Significance
We demonstrate a high-accuracy method for predicting enhancers genome-wide with >85% precision as validated by transgenic reporter assays in Drosophila embryos. This accuracy in a metazoan system enables us to predict with high confidence 1,640 enhancers genome-wide that participate in body segmentation during early development. The predicted enhancers are demarcated by heterogeneous collections of epigenetic marks; many strong enhancers are free from classic indicators of activity, including H3K27ac, but are bound by key transcription factors.
Keywords: enhancers, embryo development, machine learning, random forests, Drosophila
Abstract
Identifying functional enhancer elements in metazoan systems is a major challenge. Large-scale validation of enhancers predicted by ENCODE reveal false-positive rates of at least 70%. We used the pregrastrula-patterning network of Drosophila melanogaster to demonstrate that loss in accuracy in held-out data results from heterogeneity of functional signatures in enhancer elements. We show that at least two classes of enhancers are active during early Drosophila embryogenesis and that by focusing on a single, relatively homogeneous class of elements, greater than 98% prediction accuracy can be achieved in a balanced, completely held-out test set. The class of well-predicted elements is composed predominantly of enhancers driving multistage segmentation patterns, which we designate segmentation driving enhancers (SDE). Prediction is driven by the DNA occupancy of early developmental transcription factors, with almost no additional power derived from histone modifications. We further show that improved accuracy is not a property of a particular prediction method: after conditioning on the SDE set, naïve Bayes and logistic regression perform as well as more sophisticated tools. Applying this method to a genome-wide scan, we predict 1,640 SDEs that cover 1.6% of the genome. An analysis of 32 SDEs using whole-mount embryonic imaging of stably integrated reporter constructs chosen throughout our prediction rank-list showed >90% drove expression patterns. We achieved 86.7% precision on a genome-wide scan, with an estimated recall of at least 98%, indicating high accuracy and completeness in annotating this class of functional elements.
Enhancers are ∼100- to 1,000-bp cis-regulatory elements that direct spatial and temporal pattern transcription in metazoans. Definitive epigenetic signatures of enhancer elements have been challenging to identify. A number of computational tools have been developed to predict enhancer elements from chromatin state and transcription factor in vivo DNA binding information (1–12). Tools that attempt to measure predictive accuracy using only indirect evidence of enhancer activity [e.g., enrichment in H3K27 acetylation (H3K27ac) or histone acetyltransferase p300 (EP300)] often display excellent accuracy by these limited criteria (1, 3, 13, 14). When algorithms are benchmarked on held-out in vivo tests of functional enhancer activity, however, positive predictive power on genome-wide scans in metazoan systems has been lower than expected. In most cases, precision does not exceed 40% (13–15). However, by targeting transcription factors (TF) that function in a specific biological process, a higher precision of 56% was achieved in a randomly selected validation sample through transient transfection (16). Higher precision has also been reported when tests were confined to the top of the prediction rank list (17), but such numbers are unlikely to represent the precision of the prediction set as a whole.
There are several possible explanations for the relatively low accuracy of current enhancer prediction algorithms. The transient in vivo enhancer assays often employed to test predictions may suffer a high false-negative rate due to the loss of local chromatin context. Alternatively, the data provided to the prediction algorithms might be insufficient. Features such as H3K27ac and EP300 can partially distinguish active enhancers (18, 19), but it remains unclear whether any chromatin mark or combination of chromatin marks and EP300 uniquely identifies enhancers among all sequences in a genome (16, 20). Indeed, enhancers that lack H3K27ac yet have patterns of DNA hypermethylation are essential during early vertebrate development (21). Hence, there may be more than a single class of genomic element that drives patterned expression or, more precisely, the term “enhancer” may encapsulate a mechanistically diverse class of functional elements. TF occupancy is a better predictor of enhancer activity than canonical chromatin marks (including H3K27ac, H3K4me1, and H3K4me3) in mouse and humans (16). Thus, mechanistic subtypes of functional enhancer elements may emerge from distinct patterns of TF occupancy and chromatin context.
To test the possibility that heterogeneity among enhancers is a major reason for the difficulty in predicting enhancers, we have exploited the pregastrula Drosophila embryo network. A cohort of ∼30 spatially patterned TFs drive body patterning in concert with another 30 or so ubiquitously expressed sequence-specific TFs (22–32). Embryonic patterning is established along the anterior–posterior (A-P) axis and dorsal–ventral (D-V) axis by two separate sets of maternally deposited TFs. Over a 90-min period corresponding to developmental stages 4 and 5, these proteins act in concert with zygotically expressed A-P and D-V TFs to refine initially broad patterns of transcription into narrower striped patterns that define the basic segmental body plan of the fruit fly (33). The pregastrula fly network is thus a particularly well-defined model system for studying the relationship between TF DNA binding and spatially patterned enhancer activity.
We have tested the utility of a wide range of data for predicting enhancers, including in vivo DNA binding patterns for 22 pregastrula TFs, a variety chromatin marks, evolutionary conservation, whole-embryo mRNA sequencing (mRNA-seq), and RNA polymerase II (Pol II) location. Using a test set of nearly 8,000 genomic regions whose enhancer activity had been determined in transgenic assays in whole embryos (34, 35), we applied supervised machine learning to identify enhancer sequences active in pregastrula embryos. Verified enhancers were separated into two approximately equally sized groups based on the reproducibility with which they were correctly predicted in multiple runs of a random forest (RF). A model trained using the set of enhancers that were reproducibly classified correctly has >98% predictive accuracy when tested on a balanced set of known enhancer positive and negative genomic regions. In contrast, the other set of training enhancers generated models that predicted no better than random. Subsequent analyses revealed that the well-predicted class of enhancers are near genes that show a strong tendency to be involved in controlling segmentation and other developmental processes, and to be expressed in many cells of the embryo. The poorly predicted enhancers are without obvious ties to the control of segmentation and tend to be expressed in less than 15% of cells. By focusing on the well-predicted class of enhancers, which we term segmentation-driving enhancers (SDEs), we find that TF DNA binding is highly predictive, whereas histone modifications and the remaining features tested have little or no additional predictive power.
In a de novo, genome-wide prediction, we predict ∼1,640 SDEs in the early embryo that cover 1.6% of the euchromatic genome. As validated by an in vivo transgenic reporter gene assay, this set is predicted with 98% estimated recall and 95% precision, as validated in an in vivo transgenic assay. Unlike most previous studies, we concentrated validation away from the top of our rank list to increase the likelihood of identifying false-positives and to improve our power to compute accurate error rates. Importantly, we show that our model performance is driven by the need to treat SDEs separately from other enhancer elements, rather than the properties of a specific computational method: naïve Bayes and logistic regression perform as well as more complex models after conditioning on the SDE set. This demonstrates the prediction of a specific class of enhancers with sufficient precision to enable their identification genome-wide.
Results
Data, Feature, and Feature Selection.
Transgenic reporter data for enhancer activity in Drosophila embryos were combined from two sources. Kvon et al. (34) conducted a semiautomated screen of the reporter gene-expression patterns driven by 7,705 genomic regions (enhancers.starklab.org/) at multiple stages throughout embryogenesis. While this high-throughput assay allowed an unprecedented number of genomic areas to be tested, the small number of embryos per collection plate led to increased misclassifications in the data. The activity of an additional 282 genomic segments was determined by the Berkeley Drosophila Transcription Network Project (BDTNP) (35). Altogether, 7,987 genomic regions were examined and 731 were experimentally found to drive reporter gene expression in Drosophila embryonic stages 4–6 (36) (Dataset S1). By manually comparing the activity of overlapping genomic regions in the BDTNP database with the larger data from Kvon et al. (34), we estimate a 10% false-negative rate in the latter.
Features used in the initial model included ChIP-chip data for 20 of the TFs that pattern transcription along the A-P and D-V axes of the embryo (37–39), chromatin immunoprecipitation-sequencing (ChIP-seq) data for the ubiquitous TFs Zelda (ZLD) and Zeste (Z), 45 chromatin proteins and histone modifications (40), DNase accessibility data (41–43), and evolutionary conservation scores (44–46). Also considered were the presence of: bidirectional RNA transcripts, exon and intron coverage, distance to RNA Polymerase II ChIP-chip binding peaks, and distance to transcription start sites. A summarized list of features is presented in Table 1. For a full list and description, see SI Appendix, Table S1 and Materials and Methods, respectively.
Table 1.
Summary of features used for prediction
| Category | Features included |
| Histone and histone modifications | H3, H3K18ac, H3K27ac, H3K27me3, H3K36me3, H3K4me1, H3K4me3, H3K9ac, H4K5ac, H4K8ac |
| AP regulatory transcription on factors | BCD, CAD, GT, HB, KNI, KR, HKB, TLL, D, FTZ, PRD, RUN, SLP |
| DV regulatory transcription on Factors | DA, DL, MAD, MED SHN, SNA, TWI |
| Ubiquitous transcription on factors | Z, ZLD, sum of all transcription factor scores |
| DNA data | Conservation, DNA accessibility, distance to Pol. II, distance to TSS, bidirectional-RNA transcription |
| Exon/intron data | Exons, coding exons and introns coverage/presence |
With these data we trained and tested RF, a supervised machine-learning approach based on an ensemble of decision trees (47–49). To reduce parameter number and prevent overfitting, we culled input features (Materials and Methods). We found that TFs and histone modification data were sufficient to minimize the error rate. We note that DNase accessibility did not contribute to RF predictive power in the presence of TF binding data, nor did it significantly improve performance in the presence of histone data, and it adds only modest predictive power when it is used as the sole feature for prediction (SI Appendix, Fig. S1). Conservation scores (Materials and Methods) did not contribute to the predictive power in any fitted model, and the error rate utilizing solely conservation scores was ∼50%, suggesting that conservation is not a distinguishing feature of enhancers in the Drosophila embryo in the absence of other genomic context.
Heterogeneity Among Enhancer Elements.
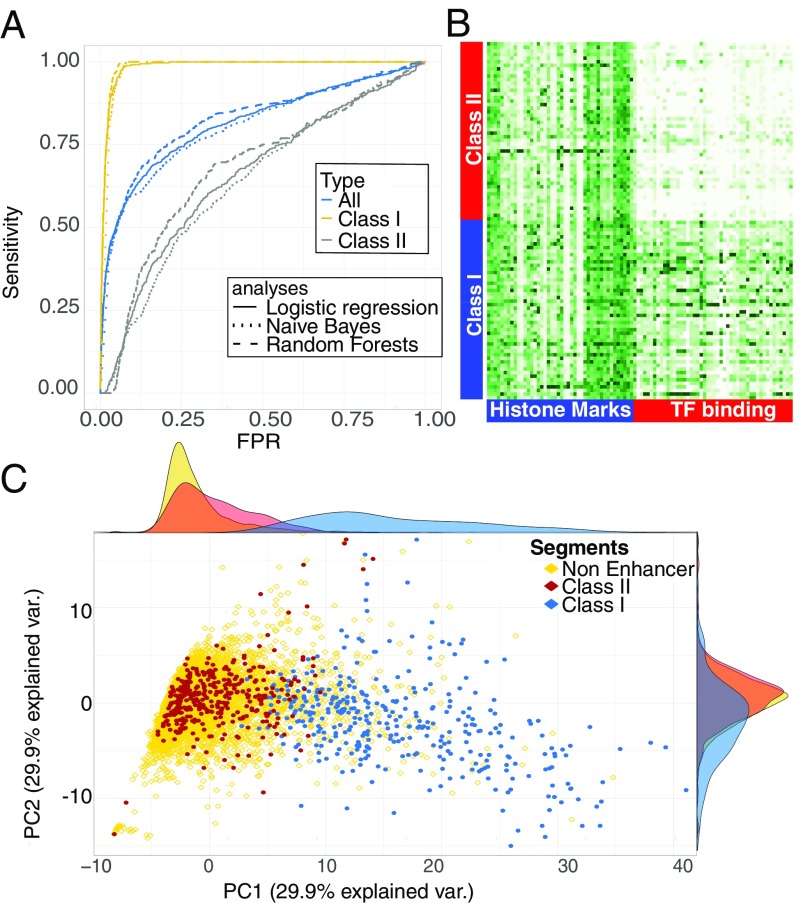
With our optimal feature set, our error rate in a single forest as defined by misclassification was nearly 30%. The performance of the forest voting probabilities as indicated by the area under the receiver operating characteristic (ROC) curve, AUC = 0.82 (Fig. 1A), is very similar to that in previously published work (16, 17), implying a similarly modest success rate. However, while this overall predictive power falls short of that required for predicting enhancers genome-wide, we noticed that some enhancers were consistently correctly classified while others were consistently misclassified. Hypothesizing that the model’s poor performance may be due to heterogeneity in the enhancer set, enhancers were separated into two classes. Class I contained the 358 enhancer segments that were correctly classified at least 75% of the time and class II contained the 373 that were not. When class II enhancers were excluded from the test sample, the single forest error rate drops to ∼3%, and the area under the ROC curve is ∼0.99 (Fig. 1A). When class I enhancers were excluded from the test sample instead, errors of a single forest are ∼40%, and the ROC curve indicated performance only marginally better than random guessing (Fig. 1A). To establish that enhancer heterogeneity is data-driven and not an artifact of our choice of method, logistic regression and naïve Bayes models of the data were also constructed. In both cases the removal of the class II enhancer set significantly improves the model’s predictive power (Fig. 1A). Interestingly, the effect of retaining and removing class I and class II enhancers appears to have almost identical effect on recall regardless of the method, and indeed the ROC curves are nearly overlapping (Fig. 1A). This is particularly noteworthy as the underlying assumption of both models—primarily, feature additivity and independence—are unlikely to be present in the data, yet both perform as well as RFs, which do not require such assumptions. Precision-recall (PR) curves also show that logistic regression performance closely matches that of RFs, although naïve Bayes precision is poor (SI Appendix, Fig. S2). In all cases accounting for heterogeneity increases precision significantly. When a nonenhancer set is purged of a later-stage enhancer, the PR curve for RF has an AUC > 0.95, demonstrating extremely high sensitivity in the data.
Fig. 1.
(A) RF ROC curves for the complete dataset of 7,987 previously validated genomic regions (blue) shows mediocre performance, with an AUC of 0.83. When only class I enhancers and nonenhancers are used for training, the predictive power rises sharply, AUC of 0.99 (yellow). When only class II enhancers and nonenhancers are used, the result is close to a random guess (gray). When predicting the class I enhancer set the ROC curves for RFs, logistic regression, and a naïve Bayes classifier are nearly overlapping. (B) This can be explained by the colocalization of class II enhancers and nonenhancers in a PCA projection. (C) The separation is mainly driven by TFs as exemplified by the normalized ChiP strength across features of 200 randomly selected class I and class II enhancers.
This separation by the model can be understood by principal component analysis (PCA) (Fig. 1C): class II enhancers are collocated with nonenhancers while class I enhancers are separated from both. Examination of feature space statistics of the three groups shows that class II enhancers are indistinguishable from nonenhancers along our entire feature space—TF DNA binding, histone marks, conservation, and DNase accessibility—while class I enhancers segregated from both by multiple features. The separation is most notable in TF DNA binding and DNase accessibility profiles (Fig. 1B and SI Appendix, Fig. S3), where class I enhancers consistently have higher ChIP scores and are more accessible in whole-embryo average data. This indicates a possible reason and mechanism for the separation of the two classes and shows that RFs can be readily used to separate heterogeneous enhancer sets.
Excluding class II enhancers from the sampled training set gives us unprecedented prediction accuracy. On a balanced held-out test set, built from genome regions that prior studies suggested half were enhancers and half were nonfunctional, more than 98% of class I enhancers are discovered by our algorithm with better than 95% precision. This model would have much lower accuracy if used to predict enhancers genome-wide, however. As one moves away from a balanced test set by adding a more realistic number of inactive genomic regions, the false-positive rate in the test set will increase.
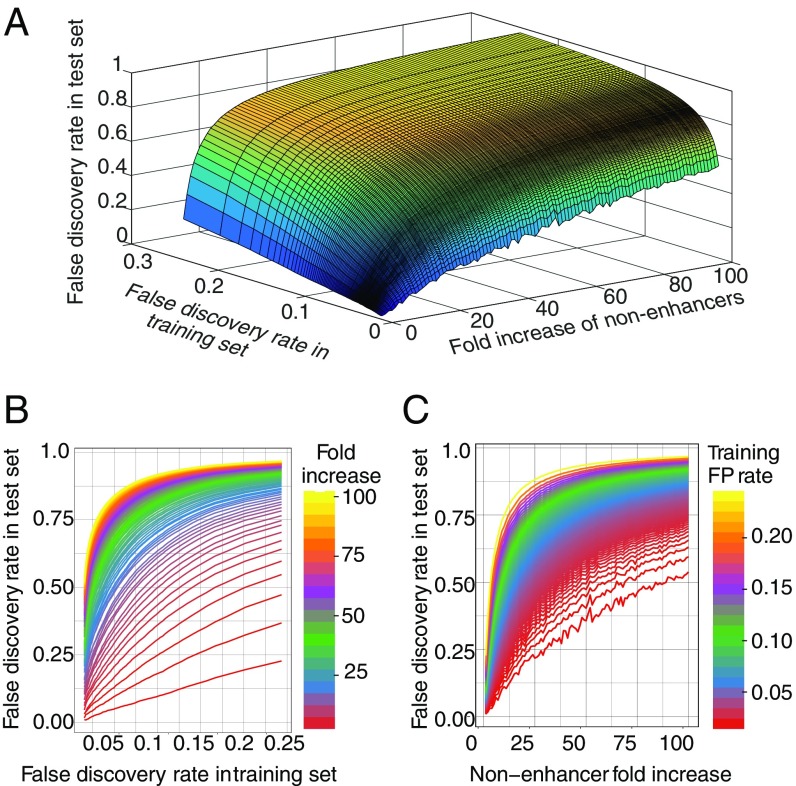
To demonstrate this point, RFs were trained on a balanced set and then tested on a series of increasingly imbalanced test sets at various degrees of stringency (Fig. 2A). The false-positive rate for test sets increases sharply as either the fraction of nonenhancers in the test set increases or as the accuracy of the model—defined during training—increases. This can also be seen in 2D plots of the same analysis (Fig. 2 B and C): unless the sample is very close to a 50%/50% balance, the rise in the false-discovery rate (FDR) in the test set is extremely sharp. Conversely, in genomic scans where nonenhancer regions are at least a 100-fold more prevalent, a precision considerably better then 95% during training (measured out of bag) (Materials and Methods) is needed to achieve a 75% FDR in the test.
Fig. 2.
False-positive rate is a function of method accuracy and imbalance in the test data. (A) A 3D surface plot shows a sharp increase in the test-set false-positive rate as either the training set false-positive rate or the fraction of nonenhancer regions in the test-set increase. This shows that in genomic settings, where the imbalance cannot be controlled, a very high degree of accuracy is required. (B and C) Two-dimensional plots of the marginals of the 3D image in A, demonstrating the sharp rise in test inaccuracy for both false-positive rate in the training set or dilution of enhancer class in the test set.
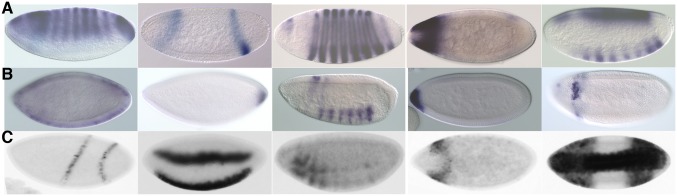
In the dataset of Kvon et al. (34) there are 20 times more annotated nonenhancers than enhancer elements. In randomly drawn test sets with only 5% true enhancers, we find that our fitted model recovers 90% of enhancers with 60% precision. However, our prediction accuracy is likely considerably higher than this analysis implies due to an abundance of false-negatives in the high-throughput Kvon et al. (34) annotations. Manual reexamination of their reporter gene-expression image data for the 100 genomic regions that our method most highly predicted to be enhancers, but which were reported as nonenhancers, revealed that only 15 were true nonenhancers, 47 were clearly enhancers, and the remainder could not be classified due to insufficient data, specifically the lack of embryos of the appropriate stage in the high-throughput images (Fig. 3C).
Fig. 3.
Examples of reporter gene-expression patterns driven by (A) class I enhancers, (B) class II enhancers, and (C) genome regions misclassified by Kvon et al. (34) as nonenhancers in stages 4–6. Magnification is 20× and the embyos are 0.5 mm in length on average.
Genome-Wide Analysis to Identify Active Enhancers in the Early Embryo.
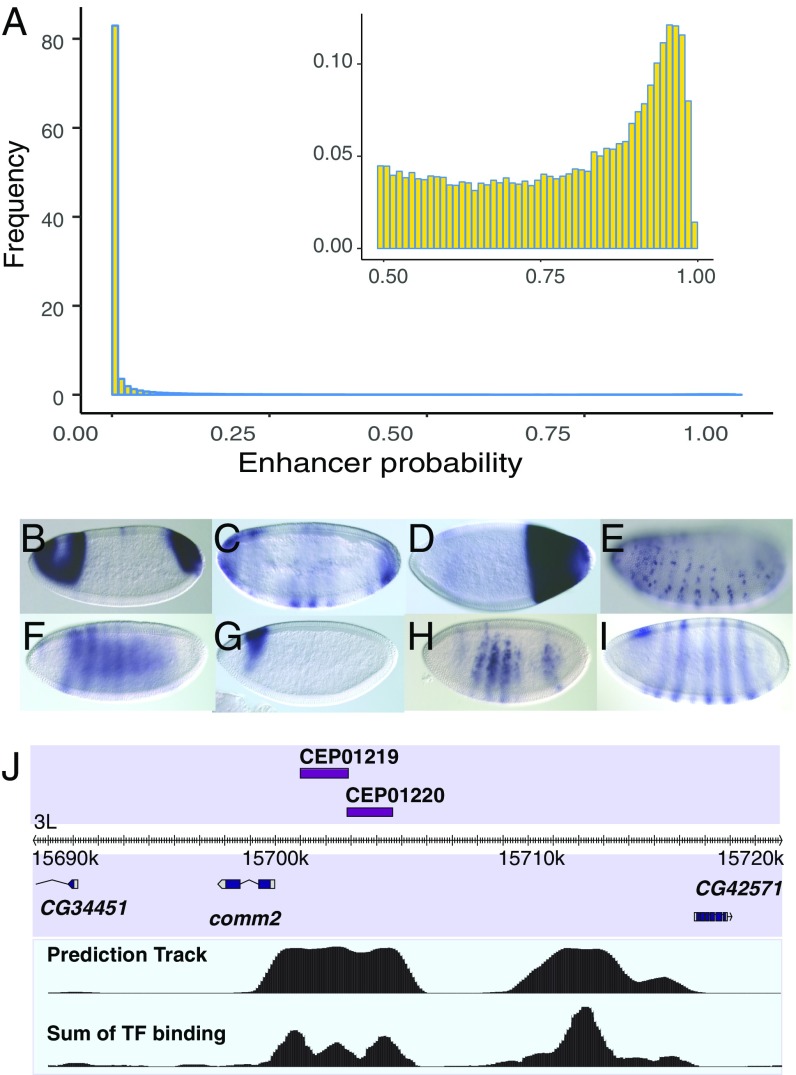
Given the high accuracy of the model on our training and held-out datasets, a genome-wide search for class I enhancers was feasible. RF was therefore used to predict enhancer probability on a computationally segmented genome (Materials and Methods). More than 82% of all segments had less than 0.01 probability of being enhancer, and more than 93% had less than 0.1 probability (Fig. 4A). While it is challenging to see initially as the histogram is dominated by a peak between probabilities 0 and 0.01, the histogram is in fact bimodal (Fig. 4A, Inset), with a secondary peak around P = 0.95. To call enhancers a threshold of P > 0.75 was established that covers ∼1.6% of the genome and rediscovers de novo 98% of the training set. Of the 1,640 class I enhancers predicted, 1,174 do not overlap with training data, 364 overlap known cis-regulatory modules identified in a database of enhancers discovered in other studies, REDfly (50–52); and 822 are completely novel. The prediction list can be viewed at genome.ucsc.edu/cgi-bin/hgTracks?db=dm3&hubUrl=https://sina.lbl.gov/seqdata/ucsc/EnhancerPrediction/hub.txt (53) and Dataset S2).
Fig. 4.
(A) Histogram of RF predicted enhancer probabilities for the entire genome. While >82% of the genome has P < 0.01, a secondary peak can be seen at P ∼ 0.95 (Inset). (B–F) As validation, predicted enhancers were inserted into the Drosophila genome and were found to drive spatial expression. (G and H) Two enhancers, CEP01219 and CEP01220, are predicted proximal to the comm2 gene. Each of their patterns is a component of the comm2 expression pattern (I). (J) The genomic region of the two predicted enhancers is shown, along with the raw prediction track showing the predicted probability of enhancer activity with 100-bp resolution and the sum of TF binding ChIP scores at the same resolution. Magnification is 20×, and the embryos are 0.5 mm in length on average.
Validation of Predicted Enhancer Set in New Experiments.
To validate the prediction precision, an in vivo reporter expression-driving test was conducted. Five, 11, and 17 genomic regions were selected with probability scores corresponding to estimated (cross-validation) FDRs of 4%, 25%, and 50%, respectively. We test down the rank list to enable estimation of the overall FDR of the entire set of predicted enhancers, not just the top predictions. Test regions were cloned into the pBPGUW expression vector, then injected into flies using the attP integration system (54) (SI Appendix, Table S2). All but three of the enhancers, including all but two of those predicted to be in the 50% FDR region, were found to be enhancers (Fig. 4 B–F and SI Appendix, Fig. S4). We thus needed to adjust our FDR estimation; assuming a Poisson distribution we obtained a maximum likelihood estimate (MLE) of 12.5% FDR at our previously cross-validation–based 50% FDR threshold (Materials and Methods). For our entire collection of 1,640 predicted class I enhancers, we estimate an overall FDR of 13.58%.
An interesting example and validation for the use of transcription factors to separate proximal enhancers (Materials and Methods) can be seen in two predicted segments (CEP01219 and CEP01220) proximal to the comm2 gene (Fig. 4J). comm2 encodes an important protein required for proper axon guidance across the embryonic midline (55, 56). The two predicted enhancer-combined expression patterns (Fig. 4 G and H) match the more complicated expression pattern of comm2 (Fig. 4J).
Segmentation Driving Enhancers.
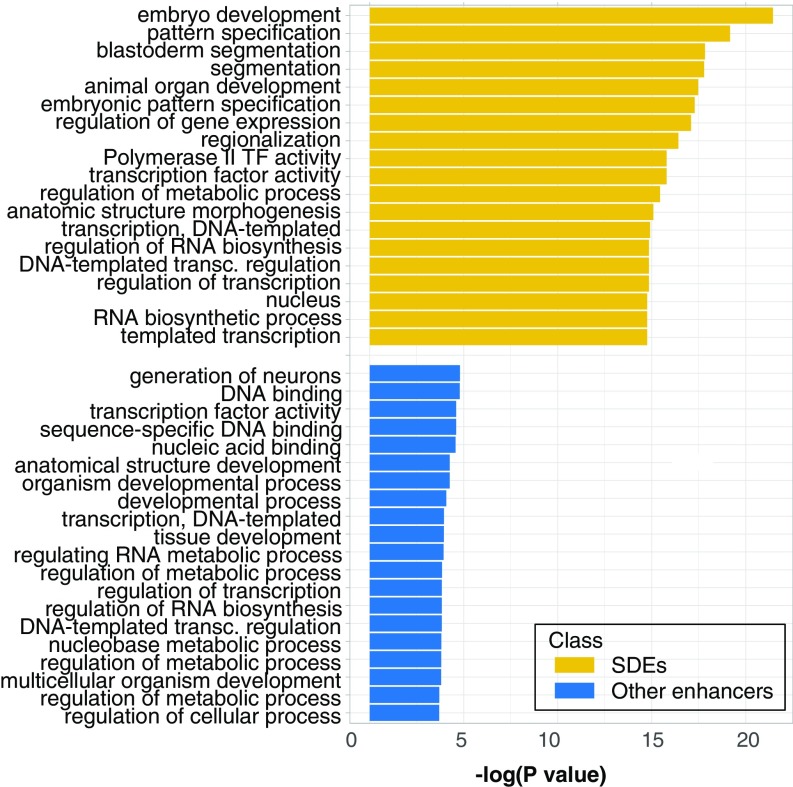
We next sought to understand if the separation of the enhancers into two classes in our feature space is related to their biology. In a detailed quantification of images of embryonic reporter gene-expression patterns for 85 randomly selected class I and 82 randomly selected class II enhancers, class II enhancers tend to be expressed in a smaller percent of nuclei. Of class II enhancers, 74% are expressed in ≤15% of cells versus only 33% of class I (SI Appendix, Fig. S3). While separation by this criterion is not complete, it is unlikely that these differences in expression are due to chance (P < 10−7). In addition, we find that class I enhancers are more likely to remain active throughout embryogenesis and show a significant enrichment for the expression in A-P stripes, posterior, or gap gene-like patterns (P < 10−4). Gene ontology (GO)-term analysis of the genes proximal to class I enhancers also showed a highly significant enrichment of terms related to segmentation (Fig. 5), while those of class II enhancers showed much lower enrichment for any GO terms and no significant enrichment for any particular pathway (Fig. 5). We therefore hypothesize that class I enhancers are likely to drive expression patterns needed for establishing the segmented body plan. We thus term class I and class II enhancers SDEs and non-SDEs respectively. We note that while the differences between these two classes are significant, there is not a clear separation in function because a minority of non-SDE direct patterns of expression resemble those of SDEs (Fig. 3 A and B).
Fig. 5.
The significance (measured as the negative log of the P value) of GO-term enrichment in genes proximal to class I enhancers is very high in terms associated with development and segmentation (SDEs, yellow). For class II enhancers, no significant GO-term enrichment (P value below 10−5) is found (non-SDEs, blue).
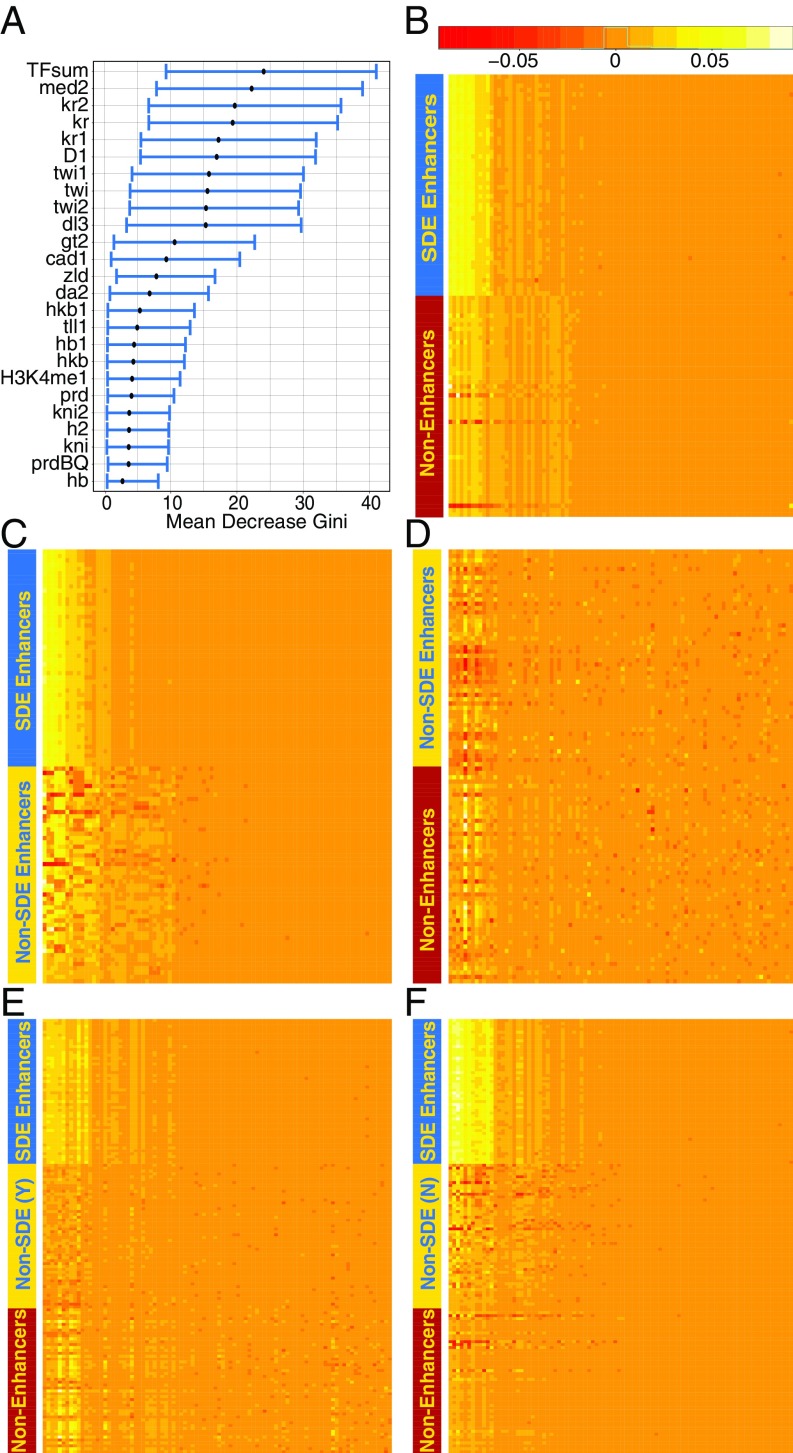
Feature Importance Is Dominated by TFs.
The RF importance measures “mean decrease accuracy” and “mean decrease Gini” (49) varied between bootstrap repetitions, but in all cases a small set of TFs were found near the top of the importance ranking list. This can be seen by the spread of the bootstrap confidence interval of these two importance measures calculated in 50,000 trees (Fig. 6A and SI Appendix, Fig. S5A). ChIP binding scores for several TFs (KR, MED, TWI, DL, D) were among the most important predictors by both measures (SI Appendix, Fig. S5B). The sum of all TF binding scores, or TFsum, was also an important predictor. Other TFs, such as BCD and FTZ (33), were uninformative despite their importance for embryo segmentation. This can be at least partially explained by low coverage in the ChIP-chip data, as there is a clear correlation (r = 0.7) between coverage and importance measure (SI Appendix, Fig. S6A). The only histone mark to have an importance above random noise was H3K4 monomethylation (H3K4me1). The histone and histone modifications we measured, including H3K27ac, which has been widely regarded as a key indicator of enhancer regions (18, 57), were found uninformative by the model in the presence of the TF data.
Fig. 6.
(A) Feature importance is dominated with transcription factors, with the H3K4me1 the only histone mark in the top 25. (B–F) “Local importance” measurements of randomly selected segments indicting how important each feature was in the segment classification when the forest was trained on (B) SDEs vs. nonenhancers, (C) SDE vs. non-SDEs, (D) non-SDEs vs. nonenhancers, (E) SDEs and non-SDEs vs. nonenhancers, and (F) SDEs vs. non-SDEs and nonenhancers. Feature order (x axis) can be found in SI Appendix.
Localized Feature-Importance Measures Support Two-Class Structure.
RF’s “local importance” (49) is a third measure that provides a detailed determination of the importance of each feature in classifying each instance, allowing a more direct understanding on the RF decision-making process (Fig. 6 B–F). This measure shows that the same small set of features are used to distinguish SDE and nonenhancers (Fig. 6B) as are used to distinguish SDEs from non-SDEs (Fig. 6C), while an attempt to separate non-SDEs and nonenhancers (Fig. 6D) shows that no variable can consistently be used and that many more parameters are employed. The increase in features used and the blurring of decision criteria is also seen when non-SDEs are presented to RF as enhancers (Fig. 6E) rather than as nonenhancers (Fig. 6F). Spectral clustering is a technique that relies on the eigenvectors of the similarity matrix or the Laplacian thereof, usually followed by k-nearest neighbors or k-means clustering (58). It is an efficient way of dimension reduction, and the number of clusters in the data can often be inferred by the eigenvalues. Applying spectral clustering to an affinity matrix computed from the local importance values (seventh-nearest neighbor of a Euclidian distance matrix calculated with a Gaussian kernel) yields a good separation of the data, however, with a sharp jump after the second eigenvalue (SI Appendix, Fig. S6B), consistent with the presence of a two-class structure.
Discussion
The identification of enhancer elements from genomics data has remained a challenging problem, in part due to the relative scarcity of enhancers in genome sequences versus nonenhancer sequences. As illustrated in Fig. 2, even an incisive enhancer prediction algorithm fitted on balanced training data (i.e., a training set with nearly equal numbers of positive and negative elements) is likely to generate high FDRs when tested on a genome-wide scan. Hence, to accurately discover enhancer elements using in silico techniques, extremely high-fidelity models are needed.
Although high-precision predictions were reported previously, the validation methods and measures used in the literature varied greatly. Many papers defined success as the colocation of data for epigenetic marks, such as EP300 and H3K27ac, but it is yet to be established that these marks are exclusive to enhancers or that all enhancers possess them. Indeed, we report here a class of H3K27ac-free enhancers (SI Appendix, Table S3). Other reports tested for functional enhancer activity of genomics regions from the top of a rank list (17), which gives a biased estimate of the overall prediction accuracy. We suggest that precision must be measured by testing throughout the prediction rank list to establish a uniform, unbiased measure of success for entire prediction sets.
We found that the prediction of enhancer elements en masse was vexed by heterogeneity among enhancer elements. For about half of previously validated enhancer elements, strong TF in vivo DNA binding signals for multiple factors is indicative of enhancer activity. The remaining half of validated elements is typically bound more weakly by fewer TFs (Fig. 1B and SI Appendix, Fig. S1). For this latter set, the residual TF binding signal is only weakly associated with enhancer activity. That is, a prediction engine that works extremely well on one class tends to fail on the other. We posit that this challenge—heterogeneity in element classes—is a widespread and foundational challenge in genomics. For example, the emerging literature on “chromatin priming elements” (59) demonstrates the existence of “enhancer-like” functional elements that, while they share chromatin structure and similar patterns of TF occupancies with enhancers, do not themselves drive patterned expression; rather, they establish chromatin context that subsequently gives rise to enhancer activity for proximal elements. It may be that the class of elements we presently denote “enhancers” is in fact diverse, admitting elements that exert regulatory effects through a variety of underlying molecular mechanisms. Indeed, it remains unclear what fraction of enhancers require eRNAs for their activity (59), or whether priming elements are transcribed like many enhancers.
It may also be that the non-SDEs or class II enhancers we studied here are simply regulated by cohorts of TFs we have yet to survey. These enhancers, however, are often expressed in a smaller fraction of the embryo than SDEs and have lower whole-embryo average DNase I-seq and TF ChIP binding scores (SI Appendix, Fig. S1), consistent with them being accessible in only a small subset of cells. This would thus make non-SDEs less amenable to interrogation through whole-animal ChIP-seq. The differences between the two enhancer classes are statistical, although not categorical. While there is a significant enrichment in segmentation GO-terms in SDEs compared with non-SDEs, some non-SDEs also display segmentally repeating expression patterns similar to those of SDEs, and many (20%) show activity above the 15% expression area threshold. Finally, 8.5% of non-SDEs were active in as much or more of the embryo as the median for SDEs.
Our validation assays revealed that cross-validation had led to significant overestimation of the FDR for SDEs. We attribute this to an abundance of false-negatives in our training set as our analysis indicated that ∼10% of negatives are erroneously labeled, which would double the number of positives and explain the significant increase in validated FDRs we observed. An alternative explanation for the discrepancy is selection bias in our training set as the genomic regions tested in the previous studies were not selected at random. Thus, it is possible that there is a stronger separation of features when the complete set of genomic enhancers is considered. Overall, we recover 98% of the training-set SDEs with an estimated FDR of less than 15%, indicating that our genome-wide predicted catalog of these elements may be close to comprehensive. Further experiments, particularly concentrated at high FDRs, are needed to better assess the boundary between functional and nonfunctional elements. It will also be important to assess the impact of the minimal promoter elements selected for these screens to study promoter–enhancer interactions, as was recently done in Arnold et al. (60), who found that different minimal promoters respond differently to the same enhancer elements. It may be that many of our remaining false-positives are in fact false-negatives in the enhancer screens due to mismatch between the putative enhancer element and our minimal promoter. At this time, it appears that at least 1,600 elements, composing more than 1.6% of the Drosophila genome, are involved in establishing early body patterning in the blastoderm.
Materials and Methods
Data Acquisition and Processing: TF Binding Data and Pol II Data.
Twenty-five percent of FDR Transcription factor ChIP-chip data were taken from the Drosophila TF network project (available at genome.ucsc.edu/cgi-bin/hgTracks?db=dm3&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr2L%3A826001-851000&hgsid=699690721_K8EWMPlLw7903qNMAaAqfOfBnHcn), containing data for 22 transcription factors: BCD, CAD, D, DA, DL, FTZ, GT, H, HB, HKB, KNI, KR, MAD, MED, PRD, RUN, SHN, SLP, SNA, TLL, TWI, Z, some with biological duplicates to give 34 tracks. Of FDR ChiP-chip data for Pol II binding, 1% was also taken from BDTNP (37–39).
Data Acquisition and Processing: Histone Data.
Histone modification and ZLD ChIP-seq data were retrieved from the University of California, Santa Cruz (UCSC) genome browser track provided by Li et al. (40). Histone modifications data collected in the Zld mutant strain were not used; all other tracks were included in the analysis.
DNase Accessibility, Conservation Scores, and Gene Annotations.
DNase accessibility data and 12-fly conservation phastCons scores were obtained from the UCSC genome browser (61, 62) and conservation scores were computed as the maximum of the mean conservation score over 200-nt windows across the element. Note that the study was repeated varying the window size from 1 nt (simple maximum over the enhancer) to 300 nt (in 50-nt steps), but our analysis was insensitive to how the conservation data were processed. Conservation scores did not constitute an important predictor under any scoring protocol that we attempted. FlyBase gene data for exon, coding exons, and intron location data (63) we also downloaded from the UCSC. Bidirectional RNA transcript data were obtained from Nechaev et al. (64) and analyzed as described in Andersson et al. (65). The transcription start site (TSS) was taken as the start of the first exon in FlyBase’s mRNA data, described above.
Defining Predictors.
Although 80% of the DNA segments in the training set were between 2- and 2.5-kb long, segment sizes varied from 100 bp to 4.5 kb in the set, and the percent of enhancer region contained by each segment is unknown, making averages a biased estimator. Thus, the maximum of ChiP data were calculated over every segment in the training set and the segmented genome using bedtools and the UCSC genome browser utilities for TF data, histones, conservation score, and DNase accessibility. In addition, the sum of TF biological replicas and the sum of all TF tracks was also calculated and included as features in the model. In addition to the maximum score, for ZLD higher-resolution ChiP-seq data and for the conservation phastCons conservation scores we also calculated the average over the segment, maximal score over a sliding window of 200, 500, and 1000 bp, and the longest continuous stretch of scores above the 0.85 quantile. For the gene data, bedtools coverage was used to calculate percent of segment covered by exons, coding exons, or introns and three binary tracks indicate the presence or absence of intron and exons. Bedtools was used to calculate distance to the closest TSS and to pol II binding peak.
Modeling.
RFs were modeled in R (66) using RandomForest (48). Initial feature-set culling was done through error rates average of 1,000 forests of 500 trees when excluding/adding one feature at a time. Our training data are highly unbalanced, with only 10% of segments being enhancers. To improve RF performance, balanced samples were used as a training set. To improve stability of the prediction, and counteract the sampling process employed by balancing the training set, we relied on forest voting. One-thousand forests of 50 trees each were trained on randomly selected sets of 300 enhancer and 300 nonenhancers with 10% of the data held out of the samples and used as test sets. Then we estimated the probability that a given segment will function as an enhancer in our assay as the fraction of trees predicting that the segment is an enhancer. This was repeated until such score was computed for each segment in the set. The same sampling and testing scheme was employed for logistic regression and naïve Bayes (67).
Importance measures varied from sample to sample and averages required 10,000 forests of 50 trees to converge. To increase stability of the importance measure, the average of 50,000 RFs mean decrease in accuracy and mean decrease in Gini index were used to find the importance RF confidence intervals. For local importance calculations, we used a single forest of 50,000 trees produced using all enhancers and a balanced nonenhancer subsample.
Analyses.
ROC curve areas were calculated with R package PRROC (68). PCA was done using prcomp (66). GO term analysis used bedtools (69) to find FlyBase genes located inside training enhancer regions, or to identify the closest genes if none are overlapping. The DAVID bioinformatics resource (70, 71) was used to find and quantify GO term and GO-term enrichment, with the full set of ∼8,000 genomic regions as the genomic background. To find the Affinity matrix of the data, we converted Euclidian distance into a similarity matrix, and calculated seven nearest neighbors for each segment. Spectral clustering and eigenvalue extraction was done using kknn (72) with default settings. We used a masked strategy to assess expression size and pattern on an unannotated randomly ordered set of both enhancer classes.
Genome-Wide Prediction.
A sliding window of 1,000 bp with 100-bp distance was used to create overlapping bins across the entire Drosophila genome. As above, we used an ensemble of RFs (1,000 forests each composed of 500 trees) trained on SDE and nonenhancers only. As above, training sets included a random sample of 300 SDEs and 300 nonenhancers. We then generated genome-wide predictions as follows: for each 1,000-bp segment in the genome, we computed the percent trees (across all forests) identifying the segment as an SDE, a number that we interpret as an estimate of the probability that the given segment is an SDE. Note that each 100-bp segment in the genome occurs in nine 1,000-bp windows. Hence, for each 100-bp segment in the genome, we have nine predicted probabilities corresponding to each of the 1,000-bp windows in which it is included. We define our estimate of the probability that a given 100-bp segment is part of an SDE as the mean of the estimated SDE probabilities for each of the overlapping 1,000-bp windows. We defined a threshold of 0.75 for 100-bp segments; all segments with predicted probabilities greater than 0.75 were labeled as part of enhancers, and all other segments were labeled as nonenhancer regions. Adjacent 100-bp windows above this threshold were merged into larger enhancer elements. For predicted SDEs longer than 1.5 kb, we attempted to refine our resolution by leveraging the TF binding data. Specifically, we looked for distinct peaks in the TFsum predictor as follows: the mean of the TFsum track was calculated for each 100-bp window; we then computed the numerical second derivative along the SDE to find extremum points and thus call peaks in the data. Peaks below noise threshold were removed, and peaks closer than 200 bp were merged. If more than one peak remained, the minimum between adjacent peaks was used to separate the longer predicted enhancers. We call this set of elements our “preliminary predicted SDE” (PPSDE) set. Finally, we reran the ensemble of RFs across all PPSDEs and computed estimates of the probability that each PPSDE is an SDE. To estimate a new FDR MLE and confidence interval, we considered the probability of being an enhancer calculated on our training set. By considering the FDR in each of several short probability threshold regions in the training set, and assuming a Poisson distribution for the false discovery, we calculated the MLE of FDR in those regions. The center of the probability threshold region points was taken to have that FDR, and we further considered 100% FDR at 0 probability as an additional data point. A second-order polynomial was fitted to these data points, so that an FDR can be calculated at each probability level. The 1,640 predicted enhancers were fitted to the polynomial, with the average taken as the predicted and confidence interval FDR score.
PCR of Fragments from Genomic DNA and Cloning into the Gateway Vector.
PfuUltra High-Fidelity DNA Polymerase (Stratagene) or EASYA DNA polymerase (Agilent) was used to amplify selected fragments (see above) by using isogenic genomic DNA from y; cn bw sp (73) as a template. The PCR products were confirmed by agarose gel analysis, purified by using the QIA-quick PCR Purification Kit (Qiagen). PCR fragment cloning was performed by adding three A-overhangs to the PCR products produced using the PfuUltra High-Fidelity DNA Polymerase (A-overhangs were not added to the products produced using the EasyA DNA Polymerase) with the addition of dATP and Taq polymerase in a 10-min incubation at 72 °C before Qiagen purification. The products (9.5 μL of each) were used in a TA TOPO cloning reaction with pCR8/GW/TOPO, as described by the manufacturer (Invitrogen). Cloning reactions were allowed to proceed for 30 min at room temperature, and then 2 μL of each reaction was used to transform Mach1 cells (Invitrogen). For each cloning reaction, two isolates were picked, purified, and confirmed by sequence verification.
Sequence Verification of Clones.
Two Gateway clones were picked for each enhancer fragment, for a total of 78 processed clones. Sequencing primers M13 forward -20 (5′ GTAAAACGACGGCCA 3′; Invitrogen) and M13 reverse (5′ GGAAACAGCTATGACCATG 3′; Invitrogen) were used to generate sequences to verify targets. One clone was identified and selected for future studies.
Transfer of Gateway Clones into Integration Vectors.
Thirty-seven nanograms of the destination vector, pBPGUw, were combined with 37.5 ng of DNA carrying a PCR fragment cloned in the Gateway vector in a LR reaction (Invitrogen) and incubated overnight at room temperature. TAM1 cells (Invitrogen) were transformed with 2.5 μL of the LR reaction and plated. A single isolate from each reaction was picked into a 96-well Beckman Deepwell block, allowed to grow overnight at 37 °C, and DNA was prepared by using the PerfectPrep kit (5 PRIME). The constructs were verified by analysis of restriction enzyme digests. A second isolate was picked in cases where there was a discrepancy between the observed and expected results. DNA for injection was prepared from 7 mL of overnight culture for production of transgenic flies.
Drosophila Genetics.
DNA constructs (100–200 ng/μL) were microinjected into embryos derived from parents homozygous for both the attP2 integration site (74) and a fusion gene encoding the PhiC31 integrase under the control of the nanos promoter (nos–integ), which provides a maternal source of integrase (75). Single males derived from these embryos were crossed to y w;Sco/CyO females, and males carrying the inserted construct (identified by their w+ eye color) were selected; integrase is removed in this step. These males were crossed to w[1118]/Dp(1;Y)y[+]; TM2/TM6C, Sb[1] (Bl stock #5906) females to establish balanced, homozygous stocks. We obtained 48 transgenic lines containing predicted enhancer elements, called CEPs.
Verification of Insertion Site and Fragment Identity by Genomic PCR.
To verify the identity of transformant flies and to confirm that all integration events occurred at the attP2 site, we performed genomic PCR on DNA isolated from homozygous transformant flies. Twenty flies were homogenized and genomic DNA isolated by using the ZR Genomic DNA II Kit (Zymo Research). To assay proper integration in the attP2 landing site, PCR was performed by using a primer from the y gene marker in the attP2 genomic docking site (TCATGACTTTGTTGCCTTAGA) and a reverse primer from the w gene (CGAAAGAGACGGC- GATATT) carried in the constructs. Only proper integration events yield a product of 1,839 bp, because y and w lie more than 2 Mb away in the Drosophila genome. Fragment identity was confirmed by using a vector-specific primer (ACAAGTTTGTACAAAAAAGCAGGCT) and a reverse primer specific to the cloned fragment being tested for enhancer activity; the position of the fragment-specific primer was chosen so as to yield a PCR product of 350–400 bp.
Embryo Whole-Mount in Situ Hybridizations.
Embryos were collected directly from the homozygous stock. Embryonic whole-mount in situ RNA hybridizations were performed as described previously (76). A summary is shown in SI Appendix, Fig. S6.
Supplementary Material
Acknowledgments
This work was funded by NIH/National Institute of General Medical Sciences Grant 5P01GM0999655 under Department of Energy Contract DE-AC02-05CH11231. J.B.B.’s work was supported by National Human Genome Research Institute Grant R00 HG006698 and Department of Energy/Laboratory Directed Research and Directed DE-AC02-05CH11231/14-200. P.J.B. was supported by NIH Grant 1U01HG007031-01. S.E.C. was also supported by NIH Grant R01-GM076655.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808833115/-/DCSupplemental.
References
- 1.Fernández M, Miranda-Saavedra D. Genome-wide enhancer prediction from epigenetic signatures using genetic algorithm-optimized support vector machines. Nucleic Acids Res. 2012;40:e77. doi: 10.1093/nar/gks149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F, Li H, Ren C, Bo X, Shu W. PEDLA: Predicting enhancers with a deep learning-based algorithmic framework. Sci Rep. 2016;6:28517. doi: 10.1038/srep28517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajagopal N, et al. RFECS: A random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erwin GD, et al. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol. 2014;10:e1003677. doi: 10.1371/journal.pcbi.1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia C, He W. EnhancerPred: A predictor for discovering enhancers based on the combination and selection of multiple features. Sci Rep. 2016;6:38741. doi: 10.1038/srep38741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee D, Karchin R, Beer MA. Discriminative prediction of mammalian enhancers from DNA sequence. Genome Res. 2011;21:2167–2180. doi: 10.1101/gr.121905.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Qu W, Shan G, Zhang C. DELTA: A distal enhancer locating tool based on AdaBoost algorithm and shape features of chromatin modifications. PLoS One. 2015;10:e0130622. doi: 10.1371/journal.pone.0130622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comin M, Antonello M. On the comparison of regulatory sequences with multiple resolution entropic profiles. BMC Bioinformatics. 2016;17:130. doi: 10.1186/s12859-016-0980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang B, et al. BiRen: Predicting enhancers with a deep-learning-based model using the DNA sequence alone. Bioinformatics. 2017;33:1930–1936. doi: 10.1093/bioinformatics/btx105. [DOI] [PubMed] [Google Scholar]
- 10.Liu B, Fang L, Long R, Lan X, Chou KC. iEnhancer-2L: A two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinformatics. 2016;32:362–369. doi: 10.1093/bioinformatics/btv604. [DOI] [PubMed] [Google Scholar]
- 11.Kleftogiannis D, Kalnis P, Bajic VB. DEEP: A general computational framework for predicting enhancers. Nucleic Acids Res. 2015;43:e6. doi: 10.1093/nar/gku1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imrichová H, Hulselmans G, Atak ZK, Potier D, Aerts S. i-cisTarget 2015 update: Generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res. 2015;43:W57–W64. doi: 10.1093/nar/gkv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firpi HA, Ucar D, Tan K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics. 2010;26:1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dogan N, et al. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than histone modifications or chromatin accessibility. Epigenetics Chromatin. 2015;8:16. doi: 10.1186/s13072-015-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 18.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilton IB, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kellis M, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaaij LJ, et al. Enhancers reside in a unique epigenetic environment during early zebrafish development. Genome Biol. 2016;17:146. doi: 10.1186/s13059-016-1013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiménez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liaw GJ, et al. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 1995;9:3163–3176. doi: 10.1101/gad.9.24.3163. [DOI] [PubMed] [Google Scholar]
- 25.Guichet A, et al. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 26.Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10:1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- 27.Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson P. Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embryos. Genetics. 1983;105:615–632. doi: 10.1093/genetics/105.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence PA. The Making of a Fly: The Genetics of Animal Design. Blackwell Scientific; Oxford: 1992. [Google Scholar]
- 30.Parkhurst SM, Meneely PM. Sex determination and dosage compensation: Lessons from flies and worms. Science. 1994;264:924–932. doi: 10.1126/science.8178152. [DOI] [PubMed] [Google Scholar]
- 31.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 32.Mahowald AP, Hardy PA. Genetics of Drosophila embryogenesis. Annu Rev Genet. 1985;19:149–177. doi: 10.1146/annurev.ge.19.120185.001053. [DOI] [PubMed] [Google Scholar]
- 33.St Johnston D, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 34.Kvon EZ, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512:91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 35.Fisher WW, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci USA. 2012;109:21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer; Berlin: 2013. [Google Scholar]
- 37.Moses AM, et al. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLOS Comput Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacArthur S, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li XY, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife. 2014;3:e03737. doi: 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas S, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplan T, et al. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 2011;7:e1001290. doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li XY, et al. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z. A space-time process model for the evolution of DNA sequences. Genetics. 1995;139:993–1005. doi: 10.1093/genetics/139.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felsenstein J, Churchill GA. A Hidden Markov model approach to variation among sites in rate of evolution. Mol Biol Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- 47.Biau G, Scornet E. A random forest guided tour. TEST. 2016;25:197–227. [Google Scholar]
- 48.Andy Liaw MW. Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 49.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 50.Halfon MS, Gallo SM, Bergman CM. REDfly 2.0: An integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res. 2008;36:D594–D598. doi: 10.1093/nar/gkm876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallo SM, Li L, Hu Z, Halfon MS. REDfly: A regulatory element database for Drosophila. Bioinformatics. 2006;22:381–383. doi: 10.1093/bioinformatics/bti794. [DOI] [PubMed] [Google Scholar]
- 52.Gallo SM, et al. REDfly v3.0: Toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 2011;39:D118–D123. doi: 10.1093/nar/gkq999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raney BJ, et al. Track Data Hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Brink DM, Banerji O, Tear G. Commissureless regulation of axon outgrowth across the midline is independent of Rab function. PLoS One. 2013;8:e64427. doi: 10.1371/journal.pone.0064427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keleman K, et al. Comm sorts robo to control axon guidance at the Drosophila midline. Cell. 2002;110:415–427. doi: 10.1016/s0092-8674(02)00901-7. [DOI] [PubMed] [Google Scholar]
- 57.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng AY, Jordan MI, Weiss Y. On spectral clustering: Analysis and an algorithm. Adv Neur In. 2002;14:849–856. [Google Scholar]
- 59.Bevington SL, et al. Inducible chromatin priming is associated with the establishment of immunological memory in T cells. EMBO J. 2016;35:515–535. doi: 10.15252/embj.201592534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnold CD, et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol. 2017;35:136–144. doi: 10.1038/nbt.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karolchik D, et al. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. FlyBase Consortium FlyBase: Genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Core Team 2014 R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna). Available at https://www.r-project.org/. Accessed December 18, 2018.
- 67.Lewis DD. European Conference on Machine Learning. Springer; Berlin: 1998. Naive (Bayes) at forty: The independence assumption in information retrieval; pp. 4–15. [Google Scholar]
- 68.Keilwagen J, Grosse I, Grau J. Area under precision-recall curves for weighted and unweighted data. PLoS One. 2014;9:e92209. doi: 10.1371/journal.pone.0092209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 72.Hechenbichler K, Schliep K. 2004 doi: 10.5282/ubm/epub.1769. Weighted k-Nearest-Neighbor Techniques and Ordinal Classification. Available at . . Accessed December 10, 2018. [DOI]
- 73.Hoskins RA, et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015;25:445–458. doi: 10.1101/gr.185579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weiszmann R, Hammonds AS, Celniker SE. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nat Protoc. 2009;4:605–618. doi: 10.1038/nprot.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.