Significance
Many groups of flowering plants have become parasites and have lost the capacity to carry out photosynthesis. The plastid genomes of these parasitic plants are often highly reduced in size and gene content and are divergent in other ways too. Here we report, to our knowledge, unprecedented levels and novel types of plastid-genome divergence in the bizarre, mushroom-like parasitic plant Balanophora. The miniaturized plastid genome in Balanophora is exceptionally streamlined, has evolved a novel genetic code, and possesses the most A+T-rich protein-coding genes known. These findings extend our understanding of the lower limits of genome complexity and offer exciting opportunities to explore the mutational and selective forces that drive radical genome evolution.
Keywords: parasitic plants, AT-biased base composition, genome reduction, genetic code change, overlapping genes
Abstract
Plastid genomes (plastomes) vary enormously in size and gene content among the many lineages of nonphotosynthetic plants, but key lineages remain unexplored. We therefore investigated plastome sequence and expression in the holoparasitic and morphologically bizarre Balanophoraceae. The two Balanophora plastomes examined are remarkable, exhibiting features rarely if ever seen before in plastomes or in any other genomes. At 15.5 kb in size and with only 19 genes, they are among the most reduced plastomes known. They have no tRNA genes for protein synthesis, a trait found in only three other plastid lineages, and thus Balanophora plastids must import all tRNAs needed for translation. Balanophora plastomes are exceptionally compact, with numerous overlapping genes, highly reduced spacers, loss of all cis-spliced introns, and shrunken protein genes. With A+T contents of 87.8% and 88.4%, the Balanophora genomes are the most AT-rich genomes known save for a single mitochondrial genome that is merely bloated with AT-rich spacer DNA. Most plastid protein genes in Balanophora consist of ≥90% AT, with several between 95% and 98% AT, resulting in the most biased codon usage in any genome described to date. A potential consequence of its radical compositional evolution is the novel genetic code used by Balanophora plastids, in which TAG has been reassigned from stop to tryptophan. Despite its many exceptional properties, the Balanophora plastome must be functional because all examined genes are transcribed, its only intron is correctly trans-spliced, and its protein genes, although highly divergent, are evolving under various degrees of selective constraint.
Parasitic plants, which penetrate their host plant tissues to form a vascular connection to acquire water, carbohydrates, and mineral nutrients, have arisen at least 12 times during angiosperm evolution (1, 2). Although ∼90% of parasitic angiosperms are hemiparasites, ranging from fully to minimally photosynthetic, 10 groups of parasitic angiosperms contain holoparasites, i.e., completely nonphotosynthetic plants. Mycoheterotrophs, the term used for nonphotosynthetic plants that parasitize fungi, have arisen in 10 angiosperm families (3). The many independent lineages of holoparasitic and mycoheterotrophic angiosperms provide numerous test cases to explore the limits and potential outcomes of plastid genome (plastome) reduction in heterotrophic plants and the extent to which parallel evolution of gene content occurs. Indeed, these many independent transitions from autotrophy to heterotrophy show sufficiently similar trends in plastid gene evolution that several similar models have been erected proposing the progressive loss and diversification of different classes of plastid genes (4–9).
The >2,000 sequenced plastomes of photosynthetic angiosperms (10) are highly conserved in size, gene content, and base composition; the great majority are 135–165 kb in size (11), contain 112 or 113 different genes, and are 35–40% G+C (12). In sharp contrast, the much smaller number (∼60) of sequenced plastomes of nonphotosynthetic angiosperms vary substantially in size (11–157 kb), gene content (5–101 genes), and GC base composition (23–40%) (13, 14). Many, if not all, plastid genes encoding proteins involved in photosynthesis, ATP synthesis, and transcription have been lost or become pseudogenes in most examined nonphotosynthetic plants (13, 14). The great majority of the intact plastid genes retained by these nonphotosynthetic plants function solely in protein synthesis; these genes include, depending on the plastome, 2–21 ribosomal protein genes, 2–4 rRNA genes, and 0–30 tRNA genes (13, 14). As opposed to the supporting process of gene expression, plastomes of photosynthetic and nonphotosynthetic angiosperms contain only one to four protein genes thought to be involved in core plastid processes other than photosynthesis and ATP synthesis. One or more of these few genes, plus the dual-functional trnE gene (15, 16), are presumed to serve as the driving force—the raison d’être (15–17)—for retention of the plastome in nonphotosynthetic angiosperms, as well as retention of the hundreds of mostly nuclear genes required for its replication, repair, recombination, and expression (18).
The smallest angiosperm plastome described to date belongs to the endoparasite Pilostyles aethiopica; this 11-kb genome contains only five identified genes, only one of which does not function in protein synthesis (19). In Rafflesia lagascae, another endoparasitic holoparasite, the plastome is thought to have been lost entirely (20), although the possibility that an extremely small and divergent plastome escaped detection cannot be ruled out (9, 19). Extreme plastome reduction has also occurred in several lineages of mycoheterotrophic plants (13), most notably in Sciaphila thaidanica, whose 13-kb genome is stuffed with 20 genes as a result of severe compaction pressure (21), and Thismia tentaculata, whose 16-kb genome contains only 12 genes (22).
Of the various parasitic and mycoheterotrophic lineages of angiosperms for which no plastome sequences are available, the holoparasitic Balanophoraceae are one of the most biologically interesting. This family includes some of the most bizarre plants known, with some species mistaken for fungi because of their fleshy, nongreen, club-shaped inflorescences (23). Probably derived from the largely hemiparasitic Santalales (24), Balanophoraceae contain 17 genera and 44 species (25), with the largest genus being Balanophora (Fig. 1 A and B) with approximately 16 species (24). Balanophora are morphologically extreme because they possess some of the smallest flowers in the world, with as many as 1 million produced per female inflorescence (23, 26). To determine whether the plastome in Balanophoraceae is also bizarre, we sequenced and examined the expression of the plastid sequences of two Balanophora species. Their plastomes are indeed extremely unusual, and in many ways. Most notably, they possess a novel type of genetic-code change, which is also the first case of any code change in land-plant plastomes, set new records for AT-richness and codon-usage bias, and are exceptionally compact, with many overlapping and/or shrunken genes.
Fig. 1.
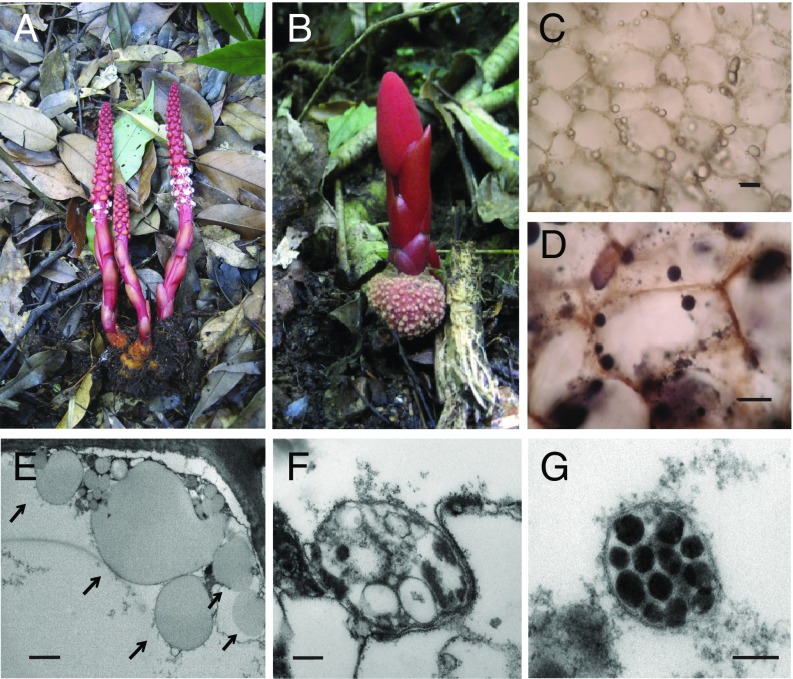
Morphology and microscopy of Balanophora. Male (A) and female (B) plants of B. laxiflora with tubers shown at the base. Light microscopy reveals that tuber cells of B. yakushimensis (unstained in C and stained with Sudan Black in D) contain numerous refractile oil droplets. (Scale bar: 50 µM.) (E) EM shows numerous lipid globules (arrows) distributed along the cell wall of bract cells of B. yakushimensis. (Scale bar: 2 µM.) (F) A plastid-like structure in bract cells of B. laxiflora. (Scale bar: 200 nm.) (G) A plastid-like structure in bract cells of B. laxiflora. (Scale bar: 100 nm.)
Results
Size, Gene Content, and Synteny of Balanophora Plastomes.
We obtained circular plastome assemblies of 15,505 bp for Balanophora laxiflora (Fig. 2) and 15,507 bp for Balanophora reflexa (GenBank accession nos. KX784265 and KX784266, respectively). The near identity in size of the two Balanophora plastomes is the fortuitous result of the accumulation of many indels in each genome that happen to virtually balance out in aggregate length; indeed, all but 2 of their 19 genes differ in length (SI Appendix, Table S1). The Balanophora genomes are the smallest known plastomes except for the 11.3- and 15.2-kb genomes of two holoparasitic species of Pilostyles (19) and the 12.8-kb genome of the mycoheterotroph S. thaidanica (21).
Fig. 2.
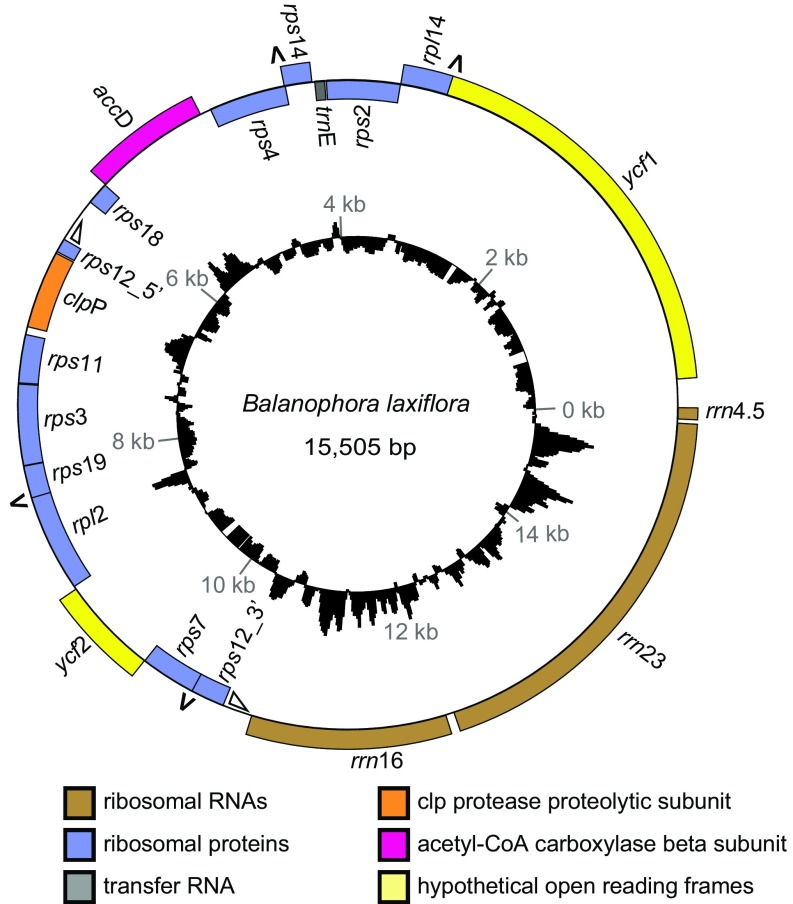
Circular map of the B. laxiflora plastome. Genes shown inside and outside the outer circle are transcribed clockwise and counterclockwise, respectively. Triangles mark the two portions, of undetermined extent, of the trans-spliced intron in rps12. The four pairs of overlapping genes are marked with inverted “V”s. The histogram shows GC content that is greater than (outside the circle) or less than (inside the circle) the plastome average of 12.2% GC.
The two Balanophora plastomes have an identical and highly reduced set of 19 putatively functional genes, consisting of three rRNA genes (rrn16, rrn23, rrn4.5), one tRNA gene (trnE), 11 ribosomal protein genes (rps2, rps3, rps4, rps7, rps11, rps12, rps14, rps18, rps19, rpl2, rpl14), and four protein genes of varying or unknown function (accD, clpP, ycf1, ycf2; Fig. 2 and SI Appendix, Fig. S1A). No traces of any photosynthetic, ATP synthase, RNA polymerase, or splicing factor genes were detected. Intergenic spacer regions in the Balanophora plastomes are exceptionally short (as detailed in the next section) and lack any ORFs of appreciable length, making it unlikely that any protein genes remain unannotated. Although the 4.5S rRNA gene was detected, albeit with difficulty, the 5S gene was not, and only a single tRNA gene was found. However, given the small size of these RNA genes and the extreme divergence and AT-richness of the Balanophora plastomes (as detailed later), we can not rule out the presence of one or a few so-far unrecognizable small RNA genes (only three spacers in both plastomes are large enough to contain a tRNA gene; Fig. 2 and next section). The most likely candidate is the 5S rRNA gene, which, by synteny, would occupy the largest intergenic spacer (between the 4.5S rRNA and ycf1 genes) in both plastomes.
Both Balanophora plastomes have a single tRNA gene (trnE) encoding tRNAGlu(UUC). In most plastid lineages, this tRNA gene is bifunctional, necessary for both protein synthesis and the biosynthesis of tetrapyrroles such as heme and, in photosynthetic plastids, chlorophyll (15, 16). The B. reflexa trnE sequence must be nonfunctional with respect to protein synthesis, as the UUC anticodon is absent (SI Appendix, Fig. S2). Although UUC is present in B. laxiflora, it is shifted by 1 nt within the 7-nt anticodon loop predicted by tRNAscan-SE (27) as a result of a 1-nt deletion in the 5′ portion of the anticodon stem (SI Appendix, Fig. S2). To the best of our knowledge, there is no precedent for an asymmetric location of a functional anticodon within an anticodon loop of a canonical length of 7 nt, even among the incredibly divergent and bizarre tRNA genes found in certain mitochondrial genomes (28–30). The tRNA structure predicted by MiTFi (28) also contains an asymmetrically positioned UUC and, in addition, an anticodon loop that is only 6 nt long, which apparently is also unprecedented for a functional tRNA. Therefore, the B. laxiflora trnE sequence is unlikely to function in protein synthesis. In contrast, it probably still functions in heme biosynthesis, as it is highly conserved (as detailed later) and contains seven of the eight sequence determinants for correct charging of tRNAGlu(UUC) in Escherichia coli that are present in Nicotiana and Schoepfia, a hemiparasitic member of the Santalales (SI Appendix, Fig. S2A) (31). Three of these determinants (UUnA) are canonically present within the anticodon loop (SI Appendix, Fig. S2A). Although the B. reflexa trnE sequence lacks the UUnA motif at its canonical position, the motif is present only a few nucleotides away (SI Appendix, Fig. S2). Therefore, the B. reflexa gene may also still function in heme biosynthesis. This heme-synthesis hypothesis (and likewise that concerning trnE functionality in B. laxiflora) is strongly supported by the fact that trnE is more highly conserved (96% identity, gaps excluded), and also more GC-rich (29%), between the two Balanophora plastomes than any of their 18 other genes (SI Appendix, Tables S1 and S2).
Taking both Balanophora plastomes together, 27 of the 30 protein genes were annotated as starting with ATG, with ATA or ATT annotated as start codons in the other three cases (SI Appendix, Table S3). All protein genes were annotated as terminating with TAA except for rpl2, which appears to terminate with TGA in both species (SI Appendix, Table S3).
The two Balanophora plastomes are colinear in gene order. Moreover, despite being an order of magnitude smaller in size and gene content than virtually all plastomes of photosynthetic land plants (e.g., SI Appendix, Fig. S1A), the two Balanophora plastomes are colinear with those of Schoepfia and Nicotiana (and most other angiosperms) except for the location of a single gene, rpl14 (SI Appendix, Fig. S1B). Unlike the great majority of angiosperm plastomes, those of Balanophora lack a large inverted repeat (Fig. 2 and SI Appendix, Fig. S3).
Extremely Compact Genes and Genomes.
The Balanophora plastomes have unusually high gene densities, with 94.2% and 95.2% of the genome assigned as genic DNA in B. reflexa and B. laxiflora, respectively. This genomic compaction is manifest in two related ways. First, Balanophora has an unusually high percentage of overlapping genes. Plastomes of photosynthetic angiosperms, with approximately 112 or 113 genes, typically have only 3 pairs of overlapping genes (32). All 6 of these genes are involved in photosynthesis and have been lost from Balanophora plastomes. However, with only 15 protein genes, the B. reflexa plastome has 5 pairs of overlapping protein genes, whereas B. laxiflora has 4 (SI Appendix, Table S4). All 9 overlaps are short, between 4 and 15 bp in length. Two of the overlaps are shared between the genomes and thus likely arose in an ancestral Balanophora plastome, whereas the others probably arose since the two diverged from a common ancestor as a result of continued genome downsizing. Second, the intergenic spacers that survive in the genome are extremely short. Excluding its 1 trans-spliced intron, B. laxiflora has 16 intergenic spacers with a median size of only 22 bp (range, 1–219 bp), while B. reflexa has 15 spacers of median size of 42 bp (range, 6–213 bp; SI Appendix, Table S4).
Plastid protein genes in Balanophora are compact too, having lost introns and sustained a net reduction in coding-sequence length. Balanophora has lost both clpP introns, the single rpl2 intron, and intron 2 of rps12, all of which are present and cis-spliced in Schoepfia and most other land-plant plastomes. The only retained intron in Balanophora (intron 1 of rps12) is trans-spliced, a configuration that makes its loss exceptionally difficult and therefore rare. The frequency of indels, especially deletions, is elevated in Balanophora protein genes and has led to a notable shortening of many genes (SI Appendix, Fig. S4 and Table S1). Only 2 of the 15 protein genes are essentially the same size as homologs in the hemiparasitic relative Schoepfia, whereas all others are ≥5% shorter, 9 are ≥10% shorter, and 5 are extremely reduced in length (32–88%; SI Appendix, Table S1).
Extreme AT Base Composition and Codon-Usage Bias.
The Balanophora plastomes are extraordinarily AT-rich, with GC content of 11.6% for B. reflexa and 12.2% for B. laxiflora (Fig. 2). Although their small-subunit and large-subunit rRNA genes are relatively GC-rich (19–24%), most protein genes are even more AT-rich than the genome as a whole, with five or six of them ≤5% GC and ycf2 an astounding 2% GC (Fig. 2 and SI Appendix, Table S1). To put these base compositional biases in perspective, we compared, in four ways, the Balanophora plastomes with 28 of the most AT-rich genomes of plastids, mitochondria, and bacteria (SI Appendix, SI Materials and Methods includes genome-selection criteria). First, the Balanophora plastomes are the most compositionally biased (toward AT or GC) plastid genomes sequenced to date and are surpassed only by the mitochondrial genome of the yeast Nakaseomyces bacillisporus (33), whose 10.9% GC content is largely the result of its high content of extremely AT-rich noncoding spacer DNA (SI Appendix, Figs. S5 and S6 and Table S5). Second, at 8.7% and 8.9% GC, the Balanophora plastid protein genes are the most AT-rich of any gene set analyzed, with the plastome of the malarial parasite Plasmodium falciparum next at 11.0% GC, followed by another apicomplexan (Babesia) at 12.1% (SI Appendix, Table S5). Third, at 1.1% and 1.2% GC, Balanophora also has the most extreme compositional bias at third-position synonymous sites (GC3), with Plasmodium next at 2.1% (SI Appendix, Table S5). Notably, Balanophora plastomes are 10-fold more biased in terms of GC3 than those of the next most AT-rich angiosperm plastomes. Fourth, Balanophora has the most unused codons (20, 21) of all examined genomes (SI Appendix, Tables S5 and S6). Furthermore, in a comparison of fewer taxa (but including the two next most GC3-biased ones; SI Appendix, Fig. S7), the B. laxiflora plastome has the most extreme amino acid bias, with only six amino acids (Asn, Ile, Leu, Lys, Phe, Tyr), each with at least one all-AT codon, comprising 80% of the plastome’s proteome. Thus, by all relevant metrics, the Balanophora plastomes have the most compositionally biased protein genes and proteins of all examined genomes.
Our results fail to support two of the most favored selective explanations for the occurrence of extreme codon-usage bias. First, selection for translationally optimal codons is often seen for highly expressed genes, including in photosynthetic plastomes (34). However, the comparable usage of A and T at third-position synonymous sites for eight of the nine codon families for which such a choice is possible (SI Appendix, Table S6) is inconsistent with translational efficiency. Also, some of the most highly used codons in Balanophora, and land plants in general, are usually translated by the relatively inefficient phenomenon of “superwobbling” (35). Therefore, selection for translational efficiency is unlikely to be a potent force in Balanophora plastomes. The one exception to the A≈T pattern, a preference for Arg-CGT over Arg-CGA (SI Appendix, Table S6), may be because there is only a single tRNA-Arg—with an anticodon of ACG—to decode both codons in other plants (35). Three lines of evidence argue against the hypothesis that the extreme codon-usage bias results from selection driven by nitrogen availability and/or energetic costs (36, 37). Although this selection hypothesis does predict the observed bias in AT over GC, it also predicts a predominance of T over A at third-position synonymous sites, which is clearly not the case in Balanophora (SI Appendix, Table S6). Also, Balanophora is not depleted for either nitrogen-rich or energetically costly-to-synthesize amino acids (SI Appendix, Table S7). Finally, one might expect selective pressure on nitrogen availability and/or energetic costs to operate on all Balanophora genomes, perhaps foremost on its vastly larger nuclear genome, yet the Balanophora nuclear genome possesses only a trivial codon-usage bias compared with the plastome (SI Appendix, Fig. S8).
An Altered Genetic Code in Balanophora Plastomes.
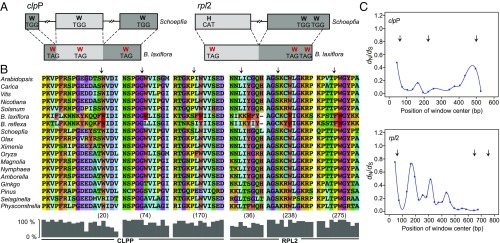
Examination of the 15 Balanophora protein genes revealed 18 internal, in-frame TAG codons in B. laxiflora and 16 in B. reflexa (Fig. 3, Table 1, and SI Appendix, Fig. S4). TAG is, of course, a stop codon in the canonical genetic code, the code that is employed by all land-plant plastomes examined heretofore. In 11 of the 18 cases for B. laxiflora and 12 of 16 for B. reflexa, these TAG codons occur at positions at which TGG (tryptophan in the canonical code) is present in most or all of the diverse photosynthetic land plants in the sequence alignments shown in SI Appendix, Fig. S4. Conversely, there is not a single TGG codon in any Balanophora plastid protein gene. Nine of these 11 and 12 cases of TAG in place of TGG are shared by the two Balanophora species (Table 1). The four partial protein-gene sequences from Balanophora fungosa (as detailed later) contain four in-frame TAG codons, two each in clpP and rpl2. All four are shared with B. laxiflora, and three are shared among all three examined Balanophora species. In both sequenced Balanophora plastomes, all occurrences of TAG are internal, and, conversely, no annotated stop codons are TAG; all are TAA or, for one gene, TGA (SI Appendix, Table S3). Collectively, these observations lead to an inescapable conclusion: a genetic-code change—the reassignment of TAG from stop codon to Trp codon, accompanied by the discontinued use of TGG—occurred in the ancestral Balanophora plastome. Sequencing the plastid proteome of Balanophora would, of course, provide even further evidence for a code change; however, such evidence is well beyond current norms for diagnosis of a genetic-code change (38, 39).
Fig. 3.
Evidence for a novel genetic-code change in Balanophora plastomes. (A) Structure of the clpP and rpl2 genes. The approximate location of six codons diagnostic of a code change are given. Exons are represented by boxes and introns by interrupted lines. (B) Partial alignments of the inferred amino acid sequences of CLPP and RPL2. The histograms indicate the percentage of the 18 sequences in the alignment that share the most common amino acid at each position. (C) Pairwise dN/dS ratios from sliding-window analysis (window size = 90 bp; step size = 30 bp) of the Balanophora clpP and rpl2 genes (SI Appendix, Fig. S9 shows the same analysis of three other Balanophora genes). Arrows mark internal TAG codons present in one or both Balanophora plastomes and inferred to encode W; note that, at five of these six positions, most or all non-Balanophora land plants contain TGG (W in the standard genetic code). The positions of these TAG codons in the complete alignments are shown in SI Appendix, Fig. S4.
Table 1.
In-frame TAG codons in Balanophora plastid genes
| Gene | Species | Positions of in-frame TAG codons in the SI Appendix, Fig. S4 alignments* | Consensus amino acid at Balanophora TAG codons in non-Balanophora sequences |
| accD | B. laxiflora | 242, 331 | W, W |
| accD | B. reflexa | 242, 331 | W, W |
| clpP | B. laxiflora | 20, 74, 170 | W, W, W |
| clpP | B. reflexa | 20 | W |
| rpl2 | B. laxiflora | 36, 238, 275 | Y, W, W |
| rpl2 | B. reflexa | 238, 275 | W, W |
| rpl14 | B. laxiflora | 107 | R |
| rps2 | B. laxiflora | 16, 200 | W, Y |
| rps2 | B. reflexa | 16, 102 | W, W/S |
| rps4 | B. laxiflora | 69 | Gap |
| rps14 | B. laxiflora | 93, 104 | C, W |
| rps14 | B. reflexa | 104 | W |
| ycf1 | B. laxiflora | 947, 1124, 1233, 1240 | Gap, R/K, W, W |
| ycf1 | B. reflexa | 13, 158, 933, 1081, 1233, 1240, 2551 | C/W, Q, H, L/M, W, W, W |
| ycf2 | B. reflexa | 693 | Y |
TAG codons shared by Balanophora species are in bold.
Two alternative hypotheses for the presence of so many in-frame internal TAG codons fail to stand up to scrutiny. One hypothesis is that these codons do in fact serve as premature stop codons, in which case 9 of the 15 protein genes (Table 1) are pseudogenes in one or both Balanophora plastomes. Although tantalizing, especially because holoparasite plastomes often contain pseudogenes with premature stops, the stop-codon hypothesis is rejected for four reasons, as follows. (i) It is entirely inconsistent with sliding-window analysis of the ratios of nonsynonymous substitution rate (dN) to synonymous substitution rate (dS) for the five longest and best-conserved genes for which the two Balanophora species share internal TAG codons (Fig. 3C and SI Appendix, Fig. S9): for all five genes, the signature of strong purifying selection is evident downstream of their internal TAG codons. (ii) As a group, these five genes (accD, clpP, rpl2, rps2, ycf1) show higher levels of purifying selection within Balanophora (mean dN/dS = 0.26, median = 0.14) than the six protein genes that lack TAGs in both plastomes (mean dN/dS = 0.33, median = 0.36; SI Appendix, Table S2). (iii) Given their extreme AT-richness and how much they have diverged since the B. laxiflora/B. reflexa divergence (SI Appendix, Table S2), one would expect these five internal-TAG presumptive pseudogenes to contain frame shifts, yet, as annotated, they have none. (iv) If all internal-TAG genes were pseudogenes, the B. reflexa plastid would be translating only ribosomal protein genes (B. laxiflora would also be translating ycf2). Such a scenario is highly implausible, as it implies maintenance of a translational apparatus merely to synthesize its own components.
The second alternative hypothesis invokes A→G RNA editing to convert the in-frame internal TAG codons to UGG at the mRNA level. However, A→G editing is unknown in land plants, for which plastid editing is exclusively C→U and U→C, and generally infrequent. More importantly, the absence of any RNA editing in the B. laxiflora cDNA sequences (as detailed later), which cover 10 of its 16 internal TAG codons, definitively rejects the editing hypothesis. In summary, there are many highly concordant lines of evidence to support a genetic-code change in the Balanophora plastome and no viable competing hypotheses.
Highly Divergent but Functional Genes.
As seen for certain other holoparasitic angiosperms, especially Hydnora and Pilostyles (9, 19), Balanophora plastid genes are, in aggregate, extremely divergent in sequence relative to a diverse range of photosynthetic land plants (SI Appendix, Fig. S10). Rapid sequence evolution in a 16-gene concatenate (3 genes present in Balanophora were excluded; Materials and Methods) is evident from both the extremely long branch leading to Balanophora and the relatively high divergence between B. laxiflora and B. reflexa. Despite the extreme divergence, phylogenetic analysis placed the Balanophora sequences within the Santalales (SI Appendix, Fig. S10), consistent with an analysis of three nuclear genes, one mitochondrial gene, and three plastid genes from a large number of relevant taxa (24).
When analyzed individually, the 14 protein genes in Fig. 4 show considerable variation within Balanophora in levels of amino acid divergence (56–90% identity, gaps excluded; SI Appendix, Table S2). Because of the highly biased nucleotide composition of these genomes, nucleotide identity is actually higher than amino acid identity for all protein genes, a situation that is rarely observed (SI Appendix, Table S2).
Fig. 4.
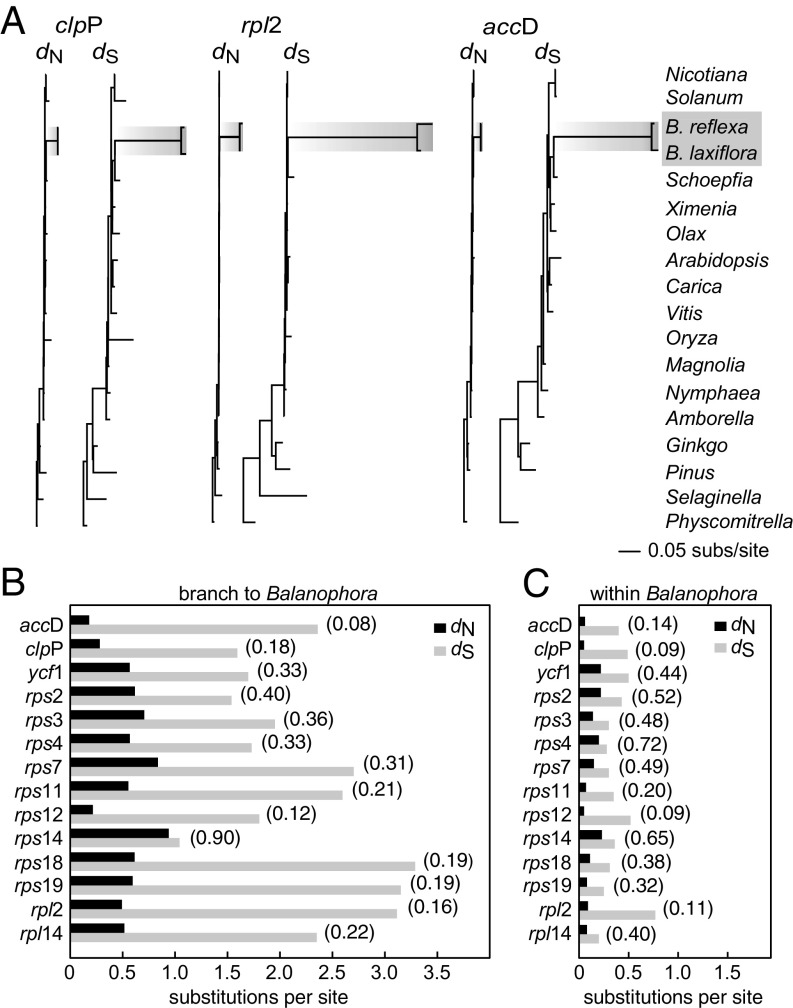
Sequence divergence of plastid genes. (A) Phylograms of nonsynonymous (dN) and synonymous (dS) site divergence for the three indicated genes. (B and C) Levels of dN (in black) and dS (in light gray) on the branch leading to Balanophora (B) and in pairwise comparisons between the two Balanophora plastomes (C). dN/dS ratios are given in parentheses.
To assess mutation and selection pressures acting on the 14 best conserved protein genes of Balanophora, we estimated dS and dN in the context of a broad sampling of land plants. The dS lengths on the branch leading to the common ancestor of the two Balanophora species are far longer than for any other branches in the three dS gene trees shown in Fig. 4A. The long dS branches for these genes, together with the high dS values shown in Fig. 4B for all 14 protein genes, indicate that a high mutation pressure is operating throughout both Balanophora plastomes. Moreover, there is clear evidence of saturation at synonymous sites on the branch leading to the Balanophora common ancestor, with dS > 1.5 for 13 of the 14 protein genes examined and >3.0 for 3 genes (Fig. 4B). The dN/dS ratio on the Balanophora stem-branch is significantly increased (branch test, P < 0.001) compared with other land plants for a concatenated alignment of the 14 genes, indicating relaxed constraint.
There is, however, clear evidence for purifying selection on the Balanophora stem-branch for 13 of the 14 protein genes, for which dN/dS is at most 0.40 (Fig. 4B). Moreover, dN/dS is below 0.20 for 6 genes and barely above it for 2 more. Pairwise comparison between the two Balanophora plastomes reveals that their genes have continued to evolve under purifying selection, albeit with what appears to be a modest overall relaxation of selective constraints (Fig. 4B and SI Appendix, Table S2). In aggregate, the preceding results constitute compelling evidence that Balanophora plastid genomes are functional. Taken individually, they indicate that all 14 genes probably encode functional proteins (a case for functionality of the ultradivergent ycf2 gene is made in SI Appendix, SI Results).
Transcription and Splicing of Balanophora Plastid Genes.
Complementary evidence for functionality comes from cDNA analysis. All 11 B. laxiflora plastid genes selected for RT-PCR amplification showed evidence of transcription, with a very strong correlation between predicted and experimentally determined cDNA product sizes (three examples are provided in SI Appendix, Fig. S11). Importantly, the only plastid intron present—the trans-spliced intron in rps12—was shown to be correctly excised in B. laxiflora because comparison of rps12 gene and cDNA sequences revealed an expected size difference as a result of splicing across sites predicted to generate a contiguous ORF. Finally, a search of the 1,000 Plants (1KP) project database (40) recovered partial transcriptome assemblies for six plastid genes (accD, clpP, rps12, rpl2, rrn16, and rrn23) from B. fungosa, indicating that its plastid genome is also expressed.
A total of 7,261 bp of the 11,165 bp covered by the B. laxiflora RT-PCR products was sequenced. All cDNA sequences are identical to the genome sequence. Therefore, there is no evidence of RNA editing in the B. laxiflora plastid. A lack of editing is not surprising because, in those plants for which plastid editing has been comprehensively determined (41), there are only very few edit sites in the protein genes present in Balanophora plastomes, but it does provide important evidence that the observed internal TAG codons are not altered by editing to a sense codon in the extremely divergent Balanophora plastome.
Abundance of Oil Droplets and Presence of Elaioplasts in Balanophora Tissues.
To investigate if Balanophora, like many parasitic plants (9, 42, 43) and most photosynthetic plants, uses plastids for starch storage, we stained three tissues (tubers, inflorescence stalks, and scale-like leaf remnants known as basal bracts) from Balanophora yakushimensis with iodine to test for the presence of starch grains. No staining was observed. Instead, Sudan Black staining, which tests for lipids, revealed numerous oil bodies in all three tissues (Fig. 1 C and D). These oil droplets (Fig. 1 C–E) could be formed, at least partly, from fatty acids synthesized in plastids, as in photosynthetic plants. To search for plastids, we performed transmission EM on thin sections of bract tissues from B. laxiflora and B. yakushimensis. Fig. 1 F and G show the presence of organelles that are ovoid/spherical in shape with a bounding double membrane. These organelles are within the size range for plastids and contain numerous conspicuous internal globules that appear to possess a bounding membrane, as shown for elaioplasts in anthers of Arabidopsis (44).
Discussion
Radical Evolution of the Balanophora Plastome.
Radical evolution is practically the norm for nonphotosynthetic plastomes (14), but the Balanophora plastid genome sets new benchmarks in several respects. Although many lineages of holoparasitic and fully mycoheterotrophic plants share with Balanophora a highly reduced plastid genome and gene set, as well as highly elevated substitution rates (9, 13, 19, 22), only Pilostyles (19) must also import all tRNAs for plastid protein synthesis and only S. thaidanica (21) has a comparably compact genome. However, none approach Balanophora in AT-richness and codon-usage bias or have evolved a noncanonical genetic code, much less a novel one (https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi).
The radicalism of Balanophora plastomes is, in several respects, reminiscent of that found in “apicoplasts” of Apicomplexans, which are also extremely AT-rich and codon-biased, highly compact (including tiny spacers and many overlapping genes), and highly divergent in sequence, with a genetic-code change in a large subgroup of these pernicious parasites (45, 46). However, apicoplast genomes differ in two important ways from the highly reduced plastomes of Balanophora and many other lineages of nonphotosynthetic plants. Apicoplasts contain many more ribosomal protein and tRNA genes (approximately 25 each) and show very little gene-content variation considering their ancient origin approximately 600–800 Mya (47). These differences may largely reflect a highly reduced rate of plastid-to-nucleus gene transfer in apicomplexans and other organisms with a single plastid per cell compared with land plants and other multiplastidic organisms (16).
In the following sections, we place the most exceptional features of the Balanophora plastid genome in perspective and also discuss the function of this most unusual genome and how its ultimate fate may, ironically, be constrained by two of these features.
Novel Genetic Code in Balanophora Plastid DNA.
The GC content of Balanophora plastomes is extremely reduced, to just 11.6% and 12.2%, making them the most AT-rich plastid genomes found to date. Such highly biased base composition and the consequent bias in codon usage may be associated with codon reassignment in Balanophora, as has been hypothesized generally (48). As our data show, there is compelling evidence that tryptophan is encoded by TAG instead of TGG in Balanophora plastomes, with termination restricted to TAA (mostly) or TGA (rarely).
Deviations from the canonical genetic code have been found in many lineages, especially in mitochondrial genomes (49). These deviations include reassignments of each of the three standard stop codons to sense codons. Most commonly, TGA has been reassigned from stop to tryptophan codon; this reassignment has been reported in many mitochondrial lineages (50), two bacterial lineages (51, 52), and two plastid lineages [a subset of apicomplexans (53, 54) and Chromera velia, a photosynthetic relative of apicomplexans (55)]. Also, two unrelated green algal plastid genomes were recently reported to use TGA as stop and sense codons (56, 57). TAG occasionally encodes leucine, tyrosine, or glutamic acid (58, 59), but its use for tryptophan is a novel code variant that makes the Balanophora plastome unique. The Balanophora code change is also the first code change of any type discovered in the >2,000 sequenced plastomes of land plants.
Reassignment of TAG from stop to a tryptophan codon in Balanophora may be explained by the codon-capture hypothesis (60). The first prediction under the codon-capture hypothesis is loss of all TAG stops by mutation to TAA stops in response to increasing AT bias of the genome. Duplication of the gene for plastid tRNATrp would allow one copy to mutate such that its product recognizes UAG. Recognition of UAG would then allow for mutation of TGG codons to TAG, with both deciphered as tryptophan. Because of increasing AT content, most or all TGG codons would eventually be replaced by TAG. Indeed, UGG is probably no longer deciphered as tryptophan in Balanophora, as no TGG codons were found in the 15 protein genes of either plastome. Other AT-rich holoparasite genomes could experience codon capture of TAG (or TGA) as well. For example, there are no TAG- or TGA-encoded stops for the 16 protein genes in the Cytinus plastome (SI Appendix, Table S8) (61). In Cytinus, TGG still codes for tryptophan (SI Appendix, SI Results and Table S8), but the requisite antecedent condition appears to be in place for a codon reassignment. Note that, for Balanophora plastids, the codon-capture hypothesis also requires the loss of release factor pRF1 binding to UAG. It is also possible that an ancestral UAG codon was read in two different ways as postulated under the ambiguous-translation hypothesis (62).
The mitochondrial and nuclear genomes of Balanophora still use UGG for Trp and UAG as a stop codon (SI Appendix, Fig. S8). We presume that the plastid tRNATrp and accompanying tryptophanyl-tRNA synthetase that charge it are nuclear encoded, in which case a complicated translational scenario must have arisen during the evolution of the novel use of UAG for Trp in Balanophora. Specifically, if plastid-targeted tRNATrp were to be charged in the cytosol, read-through of the UAG stop codon of nuclear transcripts could occur and be deleterious. Therefore, we posit that a novel tRNATrp that binds to UAG is targeted solely to the plastid and can be charged only there; we call this proposition the “plastid-specific tRNATrp(CUA)” scenario. Perhaps the simplest possibility is that a nuclear copy of the ancestrally plastid-encoded tRNATrp(CCA) evolved a CUA anticodon that allowed its product to bind to UAG. A CCA-to-CUA anticodon mutation has been shown to allow for decoding of UAG as Trp in experimental systems (63). Further complexity arises because the same tryptophanyl-tRNA synthetase is dual-targeted to the plastid and mitochondrion in most plants (64). Therefore, the above scenario predicts that Balanophora has a unique plastid-targeted tryptophanyl-tRNA synthetase that charges the putative plastid-specific tRNATrp(CUA). Because tRNA anticodons are usually important specificity determinants for these enzymes (31, 65), the novel tryptophanyl-tRNA synthetase would likely have evolved to bind and charge the derived tRNATrp and/or additional mutations occurred to the novel trnW to allow for proper charging with tryptophan. Unfortunately, no Balanophora assembly containing a plastid-like trnW (or any other plastid tRNA gene) could be found by using BLAST against the 1KP database (34) to shed light on these hypotheses. One partial Balanophora tryptophanyl-tRNA synthetase assembly was found by BLAST analyses, but it appears to be orthologous to the cytosolic protein of other plants and, as such, would not be predicted to participate in plastid tRNA charging. Deeper transcriptome or genome sequencing has the potential to uncover the candidate translational components under the intriguingly complicated plastid-specific-tRNATrp(CUA) scenario. Other mechanisms could also allow for decoding of UAG as Trp in Balanophora, such as read-through by plastid ribosomes using a “near-cognate” tRNA (a tRNA that can pair with stop codons at two of the three positions of a codon–anticodon sequence); such read-through has been shown to occur in eukaryotes (66). However, given that the plastid-encoded ribosomal sequences are so divergent in Balanophora, it is difficult to extrapolate from other studies.
Stop-codon reassignment is the easiest type of code change to recognize (39, 49), and therefore we cannot rule out the possibility of additional reassignments in these two Balanophora plastomes. In addition, if other Balanophoraceae plastomes are also extremely AT-rich, they may have traveled a different pathway of genetic-code evolution, e.g., instead of the novel TAG-for-Trp change, they may have sustained the relatively common TGA-for-Trp change.
Extraordinary AT Content and Codon-Usage Bias.
At 91% AT in coding regions, and 99% AT at third-position synonymous sites, the Balanophora plastome is, in these respects, the most extreme genome known in any organism or genetic compartment. Because nuclear and mitochondrial genes in Balanophora possess relatively modest codon-usage bias and AT richness (SI Appendix, Fig. S8), the forces responsible for the extreme plastid biases are presumably compartment-specific. However, it may nonetheless be revealing to fully examine the Balanophora mitochondrial genome, as angiosperm plastids and mitochondria share a number of dual-targeted nuclear genes for nucleotide metabolism and DNA replication, recombination, and repair (67).
Codon-usage bias is generally explained by some combination of neutral mutational forces and natural selection (68). Mutational forces are thought to involve nucleotide substitution biases (most often, G/C-to-A/T transition biases) caused by defects in DNA replication and/or repair. When codon-usage bias occurs across all genes and all codon families and comparably on A or T (or G or C) at third-position synonymous sites, and when compositional bias occurs across the entire genome, these biases are best explained by a mutation/drift model (68). These patterns are clearly evident in Balanophora plastomes, and thus their unprecedented codon-usage bias is probably driven by a genome-wide AT mutation pressure, and possibly also AT-biased gene conversion (69), operating largely free of natural selection (70, 71). Although we cannot rule out some contribution from selective forces, a number of which have been considered in the literature (36, 37, 68), the findings presented in Results fail to support two of the most widely documented adaptive explanations, selection for translation efficiency and selection driven by nitrogen availability and/or energetic costs.
Balanophora plastomes exhibit numerous indels, principally deletions, in both coding and noncoding regions, as has been shown to be correlated with AT mutation bias in the P. falciparum nuclear genome (72). Furthermore, most spontaneous plastome mutations in the one angiosperm examined are indels occurring by DNA replication slippage at short, often AT-rich repetitive sequences (73). We can thus imagine a synergistic spiral in Balanophora plastomes, with a Balanophora-specific AT-mutation pressure driving ever higher levels of replication slippage-induced indels, and these indels, depending on their context, driving AT content even higher. On top of these forces, there is the evident pressure toward genome streamlining in Balanophora plastid DNA (as detailed in the next section), as well as a highly elevated rate of synonymous substitutions. At their zenith, these forces have combined, in the weakly selected ycf2 gene, to create a barely recognizable form of the gene in Balanophora, one that has been whittled by countless deletions to one eighth its normally large size (approximately 6 kb), and which is nearly 98% AT across all codon positions (detailed in SI Appendix, SI Results and Figs. S4, S12, and S13). The extraordinary divergence—in length, in primary sequence, and in base composition and codon usage—of ycf2 and, to a somewhat lesser extent, several other plastid protein genes in Balanophora, may make them useful models to elucidate the limits of overall change and the nature of specific alterations that these proteins can sustain and still be functional.
Extreme Compaction of the Balanophora Plastome.
The high fraction of plastid gene overlaps in Balanophora and their recent origin (most are species-specific)—together with the highly reduced lengths of the intergenic spacers that remain, the loss of all cis-spliced introns, and the overall reduction in the size of plastid-encoded proteins—speak to an exceptionally high pressure to minimize genome size in Balanophora plastids. We hypothesize that the intense streamlining in Balanophora is, as with its AT content and codon-usage bias, driven predominantly by neutral, runaway mutational forces, in this case by exceptionally high deletion rates.
With the glaring exception of the bloated, spacer-rich plastome of the nonphotosynthetic alga Polytoma uvella (74), there is a general trend toward compaction of nonphotosynthetic plastomes relative to their photosynthetic relatives. However, among the many independent lineages of nonphotosynthetic land-plant plastomes examined to date, only a single genome, that of S. thaidanica (21), approaches the Balanophora plastome in extent of compaction. Moreover, looking across all plastid genomes, the Balanophora situation may be the most extreme case known in terms of percentage of overlapping genes and spacer size, rivaled only by the plastomes of apicomplexan parasites, the green algal parasite Helicosporidium spp., and the photosynthetic red alga Cyanidioschyzon merolae (SI Appendix, Table S9). Data on protein size are unavailable for most of the highly compact plastomes analyzed in SI Appendix, Table S9, and therefore an outstanding question is whether, as in Balanophora and Sciaphila, genome compaction pressures generally lead to shrinkage of plastid-encoded proteins.
Most highly compact lineages of plastomes are, unlike Balanophora and apicomplexans, not particularly AT-rich in base composition (SI Appendix, Table S9). Therefore, the forces leading to genome compaction are probably largely distinct from those leading to AT-richness, although whether distinct forces are at play in Balanophora is an open question. In contrast, as noted here earlier, there may be a connection between AT-richness and genetic code changes in Balanophora and apicomplexans.
Function of the Balanophora Plastome.
The Balanophora plastome is clearly still functional, as its 11 examined genes are all transcribed, its only intron is correctly spliced, most if not all of its 15 protein genes are evolving under selective constraint, and none of these highly divergent genes are riddled with TAA stop codons as would be the case if they were nonfunctional.
Although 14 of the 19 genes in the Balanophora plastome are devoted to protein synthesis, its most interesting translational feature is what it lacks, which is any tRNA genes capable of serving in protein synthesis. Balanophora plastids must therefore import the full suite of tRNAs required for plastid protein synthesis. Import of some or most plastid tRNAs has been postulated for many nonphotosynthetic plants (9, 13, 17), but import of all tRNAs for plastid protein synthesis need be invoked only for one other land plant, the holoparasite Pilostyles (19), the dinoflagellate Symbiodinium (75, 76), and the green alga Boodlea (56). A key difference between the plastomes of Balanophora and these other three lineages is that only Balanophora has retained trnE, but apparently solely on account of its function in heme biosynthesis. To our knowledge, such retention is the first evidence that these two functions of plastid trnE can be effectively separated, making Balanophora an important evolutionary genetic mutant in this respect. Furthermore, if we are correct that Balanophora trnE still functions in heme synthesis, it nicely illustrates some of the divergence that this gene tolerates when selection on protein synthesis has been lost (SI Appendix, Fig. S2). These considerations emphasize the importance of confirming, experimentally and by sampling more Balanophoraceae species, that Balanophora trnE does indeed function in heme biosynthesis and is not merely a very recently evolved pseudogene in one or both Balanophora species.
Functions for three of the five raison d’être (i.e., nontranslational) genes in Balanophora plastomes are known: clpP encodes part of the Clp protease complex involved in protein degradation and import (77); accD encodes a subunit of acetyl-CoA carboxylase (ACCase), which is necessary for fatty-acid biosynthesis in most plants (78); and, as already emphasized, trnE plays an essential role in heme biosynthesis (15, 16). The function of ycf2 is unknown, whereas ycf1 may have multiple roles; it has been suggested to be involved in thylakoid membrane biogenesis in photosynthetic plants, plastid protein import, and ACCase assembly (79). The presence in Balanophora of an accD gene evolving under strong purifying selection and the potential role of ycf1 in ACCase assembly suggest that one of the primary functions of its plastome is biosynthesis of lipids. A key plastome role in lipid synthesis fits with the unusual abundance of oil/lipid droplets in Balanophora tissues, at least some of which appear to be located within plastids (Fig. 1).
Fate of the Balanophora Plastome.
Plastome loss in the context of plastid retention has recently been revealed in several lineages of protists, predominantly dinoflagellates and other Myzozoans in which loss was triggered by transfer of the last remaining raison d’être plastid gene to the nucleus (80–82). The holoparasitic angiosperm Rafflesia may have also lost its plastome (20), although, if so, this loss might be the consequence of loss of the plastid itself (83). Close to extinction are the extremely gene-reduced plastomes of the holoparasite Pilostyles and the mycoheterotrophs Thismia and S. thaidanica, which appear to contain only a single raison d’être gene, accD (19, 21, 22). Loss of these genomes sparked by accD transfer to the nucleus would hardly be surprising given the gene’s functional transfer to the nucleus in multiple lineages of seed plants (84–86).
Ironically, two of the most notable features of the Balanophora plastome likely preclude its loss via gene transfer to the nucleus. The novel genetic code and extreme AT-richness make efficient translation of full-length protein in the cytosol unlikely. The presence of even a single TAG/Trp codon in a plastid gene would result in a premature translation product upon transfer to the nucleus and gain of expression. Most of the four raison d’être protein genes in Balanophora plastomes contain multiple TAG/Trp codons; for a transferred copy of such a gene to become functional, all of its TAG codons would have to revert to TGG before the copy sustains any disabling mutations. In contrast to the extreme codon-usage bias of plastid genes in Balanophora, its nuclear genes have little bias (SI Appendix, Fig. S8); this disparity may render the Balanophora cytoplasm poorly suited to efficient translation of plastid genes transferred to the nucleus (81, 82, 87–89). In combination, these two standout features of Balanophora plastid DNA may permanently lock in some of its remaining genes, and thus the genome itself.
Prospectus.
The discovery of radical plastome evolution in Balanophora should hasten exploration of the 16 other genera of Balanophoraceae. Balanophora is one of the “fast-evolving” lineages in the family (24). Slowly evolving lineages could reveal important antecedent conditions to the genetic-code switch, the increase in AT content, and other unusual features of the Balanophora plastome. Other fast-evolving lineages should be investigated too, for features potentially shared—through convergence or common ancestry—with the divergent plastomes reported here, for new categories of extreme plastome evolution, or for even more radical cases of those already identified in Balanophora, such as further departures from the canonical genetic code.
Materials and Methods
Genome Assembly and Validation.
A plastid DNA preparation was prepared from B. laxiflora inflorescences as in ref. 90, and a library was generated by using 250-bp paired-end Illumina MiSeq reads with 350–400-bp fragment sizes. Total DNA was prepared from B. reflexa as in ref. 91, and a library was constructed by using 100-bp paired-end Illumina HiSeq2000 reads with 800-bp fragment sizes. Totals of 4.6 Gb and 8.1 Gb of raw sequence reads were obtained from B. laxiflora and B. reflexa, respectively. These reads were trimmed to the first position at the 5′ end that had a quality score of <20. Reads of <50 bp in length after trimming were discarded.
For B. laxiflora, de novo assemblies were obtained by using Velvet version 1.2.07 (92) and CLC Assembly cell 4.2.2 (CLC Bio). Based on optimization tests, assembly parameters for Velvet were set to k = 165 and expected coverage set to auto, with minimum contig length of 200. Eighty-nine Velvet contigs that used 3.2% of input reads were obtained with half of the total assembly in contigs of at least 2,155 bp and a maximum contig size of 14,160 bp. BLASTn and BLASTx searches, using both default settings and e-value = 1e−5, of all 89 contigs against the National Center for Biotechnology Information nonredundant (NCBI nr) database identified four contigs of total length 12,001 bp that contained plastid genes. These four contigs were used to build an initial assembly that was improved iteratively by using PAGIT (93). In each iteration, the raw reads were mapped to the contigs by using BWA version 0.6.2 (94), and polymorphisms were checked by IGV (95) and corrected by using SAMtools (96). The corrected and extended contigs were then BLASTed against each other and joined if their ends overlapped via paired-end reads. This process was repeated until a final, circular plastome assembly of 15,505 bp was obtained. This assembly has an average of 2,946-fold coverage of paired-end reads with a mapping quality of 60, with 843-fold coverage in the region of lowest coverage (SI Appendix, Fig. S14A).
CLC assembly of the B. laxiflora cleaned reads used default settings and produced 284,561 contigs that used 88.5% of the reads, with half of the total assembly in contigs of at least 422 bp and a maximum contig size of 43,002 bp. All contigs were used to search a set of 624 plastomes by using BLASTn (e-value = 1e−10) and BLASTx (e-value = 1e−10). This search yielded only four contigs with plastid genes (SI Appendix, Fig. S14B). The largest contig (15,505 bp in size, circular in structure, and 2,868-fold in coverage) covered the full length of the Velvet assembly, whereas the three small contigs (0.4–2.2 kb) have little (<100 bp) or no similarity to the 15.5-kb contig and less sequence depth (3–192-fold coverage). These three contigs likely represent nuclear or mitochondrial integrants and were not used for further analysis.
The plastome assembly of B. laxiflora was validated by PCR amplification of total DNA. The entire B. laxiflora plastome was covered by 13 large PCR products (1,446–6,868 bp in predicted size; mean = 3,814 bp; median = 3,548 bp) with substantial overlap and of the sizes expected according to the assembly. PCR was performed using DreamTaq Green PCR Master Mix (Thermo Fisher Scientific) and the primers and conditions in SI Appendix, Tables S10A and S11A.
We mapped the cleaned B. reflexa reads onto the B. laxiflora assembly by using BWA version 0.6.2 (94). B. reflexa plastid reads thus identified were used to design primers in an iterative manner, i.e., to primer walk, in conjunction with Sanger sequencing of the PCR products or the cloned fragments (SI Appendix, Tables S10B and S11B include primers and PCR conditions). This primer walk produced a single circular assembly of 15,507 bp that is colinear with the B. laxiflora plastome. The final assembly included many additional Illumina reads that were identified as plastomic by mapping the total B. reflexa reads onto the primer-walk assembly.
Genome Annotation and Mapping.
The plastome assemblies were analyzed by using ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to detect ORFs that were then searched by Position-Specific Iterated (PSI)-BLAST (97) for gene prediction. The B. laxiflora plastome was also analyzed by the plastome gene-prediction program DOGMA (98) using sensitive parameters (e-value = 1e−5 and match-identity threshold of 25%). Start and stop codons were assigned based on proximity to those annotated in the conserved regions found by BLAST. Transfer RNA genes were identified by tRNAscan-SE (27) with a threshold cove score of ≥20 bits and MiTFi (28) with a BLAST e-value of ≥1e−2. Ribosomal RNA genes were identified by RNAmmer (99) and annotated via alignment with homologs from other plants. All intergenic sequences were translated in all six potential reading frames and searched by PSI-BLAST (e-value = 1e−5) and BLASTn (e-value = 1e−5) against the NCBI nr database to make sure all potential protein-genes were annotated. The circular map of the B. laxiflora plastome was produced by GenomeVx (100). The GC-content plot was created by DNAPlotter (101) with a window size of 100 bp and step size of 20 bp.
GC Content, Codon Usage, and Amino Acid Usage.
CodonW 1.4 (codonw.sourceforge.net/index.html) was used to analyze GC content. Codon and amino acid usage was determined by using ENCprime (102) with genome-appropriate translation tables. Thirty taxa were selected for the gene-by-gene GC-content analyses of SI Appendix, Fig. S5 (SI Appendix, Table S12 provides full names and accession numbers, and SI Appendix, SI Materials and Methods details taxon-selection criteria). To examine nuclear codon usage, we collected from GenBank the five available nuclear protein-gene sequences for B. fungosa and randomly chose 400 nuclear cDNA sequences from the B. fungosa transcriptome assemblies of the 1KP project (40). These sequences with aligned with orthologs from 10 other angiosperms by using MAFFT (103). Maximum-likelihood gene trees were then generated by using RAxML version 8.1.17 (104) with the generalized time-reversible + proportion of invariable sites + gamma distribution (GTR+I+G) model. The 56 cDNA sequences (27,682 codons) that grouped with other Santalales in the gene trees and the aforementioned five genes were used for codon-usage analyses. To examine mitochondrial codon usage, we constructed full-length assemblies for 10 B. laxiflora mitochondrial genes from the reads used to assemble its plastid genome. GenBank accession numbers for the mitochondrial and nuclear sequences are given in SI Appendix, SI Materials and Methods.
Phylogenomic Analysis.
Maximum-likelihood analysis of both Balanophora species, 15 other vascular plants, and the moss Physcomitrella was performed by using RAxML version 8.1.17 (104) with the GTR+I+G model and 1,000 bootstrap replicates (SI Appendix, Table S12 provides accession numbers). Fourteen protein genes and the small- and large-subunit rRNA genes were clustered into ortholog groups by using OrthoMCL (105). The protein genes were aligned based on amino acid alignments constructed by using MAFFT (103) with manual adjustments. The rRNA genes were aligned by using MUSCLE version 3.8.31 (106) with default settings. A concatenate of the 16 genes, with all codon positions included for the protein genes, was used to infer a maximum-likelihood tree. Three genes (rrn4.5, trnE, and ycf2) were excluded from the phylogenomic analysis and the rate analyses described in Evolutionary Rate Estimation. The RNA genes were excluded because of their short lengths, and ycf2 was excluded because of its high sequence divergence, its extraordinary approximately eightfold reduction in size, and its problematic alignment. A detailed treatment of the ycf2 issues and why we are nonetheless confident that the Balanophora ORF we have annotated as ycf2 is functional and is in fact ycf2 is provided in SI Appendix, SI Results and Figs. S4, S12, and S13.
Evolutionary Rate Estimation.
We used codeml in the PAML version 4.8 package (107) to estimate dN and dS by using the tree topology of SI Appendix, Fig. S10. Gapped regions were excluded by using cleandata = 1, and codon frequencies were set to model F3 X 4. To test if the Balanophora clade has a different level of selective constraint than other land plants, the method described by Yang (108) was used to estimate dN/dS. A null model (H0; branch model = 0), in which one dN/dS ratio was fixed across land plants, was compared with an alternative model (HA; branch model = 2) in which the Balanophora clade was allowed to have a different dN/dS ratio. Likelihood-ratio tests were used to test if the HA model was a significant improvement over the null model. In addition, to compare dN and dS between Balanophora and Schoepfia, we allowed three sets of branches to have different dN/dS values, including the Balanophora branches, the Schoepfia branch, and all remaining branches. PAML’s yn00 program (107) was used to estimate dN and dS within Balanophora. To detect variation in selective constraints across clpP and rpl2, we calculated dN/dS by using a sliding-window approach. Sequences with a window length of 90 nt and a step size of 30 nt were generated by using a custom Perl script, and pairwise dN/dS ratios were calculated by using the yn00 program. All extreme values of dS (i.e., dS < 0.01 or dS = 99) were excluded from the sliding-window analyses shown in Fig. 3C. These extreme values correspond to sliding-window midpoints of 285, 315, and 465 bp for clpP and 105, 615, 705, and 735 bp for rpl2.
Transcript Analysis and Microscopy.
Transcript analysis and microscopy procedures followed standard protocols as described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Wann-Neng Jane and Dr. Ern-Ming Lai for help with EM; Technology Commons, College of Life Science, National Taiwan University for carrying out the B. laxiflora genome sequencing; Rimi Repin and Dr. Jamili Nais (Sabah Parks) for permission to collect Balanophora material from Sabah, Malaysia; Dr. Eva-Mari Aro for expert advice; and the three reviewers for their critical and close reading of the manuscript. This project was supported by Ministry of Science and Technology (MOST), Taiwan, Research Grant 106-2311-B-845-001-MY3 (to H.-J.S.) and US National Science Foundation Awards IOS 1027529 (to J.D.P.) and IOS 1238057 (to C.W.d.).
Footnotes
Conflict of interest statement: E.K.W. and C.W.d. are coauthors on a 2018 paper with reviewer I.D.S. This paper had 42 authors with E.K.W., C.W.d., and I.D.S. all middle authors. E.K.W. and C.W.d. had no significant interaction with I.D.S. on the project or paper.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KX784265, KX784266, and MK144465–MK144474).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816822116/-/DCSupplemental.
References
- 1.Barkman TJ, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Leake JR. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994;127:171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x. [DOI] [PubMed] [Google Scholar]
- 4.Krause K. From chloroplasts to “cryptic” plastids: Evolution of plastid genomes in parasitic plants. Curr Genet. 2008;54:111–121. doi: 10.1007/s00294-008-0208-8. [DOI] [PubMed] [Google Scholar]
- 5.Barrett CF, Davis JI. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 2012;99:1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- 6.Wicke S, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett CF, et al. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 2014;31:3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- 8.Wicke S, et al. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci USA. 2016;113:9045–9050. doi: 10.1073/pnas.1607576113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naumann J, et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae) Genome Biol Evol. 2016;8:345–363. doi: 10.1093/gbe/evv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mower JP, Vickrey TL. Structural diversity among plastid genomes of land plants. Adv Bot Res. 2018;85:263–292. [Google Scholar]
- 11.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Z, et al. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: Implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 2006;6:77. doi: 10.1186/1471-2148-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham SW, Lam VK, Merckx VS. Plastomes on the edge: The evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 2017;214:48–55. doi: 10.1111/nph.14398. [DOI] [PubMed] [Google Scholar]
- 14.Wicke S, Naumann J. Molecular evolution of plastid genomes in parasitic flowering plants. Adv Bot Res. 2018;85:315–347. [Google Scholar]
- 15.Howe CJ, Smith A. Plants without chlorophyll. Nature. 1991;349:109. [Google Scholar]
- 16.Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiura M. Plastid mRNA translation. Methods Mol Biol. 2014;1132:73–91. doi: 10.1007/978-1-62703-995-6_4. [DOI] [PubMed] [Google Scholar]
- 19.Bellot S, Renner SS. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol Evol. 2015;8:189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molina J, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol Biol Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen G, Zervas A, Pedersen HAE, Seberg O. Genome reports: Contracted genes and dwarfed plastome in mycoheterotrophic Sciaphila thaidanica (Triuridaceae, Pandanales) Genome Biol Evol. 2018;10:976–981. doi: 10.1093/gbe/evy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim GS, Barrett CF, Pang CC, Davis JI. Drastic reduction of plastome size in the mycoheterotrophic Thismia tentaculata relative to that of its autotrophic relative Tacca chantrieri. Am J Bot. 2016;103:1129–1137. doi: 10.3732/ajb.1600042. [DOI] [PubMed] [Google Scholar]
- 23.Kuijt J. The Biology of Parasitic Flowering Plants. Univ California Press; Berkeley: 1969. [Google Scholar]
- 24.Su H-J, Hu J-M, Anderson FE, Der JP, Nickrent DL. Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon. 2015;64:491–506. [Google Scholar]
- 25.Nickrent DL, Der JP, Anderson FE. Discovery of the photosynthetic relatives of the “Maltese mushroom” Cynomorium. BMC Evol Biol. 2005;5:38. doi: 10.1186/1471-2148-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen B. The genus Balanophora, a taxonomic monogragh. Dansk Botanisk Arkiv. 1972;28:1–188. [Google Scholar]
- 27.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jühling F, et al. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012;40:2833–2845. doi: 10.1093/nar/gkr1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salinas-Giegé T, Giegé R, Giegé P. tRNA biology in mitochondria. Int J Mol Sci. 2015;16:4518–4559. doi: 10.3390/ijms16034518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang BF, Lavrov D, Beck N, Steinberg SV. Mitochondrial tRNA structure, identity, and evolution of the genetic code. In: Bullerwell CE, editor. Organelle Genetics. Springer; Berlin: 2012. pp. 431–474. [Google Scholar]
- 31.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nickelsen J, Bohne A-V, Westhoff P. Chloroplast gene expression—translation. In: Theg S, Wollman F, editors. Plastid Biology. Springer; New York: 2014. pp. 49–78. [Google Scholar]
- 33.Bouchier C, Ma L, Créno S, Dujon B, Fairhead C. Complete mitochondrial genome sequences of three Nakaseomyces species reveal invasion by palindromic GC clusters and considerable size expansion. FEMS Yeast Res. 2009;9:1283–1292. doi: 10.1111/j.1567-1364.2009.00551.x. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki H, Morton BR. Codon adaptation of plastid genes. PLoS One. 2016;11:e0154306. doi: 10.1371/journal.pone.0154306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alkatib S, et al. The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet. 2012;8:e1003076. doi: 10.1371/journal.pgen.1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen WH, Lu G, Bork P, Hu S, Lercher MJ. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat Commun. 2016;7:11334. doi: 10.1038/ncomms11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seward EA, Kelly S. Dietary nitrogen alters codon bias and genome composition in parasitic microorganisms. Genome Biol. 2016;17:226. doi: 10.1186/s13059-016-1087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanova NN, et al. Stop codon reassignments in the wild. Science. 2014;344:909–913. doi: 10.1126/science.1250691. [DOI] [PubMed] [Google Scholar]
- 39.Keeling PJ. Genomics: Evolution of the genetic code. Curr Biol. 2016;26:R851–R853. doi: 10.1016/j.cub.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Matasci N, et al. Data access for the 1,000 plants (1KP) project. Gigascience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tillich M, Lehwark P, Morton BR, Maier UG. The evolution of chloroplast RNA editing. Mol Biol Evol. 2006;23:1912–1921. doi: 10.1093/molbev/msl054. [DOI] [PubMed] [Google Scholar]
- 42.Singh M, Singh DV, Misra PC, Tewari KK, Krishnan PS. Biochemical aspects of parasitism by the angiosperm parasites: Starch accumulation. Physiol Plant. 1968;21:525–538. [Google Scholar]
- 43.Walsh MA, Rechel EA, Popovich TM. Observations on plastid fine-structure in the holoparasitic angiosperm Epifagus virginiana. Am J Bot. 1980;67:833–837. [Google Scholar]
- 44.Suzuki T, et al. Development and disintegration of tapetum-specific lipid-accumulating organelles, elaioplasts and tapetosomes, in Arabidopsis thaliana and Brassica napus. Plant Sci. 2013;207:25–36. doi: 10.1016/j.plantsci.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Cai X, Fuller AL, McDougald LR, Zhu G. Apicoplast genome of the coccidian Eimeria tenella. Gene. 2003;321:39–46. doi: 10.1016/j.gene.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Wilson RJM, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- 47.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson GE, Kurland CG. An extreme codon preference strategy: Codon reassignment. Mol Biol Evol. 1991;8:530–544. doi: 10.1093/oxfordjournals.molbev.a040666. [DOI] [PubMed] [Google Scholar]
- 49.Knight RD, Freeland SJ, Landweber LF. Rewiring the keyboard: Evolvability of the genetic code. Nat Rev Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 50.Burger G, Lang BF. Parallels in genome evolution in mitochondria and bacterial symbionts. IUBMB Life. 2003;55:205–212. doi: 10.1080/1521654031000137380. [DOI] [PubMed] [Google Scholar]
- 51.Yamao F, et al. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci USA. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lang-Unnasch N, Aiello DP. Sequence evidence for an altered genetic code in the Neospora caninum plastid. Int J Parasitol. 1999;29:1557–1562. doi: 10.1016/s0020-7519(99)00119-8. [DOI] [PubMed] [Google Scholar]
- 54.Tang K, et al. Genetic similarities between Cyclospora cayetanensis and cecum-infecting avian Eimeria spp. in apicoplast and mitochondrial genomes. Parasit Vectors. 2015;8:358. doi: 10.1186/s13071-015-0966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 56.Del Cortona A, et al. The plastid genome in cladophorales green algae is encoded by hairpin chromosomes. Curr Biol. 2017;27:3771–3782.e6. doi: 10.1016/j.cub.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Turmel M, Lemieux C. Evolution of the plastid genome in green algae. Adv Bot Res. 2018;85:157–193. [Google Scholar]
- 58.Nedelcu AM, Lee RW, Lemieux C, Gray MW, Burger G. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 2000;10:819–831. doi: 10.1101/gr.10.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in Condylostoma magnum. Mol Biol Evol. 2016;33:2885–2889. doi: 10.1093/molbev/msw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osawa S, Jukes TH. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989;28:271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 61.Roquet C, et al. Understanding the evolution of holoparasitic plants: The complete plastid genome of the holoparasite Cytinus hypocistis (Cytinaceae) Ann Bot. 2016;118:885–896. doi: 10.1093/aob/mcw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schultz DW, Yarus M. On malleability in the genetic code. J Mol Evol. 1996;42:597–601. doi: 10.1007/BF02352290. [DOI] [PubMed] [Google Scholar]
- 63.Hughes RA, Ellington AD. Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 2010;38:6813–6830. doi: 10.1093/nar/gkq521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duchêne AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 66.Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes–The impact of RNA sequence. RNA Biol. 2015;12:950–958. doi: 10.1080/15476286.2015.1068497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrie C, Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:253–259. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 68.Plotkin JB, Kudla G. Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khakhlova O, Bock R. Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 2006;46:85–94. doi: 10.1111/j.1365-313X.2006.02673.x. [DOI] [PubMed] [Google Scholar]
- 70.Smith DR. The mutational hazard hypothesis of organelle genome evolution: 10 years on. Mol Ecol. 2016;25:3769–3775. doi: 10.1111/mec.13742. [DOI] [PubMed] [Google Scholar]
- 71.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 2015;112:10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamilton WL, et al. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 2017;45:1889–1901. doi: 10.1093/nar/gkw1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Massouh A, et al. Spontaneous chloroplast mutants mostly occur by replication slippage and show a biased pattern in the plastome of Oenothera. Plant Cell. 2016;28:911–929. doi: 10.1105/tpc.15.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Figueroa-Martinez F, Nedelcu AM, Smith DR, Reyes-Prieto A. The plastid genome of Polytoma uvella is the largest known among colorless algae and plants and reflects contrasting evolutionary paths to nonphotosynthetic lifestyles. Plant Physiol. 2017;173:932–943. doi: 10.1104/pp.16.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mungpakdee S, et al. Massive gene transfer and extensive RNA editing of a symbiotic dinoflagellate plastid genome. Genome Biol Evol. 2014;6:1408–1422. doi: 10.1093/gbe/evu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barbrook AC, Voolstra CR, Howe CJ. The chloroplast genome of a Symbiodinium sp. clade C3 isolate. Protist. 2014;165:1–13. doi: 10.1016/j.protis.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 77.Olinares PD, Kim J, van Wijk KJ. The Clp protease system; a central component of the chloroplast protease network. Biochim Biophys Acta. 2011;1807:999–1011. doi: 10.1016/j.bbabio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 78.Nishida I. Plastid metabolic pathways for fatty acid metabolism. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles. Springer; Dordrecht, The Netherlands: 2004. pp. 543–564. [Google Scholar]
- 79.Sjuts I, Soll J, Bölter B. Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front Plant Sci. 2017;8:168. doi: 10.3389/fpls.2017.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith DR, Lee RW. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164:1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Janouškovec J, et al. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA. 2015;112:10200–10207. doi: 10.1073/pnas.1423790112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janouškovec J, et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc Natl Acad Sci USA. 2017;114:E171–E180. doi: 10.1073/pnas.1614842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krause K. Grand-scale theft: Kleptoplasty in parasitic plants? Trends Plant Sci. 2015;20:196–198. doi: 10.1016/j.tplants.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 84.Liu TJ, et al. Complete plastid genome sequence of Primula sinensis (Primulaceae): Structure comparison, sequence variation and evidence for accD transfer to nucleus. Peer J. 2016;4:e2101. doi: 10.7717/peerj.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rousseau-Gueutin M, et al. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 2013;161:1918–1929. doi: 10.1104/pp.113.214528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li J, et al. Evolution of short inverted repeat in cupressophytes, transfer of accD to nucleus in Sciadopitys verticillata and phylogenetic position of Sciadopityaceae. Sci Rep. 2016;6:20934. doi: 10.1038/srep20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evol. 2013;28:489–495. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 88.Heitzer M, Eckert A, Fuhrmann M, Griesbeck C. Influence of codon bias on the expression of foreign genes in microalgae. Adv Exp Med Biol. 2007;616:46–53. doi: 10.1007/978-0-387-75532-8_5. [DOI] [PubMed] [Google Scholar]
- 89.Tuller T, et al. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 2011;39:4743–4755. doi: 10.1093/nar/gkr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shi C, et al. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One. 2012;7:e31468. doi: 10.1371/journal.pone.0031468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 92.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swain MT, et al. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc. 2012;7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 99.Lagesen K, et al. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Conant GC, Wolfe KH. GenomeVx: Simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24:861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- 101.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Novembre JA. Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol. 2002;19:1390–1394. doi: 10.1093/oxfordjournals.molbev.a004201. [DOI] [PubMed] [Google Scholar]
- 103.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 108.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.