Significance
Herpes simplex virus (HSV) is a ubiquitous human pathogen. The mechanism of HSV DNA replication is poorly understood. ICP8, the multifunctional HSV single-strand DNA binding protein (SSB), is essential for viral growth and DNA replication. ICP8 is also capable of efficiently annealing complementary ssDNA and has been classified as a single-strand annealing protein (SSAP); however, the role of annealing during infection is still unclear. In this paper, we characterize a mutant of ICP8 that retains many of its functions as a replicative SSB but lacks annealing activity. We show that ICP8 annealing activity is essential for viral DNA replication. These findings support our hypothesis that HSV-1 uses recombination-dependent mechanisms to replicate its DNA.
Keywords: ICP8, annealing, HSV-1, DNA replication, recombination
Abstract
Most DNA viruses that use recombination-dependent mechanisms to replicate their DNA encode a single-strand annealing protein (SSAP). The herpes simplex virus (HSV) single-strand DNA binding protein (SSB), ICP8, is the central player in all stages of DNA replication. ICP8 is a classical replicative SSB and interacts physically and/or functionally with the other viral replication proteins. Additionally, ICP8 can promote efficient annealing of complementary ssDNA and is thus considered to be a member of the SSAP family. The role of annealing during HSV infection has been difficult to assess in part, because it has not been possible to distinguish between the role of ICP8 as an SSAP from its role as a replicative SSB during viral replication. In this paper, we have characterized an ICP8 mutant, Q706A/F707A (QF), that lacks annealing activity but retains many other functions characteristic of replicative SSBs. Like WT ICP8, the QF mutant protein forms filaments in vitro, binds ssDNA cooperatively, and stimulates the activities of other replication proteins including the viral polymerase, helicase–primase complex, and the origin binding protein. Interestingly, the QF mutant does not complement an ICP8-null virus for viral growth, replication compartment formation, or DNA replication. Thus, we have been able to separate the activities of ICP8 as a replicative SSB from its annealing activity. Taken together, our data indicate that the annealing activity of ICP8 is essential for viral DNA replication in the context of infection and support the notion that HSV-1 uses recombination-dependent mechanisms during DNA replication.
Recombination is a universal genetic process in all organisms and is essential for DNA replication, repair of DNA damage, and genetic diversification. dsDNA viruses of bacteria, protozoa, plants, insects, and mammals utilize recombination-dependent mechanisms during DNA replication to produce longer-than-unit-length genomes (concatemers) (1–3). The recombination machinery encoded by many dsDNA viruses is composed of a 5′-to-3′ exonuclease capable of catalyzing end resection and a single-strand annealing protein (SSAP) (4, 5). The best-characterized example of an exo/SSAP complex is phage λ Redα/β, which in addition to its role in viral replication has been used to promote in vivo recombination-mediated genetic engineering or recombineering (6–9). Interestingly, herpes simplex virus (HSV) also encodes an exo/SSAP and, like other dsDNA viruses, must generate concatemers to produce viral DNA that can be packaged into infectious virions (3, 10, 11).
Several lines of evidence support the notion that HSV performs recombination-dependent replication (RDR). The HSV genome consists of two unique regions, UL and US, flanked by inverted repeats. During DNA replication, UL and US invert relative to each other, resulting in the formation of four different isomeric viral genomes (12, 13). Genomic inversions occur as soon as DNA synthesis can be detected (14–16) and are dependent upon the presence of the seven essential viral replication proteins, suggesting that HSV-1 DNA synthesis is inherently recombinogenic (17). Evidence for high rates of recombination between coinfecting HSV genomes comes from cell culture and animal infection models as well as from analysis of viruses circulating in patient populations (18–22). Additionally, it has long been recognized that viral replication intermediates are composed of complex X- and Y-branched structures as evidenced by electron microscopy (10, 16) and pulsed-field gel electrophoresis (15, 23, 24).
The HSV exo/SSAP, composed of a 5′-to-3′ exonuclease (UL12) and an SSAP (ICP8), is capable of promoting strand exchange in vitro (25, 26). More recently, we have shown that HSV infection stimulates single-strand annealing (27), and ICP8 has been reported to promote recombineering in transfected cells (28). However, these assays do not directly address the importance of the exo/SSAP during viral replication. We have recently demonstrated that the exonuclease activity of UL12 is essential for the production infectious virus (29); however, the role of ICP8 as an annealing protein has not been explored.
ICP8 is a multifunctional 128-kDa single-stranded DNA (ssDNA) binding protein (SSB) that is essential for viral DNA synthesis and viral growth (30, 31). ICP8 participates in multiple steps of the HSV life cycle, including gene expression, DNA replication, and replication compartment (RC) formation (31–33). During DNA replication, ICP8 functions as a replicative SSB, interacting with and stimulating other HSV-1 replication proteins (34–40). ICP8 preferentially binds ssDNA in a nonspecific and cooperative manner and holds it in an extended conformation, resulting in the formation of a nucleoprotein filament that resembles a beaded necklace (39, 41). Interestingly, ICP8 can stimulate the annealing of complementary ssDNA (42, 43) and has, thus, been classified as a member of the SSAP family (3). Due to the multifunctional nature of this protein, it has been difficult to tease out the role of ICP8 as an SSAP from the other roles that it plays during viral replication. In this paper, we describe a mutant of ICP8, Q706A/F707A, that lacked annealing activity but retained most if not all of the other functions associated with replicative SSBs. The QF mutant was not able to complement an ICP8-null virus for viral growth, RC formation, or DNA replication. Taken together, these results suggest that ICP8 annealing activity is essential for HSV-1 viral replication.
Results
ICP8 Residues Q706 and F707 Are Essential for Viral Growth.
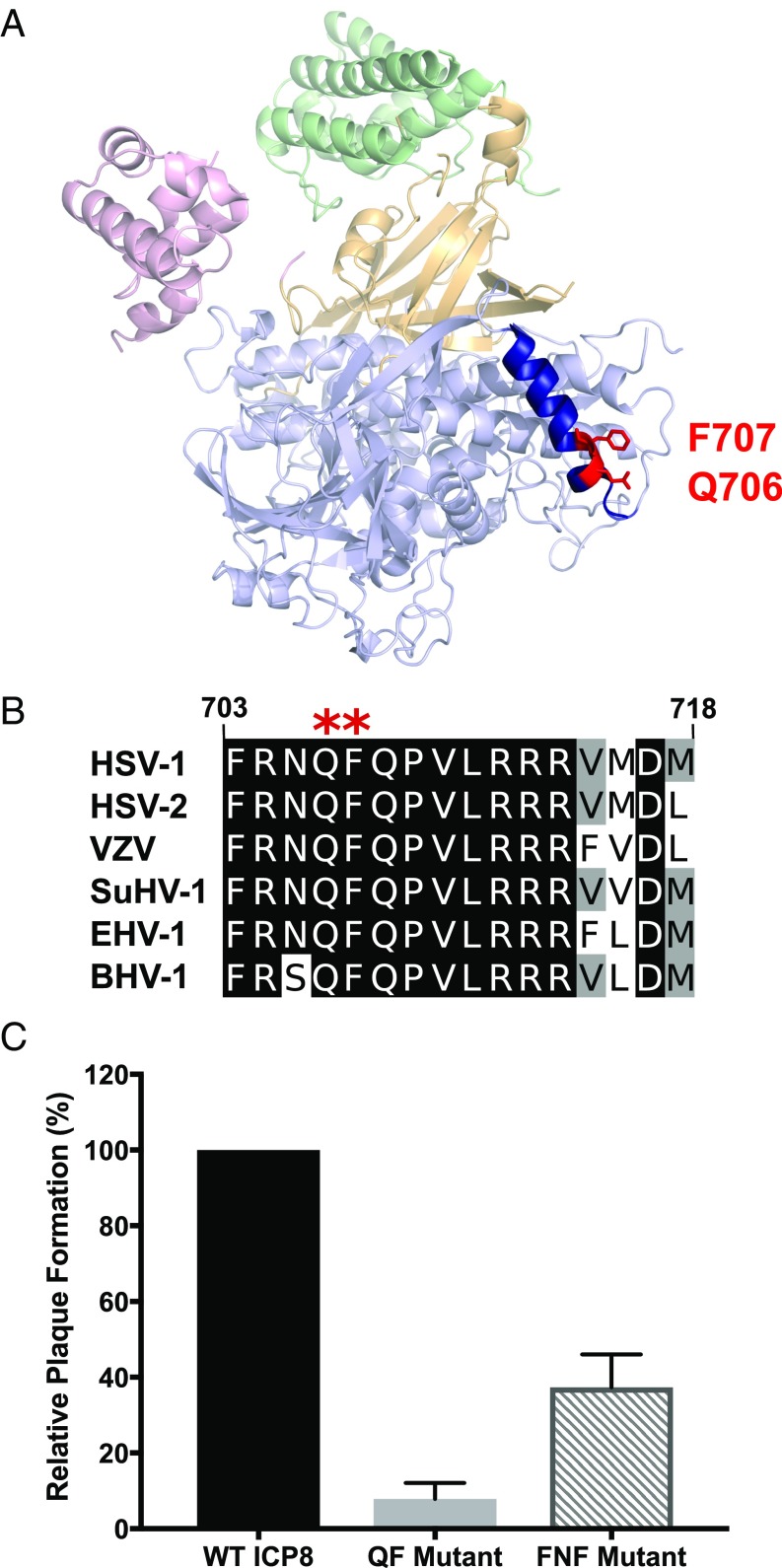
The structure of ICP8 reveals a large N-terminal domain (amino acids 9–1038) consisting of head, neck, and shoulder regions and a much smaller C-terminal domain composed of two elements, a compact C-terminal helical region (1049–1129), and a largely disordered region at the extreme C terminus (1127–1187) (Fig. 1A) (44). The N-terminal domain exhibits an unusual structure in that it is made up of noncontiguous polypeptide chains. This structure has made it difficult to express individual functional subdomains and has hampered efforts to map regions of ICP8 responsible for its many functions. To generate mutations that would not disrupt global protein folding and would allow us to genetically separate ICP8 functions, we used a targeted mutagenic approach. A bioinformatic analysis was performed, resulting in the identification of conserved surface-exposed residues. Five clusters of surface-exposed residues were identified: R644–D645, N882–L883, and D905–Y909 in the head region, and R262–H266 and Q706–F707 in the shoulder region. Additionally, we included two clusters within the shoulder region, Y20–F61–Y90 and C116–R120, that were previously identified by Mapelli et al. (44). Alanine substitution mutations in the residues shown in Table 1 were introduced into an ICP8 mammalian expression vector, and mutant proteins were examined for expression, localization, and ability to complement the ICP8-null virus (HD-2) for viral growth. Mutants that failed to express stable protein or to localize to the nucleus were excluded from further study. The Q706A–F707A (QF) mutant showed the most severe defect in its ability to complement HD-2 for viral growth. Q706 and F707 are located in the shoulder region of ICP8 within a surface-exposed α-helix that is highly conserved among alphaherpesviruses (Fig. 1 A and B).
Fig. 1.
Residues Q706 and F707 are essential for virus growth, and alanine substitution mutants in these positions have a transdominant phenotype. (A) Ribbon diagram of ICP8 (PDB ID code 1URJ) illustrating major structural domains: head (green), neck (yellow), shoulder (light blue), and C terminus (pink). Q706 and F707 residues (red) lie within a conserved α-helix (amino acids 703–718; dark blue) on the surface of the ICP8 shoulder domain. (B) Conservation of residues 703–718 among alphaherpesviruses [HSV type 1 (P17479), HSV type 2 (P36384), varicella zoster virus (P09245), Suid herpes virus type 1 (P11870), equine herpes virus type 1 (Q6S6PO), and bovine herpes virus type 1 (P12639)]. Residues Q706 and F707 are marked with a red asterisk. Alignment was performed using T-Coffee Expresso and edited with Jalview. (C) A plaque reduction assay was performed by transfecting Vero Cells with 50 ng of pSAK plasmid containing WT ICP8, QF mutant, or FNF mutant along with 1:1 molar ratio of infectious KOS DNA. Plaques were counted 3–4 d posttransfection. The graph represents percent plaque formation relative to the cells transfected with WT ICP8. Columns represent the average of three independent experiments. Error bars signify SD.
Table 1.
Analysis of ICP8 alanine substitution mutants
| ICP8 mutant | Protein expression | Protein localization | % Complementation |
| ICP8 WT | Yes | Nuclear | 100 |
| Y20A–F61A–Y90A | Yes | Cytoplasmic | 0 |
| C116A–R120A | Yes | Nuclear | 70 |
| R262A–H266A | Yes | Nuclear | 14 |
| R644A–D645A | Yes | Nuclear | 54 |
| Q706A–F707A | Yes | Nuclear | 0 |
| N882A–L883A | Yes | Nuclear | 14 |
| D905A–Y909A | No | N/A | N/A |
Table showing the analysis of alanine substitution mutants assessed in this study. Vero cells were transiently transfected with 50 ng of mammalian expression plasmids containing ether WT or mutant versions of ICP8 and analyzed by Western blot and IF with anti-ICP8 antibody for protein expression and localization, respectively. For complementation analysis, transfected Vero cells were superinfected with HD-2 at an MOI of 3 for 24 h. Virus was harvested and titrated on the ICP8-complementing cell line (S2). Viral plaques were counted and reported as a percentage of WT ICP8. Results for Q706A and F707A mutant are in bold.
The Q706A F707A Mutant Shows a Transdominant Phenotype.
Mutations of multifunctional proteins such as ICP8 often result in a transdominant or dominant-negative phenotype in which the mutant protein antagonizes the function of its WT counterpart. This phenotype results from the ability of the mutant protein to perform some of its essential functions while being deficient in others and is often associated with mutants of proteins that oligomerize or form filaments. In addition to nucleoprotein filaments on ssDNA, ICP8 forms double-helical filaments in the absence of DNA (34). A transdominant mutant, containing alanine substitution mutations of a linear FNF motif within the C terminus of ICP8, was shown to be defective for viral growth and filament formation, indicating a defect in oligomerization (38). A plaque reduction assay was performed as described in Materials and Methods. The QF mutant exhibited a strong transdominant effect compared with the FNF mutant, reducing plaque formation by ∼92% (Fig. 1C), suggesting that mutation of the QF residues disrupts an essential function of the protein while preserving others.
The QF Mutant Forms Double-Helical Filaments.
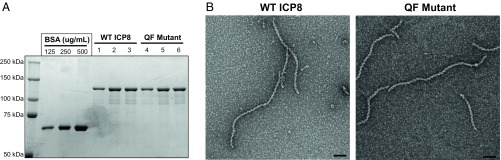
To determine whether the transdominant phenotype of the QF mutant is associated with a defect in oligomerization, we assessed the ability of the QF mutant protein to form filaments by negative-stain EM. WT and QF mutant proteins were expressed and purified from Sf9 insect cells infected with recombinant baculovirus. The yield and purity of the QF mutant protein were similar to WT (Fig. 2A). The filaments formed by the mutant protein are similar in appearance to those formed by WT, suggesting that there is no defect in filament formation (Fig. 2B). If anything, the QF mutant protein may have a slightly higher propensity to form filaments. Further experimentation will be required to determine whether this property is biologically significant.
Fig. 2.
The QF mutant forms double-helical protein filaments. (A) WT ICP8 and QF mutant proteins were expressed and purified from Sf9 insect cells infected with recombinant baculovirus as described in Materials and Methods. Purified proteins were resolved on 8% SDS/PAGE gel and stained with Coomassie blue. A BSA standard was used to confirm protein concentration determined by Bradford assay. Molecular weight markers (in kilodaltons) are shown on the Left. (B) Both WT ICP8 and QF mutant formed double-helical filaments after overnight incubation at 4 °C in the presence of a buffer containing 5 mM Mg2+. Samples were prepared for TEM at a magnification of 40,000×. (Scale bar: 100 nm.)
QF Mutant Protein Binds to ssDNA.
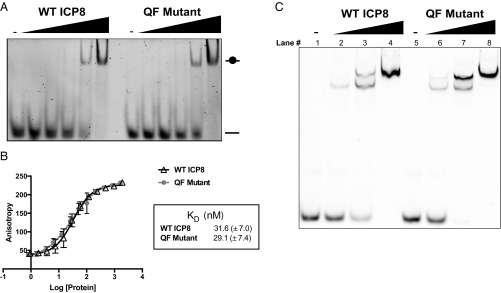
The ability of the QF mutant protein to bind ssDNA was measured using a 5′-fluorescein–tagged 14-mer oligonucleotide that is capable of binding a single molecule of ICP8. Fig. 3A shows an electrophoretic mobility shift assay (EMSA) indicating that WT and QF mutant proteins exhibited similar ssDNA binding abilities. To more precisely quantify binding affinity, fluorescence anisotropy was performed using this oligonucleotide. Fig. 3B shows nearly identical binding curves for WT and QF mutant proteins, with dissociation constants (Kd) of 31.6 (±7.0) nM and 29.1 (±7.4) nM, respectively. To assess whether the DNA binding activity of the QF mutant protein is cooperative, an EMSA was performed using a 5′-Cy3–labeled 50-mer oligonucleotide that is predicted to bind three to five molecules of ICP8 per oligo. Multiple shifted bands were observed in Fig. 3C, indicative of cooperative binding for both WT and QF mutant proteins. These data demonstrate that the QF protein binds to DNA with similar or slightly higher cooperativity compared with WT (Fig. 3C, compare lanes 7 and 3).
Fig. 3.
The QF mutant binds to ssDNA. (A) EMSA showing the ability of WT ICP8 and QF mutant proteins to bind to ssDNA. Proteins were titrated (0–300 nM, twofold serial dilution) against 100 nM fluorescein-labeled dT-14-mer ssDNA oligo in DNA binding buffer. Samples were incubated for 30 min at room temperature. Bound and unbound species were separated on 5% nondenaturing polyacrylamide gel in 1× TBE and imaged using a fluorescence imager. (B) Purified WT or QF mutant proteins were titrated (∼0.9–2 µM) against 2.5 nM of the same fluorescein-labeled dT-14-mer in the DNA binding buffer used above. FP measurements were taken on TECAN M1000 Pro plate reader with excitation wavelength of 470 nm and emission wavelength of 521 nm. Triplicate readings were averaged and the data from three independent experiments were fit to a 1:1 binding model from which dissociation constants (Kd) were determined. (C) A gel shift assay was performed with a Cy3-labeled 50-mer capable of binding multiple ICP8 monomers. Proteins were titrated (0, 50, 200, 400 nM) against 100 nM oligo.
The QF Mutant Protein Stimulates HSV-1 Replication Proteins in Vitro.
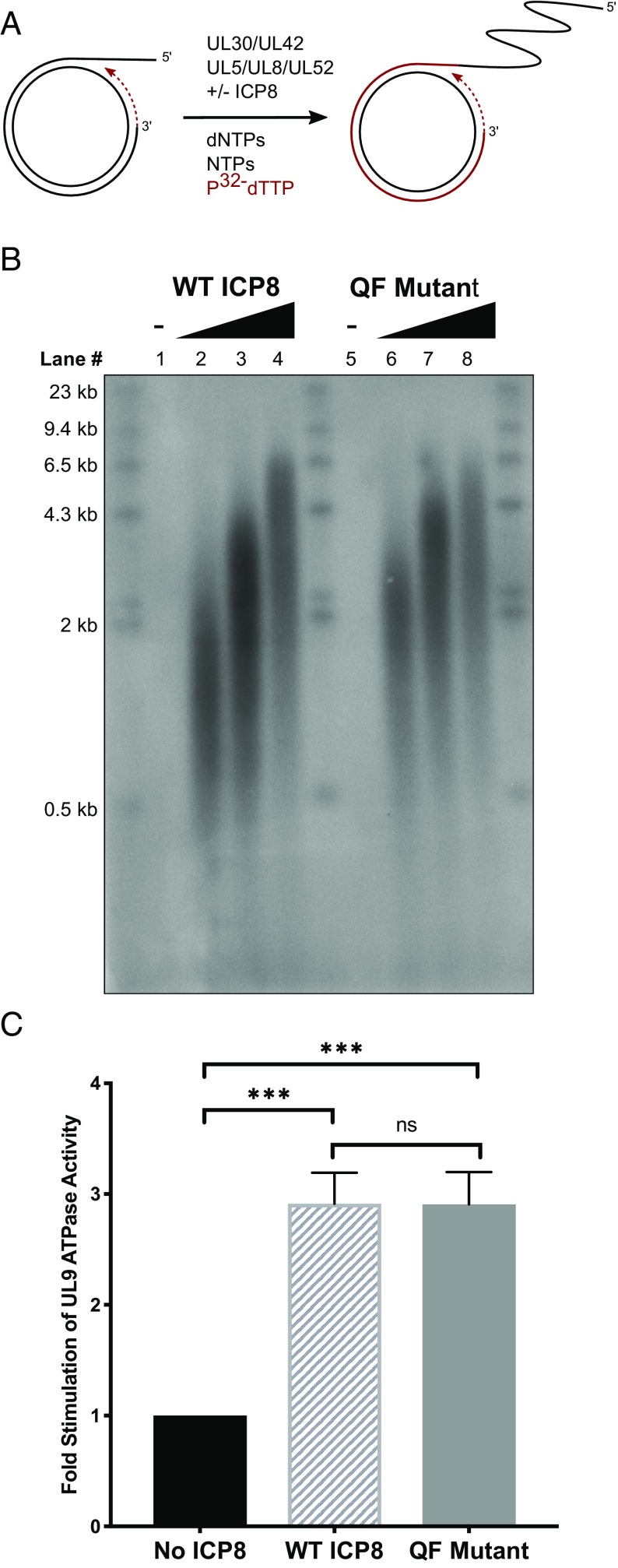
The QF mutant can form filaments and binds to ssDNA with a similar affinity to WT ICP8; therefore, we asked whether it retained other functions associated with replicative SSBs. ICP8 has been reported to stimulate the activities of other core HSV-1 replication proteins involved in leading- and lagging-strand DNA synthesis: HSV polymerases (UL30/UL42) and helicase–primase (UL5/8/52) (34, 36, 37, 39, 45–47). To assess the ability of WT and QF mutant proteins to stimulate these activities, we have employed an in vitro minicircle replication assay using a DNA substrate consisting of a 90-mer linear oligo annealed to a 70-bp minicircle (Fig. 4A) (46, 47). In this assay, the minicircle is the template for leading-strand synthesis and lacks thymidine residues. Thus, leading-strand synthesis can be detected by incorporation of [α-32P]dTTP. In the absence of ICP8, no significant incorporation of dTTP into the leading strand was detected (Fig. 4B, lanes 1 and 5). However, in the presence of either WT or QF mutant protein, incorporation of dTTP was detected, and the length of products generated increased in an ICP8 concentration-dependent fashion (Fig. 4B, lanes 2–4 and 6–8). Thus, the QF mutant protein can stimulate the HSV polymerase and helicase–primase to promote leading-strand DNA synthesis in vitro similar to WT.
Fig. 4.
The QF mutant stimulates replication proteins in vitro. (A) Schematic of rolling-circle replication on a minicircle substrate comprised of a 70-mer ssDNA circle annealed to a 90-mer linear oligo. The 3′ end of the 90-mer acts as the primer for leading-strand synthesis. Leading-strand DNA synthesis is detected by incorporation of [α-32P]dTTP. Newly synthesized DNA is shown in red. (B) An alkaline agarose gel showing stimulation of in vitro leading-strand DNA synthesis by WT and QF mutant proteins. The minicircle assay was performed using increasing concentrations of either WT ICP8 or mutant QF protein (0, 190, 380, and 760 nM). After 60-min incubation at 37 °C, reactions were quenched, and replication products were separated by 1% alkaline agarose gel electrophoresis and imaged with a phosphorimager. (C) ICP8 stimulation of UL9 ssDNA-dependent ATPase activity. WT or QF mutant proteins (700 nM) were mixed with 100 nM UL9 in the presence of 5 mM ATP and 10 µM ssDNA. Reactions were incubated for 1 h at room temperature and quenched with EDTA. ATPase activity was measured by the release of free phosphate as described in Materials and Methods. Fold stimulation relative to the control lacking ICP8 is plotted on the graph. Error bars represent SD. P values were determined by ordinary one-way ANOVA with Tukey’s multiple-comparisons test (***P = 0.0001 and nsP = 0.9993).
Another function of ICP8 consistent with its role as an SSB is its ability to interact with the origin binding protein, UL9, and to stimulate its ssDNA-dependent ATPase and helicase activities in vitro (48). Stimulation of UL9 activity is thought to be dependent on the physical interaction between ICP8 and UL9 (49). We examined the ability of WT and QF mutant proteins to stimulate the ATPase activity of UL9 in the presence of ssDNA and ATP (Materials and Methods). Fig. 4C shows that both WT and the QF mutant proteins were able to stimulate UL9 ATPase activity to a similar degree (approximately threefold; Fig. 4C), suggesting that the QF mutant still interacts with UL9 and would be expected to be competent for initiation of DNA replication.
QF Mutant Is Unable to Complement the ICP8-Null Virus for Viral DNA Replication and Replication Compartment Formation.
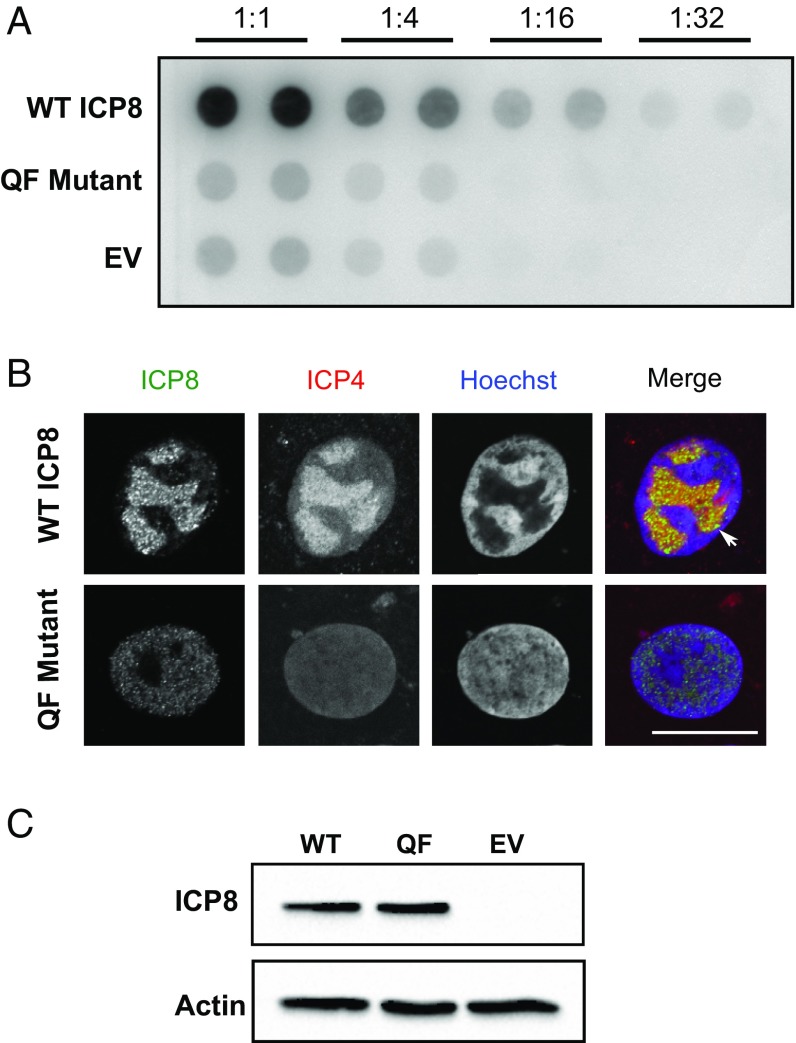
The experiments shown in Figs. 2–4 indicate that the QF mutant is capable of all of the in vitro functions classically associated with an SSB, including ssDNA binding, filament formation, and stimulation of other HSV replication proteins. However, the inability of the QF mutant to complement HD-2 for viral growth suggests that this mutant lacks a function essential in the context of viral infection. We next asked whether the QF mutant can complement HD-2 for DNA replication in infected cells. Vero cells were transfected with plasmids expressing WT or QF mutant protein or with an empty vector (EV) and subsequently superinfected with HD-2. At 24 h postinfection, total DNA was extracted, and the amount of replicated viral DNA was assessed by dot blot analysis. Fig. 5A shows that WT was able to efficiently complement HD-2 for DNA replication. In stark contrast, the amount of DNA replicated in cells transfected with the QF mutant are similar to those seen in the EV-transfected sample, indicating that the QF mutant was unable to complement HD-2 for DNA replication.
Fig. 5.
The QF mutant is unable to complement HD-2 for DNA replication and RC formation. (A) A dot blot showing the ability of WT ICP8 or the QF mutant to complement HD-2 for DNA replication. Vero cells were transfected with WT ICP8, QF mutant, or EV construct and subsequently infected with HD-2 at an MOI of 5 PFU/cell. Samples were harvested at 24 h postinfection, and total DNA was extracted. Serial dilutions were prepared starting with the same amount of total DNA for each sample. Southern blot analysis was performed using α-32P–labeled random probes generated from a PCR-derived UL30 gene template. Duplicate samples for each dilution are shown. In addition, two biological replicates were performed, and a representative image is shown. (B) IF showing the ability of WT ICP8 or the QF mutant to complement HD-2 for RC formation. Vero cells grown on coverslips were transfected with a plasmids expressing WT ICP8 or the QF mutant and subsequently infected with HD-2 at a MOI of 20 PFU/cell. Six hours postinfection, cells were fixed and stained with monoclonal mouse anti-ICP4 and polyclonal rabbit anti-ICP8 primary antibodies followed by Alexa Fluor-labeled secondary antibodies. The white arrowhead indicates a mature RC. DNA was visualized by Hoechst stain. Images were taken using Zeiss LSM 880 Meta confocal microscope. (Scale bar: 3.6 µm.) (C) A Western blot showing equal expression of WT ICP8 and QF mutant protein in transfected Vero cells.
HSV-1 DNA replication occurs within membraneless subdomains in the nucleus known as RCs (50). RC formation and maturation is believed to be dependent on active DNA synthesis, as the assembly of large mature RCs is blocked by addition of the viral polymerase inhibitor phosphonoacetic acid or by omission of any of the seven essential viral replication proteins during infection (33, 50–52). Because the QF mutant is unable to complement HD-2 for DNA replication, we asked whether RCs could be detected in Vero cells transfected with plasmids expressing WT or QF mutant protein and superinfected with HD-2. Infected cells were identified using an antibody against the immediate early protein, ICP4, and transfected cells were identified by ICP8 staining. Fig. 5B shows that in cells that were positive for both ICP4 and ICP8 staining, RCs could be observed in cells expressing WT but not the QF mutant protein. Fig. 5C shows that similar levels of ICP8 were observed in cells transfected with plasmids expressing WT and QF mutant proteins. Taken together, the results shown in Fig. 5 indicate that the QF mutant is defective in a function required for viral DNA replication and RC formation in the context of infection.
The QF Mutant Is Unable to Promote Annealing of Complementary ssDNA.
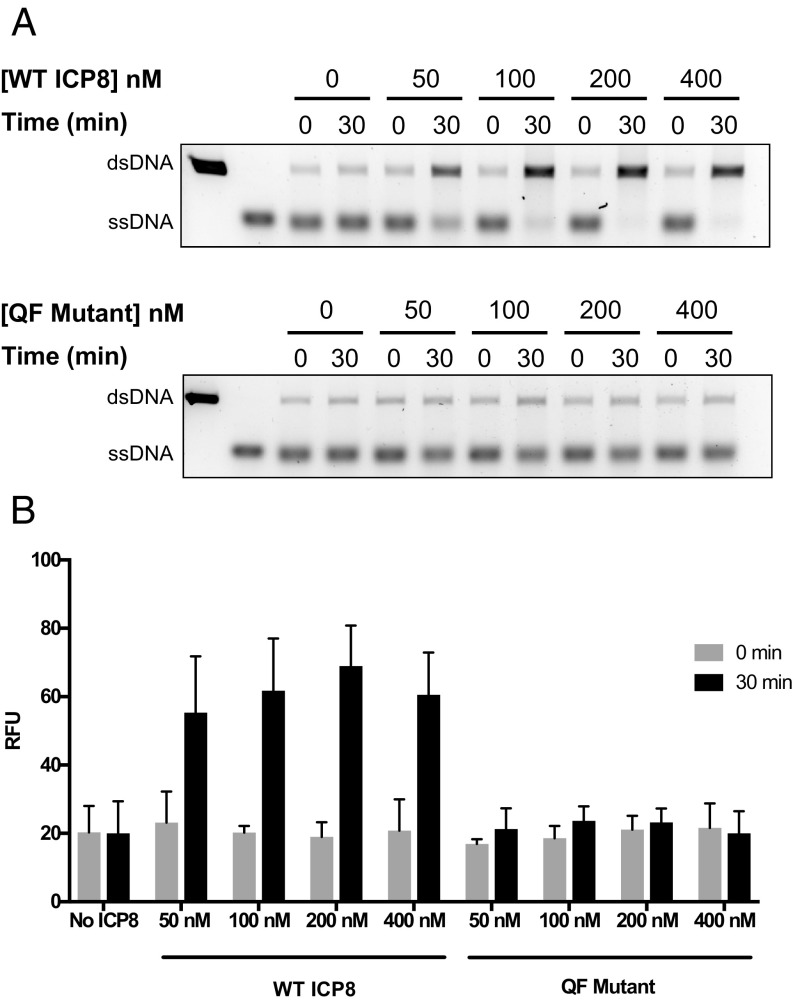
As described in the Introduction, ICP8 has been shown to efficiently promote single-strand annealing in vitro (42, 43), resulting in its characterization as an SSAP. Therefore, we asked whether the QF mutant protein can promote the annealing of complementary ssDNA. Annealed products were either visualized by gel electrophoresis (Fig. 6A) or quantified by incubation with PicoGreen, a fluorescent sensor specific for dsDNA (Fig. 6B). WT protein was able to promote efficient annealing in a concentration-dependent fashion (Fig. 6 A and B). In stark contrast, the QF mutant protein was unable to promote annealing even at the highest concentration tested (Fig. 6). These results indicate that the QF residues are essential for ICP8-mediated annealing.
Fig. 6.
The QF mutant is defective for DNA annealing. (A) Gel-based assay to determine the ability of WT ICP8 and QF mutant to promote annealing. A linearized heat-denatured plasmid was incubated in DNA annealing buffer with varying amounts of either WT or QF mutant proteins (0, 50, 100, 200, and 400 nM). The reactions were initiated by adding ssDNA and incubated for 30 min at 37 °C. Samples were collected at 0 and 30 min and subjected to proteinase K digestion. The products were analyzed by 1% agarose gel electrophoresis followed by staining with SYBR Gold. The migration of the ssDNA and the annealed products are denoted on the Left. A small amount of background annealing was observed in all of the reactions due to self-annealing of the complementary strands. (B) Annealing products were quantified by Quant-iT PicoGreen. The gray colored bars represent fluorescence due to self-annealing of complementary strands at 0 min, while the black bars represent annealing after 30-min incubation in the presence or absence of ICP8. Three independent experiments were performed, and the average values were obtained to determine annealing efficiency. Error bars indicate SD.
The QF Mutant Is Unable to Form Annealing Intermediates as Assessed by EM.
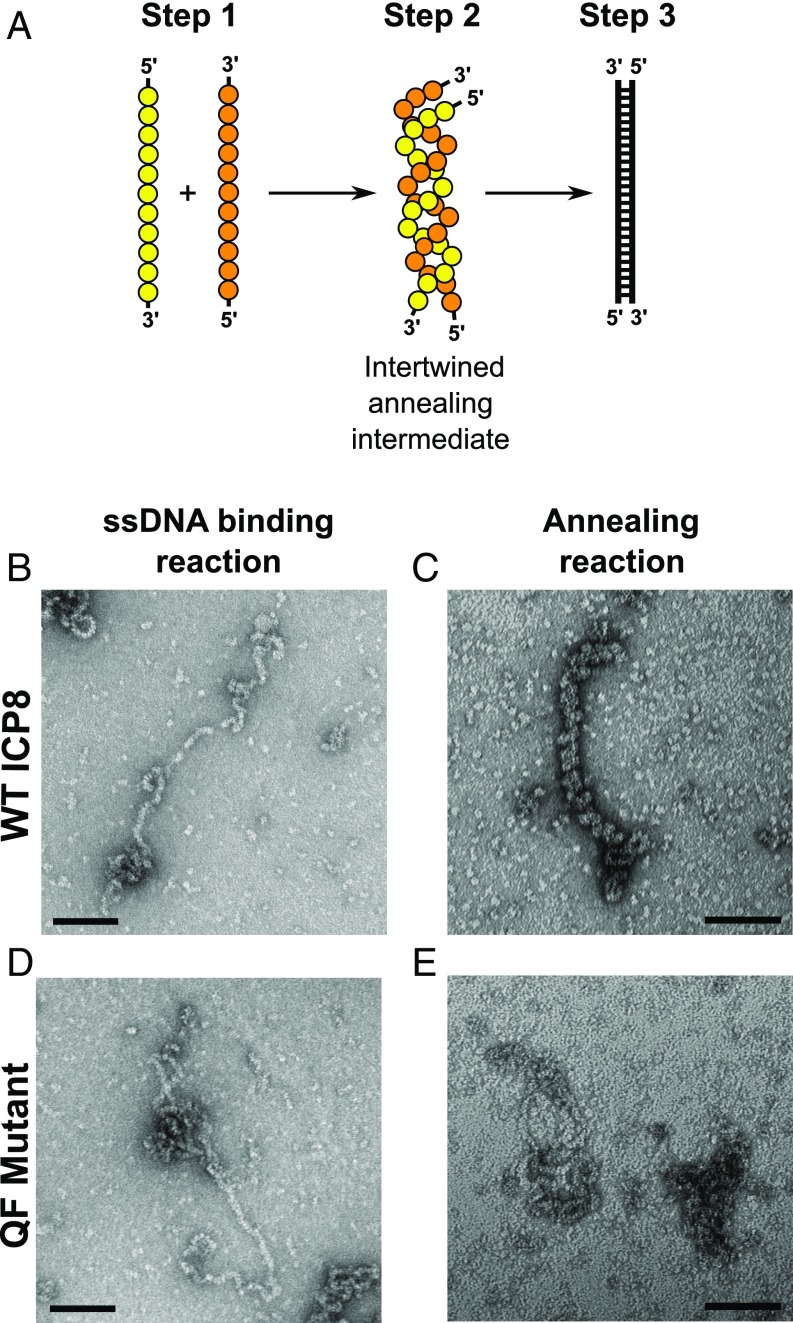
A simplified model showing steps involved in the ICP8-induced annealing reaction is depicted in Fig. 7A. This model is based on the electron-microscopic characterization of the annealing reaction describing the formation of nucleoprotein filaments on ssDNA and the demonstration that two such nucleoprotein filaments on complementary ssDNA come together to form an intertwined annealing intermediate (53). Fig. 7 B and D shows that WT ICP8 and QF mutant proteins both formed similar nucleoprotein filaments on ssDNA, consistent with the fact that the QF mutant did not exhibit a defect in ssDNA binding. Fig. 7C shows that, in the presence of complementary DNA, the WT protein was able to promote the formation of an annealing intermediate with an intertwined coiled-coil structure (53). In contrast, the QF mutant could bind to ssDNA but was unable to form this intertwined annealing intermediate (Fig. 7E). These observations are consistent with its defect in the annealing reaction.
Fig. 7.
Electron-microscopic analysis of WT and QF mutant binding to ssDNA and annealing intermediates. (A) A simplified model showing steps involved in the ICP8-induced annealing reaction. In step 1, ICP8 binds to ssDNA to form a nucleoprotein filament. In step 2, two nucleoprotein filaments containing complementary ssDNA come together to form an intertwined annealing intermediate. In step 3, dsDNA is formed. ICP8 monomers are represented by yellow circles and orange, and DNA is depicted by solid black lines. (B and D) Visualization of ICP8-ssDNA nucleoprotein filaments by EM. Linearized M13 ssDNA was incubated with 1 µg of either WT ICP8 or QF mutant in 20-µL final volume in DNA binding buffer. Samples were incubated for 20 min at room temperature and prepared for TEM at a magnification of 60,000×. (Scale bar: 100 nm.) (C and E) Visualization of intermediates performed during ICP8-induced annealing reaction. WT ICP8 or QF mutant protein (312 nM) was incubated with heat-denatured plasmid in a 20-µL volume reaction in annealing buffer. Samples were incubated at 37 °C for 15–30 min and processed for negative-stain EM. (Scale bar: 100 nm.)
Taken together, our results indicate that the QF mutant can perform biochemical activities that are consistent with its role as an SSB; however, it is unable to carry out annealing of complementary ssDNA. The defect in annealing correlates with the inability of the QF mutant to carry out viral DNA replication and RC formation. This surprising result suggests that the annealing activity of ICP8 is essential for viral DNA replication.
Discussion
Production of HSV concatemeric DNA is an essential step for the generation of progeny virus, as the packaging machinery must recognize longer-than-unit-length concatemers during encapsidation (3, 10, 11); however, the mechanism by which concatemers are formed has not been resolved. It has been proposed that the viral genome circularizes, and subsequent replication proceeds by a rolling circle mechanism (54). Evidence for this model includes the loss of genome free-ends and the appearance of junctional fragments at an early time during infection (55–58). On the other hand, several lines of evidence suggest that HSV DNA replication is more complex (59, 60). For example, it has not been possible to detect circular genomes at early times during lytic infection in cells infected with WT virus (61). Many dsDNA viruses including bacteriophages encode recombination machinery and utilize RDR. In bacteriophage λ, concatemeric DNA can be generated by either rolling-circle replication or RDR (3). Evidence supporting a role for recombination in HSV DNA replication was presented in the Introduction; however, no direct evidence for a role of annealing in this process had been reported. In this paper, we describe an ICP8 mutant, Q706A–F707A (QF), that is defective in annealing but retains the other known functions of a replicative SSB, such as the ability to form filaments, bind ssDNA, and stimulate other replication proteins. Despite its ability to carry out these replication functions, the QF mutant was not able to complement an ICP8-null virus for virus growth, DNA replication, or RC formation. These results suggest that the annealing activity of ICP8 is essential for viral DNA replication and support the notion that RDR is required during HSV DNA replication.
The observation that the QF mutant retained several replicative functions but was unable to promote viral DNA replication in the context of infection was intriguing, and we considered several explanations including the possibility that the QF mutant was unable to support initiation of viral DNA replication. During initiation, ICP8 and UL9 have been proposed to work together to distort and unwind an origin of replication followed by ssDNA binding and helix destabilization by ICP8 (48, 62–70). In this paper, we showed that the QF mutant was able to stimulate ssDNA-dependent UL9 ATPase activity in vitro (Fig. 4C) and bind cooperatively to ssDNA (Fig. 3). Although it has not been possible to measure initiation directly, the QF mutant protein can promote two of the known ICP8 functions associated with initiation. However, we cannot rule out the possibility that annealing may play an unexpected role in initiation. Another explanation for the inability of QF to perform DNA replication would be the failure to stimulate replication proteins such as helicase–primase and DNA polymerase. Fig. 4B shows that the QF mutant is capable of stimulating leading-strand DNA synthesis using an in vitro minicircle assay. Interestingly, this assay mimics rolling-circle replication. Thus, the QF mutant seems to possess the necessary functions to carry out rolling-circle DNA replication. Taken together, these findings support the hypothesis that rolling-circle replication may not be the predominant mode of replication during infection. The inability of the QF mutant to perform HSV DNA replication correlated with its inability to anneal complementary ssDNA, suggesting that annealing plays a previously unappreciated role during replication. The lack of detectable DNA replication and the inability to form RCs suggests that the defect in QF is manifested at an early stage of DNA replication and that the processes of recombination and replication may be inseparable.
Model for the Role of ICP8 in DNA Replication and Recombination.
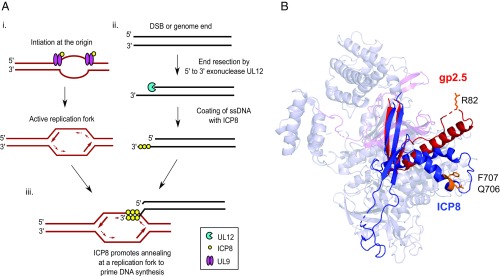
Although all dsDNA viruses that replicate by concatemer formation utilize RDR and encode recombination machinery, diverse mechanisms have evolved (2). Bacteriophage T4 encodes an ATP-dependent recombinase (UvsX), related to RecA and Rad51, that is capable of strand invasion (71–73). Most of the other bacterial viruses, however, encode SSAPs that function in an ATP-independent manner to anneal ssDNA but cannot perform strand invasion (2). Although Nimonkar and Boehmer (74, 75) reported that ICP8 could promote strand assimilation of a small ssDNA oligo into a supercoiled plasmid forming a D-loop-like structure, this reaction relied on the presence of locally denatured DNA and is therefore dependent on the activity of ICP8 as an SSAP. Many models that link replication and recombination have been proposed (1, 3, 8, 76, 77), and one common step in these models is the annealing of a ssDNA molecule to an active replication fork. In Fig. 8, we propose a simplified model for recombination-dependent HSV DNA replication. According to this model, ICP8 and UL9 promote initiation to create an active replication fork, and the other HSV replication proteins, including helicase–primase and polymerase, cooperate to promote leading- and lagging-strand synthesis (60). Previous reports from our laboratory indicate that the HSV primase is inefficient (78), and attempts to recapitulate coordinated leading- and lagging-strand synthesis in vitro with HSV 1 replication proteins required the use of high concentrations of primase (UL52) (47). These findings suggest that HSV may utilize alternative methods during infection to prime for DNA synthesis, especially on the lagging strand. Based on elegant studies in the T4 system, Luder and Mosig (79) showed that DNA synthesis can be primed by the annealing of a complementary strand of DNA at the replication fork. Nimonkar and Boehmer (80) showed that ICP8 can promote the annealing of a small oligo at a locally disrupted region of DNA and that this annealed oligo could act as a primer for in vitro DNA synthesis promoted by the HSV viral replication machinery consistent with RDR. In the model shown in Fig. 8, we suggest that ICP8 can promote the annealing of a 3′ ssDNA terminus at an HSV replication fork. The 3′ terminus could arise by resection of a dsDNA molecule, either from a free genome end or from a double-strand break generated by replication through a nick or a gap in the viral genome (81–83). We suggest that resection could be carried out by the viral alkaline nuclease, UL12 (27), and we have recently demonstrated that UL12 is also essential for the formation of concatemers that can be processed to produce infectious virus (29). However, it is also possible that end resection could be carried out by other cellular exonucleases. Priming by this annealing mechanism could generate new replication forks and lead to the formation of branched intermediates consistent with the complex replication intermediates observed during HSV infection (3, 76, 80).
Fig. 8.
Role of ICP8 annealing activity during HSV-1 DNA replication. (A) A model for HSV-1 RDR. (i) ICP8 and UL9 distort and unwind the origin to create an active replication fork. (ii) A free genome end or double-strand break created during infection is resected by a 5′-to-3′ exonuclease and the resulting 3′ overhang coated by ICP8. (iii) ICP8 promotes annealing of the resulting 3′-ssDNA overhang to an active replication fork to prime DNA synthesis. (B) Structural alignment of T7 bacteriophage SSAP gp2.5 (red; PDB ID code 1JE5; chain A) and ICP8 (blue; PDB ID code 1URJ; chain A) using jCE circular permutation algorithm. ICP8 Q706/F707 residues and gp2.5 R82 residue are illustrated in orange.
Understanding the Basic Mechanisms of How SSAPs Promote Single-Strand Annealing.
Although dsDNA viruses from bacteriophages to eukaryotic viruses encode SSAPs such as λ red beta and ICP8, the mechanisms by which they promote the annealing of complementary strands are poorly understood. A simplified model for steps in the annealing reaction is presented in Fig. 7 along with data indicating that the QF mutant was able to bind ssDNA to form a nucleoprotein filament but was unable to form an intertwined annealing intermediate. We suggest that formation of the annealing intermediate depends upon a specific protein–protein interaction between ICP8 molecules in complementary nucleoprotein filaments and that this interaction is mediated either directly or indirectly by the region of the molecule containing the QF residues (84).
Although little or no sequence conservation exists between different SSAPs, many if not all of them contain an oligonucleotide-binding fold (OB fold), composed of five antiparallel β-sheets with interspaced loops and helical regions (85). Bioinformatic analysis of ICP8 has identified an OB fold within the neck domain of the ICP8 structure (44). Interestingly, structural comparisons have revealed a remarkable level of similarity between the bacteriophage T7 SSAP gp2.5 and ICP8 (86, 87). Fig. 8B shows the structure of the T7 phage SSAP gp2.5 (in red) overlaid on the ICP8 structure (in blue). These proteins exhibit functional similarities as well. Like ICP8, gp2.5 binds to ssDNA, simulates other essential replication proteins, and promotes the annealing of complementary ssDNA (87). Similar to the QF mutant, a gp2.5 mutant (R82C) was reported to lack annealing activity while still maintaining the ability to bind to ssDNA and to stimulate other essential replication functions (88). Annealing activity of gp2.5 was shown to be essential for both viral growth and DNA replication (88). Intriguingly, R82 and Q706/F707 residues lie within a similar region of the protein located between a key α-helix (a1) and β-sheet (b3) within the OB fold. Therefore, we speculate that this region may be important for mediating the annealing of complementary strands of ssDNA (87). Additional genetic and biochemical analysis will be required to understand the precise requirements for the ability of gp2.5 and ICP8 to stimulate annealing.
In summary, analysis of the QF mutant revealed that the annealing activity of ICP8 is essential for viral DNA replication, providing direct evidence for RDR. We anticipate that further analysis of the region of ICP8 containing the QF residues will provide invaluable insight into the mechanisms by which SSAPs promote annealing.
Materials and Methods
Cell Lines.
African green monkey kidney (Vero) cells were purchased from the American Type Culture Collection and maintained as monolayers in DMEM (Invitrogen) supplemented with 5% FBS and 0.1% penicillin–streptomycin. The ICP8-complementing cell line S2 was generously provided by David Knipe (Harvard University, Boston, MA) and was maintained in DMEM supplemented with 5% FBS and 0.1% penicillin–streptomycin under G418 selection (400 µg/mL). All cell cultures were maintained in humidified atmosphere containing 5% CO2.
Viruses and Infection.
The HSV-1 KOS strain was used as the WT strain in all experiments. The ICP8-null virus, HD-2, containing a LacZ insertion mutation in the ICP8 gene was a generous gift from David Knipe (Harvard University, Boston, MA)
DNA Constructs and Mutagenesis.
The ICP8 expression vector, pSAK-ICP8, contains the full-length ICP8 gene under control of the CMV promoter. Alanine substitution mutations were made in pSAK-ICP8 and pFastBAC1-ICP8 using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s suggested protocol for use in mammalian expression and recombinant baculovirus production for protein expression, respectively.
Antibodies Used.
Monoclonal mouse anti-ICP4 [1:200; immunofluorescence (IF); US Biological], polyclonal rabbit anti-ICP8 clone 367 [1:400; IF; a gift from William Ruyechan, State University of New York at Buffalo, Buffalo, NY], monoclonal mouse anti-ICP8 (1:2,000; Western blot; Santa Cruz Biotechnology) and Alexa Fluor secondary antibodies 594 and 488 (1:200; IF; Molecular Probes).
Protein Purification.
ICP8 and UL9 protein purification was performed as previously described by our laboratory (38, 89).
Transient Complementation Assay.
The transient transfection complementation assay was performed as previously described (38). The average percent complementation was calculated using three independent experiments.
Plaque Reduction Assay.
KOS infectious DNA was prepared as described previously (83). Vero cells were grown to ∼95% confluency in 60-mm dishes. Cells were transfected with 50 ng of either pSAK-EV, pSAK-ICP8 (WT), pSAK-ICP8 FNF (positive control), or pSAK-ICP8 QF, 1:1 molar ratio of KOS infectious DNA:pSAK construct, and 1.4 µg of pUC19 carrier DNA using Lipofectamine Plus (Invitrogen) according to manufacturer’s suggested protocol. Twenty-four hours posttransfection, plates were washed with 1× PBS, overlaid with 2% methylcellulose in DMEM, and allowed to incubate at 37 °C for 3–4 d. Cells were then fixed with 8% formaldehyde and stained with crystal violet. Plaques were manually counted and the percent complementation compared with WT was calculated. Percentages calculated from three independent experiments were averaged together. Equal expression of ICP8 constructs was confirmed by Western blot.
Western Blot Analysis.
Western blot analysis for ICP8 was performed as previously described (38)
EMSA.
Purified WT or QF mutant protein (0–300 nM) was titrated against either 100 nM fluorescein-labeled dT-14-mer ssDNA oligo (IDT) or Cy3-labeled 50-mer ssDNA oligo (IDT) in DNA binding buffer (20 mM Tris⋅HCl, pH 7.5, 4% glycerol, 0.1 mg/mL BSA, 0.5 mM DTT, and 5 mM MgCl2). Samples were incubated for 30 min at room temperature and quenched with 40% sucrose solution. Bound and unbound DNA species were separated on 5% nondenaturing polyacrylamide gel in 1× TBE and imaged using ChemiDoc MP Imaging System (Bio-Rad).
Fluorescence Polarization Assay.
Purified WT or QF mutant protein was titrated (∼0.9 nM to 2 µM) against 2.5 nM fluorescein-labeled dT-14-mer ssDNA oligo (IDT) in DNA binding buffer (20 mM Tris⋅HCl, pH 7.5, 4% glycerol, 0.1 mg/mL BSA, 0.5 mM DTT, and 5 mM MgCl2). Fifteen microliters of each reaction were pipetted into Corning Low-Volume 384-Well Black Polystyrene Flat-Bottom Plates (catalog no. 3540) in triplicates. Samples were incubated for 1 h at room temperature, and fluorescence polarization (FP) measurements were taken on TECAN M1000 Pro plate reader with excitation wavelength of 470 nm and emission wavelength of 521 nm. Triplicate readings were averaged together, and results from three independent experiments were used to determine binding curve and Kd. Curve was fit to a 1:1 binding model using Prism software, and Kd values were determined for each experiment and then averaged together to obtain final values.
Annealing of Complementary ssDNA.
A linearized heat-denatured pSAK vector was used as the DNA substrate. pSAK plasmid DNA (25 µg) was linearized by digestion with excess PstI (New England Biolabs), purified, and denatured by boiling for 5 min. Unless otherwise stated, 30-µL reactions were performed in a buffer containing 20 mM Tris⋅HCl (pH 7.5), 100 nM NaCl, 1 mM DTT, 0.1 mg/mL BSA, and varying amounts of either WT or QF mutant protein (50–400 nM). The reactions were initiated by adding 53.6 ng of ssDNA, incubated for 30 min at 37 °C, and quenched by adding termination buffer [final concentration of 45 mM EDTA (pH 8) and 1 M urea]. The samples were subjected to proteinase K digestion for 10 min at 42 °C and the products generated were analyzed by 1% agarose gel electrophoresis followed by staining with SYBR Gold reagent (Invitrogen; catalog no. S11494) and imaged using ChemiDoc MP Imaging System (Bio-Rad). For quantification of annealed products, Quant-iT PicoGreen dsDNA Reagent was used according to the manufacturer’s specifications (Invitrogen; catalog no. P7589).
EM.
Annealing.
For analysis of intermediates formed during annealing, 312 nM WT or QF mutant protein was incubated with 29.5 ng of heat-denatured pSAK plasmid DNA in 20-µL final volume reaction in annealing buffer [20 mM Tris⋅HCl (pH 7.5), 100 nM NaCl, 1 mM DTT, 0.1 mg/mL BSA, 6 mM MgCl2]. Samples were incubated at 37 °C for 15–30 min and processed for EM analysis.
ssDNA binding.
To visualize ICP8-ssDNA nucleofilaments by EM, 145.6 ng of linearized M13 ssDNA was incubated with 1 µg of either WT ICP8 or QF mutant protein in 20-μL reaction in DNA binding buffer (20 mM Tris⋅HCl, pH 7.5, 4% glycerol, 0.1 mg/mL BSA, 0.5 mM DTT, and 5 mM MgCl2). Samples were incubated at room temperature for 20 min and processed for EM analysis.
Filamentation.
Samples to assess filamentation of WT ICP8 or QF mutant were prepared as previously described (38).
All samples for EM analysis were diluted 1:10 in water and absorbed onto Formvar-carbon–coated 300-mesh copper grids negatively stained with 1–2% uranyl acetate and examined using a Hitachi H-2650 transmission electron microscope (TEM) at accelerating voltage of 80 kV. Images were acquired at 40,000×, 50,000×, and 60,000×.
Minicircle Replication Assay for Stimulation of Leading-Strand DNA Synthesis by ICP8.
The 70-bp minicircle was prepared as previously described (47). The standard replication reaction (10 µL) contained 20 nM minicircle, 3.2 mM each ATP and GTP, 0.8 mM each UTP and CTP, 100 µM each dATP, dCTP, dGTP, and 40 μM dTTP, 1 μCi of [α-32P]dTTP, 50 mM Tris⋅HCl, 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/mL BSA, 1% glycerol, 300 nM UL5-UL8-UL52, 100 nM UL30-UL42, and increasing concentration (0.19 µM–0.76 µM) of either WT or QF mutant protein. After 60-min incubation at 37 °C, the reactions were quenched by adding 2 µL of 0.5 M EDTA and 3 µL of loading dye (50 mM NaOH, 1 mM EDTA, 4% glycerol, 0.03% bromocrysol green, and 0.05% xylene cyanol), and the replication products were separated by 1% alkaline agarose gel electrophoresis. The gel was fixed with 7% trichloroacetic acid, dried and imaged with phosphoimager (PerkinElmer; Cyclone Plus Storage Phosphor System). HindIII-digested radiolabeled λ-DNA was used as a molecular marker.
ICP8 Stimulation of UL9 ssDNA-Dependent ATPase Activity.
WT or QF mutant protein (700 nM) were mixed in a 20 µL with 100 nM UL9 in the presence of 5 mM ATP (Thermo Fisher Scientific) and 10 µM ssDNA (5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTAAGCTTGCATGCCTGCAGGTCGACTCTAGA-3′) in ATPase buffer (20 mM Hepes, pH 7.6, 1 mM DTT, 5 mM MgCl2, 10% glycerol, 0.1 µg/mL BSA, 200 mM NaCl). Reactions were incubated at room temperature for 1 h and quenched with 0.05 M EDTA (pH 8.0). ATPase activity was measured by the release of free phosphate using Abcam Phosphate Assay Kit (Colorimetric; ab 65622). Absolute values for the concentration of Pi/well were determined using the kit phosphate standard. Fold stimulation was calculated relative to a control lacking ICP8.
Quantification of Viral DNA Synthesis.
Vero cell were grown on 60-mm dishes to 80% confluency and transfected with 300 ng of pSAK-ICP8, pSAK-ICP8 QF, or pSAK-EV, and 1.7 µg of pUC19 as carrier DNA, per dish using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer’s suggested protocol. Sixteen- to 18-h posttransfection, cells were infected with HD-2 (ICP8-null virus) at a multiplicity of infection (MOI) of 5 PFU/cell and harvested 24 h postinfection. Cell pellets were washed once and resuspended in 200 µL of PBS. Total DNA was extracted using blood/tissue DNA extraction kit (Qiagen; catalog no. 69504) according to the manufacturer’s suggested protocol. DNA was quantified using nanodrop (Thermo Fisher Scientific), and serial dilutions were prepared starting with the same amount of initial DNA. DNA samples were transferred to nylon membrane (GeneScreen Plus; catalog no. NCF10700) using dot blot manifold (PerkinElmer) according to the manufacturer’s instructions. Southern blot was performed using α-32P–labeled random probes generated from a PCR-derived UL30 gene using a random primer labeling kit (Thermo Fisher Scientific; catalog no. 17075).
RC Assay.
Vero cells were grown to ∼80% confluency on glass coverslips in a 12-well culture plate. Cells were transfected with 50 ng of either pSAK-EV, pSAK-ICP8, or pSAK-ICP8 QF, and 450 ng of pUC19 carrier DNA using Lipofectamine Plus (Invitrogen) according to the manufacturer’s suggested protocol. Sixteen- to 18-h posttransfection, samples were infected with HD-2 (ICP8-null virus) at an MOI of 20. IF was performed as has previously been described (38). Images were taken on Zeiss LSM 880 Meta confocal microscope and a Plan-Apochromat 63× oil differential interference contrast M27 objective (numerical aperture, 1.4) at a zoom of 1.5×.
Acknowledgments
We thank members of the S.K.W. laboratory and Dr. Lee Wright, Dr. Lynn Thomason, and Dr. James Sawitzke for discussion and careful reading of the manuscript. Additionally, we thank Maya Yankova and the EM Core facility for assistance with EM, Bing Hao and Kazuya Machida for assistance with fluorescence polarization, and Mitali Adlakha for assistance with confocal microscopy. We thank the US Department of Health and Human Services, and the NIH provided funding (to S.K.W.) under Grants AI069136 and AI021747.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Mosig G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet. 1998;32:379–413. doi: 10.1146/annurev.genet.32.1.379. [DOI] [PubMed] [Google Scholar]
- 2.Lo Piano A, Martínez-Jiménez MI, Zecchi L, Ayora S. Recombination-dependent concatemeric viral DNA replication. Virus Res. 2011;160:1–14. doi: 10.1016/j.virusres.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Weller SK, Sawitzke JA. Recombination promoted by DNA viruses: Phage λ to herpes simplex virus. Annu Rev Microbiol. 2014;68:237–258. doi: 10.1146/annurev-micro-091313-103424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind L, Makarova KS, Koonin EV. Survey and summary: Holliday junction resolvases and related nucleases: Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valledor M, Hu Q, Schiller P, Myers RS. Fluorescent protein engineering by in vivo site-directed mutagenesis. IUBMB Life. 2012;64:684–689. doi: 10.1002/iub.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muyrers JPP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosberg JA, Lajoie MJ, Church GM. Lambda red recombineering in Escherichia coli occurs through a fully single-stranded intermediate. Genetics. 2010;186:791–799. doi: 10.1534/genetics.110.120782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 10.Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baines J, Weller S. Cleavage and packaging of herpes simplex virus 1 DNA, herpesvirus assembly. In: Catalano C, editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York: 2005. pp. 135–150. [Google Scholar]
- 12.Hayward GS, Jacob RJ, Wadsworth SC, Roizman B. Anatomy of herpes simplex virus DNA: Evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci USA. 1975;72:4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheldrick P, Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39:667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- 14.Lamberti C, Weller SK. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: Implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 16.Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber PC, Challberg MD, Nelson NJ, Levine M, Glorioso JC. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54:369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- 18.Bowden R, Sakaoka H, Donnelly P, Ward R. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect Genet Evol. 2004;4:115–123. doi: 10.1016/j.meegid.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Timbury MC, Subak-Sharpe JH. Genetic interactions between temperature-sensitive mutants of types 1 and 2 herpes simplex viruses. J Gen Virol. 1973;18:347–357. doi: 10.1099/0022-1317-18-3-347. [DOI] [PubMed] [Google Scholar]
- 20.Hayward GS, Frenkel N, Roizman B. Anatomy of herpes simplex virus DNA: Strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci USA. 1975;72:1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kintner RL, Allan RW, Brandt CR. Recombinants are isolated at high frequency following in vivo mixed ocular infection with two avirulent herpes simplex virus type 1 strains. Arch Virol. 1995;140:231–244. doi: 10.1007/BF01309859. [DOI] [PubMed] [Google Scholar]
- 22.Lingen M, Hengerer F, Falke D. Mixed vaginal infections of Balb/c mice with low virulent herpes simplex type 1 strains result in restoration of virulence properties: Vaginitis/vulvitis and neuroinvasiveness. Med Microbiol Immunol (Berl) 1997;185:217–222. doi: 10.1007/s004300050033. [DOI] [PubMed] [Google Scholar]
- 23.Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severini A, Morgan AR, Tovell DR, Tyrrell DLJ. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 25.Reuven NB, Staire AE, Myers RS, Weller SK. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J Virol. 2003;77:7425–7433. doi: 10.1128/JVI.77.13.7425-7433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuven NB, Weller SK. Herpes simplex virus type 1 single-strand DNA binding protein ICP8 enhances the nuclease activity of the UL12 alkaline nuclease by increasing its processivity. J Virol. 2005;79:9356–9358. doi: 10.1128/JVI.79.14.9356-9358.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher AJ, et al. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012;8:e1002862. doi: 10.1371/journal.ppat.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valledor M, Myers RS, Schiller PC. Herpes ICP8 protein stimulates homologous recombination in human cells. PLoS One. 2018;13:e0200955. doi: 10.1371/journal.pone.0200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grady LM, et al. The exonuclease activity of herpes simplex virus 1 UL12 is required for production of viral DNA that can be packaged to produce infectious virus. J Virol. 2017;91:e01380-17. doi: 10.1128/JVI.01380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conley AJ, Knipe DM, Jones PC, Roizman B. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J Virol. 1981;37:191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weller SK, Lee KJ, Sabourin DJ, Schaffer PA. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983;45:354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao M, Knipe DM. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J Virol. 1991;65:2666–2675. doi: 10.1128/jvi.65.5.2666-2675.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 34.O’Donnell ME, Elias P, Funnell BE, Lehman IR. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J Biol Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 35.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: Association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J Virol. 2004;78:5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez TR, Lehman IR. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 37.Falkenberg M, Bushnell DA, Elias P, Lehman IR. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J Biol Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 38.Darwish AS, Grady LM, Bai P, Weller SK. ICP8 filament formation is essential for replication compartment formation during herpes simplex virus infection. J Virol. 2015;90:2561–2570. doi: 10.1128/JVI.02854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruyechan WT, Weir AC. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J Virol. 1984;52:727–733. doi: 10.1128/jvi.52.3.727-733.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehmer PE, Lehman IR. Herpes simplex virus type 1 ICP8: Helix-destabilizing properties. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruyechan WT. The major herpes simplex virus DNA-binding protein holds single-stranded DNA in an extended configuration. J Virol. 1983;46:661–666. doi: 10.1128/jvi.46.2.661-666.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutch RE, Lehman IR. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J Virol. 1993;67:6945–6949. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bortner C, Hernandez TR, Lehman IR, Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J Mol Biol. 1993;231:241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 44.Mapelli M, Panjikar S, Tucker PA. The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding. J Biol Chem. 2005;280:2990–2997. doi: 10.1074/jbc.M406780200. [DOI] [PubMed] [Google Scholar]
- 45.Hamatake RK, Bifano M, Hurlburt WW, Tenney DJ. A functional interaction of ICP8, the herpes simplex virus single-stranded DNA-binding protein, and the helicase-primase complex that is dependent on the presence of the UL8 subunit. J Gen Virol. 1997;78:857–865. doi: 10.1099/0022-1317-78-4-857. [DOI] [PubMed] [Google Scholar]
- 46.Bermek O, Willcox S, Griffith JD. DNA replication catalyzed by herpes simplex virus type 1 proteins reveals trombone loops at the fork. J Biol Chem. 2015;290:2539–2545. doi: 10.1074/jbc.M114.623009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stengel G, Kuchta RD. Coordinated leading and lagging strand DNA synthesis by using the herpes simplex virus 1 replication complex and minicircle DNA templates. J Virol. 2011;85:957–967. doi: 10.1128/JVI.01688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehmer PE. The herpes simplex virus type-1 single-strand DNA-binding protein, ICP8, increases the processivity of the UL9 protein DNA helicase. J Biol Chem. 1998;273:2676–2683. doi: 10.1074/jbc.273.5.2676. [DOI] [PubMed] [Google Scholar]
- 49.Arana ME, Haq B, Tanguy Le Gac N, Boehmer PE. Modulation of the herpes simplex virus type-1 UL9 DNA helicase by its cognate single-strand DNA-binding protein, ICP8. J Biol Chem. 2001;276:6840–6845. doi: 10.1074/jbc.M007219200. [DOI] [PubMed] [Google Scholar]
- 50.Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 51.Liptak LM, Uprichard SL, Knipe DM. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukonis CJ, Weller SK. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J Virol. 1996;70:1751–1758. doi: 10.1128/jvi.70.3.1751-1758.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makhov AM, Griffith JD. Visualization of the annealing of complementary single-stranded DNA catalyzed by the herpes simplex virus type 1 ICP8 SSB/recombinase. J Mol Biol. 2006;355:911–922. doi: 10.1016/j.jmb.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 54.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16:481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 55.Efstathiou S, Minson AC, Field HJ, Anderson JR, Wildy P. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J Virol. 1986;57:446–455. doi: 10.1128/jvi.57.2.446-455.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garber DA, Beverley SM, Coen DM. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology. 1993;197:459–462. doi: 10.1006/viro.1993.1612. [DOI] [PubMed] [Google Scholar]
- 57.Strang BL, Stow ND. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J Virol. 2005;79:12487–12494. doi: 10.1128/JVI.79.19.12487-12494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poffenberger KL, Roizman B. A noninverting genome of a viable herpes simplex virus 1: Presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J Virol. 1985;53:587–595. doi: 10.1128/jvi.53.2.587-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55:451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 60.Weller SK, Coen DM. Herpes simplex viruses: Mechanisms of DNA replication. Cold Spring Harb Perspect Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson SA, DeLuca NA. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc Natl Acad Sci USA. 2003;100:7871–7876. doi: 10.1073/pnas.1230643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manolaridis I, et al. Structural and biophysical characterization of the proteins interacting with the herpes simplex virus 1 origin of replication. J Biol Chem. 2009;284:16343–16353. doi: 10.1074/jbc.M806134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boehmer PE, Dodson MS, Lehman IR. The herpes simplex virus type-1 origin binding protein. DNA helicase activity. J Biol Chem. 1993;268:1220–1225. [PubMed] [Google Scholar]
- 64.Boehmer PE, Craigie MC, Stow ND, Lehman IR. Association of origin binding protein and single strand DNA-binding protein, ICP8, during herpes simplex virus type 1 DNA replication in vivo. J Biol Chem. 1994;269:29329–29334. [PubMed] [Google Scholar]
- 65.Makhov AM, Lee SS-K, Lehman IR, Griffith JD. Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: Visualization of a specific preunwinding complex. Proc Natl Acad Sci USA. 2003;100:898–903. doi: 10.1073/pnas.0237171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olsson M, et al. Stepwise evolution of the herpes simplex virus origin binding protein and origin of replication. J Biol Chem. 2009;284:16246–16255. doi: 10.1074/jbc.M807551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gustafsson CM, Falkenberg M, Simonsson S, Valadi H, Elias P. The DNA ligands influence the interactions between the herpes simplex virus 1 origin binding protein and the single strand DNA-binding protein, ICP-8. J Biol Chem. 1995;270:19028–19034. doi: 10.1074/jbc.270.32.19028. [DOI] [PubMed] [Google Scholar]
- 68.Dodson MS, Lehman IR. The herpes simplex virus type I origin binding protein. DNA-dependent nucleoside triphosphatase activity. J Biol Chem. 1993;268:1213–1219. [PubMed] [Google Scholar]
- 69.Lee SS, Lehman IR. The interaction of herpes simplex type 1 virus origin-binding protein (UL9 protein) with Box I, the high affinity element of the viral origin of DNA replication. J Biol Chem. 1999;274:18613–18617. doi: 10.1074/jbc.274.26.18613. [DOI] [PubMed] [Google Scholar]
- 70.Aslani A, Olsson M, Elias P. ATP-dependent unwinding of a minimal origin of DNA replication by the origin-binding protein and the single-strand DNA-binding protein ICP8 from herpes simplex virus type I. J Biol Chem. 2002;277:41204–41212. doi: 10.1074/jbc.M208270200. [DOI] [PubMed] [Google Scholar]
- 71.Formosa T, Alberts BM. Purification and characterization of the T4 bacteriophage uvsX protein. J Biol Chem. 1986;261:6107–6118. [PubMed] [Google Scholar]
- 72.Hinton DM, Nossal NG. Cloning of the bacteriophage T4 uvsX gene and purification and characterization of the T4 uvsX recombination protein. J Biol Chem. 1986;261:5663–5673. [PubMed] [Google Scholar]
- 73.Yonesaki T, Minagawa T. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 1985;4:3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nimonkar AV, Boehmer PE. In vitro strand exchange promoted by the herpes simplex virus type-1 single strand DNA-binding protein (ICP8) and DNA helicase-primase. J Biol Chem. 2002;277:15182–15189. doi: 10.1074/jbc.M109988200. [DOI] [PubMed] [Google Scholar]
- 75.Nimonkar AV, Boehmer PE. On the mechanism of strand assimilation by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) Nucleic Acids Res. 2003;31:5275–5281. doi: 10.1093/nar/gkg740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murphy KC. Phage recombinases and their applications. Adv Virus Res. 2012;83:367–414. doi: 10.1016/B978-0-12-394438-2.00008-6. [DOI] [PubMed] [Google Scholar]
- 77.Maresca M, et al. Single-stranded heteroduplex intermediates in λ Red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen Y, et al. Herpes simplex virus type 1 helicase-primase: DNA binding and consequent protein oligomerization and primase activation. J Virol. 2011;85:968–978. doi: 10.1128/JVI.01690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luder A, Mosig G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: Priming by RNA polymerase and by recombination. Proc Natl Acad Sci USA. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nimonkar AV, Boehmer PE. Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1. Proc Natl Acad Sci USA. 2003;100:10201–10206. doi: 10.1073/pnas.1534569100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkie NM. The synthesis and substructure of herpesvirus DNA: The distribution of alkali-labile single strand interruptions in HSV-1 DNA. J Gen Virol. 1973;21:453–467. doi: 10.1099/0022-1317-21-3-453. [DOI] [PubMed] [Google Scholar]
- 82.Kieff ED, Bachenheimer SL, Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971;8:125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith S, Reuven N, Mohni KN, Schumacher AJ, Weller SK. Structure of the herpes simplex virus 1 genome: Manipulation of nicks and gaps can abrogate infectivity and alter the cellular DNA damage response. J Virol. 2014;88:10146–10156. doi: 10.1128/JVI.01723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tolun G, Makhov AM, Ludtke SJ, Griffith JD. Details of ssDNA annealing revealed by an HSV-1 ICP8-ssDNA binary complex. Nucleic Acids Res. 2013;41:5927–5937. doi: 10.1093/nar/gkt266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kazlauskas D, Venclovas C. Two distinct SSB protein families in nucleo-cytoplasmic large DNA viruses. Bioinformatics. 2012;28:3186–3190. doi: 10.1093/bioinformatics/bts626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hernandez AJ, Richardson CC. Gp2.5, the multifunctional bacteriophage T7 single-stranded DNA binding protein. Semin Cell Dev Biol. 2018:S1084-9521(17)30605-5. doi: 10.1016/j.semcdb.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rezende LF, Willcox S, Griffith JD, Richardson CC. A single-stranded DNA-binding protein of bacteriophage T7 defective in DNA annealing. J Biol Chem. 2003;278:29098–29105. doi: 10.1074/jbc.M303374200. [DOI] [PubMed] [Google Scholar]
- 89.Marintcheva B, Weller SK. Helicase motif Ia is involved in single-strand DNA-binding and helicase activities of the herpes simplex virus type 1 origin-binding protein, UL9. J Virol. 2003;77:2477–2488. doi: 10.1128/JVI.77.4.2477-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]