Significance
Fork regression is a way of circumventing or dealing with DNA lesions and is important to genome integrity. Fork regression is performed by double-strand DNA ATPases that initially cause newly synthesized strands to unpair from the parental strands, followed by pairing of the new strands and reversal of the fork. This study shows that Mcm10, an essential replication factor, efficiently anneals complementary strands and also inhibits fork regression by SMARCAL1. Moreover, the study localizes the Mcm10 DNA-binding domain to the N-terminal domains of the replicative CMG helicase at the forked nexus. Thus, forks that are unimpeded would contain Mcm10 at a strategic position where its DNA-binding and/or annealing function may block fork regression enzymes and thereby protect active forks from becoming reversed.
Keywords: DNA replication, fork regression, CMG helicase, SMARCAL1, Mcm10
Abstract
The 11-subunit eukaryotic replicative helicase CMG (Cdc45, Mcm2-7, GINS) tightly binds Mcm10, an essential replication protein in all eukaryotes. Here we show that Mcm10 has a potent strand-annealing activity both alone and in complex with CMG. CMG-Mcm10 unwinds and then reanneals single strands soon after they have been unwound in vitro. Given the DNA damage and replisome instability associated with loss of Mcm10 function, we examined the effect of Mcm10 on fork regression. Fork regression requires the unwinding and pairing of newly synthesized strands, performed by a specialized class of ATP-dependent DNA translocases. We show here that Mcm10 inhibits fork regression by the well-known fork reversal enzyme SMARCAL1. We propose that Mcm10 inhibits the unwinding of nascent strands to prevent fork regression at normal unperturbed replication forks, either by binding the fork junction to form a block to SMARCAL1 or by reannealing unwound nascent strands to their parental template. Analysis of the CMG-Mcm10 complex by cross-linking mass spectrometry reveals Mcm10 interacts with six CMG subunits, with the DNA-binding region of Mcm10 on the N-face of CMG. This position on CMG places Mcm10 at the fork junction, consistent with a role in regulating fork regression.
DNA replication is performed by a replisome composed of numerous proteins that work together to separate and duplicate DNA strands to form two new daughter duplexes (1–5). The replication machinery is conserved in all eukaryotes, and consists of an 11-subunit helicase known as CMG (Cdc45, Mcm2-7, GINS) (6); DNA polymerase Pol α-primase, which primes both strands; and Pols ε and δ, which duplicate the leading and lagging strands, facilitated by proliferating cell nuclear antigen (PCNA) clamps and the clamp loader, replication factor C. Numerous other factors move with replisomes, the functions of which are only now being elucidated. Most replisome accessory factors are nonessential but have roles in genome stability; however, the Mcm10 replisome accessory factor is essential in all eukaryotes tested thus far (7, 8).
Structural studies have shown that Mcm10 contains a globular “internal domain” within which are two DNA-binding elements: an oligonucleotide/oligosaccharide-binding fold and a zinc finger (9, 10). We refer to this globular internal domain as the DNA-binding region. The DNA-binding region of Mcm10 is flanked by N- and C-terminal regions. The N-terminal region of Mcm10 contains a sequence that forms a triple strand α-helical coiled coil and is proposed to mediate Mcm10 oligomerization and/or facilitate interactions with other proteins (11). The C-terminal region of Mcm10 varies in length and complexity across organisms (reviewed in ref. 12). Homology alignments indicate that the Saccharomyces cerevisiae Mcm10 N-region comprises residues 1–149, the globular DNA-binding region comprises residues 150–356, and the C-region comprises residues 357–595 (10).
Mcm10 was recently shown to tightly bind CMG, forming a CMG-Mcm10 complex (CMGM), which stimulates both CMG helicase activity and the rate of the replisome (13, 14). Mcm10 is required for CMG activation at origins during replication initiation (15–19). Along with binding CMG, Mcm10 is documented to bind Pol α and PCNA (9, 20). Studies of Mcm10 depletion or loss of function using genetics, cell biology, and cell extracts have identified Mcm10 functions in replisome stability, fork progression, and DNA repair (21–25). Despite significant advances in the understanding of Mcm10’s functions, mechanistic in vitro studies of Mcm10 in replisome and repair reactions are lacking.
The present study demonstrates that Mcm10 on its own rapidly anneals cDNA strands even in the presence of the single-strand (ss) DNA-binding protein RPA, a property previously associated with the recombination protein Rad52 (26). While CMG does not perform strand annealing, CMGM unwinds DNA and then rapidly rewinds the unwound products. Several helicases demonstrate both DNA unwinding and annealing (reviewed in ref. 27). The combined functions are proposed to be involved in such processes as double-strand (ds) break repair, fork regression, stabilization of stalled forks, transcription, and telomere metabolism (27). Roles for Mcm10 in some of these processes have been suggested by studies involving mutation or depletion of Mcm10 (21–25).
To more precisely understand the role of Mcm10 strand annealing activity, we determined the location of the DNA-binding region of Mcm10 on CMG by cross-linking mass spectrometry (CX-MS). This analysis showed that Mcm10 is in close proximity to 6 of the 11 CMG subunits and extends from the N-terminal to the C-terminal domains of CMG. The DNA-binding region of Mcm10 localizes to the N-face of CMG, which places DNA binding by Mcm10 within a moving replisome at the fork junction (28). This location suggests a possible role of Mcm10 in fork regression, important to genomic stability (29–31), and is consistent with the findings of studies of Mcm10 in metazoans (22).
Fork regression is catalyzed by a specialized class of ATP helicases that reverse forks rather than unwind them, most notably SMARCAL1, ZRANB3, and HLTF (32). In the present study, we examined the effect of S. cerevisiae Mcm10 on SMARCAL1 using previously established DNA substrates for this enzyme (33). We found that Mcm10 completely inhibits SMARCAL1 on several forked DNA structures tested here. We propose that Mcm10 is held in proximity to the fork by its known tight interaction with CMG (13). Mcm10-mediated inhibition of SMARCAL1 could be due to nonspecific binding of Mcm10 to ssDNA, or by Mcm10’s annealing activity, either of which could prevent fork regression. We propose a possible regulatory mechanism that prevents the reversal of actively replicating forks and targets fork regression only to those forks that have DNA damage.
Results
CX-MS of CMG-Mcm10.
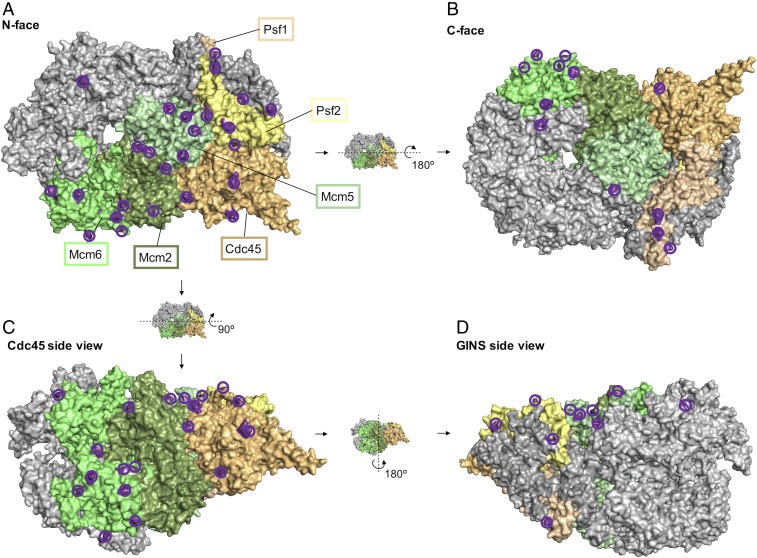
Mcm10 tightly binds CMG and greatly enhances helicase activity (13). To better understand how Mcm10 functions with CMG, we determined the location of Mcm10 on CMG. To delineate the position of Mcm10 on CMG, we reconstituted and purified CMGM then performed CX-MS on the complex. MS analysis of CMGM treated with a bifunctional lysine cross-linking agent revealed an extensive network of intermolecular and intramolecular cross-links between Mcm10 and six subunits of CMG (a total of >1,000 cross-links) (Fig. 1, SI Appendix, Fig. S1, Movie S1, and Dataset S1). We observed cross-links between the N region of Mcm10 and CMG, specifically with Mcm2, Mcm5, Mcm6, Cdc45, Psf1, and Psf2 (Fig. 1, Movie S1, and Dataset S1). The results indicate a central location of Mcm10 on the N-face of CMG near the interface between the Mcm2-7 ring and its accessory factors. In addition, some interactions extend around the outer perimeter of CMG all of the way to the C-face. These CX-MS results greatly extend earlier studies demonstrating the interaction of Mcm10 with the N-terminal regions of Mcm2 and Mcm6 (14, 34).
Fig. 1.
CX-MS proximity assays of CMG-Mcm10. Cross-links between Mcm10 and CMG on performing CX-MS on the reconstituted CMG-Mcm10 complex are colored and encircled in dark purple. Four different views of the CMG are shown: N-face (A); C-face (B); Cdc45, side view (C); and GINS, side view (D). More information is provided in Dataset S1 and Movie S1.
The globular DNA-binding region of Mcm10 contains eight residues that cross-link to CMG, and all but one of these 31 cross-links are located on the N-face of CMG (Dataset S1). The cryoEM structure of CMG on a forked DNA with ATP shows CMG tracks on DNA N-face first (28), and thus the Mcm10 DNA-binding region is located on the front edge of CMG, directly facing the fork junction. Interestingly, there are many cross-links between CMG and the N- and C-regions of Mcm10 but few cross-links to the Mcm10 DNA-binding region, suggesting that the N- and C-regions of Mcm10 may establish the main affinity of Mcm10 to CMG. This leads us to predict that Mcm10’s DNA-binding domain may be mobile on CMG. We note that most Mcm subunits contain long N-tails that are not visible in cryoEM structures, indicating mobility in these regions (35, 36), and Mcm10 may possibly dock onto these mobile Mcm N-tails.
The CX-MS results of CMG-Mcm10 revealed extensive cross-links between the C-region of Mcm10 and CMG, consistent with a previous study reporting an interaction of Mcm2 with a deletion mutant of Mcm10 that contained only the C-region (37). Our detailed CX-MS study unexpectedly revealed that the C-region of Mcm10 wraps around the entire outside surface of Mcm6 from the N-face to the C-face of CMG (Fig. 1C and Dataset S1), suggesting that the C-region of Mcm10 may be highly elongated. A few Mcm10 cross-links also extend to the C-face of CMG on the Psf1 subunit (Fig. 1D). In addition, we observed three intramolecular Mcm10 crosslinks between identical residues, consistent with an oligomeric state of Mcm10 mediated by the N-region coiled-coil domain (Dataset S1) (11).
CMG-Mcm10 Reanneals Unwound Single Strands.
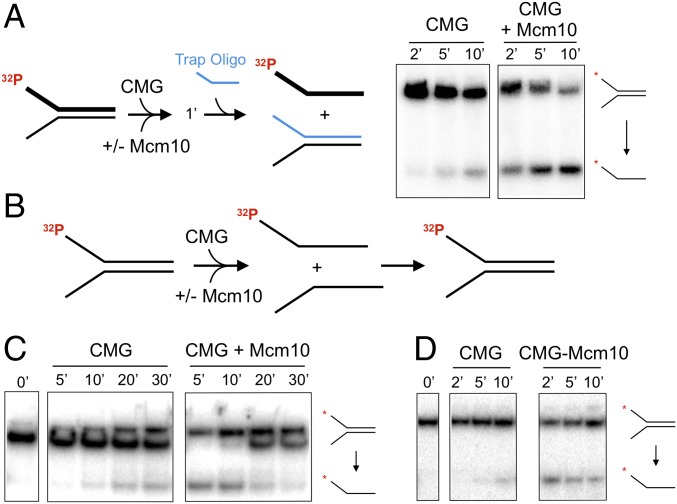
Our previous CMG-Mcm10 helicase studies were performed in the presence of a trap oligonucleotide, a standard addition to unwinding assays (13). We performed unwinding reactions using CMG with and without Mcm10 in the presence of a trap oligo (Fig. 2A). CMG helicase is greatly stimulated by Mcm10, as we reported previously (13). However, when the trap oligo is omitted from helicase assays (Fig. 2B), unwound products appear to be reannealed (Fig. 2C). CMG unwinding is initially stimulated by Mcm10, but the separated strands are rapidly converted back into the duplex substrate. We also tested purified CMGM complex compared with CMG without Mcm10 and again observed unwinding followed by reannealing when Mcm10 is bound to CMG (Fig. 2D).
Fig. 2.
Mcm10 stimulates CMG helicase, followed by reannealing of the unwound products. (A, Left) Schematic of the assay. (A, Right) Unwinding time courses of 30 nM CMG with and without mixing with 60 nM Mcm10 with an oligo trap (shown in blue), analyzed by native PAGE. (B) Schematic of the assays for C and D. (C) Unwinding time courses of 30 nM CMG with no oligo trap with and without mixing with 60 nM Mcm10. The substrate was partially shifted by Mcm10 and/or CMG, resulting in a doublet band. (D) Unwinding time courses of 50 nM CMG or 50 nM preformed CMGM complex with no oligo trap.
Mcm10 Is a Potent DNA Strand-Annealing Protein.
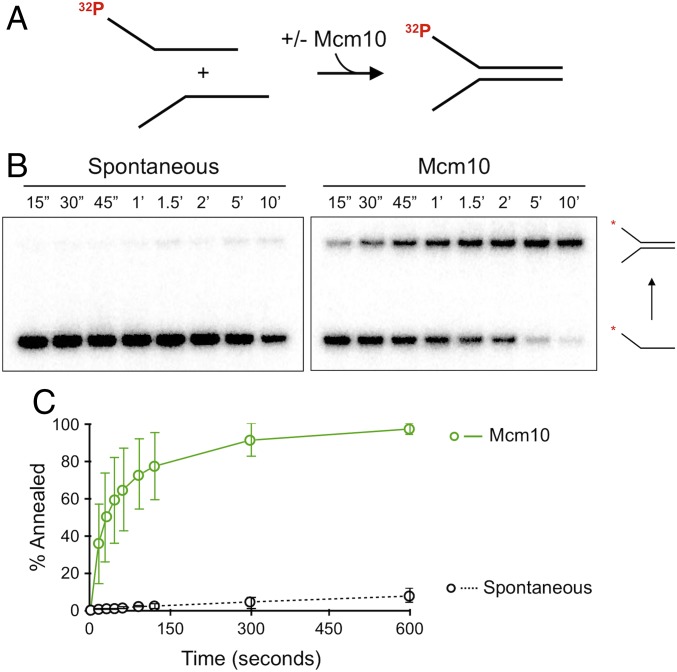
To directly test Mcm10 for annealing activity, we mixed the two individual oligos that compose the mini fork—a 32P-5′ end-labeled 5′ flap lagging strand oligo and a 3′ flap leading strand oligo at 20 nM—and incubated them with or without Mcm10 in the absence of ATP and CMG. The addition of a 20-fold molar excess of unlabeled lagging strand oligo was used to quench reactions (SI Appendix, Fig. S2). Reactions were analyzed by native PAGE to assess the rate at which complementary strands anneal with and without Mcm10 (Fig. 3A). Spontaneous annealing at 30 °C without Mcm10 was nearly undetectable, but the addition of Mcm10 resulted in rapid and nearly complete annealing under the same conditions (Fig. 3 B and C). The concentration dependence of Mcm10 annealing is shown in SI Appendix, Fig. S3. CMG alone did not demonstrate annealing activity (SI Appendix, Fig. S4). We also tested fully complementary single strands and observed comparable Mcm10 annealing activity. These experiments demonstrate a previously unreported biochemical activity of Mcm10 in which Mcm10 rapidly anneals cDNA strands.
Fig. 3.
Mcm10 promotes robust strand annealing. (A) Schematic of the assay. The lagging strand oligo of the forked DNA was 5′ end-labeled with 32P. (B) Time course of strand annealing using 60 nM Mcm10 and analyzed by native PAGE. (C) Quantitation of annealing vs. time in the presence or absence of Mcm10. Each data point is the average of three separate reactions. Error bars represent 1 SD.
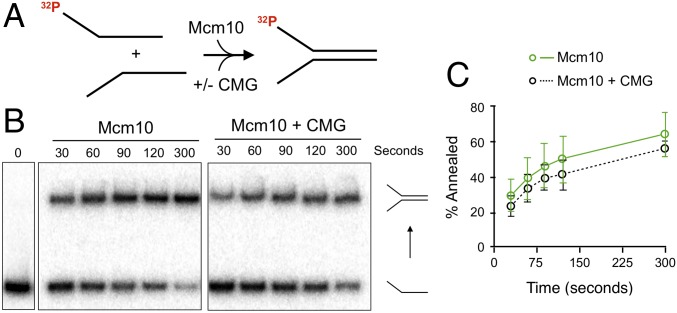
While Mcm10 has robust strand-annealing activity, it seems paradoxical that Mcm10 also stimulates CMG in DNA unwinding. To explore whether CMG affects Mcm10 annealing activity, we compared Mcm10 with and without the addition of a twofold molar excess of CMG in the annealing assay (Fig. 4 A and B). The results show no substantial effect of CMG on the rate of Mcm10 annealing, as the difference in proportion of substrate annealed is within the error of the triplicate experiments (Fig. 4C). These results are consistent with the observation that reconstituted CMGM complex also displays reannealing in the unwinding assay (Fig. 2D) and indicate that Mcm10 annealing remains active while in complex with CMG. We note that at an unperturbed replication fork in vivo, the leading strand template is readily duplicated by Pol ε, so the unwound strands would not be single-stranded and thus would not be expected to reanneal.
Fig. 4.
CMG does not inhibit Mcm10 reannealing. (A) Schematic of the reaction. (B) Mcm10 (60 nM) was added with or without CMG (120 nM) to standard annealing assays, and timed aliquots were analyzed by native PAGE. The 0 s lane is a mixture of the two oligonucleotides without protein and stopped within 30 s. (C) Quantitation of reactions. Values are the average of three experiments. Error bars represent 1 SD.
Mcm10 Catalyzes Strand Annealing in the Presence of RPA.
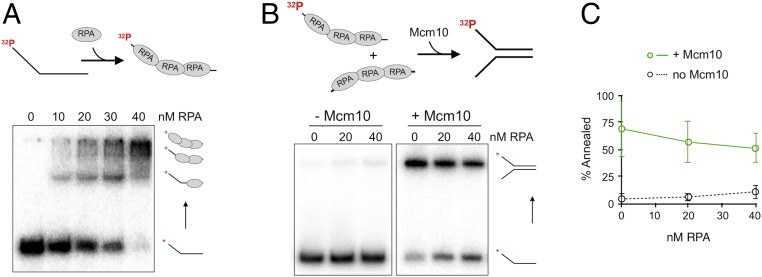
RPA binds ssDNA tightly and is presumed to prevent complementary single strands from reannealing. Several proteins have been identified that promote annealing of RPA-coated ssDNA, most notably Rad52, which is involved in homologous recombination in yeast (26). To study the capability of Mcm10 to reanneal ssDNA in the presence of RPA, we titrated RPA into one strand and empirically determined that 40 nM RPA saturates the 5′ flap lagging strand oligo as determined by electrophoretic mobility shift assay (EMSA) gel shift in a native gel (Fig. 5A). We then performed annealing assays by adding 40 nM RPA to both the leading and lagging strand oligos and then mixing them to initiate the annealing reaction. Reactions were quenched using SDS to remove protein along with a 20-fold excess of unlabeled oligo trap. Spontaneous strand annealing of the RPA-coated oligos showed very little annealed duplex (Fig. 5B). However, Mcm10 efficiently annealed the RPA coated strands (Fig. 5B), with only a 25% decrease in annealed product compared with Mcm10 activity with no RPA (Fig. 5C).
Fig. 5.
Mcm10 can anneal RPA-coated single strands. (A) Native PAGE EMSA of Mcm10 binding the radiolabeled 5′ flap lagging-strand oligonucleotides used in annealing reactions. (B) Annealing assays with and without 60 nM Mcm10, with increasing concentrations of RPA precoating the ssDNA. Gel loading buffer contains SDS to dissociate protein from DNA. (C) Quantitation of reannealing reactions. Values are the average of three experiments. Error bars represent 1 SD.
Mcm10 Inhibits Fork Regression.
Studies focusing on Mcm10 loss of function indicate that Mcm10 is important for DNA repair as well as for fork progression and stability of the replisome (21–25). Our present CX-MS findings show that the DNA-binding region of Mcm10 binds the N-terminal face of CMG (Fig. 1). In addition, our earlier cryoEM structure of CMG at a fork shows that the N-terminal face of CMG is directed at the forked junction (28). Taken together, these two structural observations indicate that Mcm10 is located at or near the fork junction. This places Mcm10 in a position where it could play a role in regulating fork regression. Recent cellular studies have demonstrated that fork regression is a primary response to replication stress or damage (29–31). A subset of SWI/SNF family dsDNA translocases– SMARCAL1, ZRANB3, and HLTF–has been shown to facilitate reversal of stalled or damaged forks to maintain genomic integrity (29–31). Fork regression involves the unwinding and subsequent pairing of nascent strands to form a four-arm Holliday junction that can be migrated to further reverse the fork.
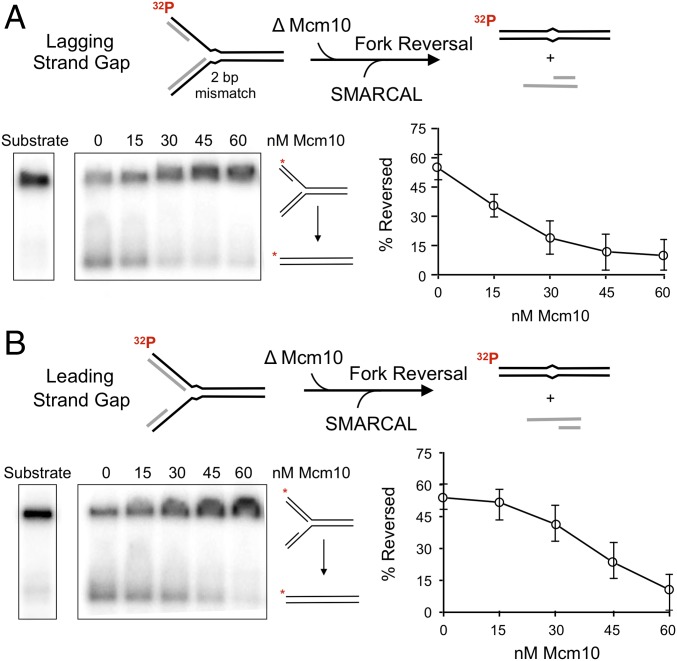
Given the phenotypic consequences of Mcm10 mutation or depletion, and considering that Mcm10 binds to ssDNA (Kd = 50 nM; SI Appendix, Fig. S5) and catalyzes strand annealing in the presence of RPA (Fig. 5), we explored the impact of Mcm10 on fork regression. To do so, we used the well-characterized fork reversal enzyme SMARCAL1 and DNA substrates used previously to study SMARCAL1: forked substrates with a single-strand gap on either the leading or the lagging strand arm (33). The fork regression substrates contain two mismatches at the base of the fork that prevent spontaneous branch migration/fork reversal. SMARCAL1 catalyzes branch migration through the mismatched region, resulting in two duplex products: a duplex of the nascent strands and a duplex of the parental strands (Fig. 6A).
Fig. 6.
Mcm10 inhibits SMARCAL1 catalyzed fork regression. Fork regression assays of SMARCAL1 with increasing Mcm10 added. (A) 2 nM SMARCAL1 on a lagging single-strand gap substrate at 2 min of reaction. (B) 5 nM SMARCAL on a leading single-strand gap substrate at 5 min of reaction. The two arms of the forked DNA are complementary, and substantial spontaneous regression is prevented by two mismatches at the fork junction, indicated by a kink in the duplex. Quantitation is the average of triplicate experiments. Error bars represent 1 SD.
We expressed and purified human SMARCAL1 in a yeast expression system (SI Appendix, Fig. S6) and tested the impact of S. cerevisiae Mcm10 on fork reversal. SMARCAL1 titrations and time courses showed that both substrates were reversed by SMARCAL1 (SI Appendix, Fig. S7), with the lagging gap substrate reversed more efficiently compared with the leading gap substrate. At 2 nM SMARCAL1, we observed >50% reversal in 2 min for the lagging gapped DNA (Fig. 6A and SI Appendix, Fig. S7). The leading gapped fork DNA required 5 nM SMARCAL1 for 5 min to yield comparable results (Fig. 6B and SI Appendix, Fig. S7). The fact that SMARCAL1 is more efficient in reversing forks having a lagging strand gap, compared with a leading strand gap, is consistent with an earlier report which used identical substrates (33). Mcm10 alone displayed no fork regression activity on either DNA substrate (SI Appendix, Fig. S8). Titration of Mcm10 into SMARCAL1 fork reversal reactions showed strong inhibition of SMARCAL1 on both substrates. We also formed a DNA fork with no ssDNA gap and found that Mcm10 still inhibited fork reversal by SMARCAL1 (SI Appendix, Fig. S9). These results are consistent with studies of Xenopus laevis Mcm10 demonstrating that Mcm10 binds ssDNA and dsDNA with similar affinities (38). Two conceivable ways in which that Mcm10 may inhibit fork regression are (i) the use of Mcm10 annealing activity in which Mcm10 reverses the initial SMARCAL1-induced unwinding of the nascent strands from the parental strands, a necessary first step in forming the four-way junction needed for fork reversal, or (ii) nonspecific binding of Mcm10 to ssDNA or dsDNA, blocking fork regression. We examine these processes in more detail below.
Discussion
The robust strand-annealing activity of Mcm10 demonstrated in this study seems to run counter to the fact that Mcm10 stimulates CMG unwinding activity (13, 14). However, during normal replication, the two daughter strands are replicated to form two duplexes, leaving no complementary single strand for Mcm10 to anneal. As has been proposed for other helicases that both unwind and anneal DNA, CMG-Mcm10 annealing activity may be involved in ds break repair, fork reversal, transcription, and/or telomere metabolism (27). In fact, Mcm10 loss of function studies indicate that Mcm10 is involved in DNA repair and replisome stability in higher eukaryotes (21–25). Below we examine a hypothesis to possibly explain how Mcm10 may regulate fork reversal, known to be required for genome integrity. We also propose a possible consequence of Mcm10’s annealing activity at origins that may necessitate the use of Pol δ in leading strand synthesis.
Mcm10 Inhibition of Fork Regression.
Our current data demonstrate that S. cerevisiae Mcm10 inhibits fork regression catalyzed by the fork reversal enzyme SMARCAL1 (human). Fork regression is thought to occur after DNA damage when the leading polymerase uncouples from the CMG helicase (29–31). Fork regression is likely regulated and prevented at active replisomes. Based on our present results demonstrating Mcm10 inhibition of fork reversal, we propose that Mcm10 may inhibit regression of active forks, helping specify fork regression to damaged forks. The physiological link of Mcm10 to fork regression is suggested by several studies correlating Mcm10 mutation or depletion to replisome stability and DNA repair (21–25) and link Mcm10 to dsDNA repair enzymes that are proposed to act during fork regression repair (25, 29–31).
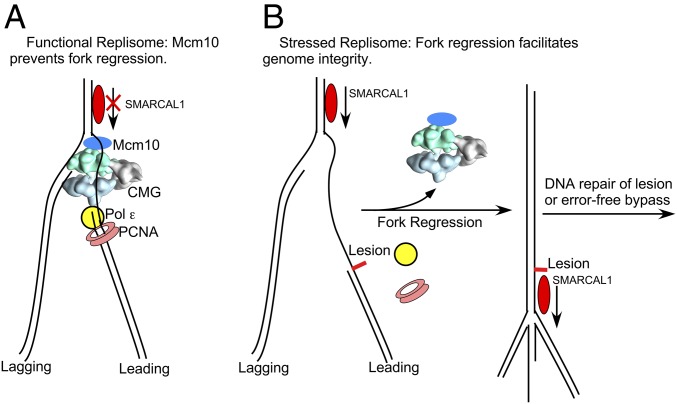
Our CX-MS analysis reveals that the DNA-binding region of Mcm10 is located at the N-face of CMG (Fig. 1, Movie S1, and Dataset S1). Combined with our earlier finding that CMG tracks first on the DNA N-face (28), this places Mcm10’s binding region at the front of CMG, facing the fork junction (Fig. 7A). This location is consistent with at least two possible mechanisms for Mcm10 inhibition of fork regression: (i) Mcm10 nonspecifically binds the ssDNA of a forked DNA that is stalled by a lesion on the leading strand, thereby inhibiting fork reversal, as observed in an earlier study using Escherichia coli ssDNA-binding protein (33), and (ii) Mcm10 prevents fork reversal by its strand-annealing activity, inhibiting the initial unpairing step in fork reversal in which newly synthesized DNA strands must unpair from their parental DNA strands before pairing to form the fourth arm. The present study demonstrating Mcm10’s robust strand-annealing activity reveals the potential for the second possibility, but we cannot distinguish between the two processes (or any others) at this time.
Fig. 7.
Putative role of Mcm10 in fork regression repair. (A) Unperturbed replication fork. Mcm10 is located at the top of the replisome and binds the forked DNA nexus, preventing fork regression by DNA translocases, such as SMARCAL1. (B, Left) Perturbed fork with a leading strand lesion. CMG and Pol ε-PCNA uncouple on encountering a leading strand lesion, and CMG may dissociate after an unspecified distance of continued unwinding and lagging strand synthesis. (B, Right) SMARCAL1 can perform fork regression for lesion repair or error-free lesion bypass.
One may question how fork regression is targeted to DNA-damaged forks but not actively replicating forks. We found that S. cerevisiae Mcm10 binds to ssDNA rather weakly (Kd = 50 nM; SI Appendix, Fig. S5), similar to the 120 nM Kd value for the more extensively characterized X. laevis Mcm10 (38). This relatively weak interaction with DNA might be harnessed for regulation. We hypothesize that when held to the fork by CMG during active replication, Mcm10 would have a high effective concentration due to proximity effects. Thus, Mcm10 could be an even more potent inhibitor of fork regression when part of an active replication fork than was observed in this study using isolated Mcm10 (Fig. 7A).
On encountering a leading strand lesion, CMG and Pol ε are thought to uncouple, which may lead to dissociation of CMG from DNA. CMG is not highly processive on its own (6, 39), and S. cerevisiae CMG-Mcm10 unwinding is 10-fold slower than when coupled to active replication forks (13). CMG dissociation would nullify the proximity effect of Mcm10 at a fork and possibly, given the low affinity of Mcm10 to DNA, no longer prevent fork regression (Fig. 7B). Experiments that enable simultaneous CMG-mediated replisome function and SMARCAL-mediated fork regression are needed to explore this hypothesis.
Mcm10 Annealing May Necessitate Pol δ Action on the Leading Strand at Some Origins.
Numerous studies have demonstrated that bulk leading synthesis is performed by Pol ε, and that Pol δ replicates the lagging strand (reviewed in ref. 3). However, Pol δ is sometimes used on the leading strand (40), and recent studies have indicated that it extends the first primer at an origin (41, 42). In E. coli, the origin is initially primed by DnaB–DnaG interactions on the lagging strands of the origin bubble, and these primers then become the two leading strands (43). This model may apply to eukaryotic origins as well. Specifically, at bidirectional origins, the two CMG helicases travel in opposite directions, forming an expanding ssDNA bubble (2, 3). Studies in yeast have demonstrated that Pol α-primase must function with CMG to form primers on the lagging strand in the presence of RPA (44). By analogy to bacterial origins, the first lagging strand primer formed by Pol α-primase at an origin would be handed off to Pol δ-PCNA (i.e., as in SV40 studies; ref. 45) and then extended to the CMG at the opposite end of the bubble, whereupon Pol ε takes over to continue the leading strand (SI Appendix, Fig. S10).
During origin ssDNA bubble formation, and before the first primers are laid down, the ssDNA is presumed to be held open by RPA. However, our findings demonstrate that Mcm10 anneals complementary strands saturated by RPA. Thus, Mcm10 may anneal the region between the bidirectional helicases, converting it to duplex DNA. In this case, the initial primers must be extended through dsDNA, thereby requiring strand displacement synthesis. Pol δ is a strong strand-displacing enzyme (3, 46), while Pol ε is nearly incapable of strand displacement (47). Thus, Mcm10 annealing may create a need for Pol δ at origins (SI Appendix, Fig. S10).
In summary, our present findings reveal that Mcm10 is a potent strand-annealing protein that inhibits fork regression by SMARCAL1. Furthermore, the CX-MS studies show that Mcm10 interacts with numerous CMG subunits on the N-surface of CMG, and even wraps to the C-face of CMG, expanding the present knowledge about Mcm10–CMG interactions. Moreover, CX-MS analysis places the DNA-binding region of Mcm10 on the N-face of CMG near the DNA fork junction, where it is poised to counteract fork regression at an active replication fork. The present study provides a conceptual framework for future studies exploring the regulatory balance of replication fork progression and fork regression.
Materials and Methods
CX-MS.
Reconstituted CMG-Mcm10 complex was cross-linked using disuccinimidyl suberate (DSS) in reactions containing ∼12 μg of CMG-Mcm10, 1 mM DSS, and 5% DMSO. Reactions were incubated for 25 min at room temperature with agitation at 1,200 rpm, then quenched with 50 mM (final concentration) NH4CO3 and methanol-precipitated. Following reduction and alkylation of cysteines, the sample was separated by SDS/PAGE in a 3–8% Tris-acetate gradient mini gel. Cross-linked products were then digested and analyzed by MS. Further details are provided in SI Appendix.
Helicase Assays.
The unwinding substrate was a fork DNA with a 32P-5′ end-labeled lagging strand. Unwinding was assayed by incubating 0.5 nM substrate with 30 or 50 nM CMG with and without 60 nM Mcm10 (as indicated in the figure legends) in the presence of 1 mM ATP at 30 °C. Timed aliquots were withdrawn, quenched, and then analyzed by native PAGE. More information is provided in SI Appendix.
Annealing Assays.
Partially complementary oligos forming a fork were combined on ice at a ratio of 10 nM leading strand to 20 nM 32P-5′ end-labeled lagging strand. Mcm10 was added immediately after combining the two oligos, and reactions were incubated at 30 °C. Reactions were stopped by adding a 10-fold excess of unlabeled lagging strand and then flash-freezing. Reactions were then analyzed by native PAGE. For experiments with RPA, both oligos were preincubated with saturating RPA for 5 min before the oligos were combined. SI Appendix provides further details.
Fork Regression Assays.
A substrate composed of four oligos was constructed by first annealing the two leading strand and two lagging strand oligos to form leading and lagging halves, which were subsequently annealed for 20 min at 30 °C to form the full substrate. Two mismatches at the fork junction prevented excessive spontaneous reversal. Fork regression activity was assayed by incubating SMARCAL1 and/or Mcm10 with 0.5 nM substrate at 37 °C with 2 mM ATP. Reactions were quenched, flash-frozen, and then analyzed by native PAGE. The product of fork regression is observed as a faster-migrating band on the gel that forms when the “nascent” strands are ejected and the 32P-labeled “parental” strands remain as a dsDNA product. More details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We appreciate the help of Dr. Grant Schauer of the M.E.O. laboratory for determining the Kd value of Mcm10 to DNA. We thank Drs. Huilin Li for suggesting the possibility that Mcm10 may bind the mobile N-tails of Mcms. This research was funded by National Institutes of Health (NIH) Grant R01 GM115809 and the Howard Hughes Medical Institute (to M.E.O.), NIH Grant R01 GM103314 (to B.T.C.), and NIH Grant T32 CA009673 (to R.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819107116/-/DCSupplemental.
References
- 1.Bai L, et al. Architecture of the Saccharomyces cerevisiae replisome. Adv Exp Med Biol. 2017;1042:207–228. doi: 10.1007/978-981-10-6955-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgers PMJ, Kunkel TA. Eukaryotic DNA replication fork. Annu Rev Biochem. 2017;86:417–438. doi: 10.1146/annurev-biochem-061516-044709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, O’Donnell ME. The eukaryotic CMG helicase at the replication fork: Emerging architecture reveals an unexpected mechanism. Bioessays. 2018;40 doi: 10.1002/bies.201700208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell ME, Li H. The ring-shaped hexameric helicases that function at DNA replication forks. Nat Struct Mol Biol. 2018;25:122–130. doi: 10.1038/s41594-018-0024-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielinsky AK. Mcm10: The glue at replication forks. Cell Cycle. 2016;15:3024–3025. doi: 10.1080/15384101.2016.1216925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thu YM, Bielinsky AK. MCM10: One tool for all—integrity, maintenance, and damage control. Semin Cell Dev Biol. 2014;30:121–130. doi: 10.1016/j.semcdb.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das-Bradoo S, Ricke RM, Bielinsky AK. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol Cell Biol. 2006;26:4806–4817. doi: 10.1128/MCB.02062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warren EM, et al. Structural basis for DNA binding by replication initiator Mcm10. Structure. 2008;16:1892–1901. doi: 10.1016/j.str.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du W, et al. Mcm10 self-association is mediated by an N-terminal coiled-coil domain. PLoS One. 2013;8:e70518. doi: 10.1371/journal.pone.0070518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du W, Stauffer ME, Eichman BF. Structural biology of replication initiation factor Mcm10. Subcell Biochem. 2012;62:197–216. doi: 10.1007/978-94-007-4572-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langston LD, et al. Mcm10 promotes rapid isomerization of CMG-DNA for replisome bypass of lagging strand DNA blocks. eLife. 2017;6:e29118. doi: 10.7554/eLife.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lõoke M, Maloney MF, Bell SP. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglas ME, Ali FA, Costa A, Diffley JFX. The mechanism of eukaryotic CMG helicase activation. Nature. 2018;555:265–268. doi: 10.1038/nature25787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31:2182–2194. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Warren EM, Huang H, Fanning E, Chazin WJ, Eichman BF. Physical interactions between Mcm10, DNA, and DNA polymerase alpha. J Biol Chem. 2009;284:24662–24672. doi: 10.1074/jbc.M109.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alver RC, et al. The N-terminus of Mcm10 is important for interaction with the 9-1-1 clamp and in resistance to DNA damage. Nucleic Acids Res. 2014;42:8389–8404. doi: 10.1093/nar/gku479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chadha GS, Gambus A, Gillespie PJ, Blow JJ. Xenopus Mcm10 is a CDK-substrate required for replication fork stability. Cell Cycle. 2016;15:2183–2195. doi: 10.1080/15384101.2016.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell. 2007;18:4085–4095. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, Bang SW, Jeon Y, Kang S, Hwang DS. Knockdown of human MCM10 exhibits delayed and incomplete chromosome replication. Biochem Biophys Res Commun. 2008;365:575–582. doi: 10.1016/j.bbrc.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Wawrousek KE, et al. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle. 2010;9:1156–1166. doi: 10.4161/cc.9.6.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y. Unwinding and rewinding: Double faces of helicase? J Nucleic Acids. 2012;2012:140601. doi: 10.1155/2012/140601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgescu R, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci USA. 2017;114:E697–E706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhat KP, Cortez D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat Struct Mol Biol. 2018;25:446–453. doi: 10.1038/s41594-018-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng W, Jasin M. Homologous recombination and replication fork protection: BRCA2 and more! Cold Spring Harb Symp Quant Biol. 2017;82:329–338. doi: 10.1101/sqb.2017.82.035006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidorova J. A game of substrates: Replication fork remodeling and its roles in genome stability and chemo-resistance. Cell Stress. 2017;1:115–133. doi: 10.15698/cst2017.12.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole LA, Cortez D. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit Rev Biochem Mol Biol. 2017;52:696–714. doi: 10.1080/10409238.2017.1380597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bétous R, et al. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 2013;3:1958–1969. doi: 10.1016/j.celrep.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douglas ME, Diffley JF. Recruitment of Mcm10 to sites of replication initiation requires direct binding to the mini chromosome maintenance (MCM) complex. J Biol Chem. 2016;291:5879–5888. doi: 10.1074/jbc.M115.707802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quan Y, et al. Cell cycle-regulated interaction between Mcm10 and double-hexameric Mcm2-7 is required for helicase splitting and activation during S phase. Cell Rep. 2015;13:2576–2586. doi: 10.1016/j.celrep.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson PD, et al. Domain architecture and biochemical characterization of vertebrate Mcm10. J Biol Chem. 2008;283:3338–3348. doi: 10.1074/jbc.M706267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci USA. 2012;109:6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garbacz MA, et al. Evidence that DNA polymerase δ contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat Commun. 2018;9:858. doi: 10.1038/s41467-018-03270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeeles JTP, Janska A, Early A, Diffley JFX. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell. 2017;65:105–116. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgescu RE, et al. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. eLife. 2015;4:e04988. doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 46.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganai RA, Zhang XP, Heyer WD, Johansson E. Strand displacement synthesis by yeast DNA polymerase ε. Nucleic Acids Res. 2016;44:8229–8240. doi: 10.1093/nar/gkw556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.