletion of arcA, which represses an sdhC–lacZ fusion but not a sucA–lacZ fusion (34, 35), increased SdhX expression in LB significantly even when grown under relatively aerobic conditions (by 12-fold) (Fig. 4C, right side of graph). Although ArcA is most active as a repressor of the sdh promoter during anaerobic growth (35), it clearly does contribute to repression under our growth conditions as well. Thus, SdhX sRNA follows the same expression pattern previously reported for the sdhCDAB-sucABCD operon (30), consistent with the majority of SdhX synthesis initiating from the sdh promoter.
SdhX Is Insulated from sRNA Regulators of the sdh-suc Genes.
The sdhCDAB-sucABCD operon is subject to posttranscriptional regulation by several Hfq-dependent sRNAs (36, 37), leading to more rapid turnover of the mRNA. Because of the effect of these sRNAs on mRNA stability, we initially expected that they would also affect accumulation of SdhX. To test this idea, we monitored the levels of native SdhX and of sdhCDAB mRNA before and after induction of each of the three known sRNA regulators, RyhB, RybB, and Spot 42. While expression of any of the three sRNAs caused the expected loss of the sdhCDAB mRNA, none had a significant effect on expression of SdhX (SI Appendix, Fig. S6D). Therefore, SdhX is insulated from the effects of these sRNAs, likely because processing separates it from the longer transcripts before it can be degraded.
SdhX-Dependent Regulation of AckA Changes with the Carbon Source.
We would expect growth conditions that lead to high levels of SdhX to affect the amounts of AckA and to a lesser extent Pta (as in Fig. 3 B and C), while lower levels of SdhX would not have major effects. We measured AckA and Pta levels in wild-type and ΔsdhX cells in exponential phase growth on some of the carbon sources (Fig. 4 C and D). AckA and Pta levels were first compared in the absence of SdhX (Fig. 4D, purple bars) to evaluate regulation by carbon source at the level of transcription, or possibly translation, but independent of SdhX sRNA. The highest levels of both proteins were observed in cells grown in pyruvate, with AckA levels three- to fourfold higher and Pta levels almost twofold higher, compared with glucose-grown cells, consistent with an effect of pyruvate on the ackA P1 promoter (SI Appendix, Fig. S4A). In SdhX+ cells, the difference in AckA levels in cells grown on pyruvate versus other carbon sources was significantly smaller (Fig. 4D, yellow bars). Presumably, this reflects the higher levels of SdhX in cells grown on pyruvate (Fig. 4C). Consistent with that finding, when AckA levels were compared in the absence and presence of SdhX, the biggest difference was observed in cells grown on pyruvate (2.5-fold), while the ratio was less with other carbon sources (1.5- to 1.8-fold). As expected, there was very little effect of SdhX on Pta expression (1- to 1.3-fold).
In the glycolytic pathway, pyruvate is the precursor of acetyl-CoA, which is a substrate for Pta and is subsequently converted to acetate by AckA (Fig. 1). Increasing expression of these genes in the presence of pyruvate might lead to more metabolizing of acetyl-CoA to AcP and acetate, diverting it from other pathways. Acetate, although the substrate for the reverse reaction catalyzed by AckA and Pta, did not lead to induction of either gene. This lack of responsiveness of ackA-pta to exogenous acetate is consistent with studies of Oh et al. (38), who found induction of acs but very little change in ackA or pta transcripts in cells grown in acetate, compared with growth in glucose.
These results suggest that both SdhX-dependent and SdhX-independent processes will regulate acetate metabolism, primarily by modulating AckA levels, and that SdhX enhances the discoordinate expression of ackA and pta genes in the presence of specific carbon and energy sources, particularly in pyruvate.
SdhX Modulates AcP and Acetate Accumulation.
We would expect that SdhX, by attenuating intracellular AckA levels, should influence phenotypes associated with AckA activity. We examined the impact of this sRNA on a number of the known functions of AckA, starting with accumulation of AcP.
Under most growth conditions, AcP is synthesized by Pta and degraded by AckA. Therefore, the balance between these two proteins could affect how much AcP accumulates. Previous studies showed that AcP levels in glucose-grown cells as sole carbon source are significantly higher in ΔackA cells, but are very low or undetectable in Δpta and Δ(ackA-pta) cells (10, 39), as expected; levels of AcP were high in cells grown on pyruvate (40). We predicted that higher SdhX levels, by reducing AckA, should increase AcP levels in vivo, and that lack of SdhX might decrease AcP levels by increasing AckA. AcP levels were determined in cells grown to midexponential phase in MOPS pyruvate, where SdhX expression is high (Fig. 4C).
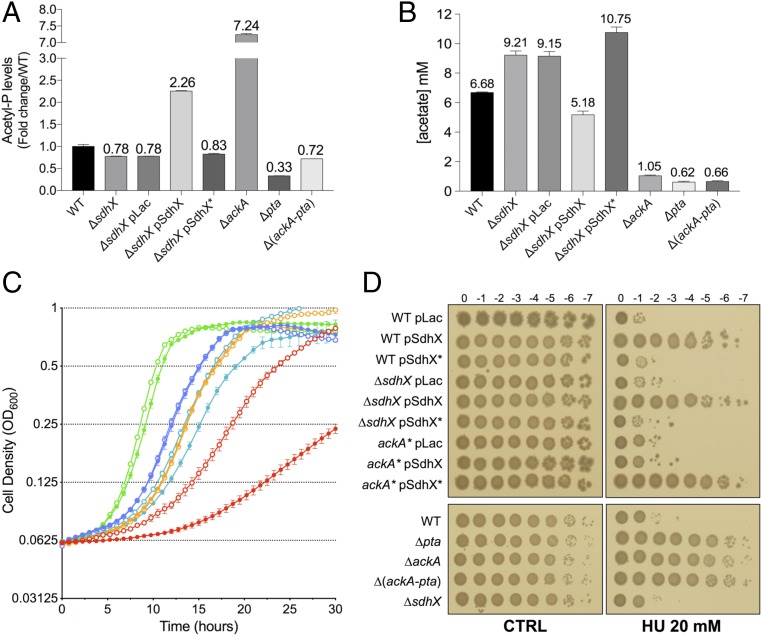
Deletion of ackA led to a dramatic increase in AcP [Fig. 5A; compare wild type (bar 1) to ΔackA (bar 6)] (41), and deletion of pta or ackA and pta lowered AcP levels (Fig. 5A, bars 7 and 8). An sdhX deletion reduced AcP levels close to that seen in the Δ(ackA-pta) cells (Fig. 5A; compare bars 2 and 3 to bar 8), although under these conditions, this level was only modestly lower than that seen in wild-type cells (Fig. 5A, compare bars 1–3 and 8). This result suggests that chromosomal levels of SdhX contribute to AcP accumulation by repressing AckA expression, thus slowing the rate at which AcP is converted to acetate. In contrast, overexpression of wild-type SdhX, which led to a dramatic reduction in AckA levels (Fig. 3B), resulted in a 2.3-fold increase in AcP levels (Fig. 5A, compare bar 4 to bar 1), while overexpression of SdhX*, which is unable to repress ackA expression (Fig. 3 A and B), had little effect on the accumulation of AcP.
Fig. 5.
Phenotypic effects of SdhX Expression. (A) Fold-change of intracellular AcP levels of bacterial strains relative to wild type [wild type: NM525; ΔsdhX: FDM1708; ΔackA: FDM1709; Δpta: FDM1711; Δ(ackA-pta): FDM1710] grown in MOPS minimal medium supplemented with sodium pyruvate 0.6% as carbon source at exponential phase (OD600 ∼ 0.4–0.5). (B) Extracellular acetate concentration in medium grown with the same set of strains and conditions as in A. Data represent the mean of three independent experiments for each strain ±SD. (C) Growth curve of SdhXseedless (FDM1752, filled circles) and its wild-type derivative (FDM1753, open circles) in MOPS minimal medium supplemented with 0.2% glucose (green lines), 0.8% Na succinate hexahydrate (blue lines), 0.5% glycerol (orange lines), 0.6% Na pyruvate (cyan lines), or 60 mM Na acetate (red lines) as carbon source. Each curve represents the mean of two independent experiments from colonies grown overnight in LB rich medium. (D) Efficiency of plating spot assays on hydroxyurea in strains listed in the legend to SI Appendix, Fig. S9A. Serial dilutions of bacterial overnight cultures were spotted on LB agar plates (control) and on LB agar plates containing 20 mM hydroxyurea (HU), supplemented with ampicillin and IPTG 1 mM when appropriate.
We also measured extracellular acetate concentrations in the medium during growth of these strains (Fig. 5B). As expected, strains in which either ackA or pta was deleted did not accumulate acetate. Overexpression of SdhX led to reduced levels of acetate, although not nearly to the extent seen in the complete absence of ackA, suggesting that even low levels of AckA are sufficient to allow acetate accumulation. Deletion of sdhX led to increased levels of acetate, consistent with more flux through the pathway in response to the increase in AckA. Thus, the effects of SdhX on acetate were inversely correlated to the AcP accumulation (Fig. 5B), providing evidence that SdhX regulates acetate metabolism and the relative abundance of AcP and acetate by modulating AckA expression. Such an inverse correlation was also seen under other growth conditions, studied by others (40). These results suggests that phosphorolysis of AcP by AckA is likely to be the rate-limiting step in this pathway under these growth conditions.
Pta and AckA control flux from acetyl-CoA to acetate, but can operate in the opposite direction when extracellular acetate concentrations are high, assimilating the acetate and metabolizing it to acetyl-CoA. AckA catalyzes the initial step by converting acetate to AcP. Strikingly, a modified endogenous SdhX sRNA without its seed region (Fig. 5C, closed circles) strongly perturbed growth on acetate as sole carbon source compared with the parental wild-type strain (Fig. 5C, open circles; acetate growth, red lines) (doubling time of ∼500 min for SdhXseedless vs. ∼250 min for SdhX wild type). The loss of the ackA pairing region had no effect on cell growth in glycerol or succinate (Fig. 5C, orange and blue lines) and modest effects on cell growth in glucose (Fig. 5C, green lines). When cells were grown in pyruvate, there was a lag in growth in the seedless mutant, but little effect on the doubling time (Fig. 5C, pink lines), indicating that the SdhXseedless primarily affects a process specifically required for growth on acetate (Fig. 5C). We separately confirmed that growth on high acetate concentrations, but not growth on pyruvate, was dependent upon AckA and Pta, but not Acs, until higher ODs, when acetate levels presumably drop (SI Appendix, Fig. S8A), consistent with previous observations (12).
SdhX- and AcP-Associated Phenotypes.
We next examined the role of SdhX under two conditions in which AcP has been implicated in regulation. RpoS degradation was suggested to be regulated in part by AcP-dependent phosphorylation of the adaptor protein RssB, based on the observation that levels of RpoS are higher in cells lacking the ackA-pta locus (42). However, we found that deletion of ackA (high AcP), pta (low AcP), or both (low AcP) all increased RpoS levels (SI Appendix, Fig. S8B), suggesting that interrupting flux through this pathway, rather than lack of AcP, leads to up-regulation of RpoS. Neither deletion of sdhX nor multicopy SdhX affected RpoS levels (SI Appendix, Fig. S8B). Because multicopy SdhX reduced AckA levels by 10-fold, these results suggest that only the full absence (or extremely low levels) of AckA generates conditions that promote higher levels of RpoS.
The RcsB response regulator has been reported to be subject to phosphorylation by AcP in the absence of the cognate histidine kinase, RcsC, or phosphorelay protein, RcsD (43). We used a highly sensitive reporter for the RcsB response regulator (44), rprA-mcherry, to test the role of AcP in promoting RcsB activity. Deleting ackA increased mCherry expression, while deleting pta alone or ackA and pta reduced expression relative to the parental ΔrcsD control (SI Appendix, Fig. S8C). Therefore, expression of this reporter correlates well with AcP levels. Surprisingly, however, even when cells were grown in pyruvate, a condition in which SdhX is highly induced, no effect of deleting sdhX was detected (SI Appendix, Fig. S8C). Multicopy expression of SdhX (which increases AcP levels, although not as much as deleting ackA) (Fig. 5A) was also not sufficient to increase the expression of the reporter (SI Appendix, Fig. S8D). Thus, under the conditions tested, we were unable to detect an SdhX phenotype attributable to changes in AcP levels.
SdhX Confers Resistance to Hydroxyurea via Repression of AckA.
To further explore the physiological roles of SdhX, we sought phenotypes characteristic of cells deleted for ackA, reasoning that these phenotypes might be mimicked by strong SdhX repression of ackA. Among the strongest phenotypes of an ackA deletion in a chemical biology screen of E. coli mutants (45), was increased resistance to hydroxyurea. Nakayashiki and Mori (46) recently reported that E. coli respiration-defective mutants were significantly more resistant to hydroxyurea, possibly because they generated less reactive oxygen species (ROS) (47). Hydroxyurea specifically inhibits class I ribonucleotide reductase, leading to depletion of dNTP pools under aerobic conditions (48, 49), and subsequently causes replication fork arrest and arrest of DNA synthesis.
In agreement with published data (46), an in-frame ackA deletion was hyperresistant to hydroxyurea (Fig. 5D and SI Appendix, Fig. S9A). Δpta and Δ(ackA-pta) strains were also resistant; thus, the phenotype is likely not dependent on AcP levels but on blocking the ackA-pta pathway, as was seen with RpoS. The wild-type strain carrying a vector control plated with an efficiency of 10−5 (Fig. 5D and SI Appendix, Fig. S9A). Overexpression of SdhX from a plasmid in the wild-type strain increased the ability to form colonies more than 10,000-fold, comparable to the ackA strain (Fig. 5D and SI Appendix, Fig. S9A). Increased resistance was not seen in wild-type cells overexpressing SdhX* from a plasmid but was restored when the SdhX* plasmid was moved to a strain expressing a chromosomal ackA* mutation, indicating that resistance to hydroxyurea was dependent on down-regulation of ackA by SdhX.
Recent work suggested that activation of the Cpx two-component system confers resistance to hydroxyurea (50). However, the hydroxyurea resistance caused by disruption of the AckA-Pta pathway does not depend upon the Cpx response, because inactivation of the response regulator for the system, CpxR, did not reverse hydroxyurea resistance (SI Appendix, Fig. S9B).
SdhX Contributes to Hydrogen Peroxide Sensitivity, Independent of AckA.
Previous studies suggested that increased resistance to hydroxyurea might reflect less endogenous ROS generation (47), leading us to examine the effect of SdhX on the ability of cells to handle ROS generated by addition of hydrogen peroxide (SI Appendix, Fig. S9C). When wild-type cells were grown in microtiter dishes in the presence of 8 mM hydrogen peroxide, they showed a long lag before growth commenced. Strikingly, deletion of sdhX consistently reduced this lag (compare orange and blue lines in SI Appendix, Fig. S9C), and increased levels of SdhX or of SdhX* (unable to regulate ackA) fully inhibited growth during the 15 h of the experiment. Thus, this phenotype, while affected by SdhX (reduced sensitivity without SdhX, increased sensitivity with more SdhX), is likely ackA-independent. We searched for other possible targets of SdhX, using the IntaRNA algorithm (51). The best predicted mRNA target for SdhX was the H2O2-induced catalase katG, which is predicted to pair in the poorly conserved 5′ region of SdhX (SI Appendix, Figs. S5C and S9C). This catalase, together with katE, is an important scavenger of high H2O2 concentrations (52) and, as expected, a katG mutant did not recover within 15 h in the presence of 8 mM H2O2 (SI Appendix, Fig. S9C). A variant of SdhX mutated in the katG pairing region (SdhX**; green line) did not block growth on H2O2 (SI Appendix, Fig. S9C), suggesting that repression of katG by SdhX is likely the basis for the increased H2O2 sensitivity.
Discussion
Enterobacteria share highly conserved central metabolic pathways and are capable of very rapid metabolic adaptation to changes in the environment. These acclimatizations depend on a variety of regulatory mechanisms, including feedback control of enzymes, transcriptional control of expression, as well as translational control, frequently mediated by sRNAs. sRNAs can refine the timing and intensity of the primary transcriptional response, as well as reinforce its function by extending the targetome (37, 53). New RNA sequencing-based approaches have established the 3′UTR of mRNAs as a previously unappreciated rich sRNA reservoir in bacteria (21, 26). Our study investigates an important function of E. coli SdhX, previously annotated as RybD (15), a 3′UTR-encoded sRNA whose expression is linked to that of the upstream sdhCDAB-sucABCD mRNA, encoding three complexes of the TCA cycle. SdhX acts to limit the expression of acetate kinase and thus helps to adjust flux through an important metabolic node highly sensitive to changes in the nutritional status of growing cells. Complementary studies of SdhX (RybD) in Salmonella and E. coli have been carried out by Miyakoshi et al. (54), highlighting some of the similarities and differences in SdhX regulation in these closely related organisms.
SdhX Is a Robust Regulator, Expressed Under Conditions of TCA Cycle Function.
SdhX is processed from the mRNA of the complex sdh-suc operon, dependent upon the activity of RNase E. In the absence of RNase E, SdhX sequences were found exclusively in the mRNA of the operon, indicating that SdhX synthesis is dependent on transcription of the upstream operon genes (SI Appendix, Fig. S6 A and B). In E. coli, the RNase E-mediated cleavage to release SdhX occurs just downstream of the sucD terminator codon (SI Appendix, Fig. S5B). The location of this AU-rich cleavage site is not conserved among other Enterobacteria; the primary cleavage site of Salmonella SdhX is within the sucD mRNA coding region (55). Nonetheless, expression of the sRNA from the upstream sdh promoters is likely to be conserved in Enterobacteria. Only a short sequence within sdhX, upstream of the Rho-independent terminator, is well conserved (SI Appendix, Fig. S5B). This sequence is the region of pairing with ackA and with most, but not all, other computationally predicted mRNA targets, and we thus identify this sequence as the seed region of SdhX.
The expression pattern of SdhX is consistent with transcription dependent on the promoter upstream of the sdh-suc operon (30, 32, 34, 35). The sdhC promoter responds to carbon availability, via activation by Crp/cAMP and to availability of oxygen, via repression by the response regulator ArcA, most active under anaerobic conditions (35, 56). As expected, SdhX levels were low in glucose relative to less-favored carbon sources, and significantly higher in cells deleted for ArcA and growing in rich media, even under aerobic conditions (Fig. 4C). Highest expression was seen when cells were grown in pyruvate, the substrate immediately upstream of acetyl-CoA (Fig. 1). Acetyl-CoA sits at the branch point between the TCA cycle and acetate metabolizing enzymes, including the ackA target of SdhX (Fig. 1).
In addition to regulation of transcription initiation, the sdhCDAB-sucABCD operon is highly regulated by several sRNAs (36). However, SdhX escapes this sRNA-dependent posttranscriptional regulation (SI Appendix, Fig. S6D), most likely because of the rapid and efficient processing of the the sdhB-sucA intergenic region (SI Appendix, Fig. S6 A and B) (31). This insulation of SdhX provides the cell the ability to regulate the synthesis of SdhCDAB-SucABCD enzyme levels without perturbing expression of SdhX, creating a hierarchical regulatory tree in which SdhX may be the most robust reporter for the sdh promoter. For example, RyhB sRNA, made when Fe2+ is limiting, down-regulates sdh, limiting the need for Fe2+ by reducing the synthesis of Sdh Fe-S clusters (57). However, excess RyhB does not reduce the accumulation of SdhX (SI Appendix, Fig. S6D), which presumably can still carry out its functions under iron-limited conditions.
Functions of SdhX in Modulating Acetate Metabolism.
Most Hfq-binding sRNAs pair with multiple targets, and the same is clearly also true for SdhX. Experiments in which sRNA/mRNA pairs were captured by ligation on Hfq (RIL-Seq) identified a range of targets for the 3′UTR of sucD (21) (here named SdhX). ackA was identified in those experiments as a partner of SdhX, and other targets have been confirmed by the work of Miyakoshi et al. (54). The region of ackA that pairs with SdhX is fully conserved in Enterobacteria, and we predict that SdhX will regulate ackA in most if not all of these species. Regions outside the SdhX seed may target mRNAs, with species specificity. Our results and those of Miyakoshi et al. are consistent with the nonconserved 5′ region in SdhX targeting katG, encoding hydroperoxidase I, explaining the contribution of SdhX to hydrogen peroxide sensitivity (SI Appendix, Fig. S9C).
SdhX affects AckA both when overproduced (leading to a fivefold decrease in AckA protein levels) (Fig. 3 A and B and SI Appendix, Fig. S3 A and B) and when expressed from the endogenous locus (deletion increases AckA significantly) (Fig. 4D). It is striking, however, that Pta is insulated from SdhX-mediated down-regulation, primarily by expression from an independent promoter (Figs. 3C and 4D and SI Appendix, Fig. S4). Thus, SdhX repression of AckA will change the ratio of AckA and Pta, likely affecting levels of AcP and, by affecting availability of AckA, may gate flux from acetyl-CoA to acetate or, in cases of high acetate, in the opposite direction, from acetate to acetyl-CoA (Fig. 1).
Under aerobic growth in the presence of glucose, the glucose is metabolized from pyruvate to acetyl-CoA; the subsequent fate of acetyl-CoA is an important node for balancing energy, redox potential, and biosynthetic capacity. Under aerobic conditions in the presence of high glucose, acetyl-CoA partially feeds into the full TCA cycle, where it is further metabolized; reduced NADH or NADPH from this cycle act as electron donors for respiration, generating ample ATP (58). However, it is clear that even under optimal growth conditions, significant amounts of acetyl-CoA are instead subject to “overflow metabolism,” the equivalent of the Warburg effect in mammalian cancer cells (7), feeding into Pta and AckA for excretion as acetate (Figs. 1 and 5B). This has been suggested to be a global cell response to balance the conflicting proteomic demands of energy biogenesis (leading to rapid growth) and biomass synthesis (59). When pta and ackA are deleted, cells grow at a modestly slower rate on pyruvate (SI Appendix, Fig. S8A), consistent with a contribution of flux through these enzymes. Our results suggest that SdhX contributes to gating overflow metabolism; cells grown in pyruvate had significantly more acetate in the medium in the absence of SdhX (Fig. 5B) and showed a significant lag in growth on pyruvate when SdhX could not pair with its major targets, including ackA (Fig. 5C). Growth on glucose also showed a modest growth defect, while growth on the gluconeogenic substrates glycerol and succinate were unaffected by the mutation in sdhX (Fig. 5C).
High levels of acetate can be taken up by cells, and metabolized by AckA and Pta back to acetyl-CoA, providing the cell with another opportunity to use this molecule. In both directions, the high-energy molecule AcP will be an intermediate. AcP itself can be used as a signaling molecule in some systems, as well as for acetylation of proteins, some of them critical for carbon metabolism (39, 60), and thus its levels probably must be tightly controlled. However, SdhX had relatively modest effects on AcP accumulation under the conditions we assayed (growth on pyruvate) (Fig. 5A), and no phenotype associated with this modest change in AcP could be detected (SI Appendix, Fig. S8 C and D).
When acetate is the sole carbon and energy source, the flux from AckA to AcP and then to acetyl-CoA should be significantly higher, and it is here that we see the most striking effects of SdhX. Deleting the seed region of SdhX led to a considerable cell growth defect on acetate (Fig. 5C). When cells were grown in acetate, AckA and Pta played a major role; at higher ODs, presumably when levels of acetate are low, Acs became critical for growth (SI Appendix, Fig. S8A). Therefore, we suggest that this brake on AckA provides a gating mechanism that may prevent excessive accumulation of AcP. If Pta levels are insufficient to handle the AcP rapidly, it may act to signal or acetylate inappropriately, or will be catabolized to acetate once again in a futile cycle.
Our study provides additional insight into regulation of the ackA-pta genes. Irrespective of SdhX, we found the highest levels of expression of AckA and Pta when cells were grown in pyruvate, although Pta was much less affected than AckA (Fig. 4 C and D). CreB, a transcriptional response regulator of the CreB-CreC two-component system, positively regulates the major promoter P1 of ackA; activity of CreB-regulated promoters also seems to be highest in pyruvate (61). The high SdhX level in pyruvate decreases AckA levels, therefore tempering the effect of pyruvate on AckA levels. We suggest that cells are poised to rapidly increase AckA when they shift from high SdhX conditions (aerobic growth, possibly) to low SdhX conditions (anaerobic growth, for example, when ArcA efficiently represses sdh-suc expression) (56). Under those conditions, the need for AckA to help promote the fermentative pathway is likely to be high.
Disruption of the AckA-Pta Pathway Has Major Effects on Cell Physiology.
We observed two major phenotypes resulting from deleting ackA, pta, or both, during our study of SdhX. The first was induction of RpoS (SI Appendix, Fig. S8B). Because blocking flux by deleting ackA (high AcP), pta (no AcP), or both (no AcP), has similar effects on induction of an rpoS–lacZ translational reporter fusion, we suggest that these mutants generates a stress response, akin to what is seen for mutants in the genes encoding pyruvate dehydrogenase (62). However, because multicopy SdhX did not mimic the effect of deleting ackA (SI Appendix, Fig. S8B), even a low level of flux through this pathway must be sufficient to avoid this stress response. The difference in levels of acetate in cells deleted for ackA compared with those overexpressing SdhX (Fig. 5B) highlights the metabolic consequences of even low levels of AckA.
The second phenotype, increased resistance to hydroxyurea, was one of the strongest phenotypes described for loss of ackA in a chemical genomics study (45). Increased hydroxyurea resistance was observed in cells with mutations in ackA or pta or both, and thus is not due to AcP (Fig. 5D and SI Appendix, Fig. S9A), and was independent of RpoS. SdhX overexpression was sufficient to make cells resistant to hydroxyurea, and this resistance was dependent upon pairing with ackA (Fig. 5D and SI Appendix, Fig. S9A). Thus, we would conclude that the remaining flux when AckA levels are only 10% of wild-type levels is insufficient to prevent hydroxyurea resistance. In addition to ackA and pta, a variety of mutants in central metabolism, including deletion of genes for ubiquinone biosynthesis, genes for NAD biosynthesis, genes for NADH dehydrogenase and succinate dehydrogenase, as well as those for pyruvate dehydrogenase have been reported to lead to hydroxyurea resistance (46, 63). While the immediate target of hydroxyurea is thought to be the essential ribonucleotide-diphosphate reductases, it has been suggested that generation of endogenous ROS is important for hydroxyurea killing and might be reduced when respiration is compromised (47). While the basis for resistance is not yet clear, the results suggest important roles for flux through the AckA and Pta pathway even when cells are growing on rich media.
Overall, our results suggest a central role for SdhX in regulating carbon usage via its modulation of AckA levels. SdhX is well expressed under any conditions that the TCA cycle enzymes are well expressed and, because of this tie to the TCA cycle, provides a direct communication between the TCA cycle and acetate metabolism. It will be of interest to see if other SdhX targets also contribute to coordinating metabolism in a similar fashion.
Materials and Methods
Bacterial Strains and Plasmids.
E. coli K-12 MG1655 was used as the wild-type strain. The strains and plasmids used in this study are listed in SI Appendix, Tables S1 and S2, and their construction is described either in the respective table or in SI Appendix, SI Materials and Methods. A number of SdhX alleles are used. SdhX* refers to the three base pair change in the seed region (nucleotides 66–68) (Fig. 2C and SI Appendix, Fig. S5C) that abolishes the ability of SdhX to pair with and regulate ackA but allows pairing to ackA* (Fig. 2 C and D); it is used in this work in the plasmid-expressed SdhX. SdhX**, also expressed from the plasmid, carries a 3-nt change in the poorly conserved 5′ portion of SdhX (nucleotides 39–41) (SI Appendix, Fig. S5C) that disrupts predicted base pairing with katG (SI Appendix, Fig. S9C). Two chromosomal mutants of SdhX were used in this work. ΔsdhX deletes nt 20–75, and carries a kanR insert in the sdhX locus. This mutation was found to have cis effects on the expression of the upstream suc genes. The SdhXseedless, a chromosomal mutation that deletes nucleotides 63–75 of sdhX (see SI Appendix, Fig. S5C for numbering), does not perturb upstream sdh and suc gene expression. It carries a zeoR gene downstream of the terminator. Experiments done with this mutant used an isogenic strain carrying the same zeoR gene, but with an intact copy of SdhX.
Plasmids (SI Appendix, Table S2) were generally introduced into strains by TSS transformation (64). Primers used for PCR, sequencing, probes, and synthetic gene fragments (Integrated DNA Technologies) are listed in SI Appendix, Table S3. All of the chromosomal modifications and derivatives of parent strains were transduced to a fresh genetic background using bacteriophage P1vir, as described by Miller (65) and verified by Sanger sequencing.
Growth Conditions.
Bacterial strains were grown in LB-Lennox medium with 250 rpm aeration at 37 °C, supplemented with standard concentrations of the appropriate antibiotic (100 μg/mL ampicillin, 25 μg/mL kanamycin, 10 μg/mL chloramphenicol, 30 μg/mL zeocin, and 50 μg/mL rifampicin). The minimal medium used was MOPS buffer (Teknova) supplemented with a single carbon source (0.4% glycerol, 0.2% d-glucose, 0.2% xylose, 0.2% d-galactose, 60 mM sodium acetate, 0.6% sodium DL-lactate, 0.6% sodium pyruvate, and 0.8% sodium succinate hexahydrate). Experiments measuring SdhX sRNA or protein levels in the presence of a specific carbon source were performed with 20 mL of cells grown in 150-mL flasks; the experiment with the ΔarcA mutant was in 250-mL flasks. Translational PBAD-ackA–lacZ fusions were induced with 0.02% arabinose and a final concentration of 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) was used to induce sRNAs on plasmids.
sRNA Library Screen.
Our library screen included 30 sRNAs that are expressed from a pBR322-derived multicopy plasmid under control of an inducible PLAC promoter. The library was introduced into strains carrying the reporter fusion of interest by TSS transformation, grown at 37 °C in LB with 100 μg/mL ampicillin, 100 μM IPTG, and the appropriate concentration of arabinose (see figure legends) until stationary phase was reached and assayed, as previously described (19). The effect of each sRNA was plotted as a function of the fold-change compared with the fusion containing the pBRPLAC control vector; fold-changes greater than two were considered significant.
β-Galactosidase Assays.
For sRNA library screens, the β-galactosidase activity of strains carrying the indicated lacZ translational fusions and transformed with pBR plasmids were assayed on a SpectraMax 250 (Molecular Devices) microtiter plate reader, as previously described (66). β-Galactosidase activities were determined as specific units by normalizing the Vmax to OD600 (machine units) (67) and were about 10-fold lower than Miller Units. All other experiments measuring lacZ expression were determined by β-galactosidase assays (Miller units) (68). Experiments were performed in biological triplicates; data were plotted as mean ± SD.
RNA Extraction and Northern Blot Analysis.
Total RNA was extracted at indicated OD600 with the hot acid phenol procedure, as previously described (69). Briefly, RNAs were transferred to a Zeta-Probe GT blotting membrane (Bio-Rad) overnight by capillary action (agarose gel) or by electro-transfer (TBE-Urea gel). Membranes were hybridized with the biotinylated probes (SI Appendix, Table S3), then further incubated with a streptavidin-conjugated alkaline phosphatase. The blot was then developed using the BrightStar BioDetect kit (Thermo Fisher Scientific), according to the manufacturer’s instructions; chemifluorescence was then captured and quantified. Further details are provided in SI Appendix, SI Materials and Methods.
Primer Extension Analysis.
Primer extension analysis was carried out as previously described (3). Total RNA samples (5 μg) were incubated with 2 pmol of 5′-32P-end-labeled primer FDM17 at 80 °C and then slow-cooled to 42 °C. Reactions were incubated with dNTP mix (1-mM each) and AMV reverse transcriptase (10 U; Life Sciences Advanced Technologies) at 42 °C for 1 h, then terminated by adding 10 μL of Gel Loading Buffer II (Thermo Fisher Scientific). The DNA sequencing ladder was generated by amplifying sdhX by PCR (oligonucleotides FDM18 and FDM17) and subjected to sequencing reaction using the 5′ end-radiolabeled FDM17 and the Thermo Sequenase Dye Primer Manual Cycle Sequencing Kit (Thermo Fisher Scientific), according to the manufacturer’s protocol. The cDNA products and DNA sequencing ladder were resolved on an 8% polyacrylamide urea sequencing gel in 1× TBE at 70 W for 80 min. Gel was vacuum-dried for 1 h at 80 °C and exposed overnight to a phosphorimager.
Western Blotting.
The analysis of endogenous AckA and Pta-FLAG protein levels was performed using standard procedures using TCA precipitation and acetone neutralization. Protein samples were separated by SDS/PAGE and transferred to nitrocellulose membrane, followed by incubation with primary and fluorescent secondary antibodies. Fluorescence signals were captured using the imaging system ChemiDoc MP (Bio-Rad) and quantified with the Image Studio software (Li-COR Biosciences). Further details are provided in SI Appendix, SI Materials and Methods.
Acetyl-Phosphate Measurements.
AcP levels were determined as previously described (39), with minor modifications (SI Appendix, SI Materials and Methods). Aliquots of cell cultures in midexponential phase (OD600 0.4–0.5) were collected and AcP was converted to ATP with purified E. coli acetate kinase (Sigma Aldrich). The ATP concentration was then quantified by luminescence using CellTiter-Glo Luminescent Cell Viability (Promega).
Acetate Measurements.
Acetate levels were determined by using the Acetate Colorimetic Assay Kit (Sigma-Aldrich). Aliquots of cell cultures in midexponential phase (OD600 0.4–0.5) were centrifuged at 10,000 × g for 10 min at 4 °C. Supernatants were isolated and subjected to quantification according to the manufacturer’s instructions. The background signal of samples was negligible.
Hydroxyurea Sensitivity Assay.
Strains were grown to stationary phase for 15 h in LB medium. Cultures were then serially diluted (from 100 to 10−7) and each dilution spotted with a volume of 4 μL on LB agar containing 20 mM HU and, when appropriate, Amp100, and 1 mM IPTG. Plates were incubated overnight at 37 °C.
Supplementary Material
Acknowledgments
We thank Michael Maurizi, Gisela Storz, Kumaran Ramamurthi, Jorg Vogel, Teppei Morita, Jiandong Chen, and Arti Tripathi for critical reading of the manuscript; J. Vogel for sharing results of their parallel studies before publication; Nadim Majdalani and Erin Wall for providing strains; Nadim Majdalani for excellent technical advice; Kyung Moon for technical guidance for the 5′ RACE; and Daniel Schu for information from initial studies on RybD. F.D.M. is a doctoral candidate with a scholarship from the Fund for Research Training in Industry and Agriculture, Belgium. This study was supported in the L.V.M. laboratory by grants from the Fonds National de la Recherche Scientifique, the Interuniversity Attraction Poles Program initiated by the Belgian Science Policy Office (MICRODEV), and the Fonds Jean Brachet and the Fondation Van Buuren. Research in the S.G. laboratory was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815288116/-/DCSupplemental.
References
- 1.Nitzan M, Rehani R, Margalit H. Integration of bacterial small RNAs in regulatory networks. Annu Rev Biophys. 2017;46:131–148. doi: 10.1146/annurev-biophys-070816-034058. [DOI] [PubMed] [Google Scholar]
- 2.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: Expanding frontiers. Mol Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schu DJ, Zhang A, Gottesman S, Storz G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015;34:2557–2573. doi: 10.15252/embj.201591569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland I, et al. Gut microbiota functions: Metabolism of nutrients and other food components. Eur J Nutr. 2018;57:1–24. doi: 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernal V, Castaño-Cerezo S, Cánovas M. Acetate metabolism regulation in Escherichia coli: Carbon overflow, pathogenicity, and beyond. Appl Microbiol Biotechnol. 2016;100:8985–9001. doi: 10.1007/s00253-016-7832-x. [DOI] [PubMed] [Google Scholar]
- 9.Enjalbert B, Millard P, Dinclaux M, Portais JC, Létisse F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci Rep. 2017;7:42135. doi: 10.1038/srep42135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J Bacteriol. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren J, Sang Y, Lu J, Yao YF. Protein acetylation and its role in bacterial virulence. Trends Microbiol. 2017;25:768–779. doi: 10.1016/j.tim.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Kumari S, Tishel R, Eisenbach M, Wolfe AJ. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumari S, et al. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:4173–4179. doi: 10.1128/jb.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esquerré T, et al. Dual role of transcription and transcript stability in the regulation of gene expression in Escherichia coli cells cultured on glucose at different growth rates. Nucleic Acids Res. 2014;42:2460–2472. doi: 10.1093/nar/gkt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 16.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmqvist E, et al. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016;35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomason MK, et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melamed S, et al. Global mapping of small RNA-target interactions in bacteria. Mol Cell. 2016;63:884–897. doi: 10.1016/j.molcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright PR, et al. CopraRNA and IntaRNA: Predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–W123. doi: 10.1093/nar/gku359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakuda H, Hosono K, Shiroishi K, Ichihara S. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem. 1994;116:916–922. doi: 10.1093/oxfordjournals.jbchem.a124616. [DOI] [PubMed] [Google Scholar]
- 25.Chao Y, et al. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell. 2017;65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyakoshi M, Chao Y, Vogel J. Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr Opin Microbiol. 2015;24:132–139. doi: 10.1016/j.mib.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Guo MS, et al. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014;28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao Y, Vogel J. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell. 2016;61:352–363. doi: 10.1016/j.molcel.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 30.Nam TW, Park YH, Jeong HJ, Ryu S, Seok YJ. Glucose repression of the Escherichia coli sdhCDAB operon, revisited: Regulation by the CRP*cAMP complex. Nucleic Acids Res. 2005;33:6712–6722. doi: 10.1093/nar/gki978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cunningham L, Georgellis D, Green J, Guest JR. Co-regulation of lipoamide dehydrogenase and 2-oxoglutarate dehydrogenase synthesis in Escherichia coli: Characterisation of an ArcA binding site in the lpd promoter. FEMS Microbiol Lett. 1998;169:403–408. doi: 10.1111/j.1574-6968.1998.tb13347.x. [DOI] [PubMed] [Google Scholar]
- 32.Cunningham L, Guest JR. Transcription and transcript processing in the sdhCDAB-sucABCD operon of Escherichia coli. Microbiology. 1998;144:2113–2123. doi: 10.1099/00221287-144-8-2113. [DOI] [PubMed] [Google Scholar]
- 33.Górna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: The structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys. 2012;45:105–145. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- 34.Park S-J, Tseng C-P, Gunsalus RP. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: Role of ArcA and Fnr. Mol Microbiol. 1995;15:473–482. doi: 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 35.Park S-J, Chao G, Gunsalus RP. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode alpha-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: Roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J Bacteriol. 1997;179:4138–4142. doi: 10.1128/jb.179.13.4138-4142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desnoyers G, Massé E. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 2012;26:726–739. doi: 10.1101/gad.182493.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. J Biol Chem. 2002;277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- 39.Weinert BT, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 40.McCleary WR, Stock JB. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 41.Prüss BM, Wolfe AJ. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 42.Bouché S, et al. Regulation of RssB-dependent proteolysis in Escherichia coli: A role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol. 1998;27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 43.Fredericks CE, Shibata S, Aizawa S, Reimann SA, Wolfe AJ. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol Microbiol. 2006;61:734–747. doi: 10.1111/j.1365-2958.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 44.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 45.Nichols RJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakayashiki T, Mori H. Genome-wide screening with hydroxyurea reveals a link between nonessential ribosomal proteins and reactive oxygen species production. J Bacteriol. 2013;195:1226–1235. doi: 10.1128/JB.02145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies BW, et al. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol Cell. 2009;36:845–860. doi: 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odsbu I, Morigen, Skarstad K. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PLoS One. 2009;4:e7617. doi: 10.1371/journal.pone.0007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eklund H, Uhlin U, Färnegårdh M, Logan DT, Nordlund P. Structure and function of the radical enzyme ribonucleotide reductase. Prog Biophys Mol Biol. 2001;77:177–268. doi: 10.1016/s0079-6107(01)00014-1. [DOI] [PubMed] [Google Scholar]
- 50.Mahoney TF, Silhavy TJ. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol. 2013;195:1869–1874. doi: 10.1128/JB.02197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mann M, Wright PR, Backofen R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyakoshi M, Matera G, Maki K, Sone Y, Vogel J. Functional expansion of a TCA cycle operon mRNA by a 3’ end-derived small RNA. Nucleic Acids Res. December 12, 2018 doi: 10.1093/nar/gky1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kröger C, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Basan M, Hui S, Williamson JR. ArcA overexpression induces fermentation and results in enhanced growth rates of E. coli. Sci Rep. 2017;7:11866. doi: 10.1038/s41598-017-12144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawers RG, Clark DP. Fermentative pyruvate and acetyl-coenzyme A metabolism. EcoSal Plus. 2004;1:1. doi: 10.1128/ecosalplus.3.5.3. [DOI] [PubMed] [Google Scholar]
- 59.Basan M, et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pisithkul T, Patel NM, Amador-Noguez D. Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr Opin Microbiol. 2015;24:29–37. doi: 10.1016/j.mib.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Avison MB, Horton RE, Walsh TR, Bennett PM. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem. 2001;276:26955–26961. doi: 10.1074/jbc.M011186200. [DOI] [PubMed] [Google Scholar]
- 62.Battesti A, Majdalani N, Gottesman S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci USA. 2015;112:5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JH. Experiments in Bacterial Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 66.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Y, Gottesman S. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J Bacteriol. 1998;180:1154–1158. doi: 10.1128/jb.180.5.1154-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
- 69.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.