Significance
The ciliary membrane-localized transmembrane Hedgehog receptor Patched1 suppresses the ciliary translocation of the transmembrane oncoprotein Smoothened and consequently Hedgehog pathway activation, but the mechanism underlying this process remains largely unknown. Here, we show that the Patched1–ArhGAP36–PKA–Inversin–Smoothened axis determines this process. We uncover that the ciliary translocation of Smoothened depends on its interaction with phosphorylated ciliary Inversin, which is sequentially achieved by the removal of centrosomal ArhGAP36 with ciliary Patched1 and the accumulation of centrosomal PKA during Hedgehog pathway activation. Our findings reveal how the Hedgehog pathway is spontaneously activated in tumor cells carrying loss-of-function mutations of Patched1 or oncogenic mutations of Smoothened.
Keywords: Sonic Hedgehog pathway, Patched1, Smoothened, Inversin, PKA
Abstract
The Sonic Hedgehog (Shh) pathway conducts primarily in the primary cilium and plays important roles in cell proliferation, individual development, and tumorigenesis. Shh ligand binding with its ciliary membrane-localized transmembrane receptor Patched1 results in the removal of Patched1 from and the translocation of the transmembrane oncoprotein Smoothened into the cilium, leading to Shh signaling activation. However, how these processes are coupled remains unknown. Here, we show that the Patched1–ArhGAP36–PKA–Inversin axis determines the ciliary translocation of Smoothened. We find that Patched1 interacts with and stabilizes the PKA negative regulator ArhGAP36 to the centrosome. Activating the Shh pathway results in the removal of ArhGAP36 from the mother centriole and the centrosomal PKA accumulation. This PKA then phosphorylates Inversin and promotes its interaction with and the ciliary translocation of Smoothened. Knockdown of Inversin disrupts the ciliary translocation of Smoothened and Shh pathway activation. These findings reveal a regulatory molecular mechanism for the initial step of Shh pathway activation.
The Sonic Hedgehog (Shh) pathway, conserved from insects to humans, plays an important role in cell proliferation, differentiation, individual development, and tumorigenesis. In most mammalian cells, it is primarily conducted in the primary cilium (1, 2). In the absence of the Shh ligand, cilium-localized Patched1 (Ptc1) suppresses the ciliary translocation of Smoothened (Smo) (3). The binding of Shh ligand to Ptc1 triggers the removal of Ptc1 from the cilium and the translocation of Smo into the cilium (3). Ciliary Smo then regulates the removal of ciliary Gpr161 (4), which negatively regulates the Shh pathway (5, 6) and promotes dissociation of Gli transcriptional factors from its inhibitory factor Sufu in the cilium (7, 8). The released Gli transcriptional factors then activate the expression of their target genes (1, 2).
The PKA kinase is one of the key players in the Hedgehog pathway (9). It in part localizes to the cilium and the basal body (10). Its activity in the cilium is correlated with the amount of ciliary cAMP under the regulation of Gpr161 (1, 6). In the absence of Shh, ciliary PKA phosphorylates Gli proteins, leading to the proteolytic processing of full-length Gli into a transcriptional repressor. Encountering Shh signaling, PKA accumulates at the centrosome (11, 12). PKA was shown to promote the translocation of Smo to the proximal region of the cilium (13, 14), although it was also shown not required for the ciliary trafficking of Smo (15). Recently, ArhGAP36 was identified as a negative regulator of PKA by blocking PKA activity and by targeting the catalytic subunit of PKA (PKAc) for degradation (16). PKA inhibition by ArhGAP36 results in activation of the Shh pathway. Up-regulation of ArhGAP36 is correlated with Shh-driven medulloblastomas (16, 17). However, besides its critical role in regulating PKA, little is known about how ArhGAP36 itself is regulated during Shh pathway activation.
The ciliary translocation of Smo is a hallmark for Shh pathway activation (18). In Shh-driven cancers, the oncogenic mutations of Smo result in its spontaneous ciliary localization and hyperactivation of the Shh pathway even in the presence of Ptc1 (19). Additionally, various posttranscriptional modifications of Smo are required for its function: the N-glycosylation modification of Smo is one of these modifications required for its activity (20), and several serine and threonine sites residing in the cytoplasmic tail of Smo are phosphorylated upon Shh signaling (9), which induces a conformational switch of Smo to facilitate its function (21). More recently, cholesterol was shown to play a critical role in the ciliary translocation of Smo and Smo-mediated Shh pathway activation (22–25). So far, the mechanism by which Ptc1 inhibits the ciliary translocation of Smo remains unknown.
Results and Discussion
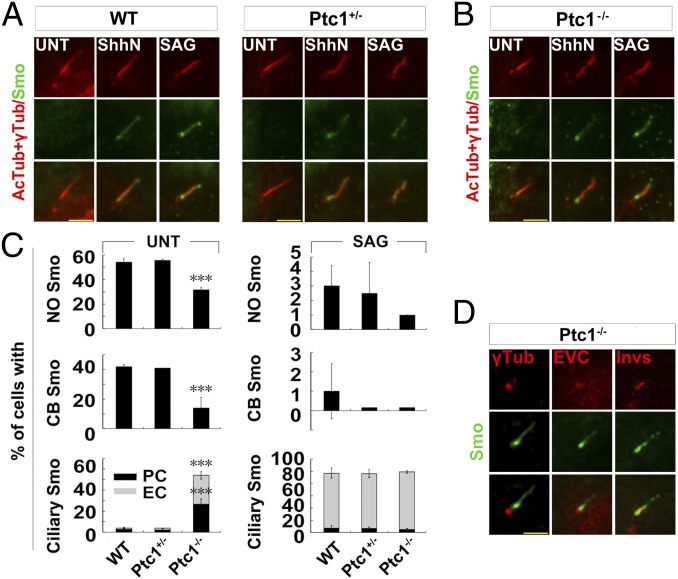
Ptc1 Knockout Results in Accumulation of Smo at the Ciliary Proximal Region.
To investigate the regulation of Smo localization by Ptc1, we carefully examined the ciliary translocation of Smo in wild-type (WT), Ptc1+/−, and Ptc1−/− mouse embryonic fibroblast (MEF) cells, in which Smo was reported localizing to the entire cilium in the absence of the Shh ligand (3). Surprisingly, while almost no ciliary Smo was observed in untreated ciliated WT and Ptc1+/− cells, Smo situated to the proximal but not the entire region of the cilium in ∼50% of untreated ciliated Ptc1−/− cells (Fig. 1 A–C), and this Smo colocalized with the marker proteins that indicate the ciliary inversin compartment, but not the basal body or the EVC zone (26–29) (Fig. 1D). Through RT-PCR and immunofluorescence labeling, we confirmed that these Ptc1−/− cells were not contaminated by Ptc1+/− or WT cells (SI Appendix, Fig. S1 A and B). Furthermore, we found besides the ciliary localization, a portion of Ptc1 localized at the centrosome (SI Appendix, Fig. S1 B and C), which was similar to the localization of Ptc1-YFP (3). Since the ciliary proximal region-localized Smo in Ptc1−/− cells could relocate to the entire cilium, like that in WT and Ptc1+/− cells, upon the treatment with the Shh ligand (ShhN) or the Smo agonist SAG (Fig. 1 A–C and SI Appendix, Fig. S2), the uneven ciliary localization of Smo in Ptc1−/− cells is not due to the defect of its transport. The ciliary translocation of Smo upon ShhN treatment in Ptc1−/− cells might be due to the presence of Ptc2 in these cells (SI Appendix, Fig. S1A), since Ptc2 could also mediate the Shh response in Ptc1−/− cells (30). And, since the protein levels of Smo in WT, Ptc1+/−, and Ptc1−/− cells were similar (SI Appendix, Fig. S1A), the ciliary localization of Smo in Ptc1−/− cells was not due to its elevated expression, which results in the spontaneous translocation of Smo into the cilium (31). Together, these results demonstrate that the depletion of Ptc1 results in the translocation of Smo to the ciliary proximal region.
Fig. 1.
Smo mainly accumulates at the ciliary proximal region in Ptc1−/− cells. (A–C) The ciliary proximal region localization of Smo in Ptc1−/− cells. G0 phase WT, Ptc1+/−, and Ptc1−/− MEF cells after the indicated treatments were immunostained for the indicated proteins. AcTub, acetylated α-tubulin; CB, ciliary base localization; EC, even ciliary localization; NO, no ciliary localization; PC, ciliary proximal region localization. In the statistics histograms for the results in A and B, the values are the mean ± SD; 50 cells per sample were randomly selected and counted in each of three independent experiments. ***P < 0.001. (D) A proportion of Smo accumulates at the Inversin compartment in Ptc1−/− cells. G0 phase Ptc1−/− cells were immunostained for the indicated proteins. Invs, Inversin; γTub, γ-tubulin. Note that only Inversin showed a partial colocalization with Smo at the ciliary proximal region. (Scale bars, 5 μm.)
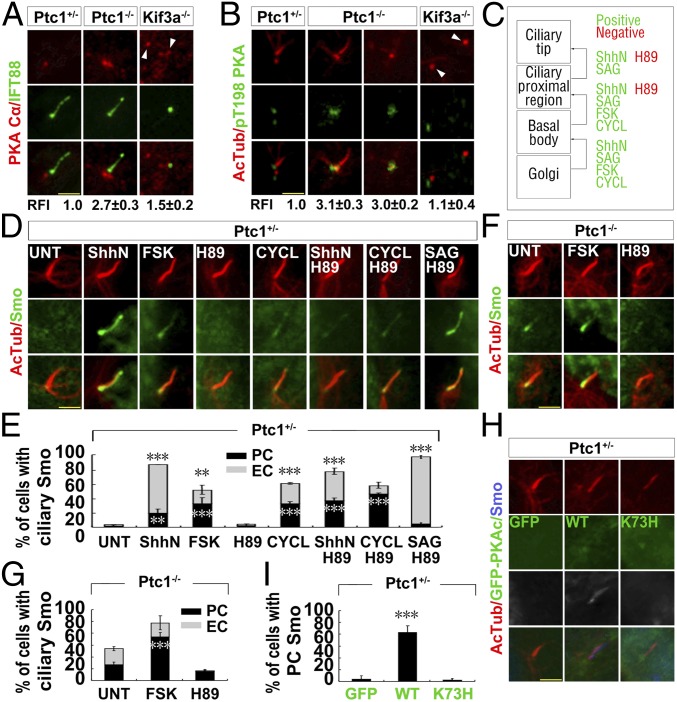
The Ciliary Proximal Region Translocation of Smo Is Promoted by PKA, Downstream of Shh Signaling.
To reveal how Smo accumulates at the ciliary proximal region in Ptc1−/− cells, we investigated several known centrosomal/ciliary proteins (SENP1, CK1γ, GRK2, and PKA) which regulate the ciliary translocation of Smo (13, 14, 32, 33). We found that the expression and centrosomal localization of GRK2, CK1γ1, and SENP1 were similar in Ptc1−/− and Ptc1+/− cells (SI Appendix, Fig. S3 A–D), but PKAc was significantly enriched at the centrosomes in Ptc1−/− cells compared with that in Ptc1+/− cells (Fig. 2A). Since the total protein level of PKAc was lower in Ptc1−/− cells than in Ptc1+/− cells (SI Appendix, Fig. S3D), the accumulation of centrosomal PKAc in Ptc1−/− cells is not due to its elevated protein amount. The staining of T198-phosphorylated PKAc (pPKAc) showed that centrosomal PKA in Ptc1−/− cells has kinase activity regardless of whether the cilium was assembled or not (Fig. 2B). To determine which domain of Ptc1 accounts for the abnormal localization of PKAc in Ptc1−/− cells, we stably expressed GFP-tagged full-length Ptc1 (Ptc1FL), N (the transmembrane domain) or C (the cytoplasmic domain) terminus truncate Ptc1 in Ptc1−/− cells (SI Appendix, Fig. S4 A and B), and found that only Ptc1FL could decrease the accumulation of PKAc at the centrosome (SI Appendix, Fig. S4 C and D). Since PKAc was not accumulated at the centrosome in Kif3a−/− cells (Fig. 2B), the inhibition of centrosomal PKAc by Ptc1 is cilium dependent.
Fig. 2.
The centrosomal accumulation of PKA results in the ciliary proximal localization of Smo in Ptc1−/− cells. (A and B) PKAc accumulates at the centrosome in Ptc1−/− cells. G0 phase Ptc1+/−, Ptc1−/−, and Kif3a−/− cells were immunostained for the indicated proteins. Arrowheads indicate the centrioles. The quantified relative fluorescent intensity (RFI) of centrosomal PKAc is shown Below each panel. The RFI results in A and B are shown as the mean ± SD; 50 cells were randomly selected for calculation. (C) A summary of the effects of the small molecules on the ciliary translocation of Smo. (D and E) PKA activity promotes Smo localization to the ciliary proximal region, downstream of Ptc1 inhibition by ShhN. G0 phase Ptc1+/− cells after the indicated treatments were immunostained for the indicated proteins. Note that PKA inhibition by H89 could abolish ShhN-induced, but had no effect on SAG- or cyclopamine-induced Smo ciliary translocation. (F and G) PKA activity accounts for Smo localization at the ciliary proximal region in Ptc1−/− cells. G0 phase Ptc1−/− cells after the indicated treatments were immunostained for the indicated proteins. (H and I) Increasing the amount of functional centrosomal PKAc promotes Smo localization at the ciliary proximal region. G0 phase Ptc1+/− cells stably expressing WT, K73H mutant GFP-PKAc, or GFP alone were immunostained for the indicated proteins. (Scale bars, 5 μm.) In the statistics histograms for results from E, G, and I, the values are the mean ± SD; 50 cells per sample were randomly selected and counted in each of three independent experiments. ***P < 0.001, **P < 0.01.
Then, through treating cells with forskolin (FSK) to activate or with H89 to inhibit PKA kinase activity, we investigated whether PKA activity controls the ciliary translocation of Smo. We showed that PKA activation in Ptc1+/− cells significantly increased the localization of Smo to the ciliary proximal region, even in the absence of Shh signaling. Importantly, the inhibition of PKA activity significantly impaired ShhN-induced ciliary translocation of Smo (Fig. 2 D and E), indicating that PKA functions downstream of Shh-induced Ptc1 removal from the cilium (Fig. 2C). Additionally, in Ptc1−/− cells, PKA activation could reinforce Smo localization at the ciliary proximal region, but not to the entire cilia; and PKA inhibition abolished this localization (Fig. 2 F and G). This indicates that PKA activity solely regulates Smo translocation to the ciliary proximal region. To further confirm the role of PKA activity in the ciliary translocation of Smo, we expressed GFP-tagged PKAcWT and its kinase-dead mutant PKAcK73H (34) in Ptc1+/− cells, and found that although both proteins accumulated at the centrosome, only PKAcWT promoted Smo translocation to the ciliary proximal region (Fig. 2 H and I). Notably, the inhibition of PKA did not affect the SAG or cyclopamine-induced Smo ciliary translocation, which binds with the transmembrane domains of Smo (Fig. 2 D and E); the inhibition of PKA also did not affect the 20-OHC–induced ciliary translocation of Smo, which binds with the cysteine-rich domain of Smo, although it decreased the intensity/amount of ciliary Smo (SI Appendix, Fig. S3E). These results demonstrate that PKA activity is solely required for Smo ciliary translocation induced by ShhN, and that the mechanism of PKA-dependent Smo ciliary translocation by ShhN is different from those governed by small molecules.
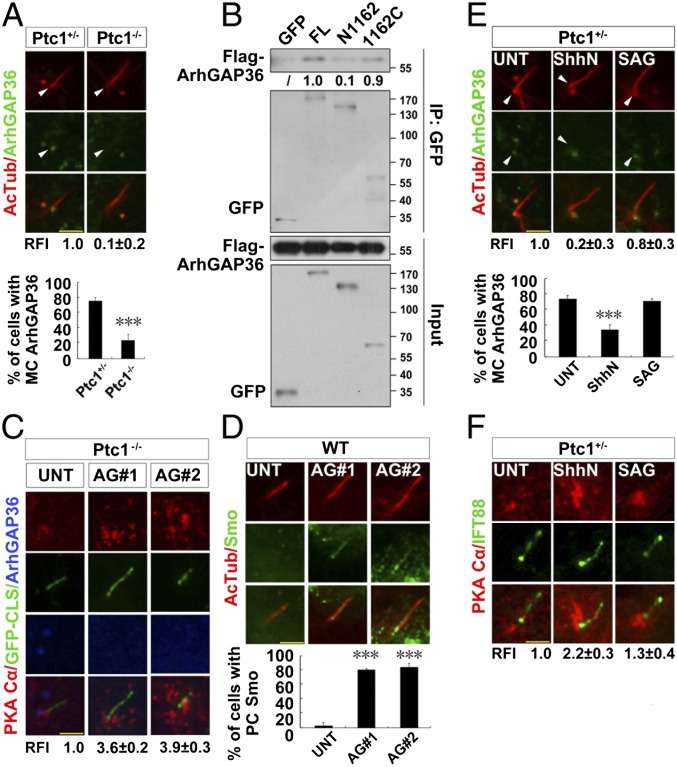
Ptc1 Interacts with and Stabilizes ArhGAP36 to the Centrosome to Inhibit Centrosomal PKA in the Absence of Shh Signaling.
To discover how PKAc is enriched at the centrosome in Ptc1−/− cells, we investigated the regulation of centrosomal PKAc. Since ArhGAP36 localizes at the centrosomal region (17), inhibits PKA activity, and promotes PKAc degradation (16), we examined its centrosomal localization. The amount of ArhGAP36 at the mother centriole (MC) and the ratio of MC-localized ArhGAP36 were both much lower in Ptc1−/− cells than in Ptc1+/− cells (Fig. 3A). When the full-length or the truncated forms of Ptc1 were each expressed in Ptc1−/− cells, the MC localization of ArhGAP36 was recovered only by Ptc1FL (SI Appendix, Fig. S4E). In comparison, the MC localization of ArhGAP36 and the centrosomal localization of PKAc was neither affected in Sufu−/− cells where the Shh pathway is activated in the presence of Ptc1 nor rescued by SANT1 treatment in Ptc1−/− cells where the Shh pathway is totally inhibited (SI Appendix, Fig. S5 A–C). This indicates that the disappearance of MC ArhGAP36 is specifically caused by the loss of function of Ptc1, but not the activation of the Shh pathway. To reveal whether Ptc1 maintains ArhGAP36 at the centrosome via interacting with it, we performed a coimmunoprecipitation assay and found that Ptc1 interacts with ArhGAP36, mainly through its C terminus (Fig. 3B). To know whether ArhGAP36 is directly involved in Smo ciliary translocation, we knocked down ArhGAP36 in Ptc1−/− cells and found PKAc was accumulated at the centrosome in these cells (Fig. 3C), with increased Smo localization to the ciliary proximal region (Fig. 3D).
Fig. 3.
Removal of ciliary Ptc1 by Shh signaling abolishes centrosomal ArhGAP36 localization and increases the amount of centrosomal PKAc. (A) The mother centriolar (MC) localization of ArhGAP36 is impaired by Ptc1 knockout. G0 phase Ptc1+/− and Ptc1−/− cells were immunostained for the indicated proteins. Arrowheads indicate the MC localization of ArhGAP36. The RFIs and the ratio of MC-localized ArhGAP36 are shown Below each panel. (B) Ptc1 interacts with ArhGAP36 mainly via its C terminus. HEK 293T cells were cotransfected with the indicated plasmids, and arrested at the G0 phase, followed by an immunoprecipitation assay. The normalized results of immunoprecipitated Flag-ArhGAP36 by the indicated proteins are shown Below each panel; Flag-ArhGAP36 immunoprecipitated by full-length Ptc1-GFP was set as 1.0. (C) Knockdown of ArhGAP36 results in PKAc accumulation at the centrosome. The Ptc1−/− cells expressing a GFP-CLS sequence of Fibrocystin (53) were knocked down for ArhGAP36, arrested at the G0 phase, and immunostained for the indicated proteins. (D) Knockdown of ArhGAP36 results in Smo translocation to the ciliary proximal region. The WT cells were knocked down for ArhGAP36, arrested at the G0 phase, and immunostained for the indicated proteins. (E and F) Ptc1 inhibition by Shh signaling causes ArhGAP36 removal from the MC and PKAc accumulation at the centrosome. G0 phase Ptc1+/− cells after the indicated treatments were immunostained for the indicated proteins. Arrowheads indicate the MC localization of ArhGAP36. (Scale bars, 5 μm.) In the statistics histograms for results from A, D, and E, the values are the mean ± SD; 50 cells per sample were randomly selected and counted in each of three independent experiments. ***P < 0.001. The RFI results in A, C, E, and F are shown as the mean ± SD; 50 cells were randomly selected for calculation.
Next, we investigated the relationship between Shh pathway activation and MC localization of ArhGAP36. When Ptc1+/− cells were treated with ShhN, the MC localization of ArhGAP36 was disappeared, and PKAc was accumulated at the centrosome (Fig. 3E and SI Appendix, Fig. S5E). Since the protein level of ArhGAP36 did not change during Shh pathway activation (SI Appendix, Fig. S5D), the removal of ArhGAP36 from the centrosome is due to its relocalization but not degradation. When Ptc1+/− cells were treated with SAG or 20-OHC, however, the MC localization of ArhGAP36 and the centrosomal accumulation of PKAc were only mildly changed (Fig. 3 E and F and SI Appendix, Figs. S5E and S6 A and B), indicating that besides the primarily Ptc1-dependent mechanism, a weaker, Ptc1-independent regulation of ArhGAP36 also exists. In Kif3a−/− cells, PKAc did not show accumulation at the centrosome regardless of whether the cells were treated with ShhN or SAG (SI Appendix, Fig. S5F). Collectively, these results indicate that Ptc1 inhibits centrosomal PKA by maintaining ArhGAP36 at the MC.
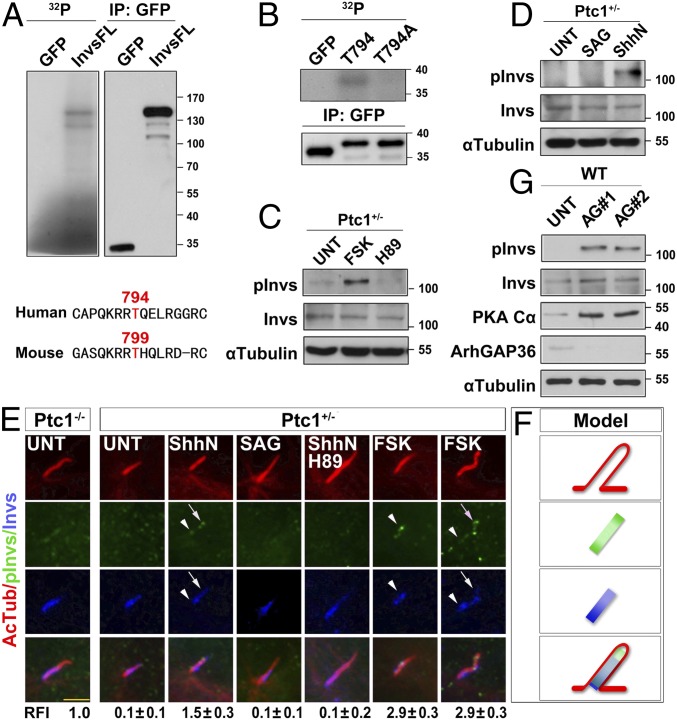
Shh Signaling Promotes Inversin Phosphorylation by PKA and Inversin–Smo Interaction.
To find out the downstream target(s) of PKA that mediates the ciliary translocation of Smo, we turned to investigate Inversin, as Smo showed a partial colocalization with Inversin in Ptc1−/− cells. We found that Inversin could be phosphorylated by PKA in vitro (Fig. 4A). Through sequence analysis, the amino acid residue T794 of human Inversin (conserved in mouse as T799) was predicted to be the phosphorylation site for PKA (Fig. 4A). Indeed, when T794 was mutated to alanine (T794A), this mutant peptide lost its phosphorylation by PKA (Fig. 4B). We further raised an antibody against phosphor-T799 of mouse Inversin and validated it by its significant increase upon forskolin treatment (Fig. 4C). With this antibody, we found that phosphor-Inversin (pInvs) was solely detected in ShhN-treated, but not SAG- or 20-OHC–treated, cells (Fig. 4D and SI Appendix, Fig. S6C), and consistently it was absent from the cilium in untreated Ptc1+/− cells. In contrast, pInvs weakly localized to the inversin compartment in Ptc1−/− cells. Importantly, upon Shh pathway activation by ShhN or PKA activation by forskolin, pInvs localized both at the ciliary proximal region and the ciliary tip in Ptc1+/− cells, whereas SAG had no such effect (Fig. 4 E and F). When PKA was inhibited by H89, both the ciliary proximal region and tip localization of pInvs disappeared (Fig. 4E). To confirm ArhGAP36 directly affects the phosphorylation of Inversin, we knocked down ArhGAP36 in WT cells and found that ArhGAP36 knockdown significantly increased the protein level of PKAc and pInvs (Fig. 4G). These results demonstrate that PKA phosphorylates Inversin at T799, under the regulation of ArhGAP36 and downstream of Shh signaling.
Fig. 4.
Shh signaling results in Inversin phosphorylation by PKA. (A and B) PKA phosphorylates Inversin at T794. HEK 293T cells were transfected with plasmids encoding the full length or fragments of human GFP-Inversin protein and arrested at the G0 phase. The GFP-tagged proteins were immunoprecipitated and subjected to in vitro kinase assay. T799 is the conserved site of the mouse Inversin protein analogous to the T794 in human protein. (C and D) Mouse Inversin is phosphorylated by PKA at T799. G0 phase Ptc1+/− cells after the indicated treatments were examined for the protein level of the indicated proteins. pInvs, phosphor-T799 Inversin. (E and F) Shh signaling leads to Inversin phosphorylation at T799 by PKA, with ciliary proximal region and tip localization. G0 phase Ptc1+/− and Ptc1−/− cells after the indicated treatments were immunostained for the indicated proteins. The localization pattern of the indicated proteins is shown in F. Arrowheads indicate the ciliary proximal region; arrows indicate the ciliary tip. (Scale bar, 5 μm.) The RFI results are shown as the mean ± SD; 50 cells were randomly selected for calculation. (G) Knockdown of ArhGAP36 results in Inversin phosphorylation at T799. WT cells were knocked down for ArhGAP36, arrested at the G0 phase, and examined for the indicated proteins.
Next, we tested whether this phosphorylation regulates Inversin–Smo interaction. Coimmunoprecipitation assays showed that Smo interacted with Inversin and that PKA activation increased this interaction (Fig. 5A). The proximal ligation assay (PLA) revealed that Inversin–Smo interaction occurred at the ciliary proximal region of untreated Ptc1−/− cells (Fig. 5B). Importantly, although Smo showed two obviously major bands in electrophoresis, Inversin solely interacted with the Smo of higher molecular weight (Fig. 5A), which is likely the glycosylated form (20, 35). Since the T794A mutation of Inversin largely reduced its interaction with Smo irrespective of whether PKA was activated (Fig. 5A), T794 phosphorylation is critical for Smo–Inversin interaction. Consistently, when the GFP-tagged WT, nonphosphorylated T794A and phosphorylation-mimicking T794D mutant Inversin were each introduced into WT cells, InversinT794D significantly increased Smo translocation to the ciliary proximal region (Fig. 5 C and D) and elevated Gli1 expression (Fig. 5E). Finally, we tested whether Shh pathway activation could induce Smo–Inversin interaction. Although promoting Smo translocation into the cilia, 20-OHC treatment did not increase the binding of Inversin and Smo (SI Appendix, Fig. S6D). However, although SAG and ShhN activate the Shh pathway with different mechanisms, they both promoted Smo–Inversin interaction (Fig. 5F). The increased binding of Smo–SAG with Inversin might be due to a specific conformation of Smo induced by SAG (36), and this changed conformation could mimic the binding mode of Smo with phosphorylated Inversin in the presence of Shh. The reinforced SmoSAG–Inversin binding helps in understanding how SAG promoted Smo ciliary translocation in PKA−/− cells (15).
Fig. 5.
Shh signaling-induced Inversin phosphorylation by PKA increases Inversin–Smo interaction. (A) Inversin interacts with Smo, and its T794 phosphorylation increases this interaction. HEK 293T cells were cotransfected with the indicated plasmids, arrested at the G0 phase, treated with or without FSK, and processed to immunoprecipitation assays. Note that losing the ability of T794 phosphorylation of Inversin by PKA impaired its binding with Smo. (B) Smo interacts with Inversin at the ciliary proximal region in Ptc1−/− cells. G0 phase Ptc1−/− cells were subjected to the PLA assay (see details in SI Appendix, Materials and Methods), singly labeled with Smo or Inversin, or colabeled with Smo and Inversin. (Scale bar, 5 μm.) (C and D) The phosphorylation-mimicking mutant of Inversin recruits Smo to the ciliary proximal region. WT cells stably expressing the WT or T794A/D mutant GFP-Inversin were arrested at the G0 phase and immunostained for the indicated proteins. The percentage of PC-patterned Smo in each cell line is shown in D. In each of three independent experiments, 50 cells per sample were randomly selected and counted. ***P < 0.001. (E) The T794D mutant Inversin results in a partial activation of Shh pathway. The parental cells (WT and Ptc1−/−) and the cells stably expressing the WT, T794A/D GFP-Inversin (labeled in green) were arrested at the G0 phase and examined for the indicated proteins. (F) Smo–Inversin interaction is increased by Shh pathway activation. HEK 293T cells were cotransfected with the indicated plasmids, arrested at the G0 phase, and processed to immunoprecipitation assays. The normalized results of total and immunoprecipitated upper band of Smo-Myc by the indicated proteins are shown Below each panel, and the amount of Smo-Myc under normal condition (UNT) was set as 1.0.
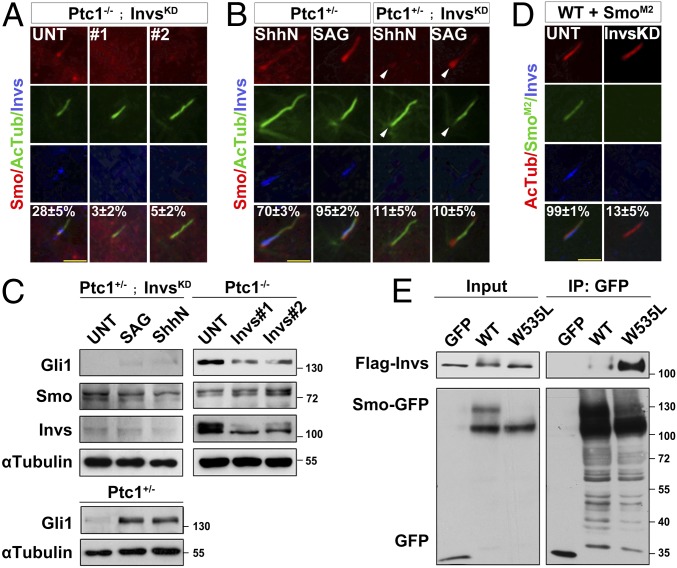
Inversin Is Required for Ciliary Translocation of Smo and Activation of the Shh Pathway.
To investigate the function of Inversin–Smo interaction, we knocked down Inversin in Ptc1−/− cells (Ptc1−/−; InvsKD), and found that Smo localization to the cilium was abolished (Fig. 6A). When Ptc1+/−; InvsKD cells were treated with ShhN or SAG, Smo was accumulated around the ciliary base, likely the EVC zone, and was almost absent from the cilium (Fig. 6B). Consistently, the protein level of Gli1 was significantly decreased by Inversin knockdown in ShhN or SAG-treated Ptc1+/− cells and impaired by Inversin knockdown in untreated Ptc1−/− cells (Fig. 6C). The failure of Shh signaling transduction in InvsKD cells could be rescued by introducing GFP-InversinWT, but not the T794A mutant (SI Appendix, Fig. S7A). Since SmoM2 localizes to the cilium independent of Shh signaling (37) and induces a hyperactivation of Shh pathway (38, 39), we examined the role of Inversin in cells stably expressing GFP-tagged SmoM2. When these cells were knocked down for Inversin, the ciliary localization of SmoM2 was abolished (Fig. 6D), and the activity of the Shh pathway in these cells was also significantly decreased (SI Appendix, Fig. S7B). These results demonstrate that Inversin determines the ciliary translocation of Smo and activation of the Shh pathway.
Fig. 6.
Inversin determines Smo ciliary translocation for Shh pathway activation. (A and B) Inversin is required for the ciliary translocation of Smo. Ptc1−/− or Ptc1+/− cells were knocked down for Inversin (InvsKD) and arrested at the G0 phase. After the indicated treatment, the cells were immunostained for the indicated proteins. Arrowheads indicate the ciliary base. The percentage of PC- and EC-localized Smo is shown. (C) Inversin is required for Shh signaling transduction. Ptc1+/−, Ptc1−/−, or Ptc1+/− cells knocked down for Inversin were arrested at the G0 phase; after the indicated treatment, the cells were lysed and the indicated proteins examined. (D) SmoM2 cannot translocate into the cilia of cells with Inversin knockdown. WT cells stably expressing SmoM2-GFP were knocked down for Inversin, arrested at the G0 phase, and immunostained for the indicated proteins. The percentage of EC-localized Smo is shown. In the statistics results from A, B, and D, the values are the mean ± SD; 50 cells per sample were randomly selected and counted in each of three independent experiments. (E) SmoM2 strongly interacts with Inversin in the absence of ShhN. HEK 293T cells were cotransfected with the indicated plasmids, arrested at the G0 phase, and processed to immunoprecipitation assays.
Finally, through establishing MEF cell lines stably expressing the oncogenic mutants of Smo (37, 40), we investigated the correlation of Smo mutations with the spontaneous activation of the Shh pathway. At the G0 phase, while the parental cells showed no expression of Gli1, SmoM2-expressing cells showed a significantly elevated protein level of Gli1 (SI Appendix, Fig. S7C). This indicates that the elevated expression of WT and mutant Smo do not always result in the activation of the Shh pathway. Since SmoM2 leads to hyperactivation of the Shh pathway (37), we examined the effect of this mutation on Smo–Inversin interaction. Coimmunoprecipitation assays showed that even in the absence of Shh, SmoM2 strongly interacted with Inversin in comparison with SmoWT (Fig. 6E), which was possibly caused by the specific, Shh-signaling activated-like conformation of SmoM2 (21, 41). However, although SmoL412F-expressing cells also showed elevated Gli1 expression (SI Appendix, Fig. S7C), it did not show an increased binding with Inversin in comparison with SmoWT (SI Appendix, Fig. S7 D and E). Consistently, Inversin knockdown had only a mild effect on the activity of the Shh pathway in SmoL412F cells (SI Appendix, Fig. S7B). These results indicate that, although the increased binding of Inversin with SmoM2 may cause the spontaneous activation of the Shh pathway in the case of SmoM2-driven tumor cells, this is not a general mechanism for the activation of the Shh pathway in other sorts of Smo mutant-driven tumors.
Based on our findings, we propose that Ptc1 stabilizes ArhGAP36, the negative regulator of PKA, to the centrosome in the absence of Shh signaling. When the amount of centrosomal PKA is low, it is unable to efficiently phosphorylate Inversin to promote Inversin–Smo interaction, and the small population of phosphorylated Inversin, if any, cannot sufficiently sequester Smo in the cilium. When Ptc1 exits from the cilium caused by either ShhN binding or loss-of-function mutations, ArhGAP36 is no longer maintained at the mother centriole, and consequently centrosomal PKAc begins to accumulate. Along with this, the phosphorylation of Inversin by centrosomal PKA increases and promotes Inversin–Smo interaction as well as the enrichment of Smo at the Inversin compartment. Once entering the cilium, either at the ciliary proximal region, the EVC zone (28), or the Inversin compartment as shown here, Smo is ready to activate the Shh pathway (SI Appendix, Fig. S8). Interestingly, although the PKA phosphorylation-mimicking mutant InversinT794D could recruit Smo to the ciliary proximal region and resulted in an elevated expression of Gli1, it could not activate the Shh pathway like that of Ptc1 knockout, indicating that additional roles of Ptc1 on Smo inhibition exist. Consistently, it was found recently that Ptc1 could inhibit Smo activity through limiting the access of cholesterol to Smo (25). Therefore, Ptc1 may adapt dual mechanisms to regulate both the localization and activity of Smo in ciliated cells.
The prevalent negative role of PKA in the Shh pathway comes from pharmacological manipulation of PKA by small molecules and genetical depletion of PKA or ArhGAP36 in cells and animals (9, 15–17). But these manipulations inevitably resulted in a universal activation or inactivation of PKA, wherein only the eventual readouts of the Shh pathway (Gli expression) could be detected, and the intermediate step(s) where PKA functions was overlooked. We propose here that the spatiotemporally restricted function of PKA in the cilium and the centrosome underlies the activation of the Smo-dependent Shh pathway: after Smo translocates into the cilium by the Ptc1–ArhGAP36–centrosomal PKA–Inversin axis, it abolishes the ciliary PKA-mediated phosphorylation of Gli (10). Our finding is therefore an important complement to the current model (9). Considering that in Drosophila, the binding of Hh ligand with Ptc1 directly induces the phosphorylation of Smo by PKA and Smo translocation to the cell membrane (42–46), our findings therefore reveal a communal positive role of PKA in the Shh pathway from insects to mammals.
Inversin plays a critical role in the establishment of the body axis and renal development. Mutations of Inversin are linked to nephronophthisis and abnormal left–right asymmetry of the body axis (47–49). It also negatively regulates the Wnt pathway (50). A potential inconsistency of our results with those Inversin mutant animals is that they do not show significant phenotypes related to Shh pathway inhibition, therefore the contribution of Inversin to Shh pathway regulation under physiological conditions remains a matter of concern. A possible cause for this is that in Inversin-null animals, the function of the Smo–EVC/EVC2 complex at the EVC zone is intact, which could maintain a basal level of Shh pathway activity required for normal development. Alternatively, a crosstalk between the Shh pathway and the Wnt pathway exists (51), where Inversin may be involved. It is therefore possible that the interplay between the two pathways in vivo could render animals that do not show Shh pathway defects.
Materials and Methods
Immunofluorescence Microscopy and the PLA.
The cells were fixed by 4% paraformaldehyde in PBS and then extracted with 0.2% Triton X-100 in PBS for detecting membrane proteins (i.e., Smo and Ptc1), or directly fixed by cold methanol for detecting centrosomal proteins (e.g., ArhGAP36, PKAc, and pPKAc). DNA was stained with DAPI. Images were captured by the immunofluorescence microscope (Axiovert 200M, Carl Zeiss). The fluorescence intensity of proteins was quantified using ImageJ. In situ interaction of Smo and Inversin was determined by using the PLA technology-based Duolink kit (cat. no. DUO92105, Sigma), following the manufacturer’s instructions.
Immunoprecipitation and Immunoblotting.
Immunoprecipitation and immunoblotting were performed as previously described (52). Briefly, MEF or HEK 293T cells transfected with the indicated plasmids were lysed using IP buffer [50 mM Hepes (pH 7.0), 125 mM NaCl, 10% glycerol, 0.1% Nonidet P-40, and 0.5 mM PMSF] on ice for 15 min. The lysates were centrifuged at 20,000 × g for 15 min, and the supernatants were incubated with the primary antibody-coated beads for 1.5 h at 4 °C on a rotator. After six washes with IP buffer, the beads were collected and the bound proteins were analyzed by Western blotting. A specific 2× loading buffer [100 mM Tris⋅HCl (pH 6.8), 6 M urea, 4% SDS, 20% glycerol, and 0.1% bromophenol blue] was used when dealing with the Smo protein. Each immunoprecipitation and Western blotting assay were independently repeated twice.
Details about cell culture, reagents, biochemistry, bioinformatics, and statistics are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Yun Zhao (Shanghai Institute of Biochemistry and Cell Biology) for a critical reading of the manuscript; Drs. William E. Moerner and Ljiljana Milenkovic (Stanford University), Rune Toftgård (Karolinska Institutet), Steven Y. Cheng (Nanjing Medical University), Oliver Rocks (Max Delbrück Center for Molecular Medicine), Gerd Walz (University of Freiburg Medical Center), and Xiaoyan Zheng (George Washington University) for cell lines and constructs; Drs. Hongxia Lv, Liying Du, and Dong Cao (Peking University) for technical support; and all the members of C.Z. laboratory for suggestions and comments. B. Yu is a visiting student from Shanghai Jiao Tong University School of Medicine. This work was supported by funds from the Ministry of Science and Technology of China and National Natural Science Foundation of China (Grants 2016YFA0100501, 2016YFA0500201, 31430051, 31520103906, 91854204, and 31571386).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804042116/-/DCSupplemental.
References
- 1.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 2.Pak E, Segal RA. Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates Hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 4.Pal K, et al. Smoothened determines β-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol. 2016;212:861–875. doi: 10.1083/jcb.201506132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachmann VA, et al. Gpr161 anchoring of PKA consolidates GPCR and cAMP signaling. Proc Natl Acad Sci USA. 2016;113:7786–7791. doi: 10.1073/pnas.1608061113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukhopadhyay S, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between suppressor of fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013;23:186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mick DU, et al. Proteomics of primary cilia by proximity labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barzi M, Berenguer J, Menendez A, Alvarez-Rodriguez R, Pons S. Sonic-hedgehog-mediated proliferation requires the localization of PKA to the cilium base. J Cell Sci. 2010;123:62–69. doi: 10.1242/jcs.060020. [DOI] [PubMed] [Google Scholar]
- 12.Saade M, Gonzalez-Gobartt E, Escalona R, Usieto S, Martí E. Shh-mediated centrosomal recruitment of PKA promotes symmetric proliferative neuroepithelial cell division. Nat Cell Biol. 2017;19:493–503. doi: 10.1038/ncb3512. [DOI] [PubMed] [Google Scholar]
- 13.Milenkovic L, Scott MP, Rohatgi R. Lateral transport of smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson CW, Chen MH, Chuang PT. Smoothened adopts multiple active and inactive conformations capable of trafficking to the primary cilium. PLoS One. 2009;4:e5182. doi: 10.1371/journal.pone.0005182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuson M, He M, Anderson KV. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011;138:4921–4930. doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eccles RL, et al. Bimodal antagonism of PKA signalling by ARHGAP36. Nat Commun. 2016;7:12963–12979. doi: 10.1038/ncomms12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rack PG, et al. Arhgap36-dependent activation of Gli transcription factors. Proc Natl Acad Sci USA. 2014;111:11061–11066. doi: 10.1073/pnas.1322362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbit KC, et al. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 19.Sharpe HJ, Wang W, Hannoush RN, de Sauvage FJ. Regulation of the oncoprotein smoothened by small molecules. Nat Chem Biol. 2015;11:246–255. doi: 10.1038/nchembio.1776. [DOI] [PubMed] [Google Scholar]
- 20.Marada S, et al. Functional divergence in the role of N-linked glycosylation in smoothened signaling. PLoS Genet. 2015;11:e1005473. doi: 10.1371/journal.pgen.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 22.Huang P, et al. Cellular cholesterol directly activates smoothened in Hedgehog signaling. Cell. 2016;166:1176–1187.e4. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luchetti G, et al. Cholesterol activates the G-protein coupled receptor smoothened to promote Hedgehog signaling. eLife. 2016;5:e20304. doi: 10.7554/eLife.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao X, et al. Cholesterol modification of smoothened is required for Hedgehog signaling. Mol Cell. 2017;66:154–162.e10. doi: 10.1016/j.molcel.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Myers BR, Neahring L, Zhang Y, Roberts KJ, Beachy PA. Rapid, direct activity assays for smoothened reveal hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci USA. 2017;114:E11141–E11150. doi: 10.1073/pnas.1717891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caparrós-Martín JA, et al. The ciliary Evc/Evc2 complex interacts with Smo and controls hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum Mol Genet. 2013;22:124–139. doi: 10.1093/hmg/dds409. [DOI] [PubMed] [Google Scholar]
- 27.Blair HJ, et al. Evc2 is a positive modulator of Hedgehog signalling that interacts with Evc at the cilia membrane and is also found in the nucleus. BMC Biol. 2011;9:14. doi: 10.1186/1741-7007-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorn KV, Hughes CE, Rohatgi R. A smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev Cell. 2012;23:823–835. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiba D, et al. Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J Cell Sci. 2009;122:44–54. doi: 10.1242/jcs.037408. [DOI] [PubMed] [Google Scholar]
- 30.Alfaro AC, Roberts B, Kwong L, Bijlsma MF, Roelink H. Ptch2 mediates the Shh response in Ptch1-/- cells. Development. 2014;141:3331–3339. doi: 10.1242/dev.110056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, et al. GSK3β-Dzip1-Rab8 cascade regulates ciliogenesis after mitosis. PLoS Biol. 2015;13:e1002129. doi: 10.1371/journal.pbio.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, et al. Sonic hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9:e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, et al. Regulation of smoothened phosphorylation and high-level Hedgehog signaling activity by a plasma membrane associated kinase. PLoS Biol. 2016;14:e1002481. doi: 10.1371/journal.pbio.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iyer GH, Moore MJ, Taylor SS. Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J Biol Chem. 2005;280:8800–8807. doi: 10.1074/jbc.M407586200. [DOI] [PubMed] [Google Scholar]
- 35.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, et al. Converse conformational control of smoothened activity by structurally related small molecules. J Biol Chem. 2009;284:20876–20884. doi: 10.1074/jbc.M807648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie J, et al. Activating smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 38.Mao J, et al. A novel somatic mouse model to survey tumorigenic potential applied to the hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong SY, et al. Primary cilia can both mediate and suppress hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atwood SX, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27:342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang P, et al. Structural basis of smoothened activation in Hedgehog signaling. Cell. 2018;175:295–297. doi: 10.1016/j.cell.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ranieri N, Thérond PP, Ruel L. Switch of PKA substrates from cubitus interruptus to smoothened in the Hedgehog signalosome complex. Nat Commun. 2014;5:5034. doi: 10.1038/ncomms6034. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Ma G, Wang B, Jiang J. Hedgehog induces formation of PKA-smoothened complexes to promote smoothened phosphorylation and pathway activation. Sci Signal. 2014;7:ra62. doi: 10.1126/scisignal.2005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 45.Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7:86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- 46.Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Extensive phosphorylation of smoothened in hedgehog pathway activation. Proc Natl Acad Sci USA. 2004;101:17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lienkamp S, Ganner A, Walz G. Inversin, Wnt signaling and primary cilia. Differentiation. 2012;83:S49–S55. doi: 10.1016/j.diff.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 48.Otto EA, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan D, et al. Inversin, a novel gene in the vertebrate left-right axis pathway, is partially deleted in the inv mouse. Nat Genet. 1998;20:149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]
- 50.Simons M, et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nat Genet. 2005;37:537–543. doi: 10.1038/ng1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takebe N, et al. Targeting Notch, hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B, et al. DAZ-interacting protein 1 (Dzip1) phosphorylation by polo-like kinase 1 (Plk1) regulates the centriolar satellite localization of the BBSome protein during the cell cycle. J Biol Chem. 2017;292:1351–1360. doi: 10.1074/jbc.M116.765438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Follit JA, Li L, Vucica Y, Pazour GJ. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol. 2010;188:21–28. doi: 10.1083/jcb.200910096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.