Significance
Active nematics are nonequilibrium fluids consisting of elongated units driven at the individual scale. They spontaneously exhibit complex spatiotemporal dynamics and have attracted the attention of scientists from many disciplines. Here, we introduce an experimental system (made of filamentous bacteria) and a type of microscopic model for active nematics. Simultaneous measurements of orientation and velocity fields yield comprehensive experimental data that can be used to identify optimal values for all important parameters in the model. At these optimal parameters, the model quantitatively reproduces all experimentally measured features. This, in turn, reveals key processes governing active nematics. Our versatile approach successfully combines quantitative experiments and data-driven modeling; it can be used to study other dense active systems.
Keywords: bacteria collective motion, active nematics, topological defects, quantitative modeling
Abstract
Active matter comprises individual units that convert energy into mechanical motion. In many examples, such as bacterial systems and biofilament assays, constituent units are elongated and can give rise to local nematic orientational order. Such “active nematics” systems have attracted much attention from both theorists and experimentalists. However, despite intense research efforts, data-driven quantitative modeling has not been achieved, a situation mainly due to the lack of systematic experimental data and to the large number of parameters of current models. Here, we introduce an active nematics system made of swarming filamentous bacteria. We simultaneously measure orientation and velocity fields and show that the complex spatiotemporal dynamics of our system can be quantitatively reproduced by a type of microscopic model for active suspensions whose important parameters are all estimated from comprehensive experimental data. This provides unprecedented access to key effective parameters and mechanisms governing active nematics. Our approach is applicable to different types of dense suspensions and shows a path toward more quantitative active matter research.
Examples of active matter can be found at diverse length scales (1–6), from animal groups (7–11) to cell colonies and tissues (12–18) to in vitro cytoskeletal extracts (19–26) and manmade microscopic objects (27–32). Energy input at the level of the individual constituents drives active matter systems out of thermal equilibrium and leads to a wide range of collective phenomena, including flocking (7, 19, 28, 29, 33, 34), swarming (12, 13), clustering (14, 27, 30, 32), 2D long-range order (15, 35), giant number fluctuations (14, 15, 33, 35, 36), spontaneous flow (21, 24, 25, 37, 38), and synchronization (16).
Active matter systems consisting of elongated particles often lead to local nematic orientational order. This important active nematics class comprises experiments with vibrating granular rods (36), crawling cells (39–41), swarming sperms (42), filamentous bacteria (15), and motor-driven microtubules (20, 21, 24), which, together with theoretical work, have shown that the interplay between orientational order, active stress, and particle and fluid flow leads to complex spatial–temporal dynamics and unusual fluctuations. The seminal work by Dogic and coworkers (21, 43) has been particularly influential. They experimentally observed spontaneous chaotic dynamics driven by topological defects, and their results triggered a large number of theoretical and modeling approaches. These are of two main types, particle-level “microscopic” models (43–45) and continuous-level “hydrodynamic” descriptions (46–53), with the latter usually written phenomenologically or by complementing equilibrium liquid crystal theories with minimal active terms. These studies provided important insights into the multifaceted dynamics of active nematics, such as hydrodynamic instabilities, long-range correlations, anomalous fluctuations, defect dynamics, and spatial and temporal chaos. However, these models generally contain a large number of parameters. This has made comparisons between models and experiments semiquantitative at best.
Bacteria are widely used as model systems to study active matter (12–16). A recent study showed that elongated E. coli cells strongly confined between two glass plates can display the long-range nematic order and anomalous fluctuations typical of dry, dilute active nematics systems (15). However, so far, almost no bacterial system has been reported to exhibit the phenomenology of dense, wet active nematics, as reported first by Dogic and coworkers (21, 43). One exception is a study of motile bacteria dispersed in a nontoxic lyotropic nematic liquid crystal (54, 55). When bacteria concentration is high enough, active stress destabilizes the ordered nematic state of this biosynthetic system, leading to a state where topological defects in the liquid crystal evolve chaotically in a manner closely resembling that of the Dogic system. Here, we show that the typical phenomenology of wet, dense, active nematics can be experimentally realized in colonies of filamentous bacteria and show how to build a data-driven quantitatively faithful theoretical description of it. To this aim, we introduce a type of microscopic model for active suspensions, and we use simultaneous experimental measurements of both orientation and velocity fields to estimate all its parameters.
Experimental Results
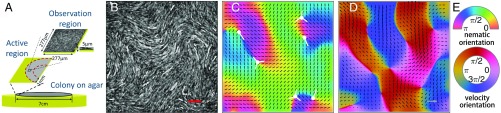
Our experiments are carried out with Serratia marcescens bacteria. At the edge of growing colonies, two to three layers of cells actively swim by rotating flagella in a micrometer-thick, millimeters-wide film of liquid on the agar surface (Fig. 1A). Apart from a narrow (m) outer ring, the thickness of this quasi-2D suspension is very constant. No obvious spatial or temporal inhomogeneity is noticeable in measured fields. A sublethal level of the antibiotic drug cephalexin is added into the growth agar medium. The drug allows bacteria to grow but not to divide, leading to long cells. By varying the drug concentration, we can change the mean cell length by a factor of 2 (SI Appendix, Fig. S1 A). Bacteria are labeled with a green fluorescent protein, which allows to record their motion under the microscope. In the dense, thin layer of interest, cells are almost always in close contact and nearly cover the whole surface. Our elongated cells are also frequently nematically aligned, as testified by the presence of charge topological defects typical of 2D nematics (Fig. 1B). (Standard cells cultivated without antibiotic drug do not give rise to any significant local order.) Our images do not allow to distinguish the current polarity of each cell, i.e., in which direction it is currently swimming with respect to the fluid. In fact, the swimming of most bacteria is strongly hampered at such high density. Nevertheless, our cells move collectively, mainly advected by the fluid they have set in motion, in a spatiotemporally chaotic manner strongly reminiscent of other active nematics systems (21, 54) (Movies S1 and S2). From each image, we extract a nematic orientation field through a gradient-based method, and we extract , the velocity field of cells in the laboratory frame, using a standard particle image velocimetry technique (Fig. 1 C and D and SI Appendix, Fig. S2). Movie S1 shows the typical evolution of the obtained coarse-grained orientation and velocity fields. This dynamics is fast. Typical correlation times are of the order of seconds (see below). In each experiment, we record images for 30 s, which is significantly shorter than the cell division time (20 min). Therefore, contributions of cell growth to active stress are negligible in our work (56, 57).
Fig. 1.
(A) Schematic illustration of a bacteria colony growing on agar; the active nematics dynamics studied here takes place in a millimeter-wide, few-micron-thick region at the edge; our observation region is typically . Mean cell length varies from 6 to , depending on antibiotic concentration. (B) Raw image of our fluorescent cells (scale bar, 30 ) in some experiment performed at 45 drug level. (C and D) nematic order and velocity fields extracted from B. Black rods in C represent unit length director vector , and length of arrows in D is proportional to the local speed (scale bar, 250 .) (E) Color maps coding the orientation of the nematic and velocity fields in C and D. The white symbols in C represent the defects (see SI Appendix, SI Text and Fig. S2 for details about their orientation).
Global Measurements.
We first measure global statistical properties of our velocity and orientation fields. The average cell speed varies between 20 and from experiment to experiment but is approximately independent of the drug concentration (SI Appendix, Fig. S1 B and C).
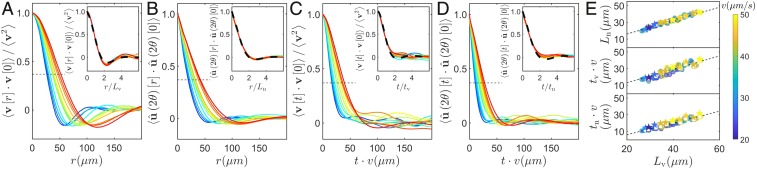
Next, we compute spatial and temporal two-point correlation functions, which are defined and shown in Fig. 2 A–D. The spatial/temporal separations corresponding to a correlation value of are identified as the correlation lengths and times. Symbols , , , and , respectively, denote velocity and orientation correlation lengths and velocity and orientation correlation times. These quantities are typically of the order of tens of and 1 s. When we increase the cell length with antibiotics, the correlation lengths and increase systematically (Fig. 2 A and B). Such a systematic variation is only observed for correlation times and if time is rescaled by the mean speed (Fig. 2 C and D). Correlation functions from various experiments with different drug concentrations collapse onto each other when space and time are rescaled by correlation lengths and times (insets in Fig. 2 A–D). Moreover, all of these quantities are linearly related to each other. Strikingly, transforming correlation times into correlation lengths using the mean speed , we find that , , and are all proportional to with approximately the same slope (Fig. 2E). This indicates that our experiments are characterized by a single lengthscale and the mean flow speed (58, 59). Because our bacteria are too closely packed to measure their length, we use and as “effective control parameters” of our experiments, with serving as a good proxy to the mean cell length (SI Appendix, Fig. S1).
Fig. 2.
Spatial (A and B) and temporal (C and D) two-point correlation functions (defined in y axis labels) from experiments with drug concentrations from 30 to 60 . The color of each curve codes for the velocity correlation length . (Insets) Correlation functions rescaled by their correlation length/time. Dashed lines indicate correlation functions calculated from numerical simulations of the model performed at the optimal-match parameter values estimated for the typical 45 experiment shown in Fig. 1, corresponding to the sixth column in SI Appendix, Table S1. (E) Variation of various correlation lengths with the velocity correlation length for both experiments (solid circles and open squares) and simulations of our model at optimal parameter values (solid stars). The experiments noted by open squares are those corresponding to the optimal model simulations noted with solid stars. Each symbol is colored according to the mean speed measured (color map at the right).
Defect Properties.
To go beyond the reduction of the complex spatiotemporal dynamics of our bacterial system to just a lengthscale and the mean speed, we now focus on the topological defects of the orientation field. Their detailed structure and their dynamics offer unique access to the coupling between nematic order and flow, all information that we will show later to be crucial to determine model parameters.
We identify the location of defects by contour integral of the director field (see Fig. 1C and Movie S1 for typical results). From the trajectories of defect cores, we measure , their velocity in the laboratory frame. We also measured the velocity of defects in the fluid frame, , where is the fluid “backflow” velocity averaged over a small region surrounding the defect core (SI Appendix, Fig. S2 D). We finally determine the intrinsic orientation of defects. This is straightforward for the comet-shaped defects. For the defects, which are not polar but have a threefold symmetry with three radial axes along which the nematic director is aligned, we choose the axis closest to the current orientation of (see SI Appendix, SI Text and Fig. S2 for details).
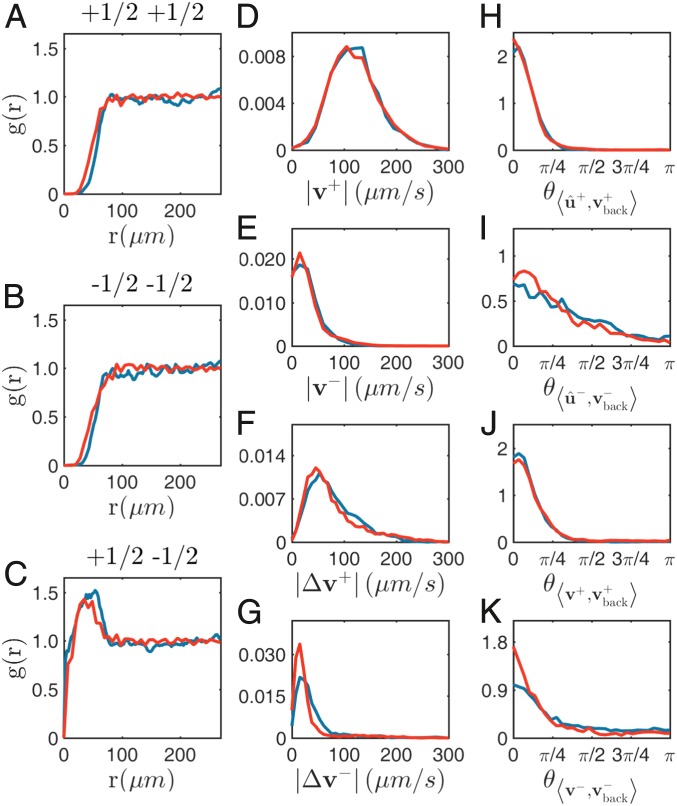
As in other active nematics systems (21, 36, 41), defects are created in pairs via the bending of ordered regions (Movie S1). Upon generation, defects typically quickly move away and less motile defects stay longer near the generation site. Pairs of defects of opposite charge may also annihilate upon encounter. In a given experiment, generation and annihilation of defects balance each other so that their total number is approximately constant in time. The radial distribution functions of defect position, , reveals that defects with the same sign repel from each other at short distances (Fig. 3 A–C). For defects of opposite sign, has a short-scale peak reflecting the fact that defects are created in pairs (43).
Fig. 3.
Statistical properties of defect cores. Experimental data extracted from the experiment at drug level 45 that gives a correlation length and a mean flow speed used in most of the text (blue curves) and from simulations of our model at parameter values optimized for that experiment (red curves), corresponding to the sixth column of SI Appendix, Table S1. (A–C) Two-point pair correlation functions for the positions of defect cores (respectively , , and pairs). (D–G) Probability distribution functions of various defect speeds (respectively speed of and defects in laboratory frame and speed of and defects in fluid frame). (H and I) Probability distribution functions of angle between defect orientations and fluid velocity at their core . (J and K) Same as H and I but for the defect core velocities in the laboratory frame .
Restricting our analysis to “isolated” defects from now on, i.e., whose distance from nearest neighbors is larger than nematic correlation length , we observe that they are essentially distributed randomly in space: no global translational nor orientational order is observed. Defect speed distributions, both in the laboratory and in the fluid frame, show that defects are more motile, but defects do not have a negligible speed, even in the fluid frame (Fig. 3 E and G). We also find that the defect orientation is strongly correlated to their velocity orientation, and to the orientation of their velocity in the fluid frame. Essentially, all three vectors are aligned, even for the defects (Fig. 3 H–K). Note that a small but finite velocity in the fluid frame is at odds with usual statements about defects in active nematics, where they are treated as symmetric, force-free, diffusive objects (44, 53, 60). We elaborate on this point under Discussion.
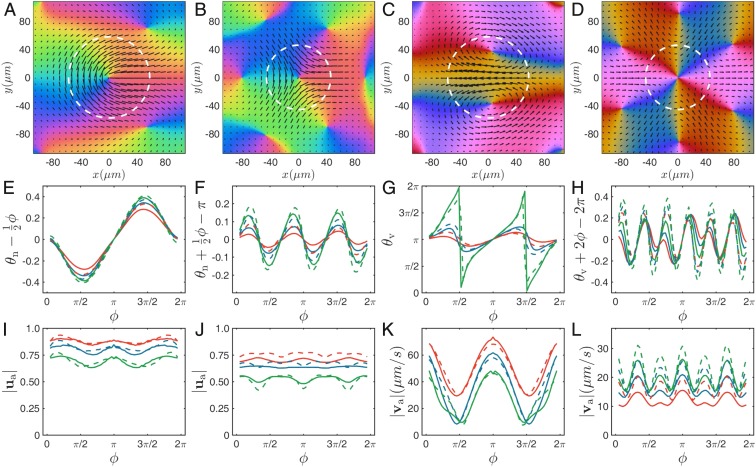
To further quantify the structure of defects, we average, over time and many defects, the orientation and velocity fields around their core, sitting in their intrinsic reference frame. The familiar mushroom-shape and threefold symmetry of, respectively, the and defect are clearly observed (Fig. 4 A and B). The flow field around the defect core shows a strong jet, while three nearly symmetric jets go through the center of the defect (Fig. 4 C and D), in agreement with previous work (53, 60).
Fig. 4.
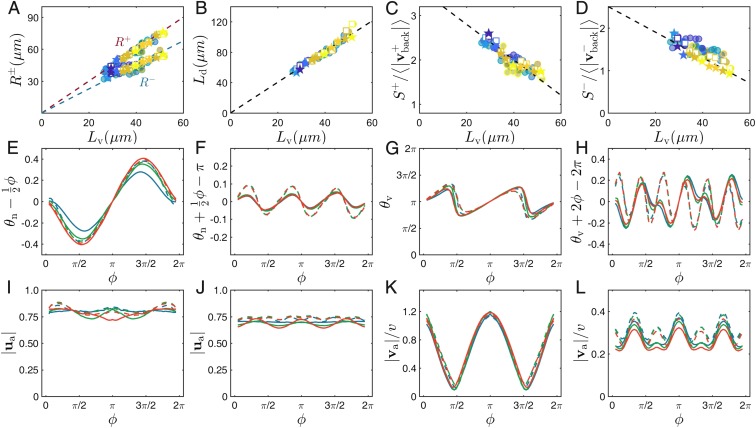
Mean nematic (first and second columns) and velocity (third and fourth columns) fields around isolated (first and third columns) and (second and fourth columns) defects. (A–D) Full 2D representation as in Fig. 1. The white circles show the defect sizes and defined in the text. Panels in the second (E–H) and third (I–L) rows contain angular profiles of the 2D fields measured at three different radii: (red), (blue), and (green). Angle is the angle depicting the circles of various radii around the defect cores. Mean director and velocity angles are defined as the angles between mean director/velocity vectors and x axis. The reference linear component ( in E, in F, and in H) has been subtracted in the orientation profiles to emphasize deviations from single-Frank constant liquid crystal theory. Solid lines indicate angular profiles from experiments. Dashed lines in E–L indicate angular profiles from simulations performed at optimal parameters.
Because of the chaotic collective dynamics, the magnitude of these averaged fields decays away from the defect core. We define defect core sizes as the radius where the magnitude of averaged director vector reaches value . For the quantitative modeling of our system, we also extracted angular profiles of orientation and velocity around defect cores from the averaged fields. In Fig. 4 E and F, we plot profiles of the angle of the nematic director calculated at three different radii around the defect cores. These profiles show clear systematic deviations from the linear variation predicted in one-constant equilibrium liquid crystals theory (61). The velocity orientation profiles, as well as the profiles of the magnitude of orientation and velocity fields, show also systematic variations reflecting the fine structure of defects (Fig. 4 I–L).
We have performed the above analysis of the dynamics and fine structure of defects on a large set of experiments. We now describe how the main defect properties vary with our two effective control parameters, the correlation length and the mean flow speed . The defect core sizes vary linearly with and are roughly independent of (Fig. 5A). In the steady state, the density of defects is statistically constant. From this steady density, one can extract an interdefect lengthscale , which behaves like all other correlation lengths, in agreement with previous work on wet active nematics (62, 63) (Fig. 5B). We also find that the speed of defects relative to the local flow speed at their core decreases with while being also roughly independent of (Fig. 5 C and D; see a discussion of this below). Remarkably, the detailed spatial structure of defects does not vary significantly between experiments with different characteristic lengths: after rescaling spatial coordinates by defect core size, or, equivalently, correlation length, averaged director and velocity fields from different datasets overlap nicely. We further confirm this by comparing defect angular profiles at 0.6 for different experiments (Fig. 5 E–L).
Fig. 5.
Dependence of defects properties on experimental conditions. (A–D) variation with nematic correlation length of defect core size (A), mean interdefect lengthscale (B), and mean of and defect speed divided by local flow speed (C and D). Experiment and simulation results are shown by open squares and solid stars, color-coded by the mean speed , following Fig. 2E. All experiments and simulations collapse onto single straight lines, indicating that does not influence the quantities represented. (E–L) Angular profiles as in Fig. 4 calculated along the circle of radius 0.6 for three experimental datasets obtained at different drug levels (blue: 30 , first column in SI Appendix, Table S1; green: 45 , sixth column in SI Appendix, Table S1; red: 60 , 12th column in SI Appendix, Table S1). Corresponding simulation results are shown using dashed lines.
Quantitative Modeling.
A microscopically faithful model of our dense, thin bacterial system where cells and their many flagella are in constant contact with each other and with the gel substrate is a formidable task well beyond current numerical power. Besides, this would require the knowledge of many specific details that are unknown. Here, we adopt a radically different approach: we treat the collisions and local interactions between cell bodies and their flagella at some effective level, where, we assume, they amount to a combination of steric repulsion and alignment. In addition, the far-field interactions and other effects due to the incompressible fluid surrounding bacteria are taken into account by solving the Stokes equation for the fluid flow. All of this also allows us to build an efficient, streamlined, but comprehensive model in two space dimensions.
Description of the Numerical Model.
Recall that most cells in our dense system are not able to swim freely, simply because nearby cells prevent them from doing so (see Movie S2). These crowded cells mostly exert force dipoles on the fluid, which is then set in motion by their collective action. Cells, in turn, are advected and rotated by the fluid. Our model thus consists of nonswimming force dipoles immersed in an incompressible fluid film and differs significantly from the common choice of using a dynamic equation for a director field (64–69). As shown by a schematic diagram in SI Appendix, Fig. S3, each dipole represents the local cell body orientation and active forcing.
The fluid flow is the solution of the (2D) Stokes equation
| [1] |
where is the fluid viscosity, is the effective friction with the substrate, is the pressure enforcing the incompressibility condition, and is the active force field exerted by dipoles on the fluid (64).
Our dipoles are point particles with position and orientation (or, equivalently, unit orientation vector . They locally align, are advected and rotated by the flow, and experience pairwise repulsion to keep their density homogeneous:
| [2] |
| [3] |
In Eq. 3, the first term on the right-hand side, with strength , codes for the nematic alignment of dipole with all neighbors currently present within distance . The next two terms govern how dipoles are rotated by the flow field , following Jeffery’s classic work: both local vorticity and local strain are playing a role, but with coefficients and taking values a priori different from the classic ones calculated for perfect ellipsoids with no-slip boundaries (70). Finally, is a unit-variance, white, angular Gaussian noise. In Eq. 2, the right-hand side term represents a unit-range pairwise soft repulsion force between dipoles of strength . Note that self-propulsion is not included in Eq. 2 because our system is crowded.
The force field in Eq. 1 is assumed to be dominated by the gradient of the active stress tensor field. (A small, residual contribution from the short-range repulsion force between neighboring dipoles exists but can usually be neglected; see SI Appendix, Eq. S2 for details about this point.) The active stress tensor is itself assumed, as usual in wet active nematics studies (50, 51), to be proportional to the gradient of the orientation field:
| [4] |
where is the typical strength of dipoles. In experiments, is the measured nematic orientation field. In the model, is the local coarse-grained orientation of our dipoles.
The full system constituted by Eqs. 1–4 can be seen as a minimal Vicsek-style model (71, 72) incorporating the main mechanisms at play in our bacterial active nematics. One thus expects a basic interplay between alignment and noise: if the alignment strength , or the alignment range , or the number density of dipoles is large enough, or if the noise strength is weak enough, local orientational (nematic) order can emerge. The global number density of dipoles and the noise strength have opposite effects. We checked that changing in the experimentally reasonable range (in simulation units) yields similar results. In the following, we fix to lighten the numerical task.
It is relatively easy to find parameter values such that the dynamics of our model closely resembles the experimental observations. As a matter of fact, the region of parameter space where spatiotemporally chaotic active nematics behavior occurs is rather large. To go beyond such qualitative agreement, we have systematically investigated the effects of parameters. We now show that for each experimental dataset, there exists a unique set of parameter values at which the model optimally matches the experiment, in the sense that all quantities studied in the previous section are in quantitative agreement.
Data-Driven Parameter Optimization.
We proceed in two steps. First, simultaneous measurements of velocity and orientation fields allow us to pinpoint the parameters in Eq. 1 without resorting to the “microscopic” part of the model, i.e., Eqs. 2 and 3.
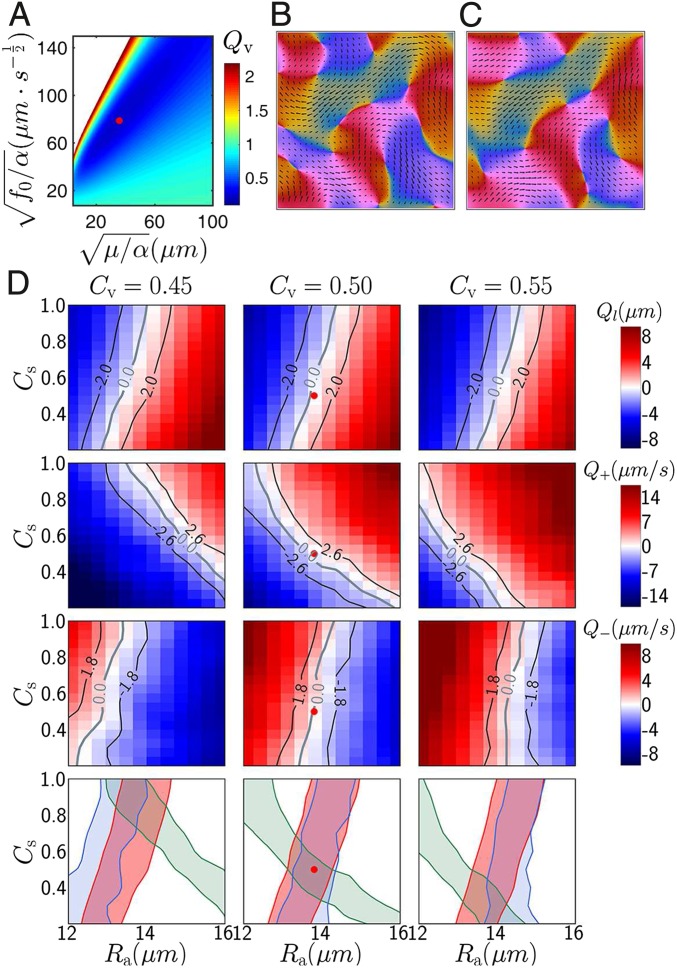
Dividing both sides of Eq. 1 by , we are left with two independent parameters, and . Therefore, for any given pair of parameters and , and a particular experimentally measured orientation field , we can compute the velocity field solution of Eq. 1. We then compare with , the velocity field measured at the same time as . Scanning the whole parameter plane, we find that there is an optimal point where the difference between and is minimal on average. Specifically, we measure the quality function where the average is carried out over both space and time. A typical result for an experiment with 45 drug concentration is in Fig. 6A, where shows a minimum for at and . Typical instantaneous velocity fields produced at these parameters compare very well to the corresponding fields (Fig. 6 B and C and Movie S3).
Fig. 6.
Quality functions and results from hydrodynamic (A–C) and full (D) model matching. Optimal parameters are marked by a red dot. (A) Quality function in the plane. (B) A typical instantaneous experimental velocity field. (C) Velocity field reconstructed from the orientation field measured at the same time as B at the optimal parameters indicated in A. The first three rows of D contain quality functions of characteristic length (), +1/2 defect speed (), and −1/2 defect speed (). Each panel represent a scan in the plane, and panels in the same column use the same value. Black contour lines mark regions of acceptable deviations. Panels in the last row contain acceptable regions of parameters extracted from quality functions: red from , green from , and blue from ; they show that yields largest overlap area for acceptable regions of parameters and a red dot in the middle panels marks the optimal choice for parameters and .
After fluid parameters are fixed, we proceed to the second step and match the full model with experiments. Eqs. 2 and 3 contain six parameters. We first evaluate their influence by varying them individually around a reference point (see SI Appendix, SI Text and Fig. S4 for details). We find that angular noise level and repulsion strength are not sensitive parameters provided local order is not destroyed by strong noise and particles do not crystallize for too-strong repulsion. We therefore fix and . Nematic alignment parameters and play a major, but similar, role, so we decide to fix and vary , mimicking the change of cell length in experiments. This leaves us with only three parameters to vary, , , and , when looking for an optimal match between model and experiment.
We performed a systematic scan of this restricted parameter space, running the model for many sets of parameter values, and extracting from each of these runs the quantities of interest, i.e., those measured also in the experiment. To quantify the match between model and experiment, we found that using three independent quality functions is sufficient. Here, we use , the difference in nematic correlation length, and , the differences in defect speed (), respectively, for the and defects. (As before, the subscript denotes quantities measured on the model.) Computed quality functions in the 3D parameter space {, , } are shown in Fig. 6D. Perfect matching () occurs for each function on a surface. These three surfaces approximately cross at a single point, as shown in the bottom row of Fig. 6D. For the particular experiment considered, we find , , and , which thus defines our optimal set of model parameters. By construction, these parameter values optimize the match between model and experiment for what concerns the quantities involved in the quality functions used. Remarkably, we observe that all other quantities not used in these functions are also quantitatively matched. This is, in particular, the case for all correlation functions in Fig. 2, all distributions of defect speed and orientation, and spatial distributions of defects in Fig. 3, all averaged angular profiles of isolated defects in Fig. 4 and defect size and speed in Fig. 5 (see also simulations of the model at optimal parameters in Movie S4).
Finally, we performed two “consistency checks.” We verified that choosing a different value of yields a different optimal value of but that all other optimal parameter values then approximately remain the same (SI Appendix, Fig. S6). In short, and are fully redundant. Next, taking our optimal parameter set, but now “freeing” the fluid parameters and from the values determined during our first step, we find that these initial values remain optimal (SI Appendix, Fig. S7). This confirms that our procedure, for a given experiment, yields a unique set of model parameters at which model dynamics optimally matches spatiotemporal data.
Variation of Model Parameters.
We have successfully applied our matching procedure to a large set of experiments with drug concentration above 15 , the level below which cells are too short to give rise to a clear local nematic orientation that can be reliably measured. For each experiment, the quality of the matching between experiments and simulations remains excellent. Corresponding orientation and velocity fields from simulations at these optimal parameters are shown in Movie S4. We thus obtained the variation of the optimal model parameter values with the two experimental effective control parameters, the correlation length (proxy for cell size) and the mean flow speed (see Fig. 7 and SI Appendix, Table S1). This provides us with a wealth of information about our experimental system.
Fig. 7.
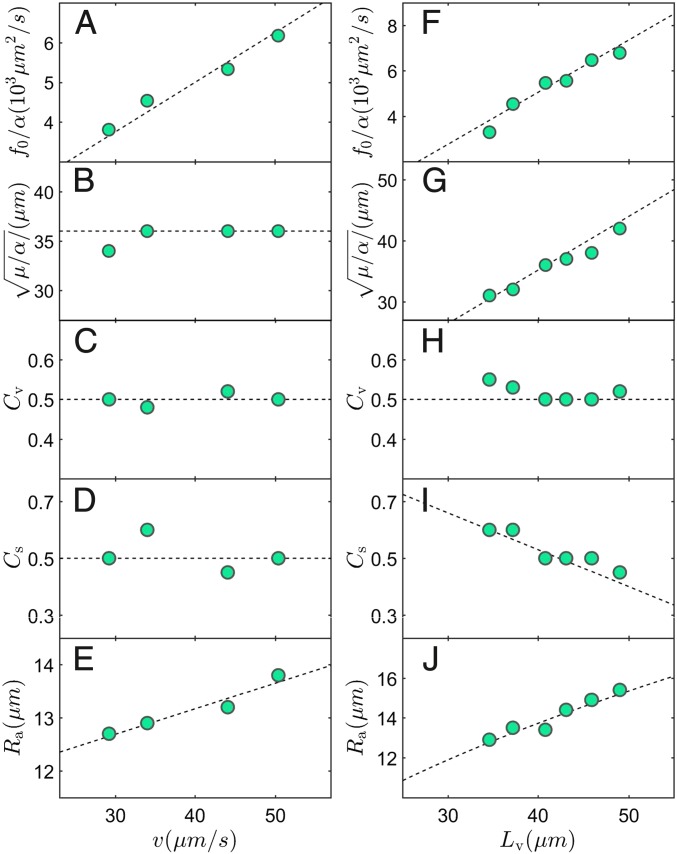
Variation of optimal model parameters with mean flow speed (A–E) and velocity correlation length (F–J). Data points in A–E have , and data points in F–J have . The numerical values of all of these parameters are listed in SI Appendix, Table S1. Dashed lines in A–I are linear fits (Variation of Model Parameters); the dashed line in J is a fit of with a .
We first discuss the effect of the mean speed at fixed correlation length. Choosing a subset of experiments yielding approximately the same correlation length, we observe that almost exclusively influences , and does so linearly (Fig. 7A). The other parameters remain constant with the exception of the interaction range , which grows slightly with (Fig. 7 B–E). The clear linear growth of confirms that, via , one has direct access to the strength of forces dipoles, which, in turn, can be interpreted to be proportional to the power developed on average by each flagellum. As for the weaker linear variation of with , we attribute it to the fact that for higher , which corresponds to higher , the fluid flow would be destabilized faster, leading to a smaller correlation length. Increasing compensates for this.
The variation of optimal model parameters with correlation length, at fixed mean speed , is presented in Fig. 7 F–J. From the extracted “fluid” parameters, we can construct two length scales that are proportional to . Balancing the active force term in Eq. 1 with the friction term , we have , which leads to . This is confirmed in Fig. 7F. We can also balance friction with the viscous force and get , as shown in Fig. 7G. (We show that the two scalings above are verified for all our data points in SI Appendix, Fig. S8.) These findings provide a physical understanding of the factors contributing to the correlation length and, in particular, of how it is connected to the fluid effective parameters. The vorticity coupling parameter is approximately a constant, (Fig. 7H). This is in agreement with Jeffery’s theory, which shows that for almost any axisymmetric shape from needles to ellipsoids to disks. Nearly constant is also consistent with observations that defect shape changes little in different experiments (Fig. 5 E–L) and that is closely connected to defect shape in our model (SI Appendix, Fig. S5). On the other hand, the strain coupling parameter decreases with (Fig. 7I), at odds with Jeffery’s results, which show that longer objects have higher . This can be understood by noticing that in the model, particles do not represent cells. Rather, over the interaction range , several dipoles stand for a cell. They react individually to the local strain, and thus their response must be weaker than that of a cell, and the longer the cell, the weaker the response. Finally, the range of nematic alignment (Fig. 7J), which shows that the correlation length increases linearly with the area where nematic alignment takes place, i.e., the number of aligning neighbors, in our model.
Discussion
To summarize, we presented a systematic study of collective motion and defect properties in a dense, wet active nematic system composed of filamentous bacteria and introduced a minimal microscopic model to account for our experiments. We have shown that using both orientation and velocity measurements enables to determine a unique, optimal set of parameter values at which our Vicsek-style model for active suspensions accounts quantitatively for many, if not all, quantities that one can extract from experimental data. Because the collective dynamics of our bacterial active nematics is always chaotic, we have used topological defects to estimate these optimal parameter values. As a matter of fact, it is sufficient to use a small subset of the various quantities we measured to determine all optimal parameter values, after which the remaining subset is “automatically” matched too. The existence of a unique optimum at which matching is nearly perfect constitutes, in retrospect, evidence of the quality of our model.
Thanks to quantitative match at a remarkable level of detail, the interplay between experiments and model provides a deeper understanding of our system. This is, in particular, the case for the dynamics and structure of topological defects. Fig. 5C demonstrates that defects move approximately twice as fast as the background flow. This acceleration can be explained by the local flow field (Fig. 4C), which shows two vortices above and below the strong jet advecting the defects. The shape of defects is essentially governed by the vorticity coupling constant [the orientation field around defects changes from arrow-like to mushroom-like shape when increasing (61, 73)]. This cannot be seen in our experiments, in which is essentially constant (Fig. 7 C and H) but is shown by simulations of our quantitatively faithful model (SI Appendix, Fig. S5). Thus, the vortices can destabilize orientational order ahead of the core, causing the defect to move faster than the background flow. The accelerating effect weakens when particles are less sensitive to flow vorticity or when nematic interaction becomes stronger, as shown in simulation (SI Appendix, Fig. S4 C) and experiments (Fig. 5 C and D).
The averaged orientation and velocity fields around defects (Fig. 4 B and D) show an approximate threefold rotational symmetry, which is consistent with the conventional, equilibrium picture: this symmetry implies that active and elastic stresses are balanced around the defect core and that defects are passive particles advected by the background flow. However, our experiments and simulations indicate that defects possess a small but significant velocity in the fluid frame (Figs. 3 E, I, G, and K and 5D). Moreover, instantaneous fields around defects often deviate significantly from threefold symmetry (SI Appendix, Fig. S9). Such deviations break stress balance around the core and give defects their velocity over the background flow. Our data (SI Appendix, Figs. S9 and S10) indeed show that the degree of deviation from threefold symmetry correlates with this velocity.
Our work also explains the multiple effects of cell length (under the influence of cephalexin). Cell length directly, and not surprisingly, governs all length scales in our system and does so nearly identically (Figs. 2E, 5 A and B, and 7 F–J). More surprising is the observation that the relative speed of defects decreases with cell length (Fig. 5 C and D) and that the strain coupling constant decreases for long cells (Fig. 7I).
These findings are just a subset of all those illustrating how, thanks to the quantitative modeling, one cannot only determine key effective parameters (such as the strength of flagella or the effective viscosity of our suspension) but also “read” important physical mechanisms from observing how model parameters change in experiments or are changed in simulations.
Our data-driven quantitative matching was made possible thanks to the relative simplicity of our Vicsek-style model: even though it deals with wet active suspensions, it possesses a relatively small number of parameters and is numerically efficient. Treating near-field interactions only effectively, it is also versatile, and we believe the same approach can be applied to other active suspensions and extended to include other effects, such as external field and polar order.
The simplicity of our model should also allow for derivation of continuous, hydrodynamic equations. Works on hydrodynamic theories of wet active nematics abound, but they typically lack a direct connection to microscopic mechanisms. Thus, deriving a faithful hydrodynamic theory from our quantitatively valid model is a very promising step. That would, in particular, allow to estimate how far our active nematics deviates from elastic theory predictions, something hinted by the structure of defects (Fig. 4 E and F).
Materials and Methods
Bacteria Strain and Colony Growth.
We use wild-type S. marcescens strain American Type Culture Collection 274 labeled with green fluorescent protein p15A-eGFP. Bacteria colonies are grown on a soft () Difco agar plate containing Luria Broth (Sigma). We mix cephalexin with molten agar at . We then pour 40 mL of molten agar into a 15-cm-diameter Petri dish, which is then dried with a lid on for 16 h ( and humidity). About of overnight bacteria culture is then inoculated on the agar. The inoculated plates are dried for another without a lid and then stored in an incubator at and humidity.
Imaging Procedure.
After a growth time of 8–9 h, collective motion is observed for as long as 2 h near the expanding edge of a colony, in an active region about wide. The colony expansion speed is approximately , i.e., much smaller than the measured bacteria flow speed. Thus, its influence on bacteria velocity measurement can be neglected. We capture bacteria motion in the central part () of this active region through a objective (Nikon S plan Fluor). S. marcescens colonies quickly change from monolayer to three-layer within from the swarming edge; thus, the thickness of swarming cells is constant in the observation region. A Nikon MBE45510 filter cube (excitation, ; emission, ) is used for fluorescent imaging. Images are acquired by a high-speed camera (Basler acA2040-180km) at for , during which bacteria motility remains unchanged. Bacteria form an immobile film in the central part of the colony. We record bacteria motion far enough from this immobile region (Movies S1–S3).
Supplementary Material
Acknowledgments
H.P.Z. thanks Julia Yeomans for useful discussions at the initial stage of this work. X.-q.S., H.C., and H.P.Z. acknowledge financial support from National Natural Science Foundation of China Grants 11422427 and 11774222 (to H.P.Z.), 11635002 (to X.-q.S. and H.C.), and 11474210 and 11674236 (to X.-q.S.). H.P.Z. thanks the Program for Professor of Special Appointment at Shanghai Institutions of Higher Learning (Grant GZ2016004). H.C. thanks the French Agence Nationale de la Recherche project “Baccterns.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812570116/-/DCSupplemental.
References
- 1.Ramaswamy S. The mechanics and statistics of active matter. Annu Rev Condens Matter Phys. 2010;1:323–345. [Google Scholar]
- 2.Vicsek T, Zafeiris A. Collective motion. Phys Rep. 2012;517:71–140. [Google Scholar]
- 3.Marchetti MC, et al. Hydrodynamics of soft active matter. Rev Mod Phys. 2013;85:1143–1189. [Google Scholar]
- 4.Bechinger C, et al. Active particles in complex and crowded environments. Rev Mod Phys. 2016;88:045006. [Google Scholar]
- 5.Elgeti J, Winkler RG, Gompper G. Physics of microswimmers-single particle motion and collective behavior: A review. Rep Prog Phys. 2015;78:056601. doi: 10.1088/0034-4885/78/5/056601. [DOI] [PubMed] [Google Scholar]
- 6.Needleman D, Dogic Z. Active matter at the interface between materials science and cell biology. Nat Rev Mater. 2017;2:17048. [Google Scholar]
- 7.Cavagna A, et al. Scale-free correlations in starling flocks. Proc Natl Acad Sci USA. 2010;107:11865–11870. doi: 10.1073/pnas.1005766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavagna A, Giardina I. Bird flocks as condensed matter. Annu Rev Condens Matter Phys. 2014;5:183–207. [Google Scholar]
- 9.Buhl J, et al. From disorder to order in marching locusts. Science. 2006;312:1402–1406. doi: 10.1126/science.1125142. [DOI] [PubMed] [Google Scholar]
- 10.Berdahl A, Torney CJ, Ioannou CC, Faria JJ, Couzin ID. Emergent sensing of complex environments by mobile animal groups. Science. 2013;339:574–576. doi: 10.1126/science.1225883. [DOI] [PubMed] [Google Scholar]
- 11.Ni R, Puckett JG, Dufresne ER, Ouellette NT. Intrinsic fluctuations and driven response of insect swarms. Phys Rev Lett. 2015;115:118104. doi: 10.1103/PhysRevLett.115.118104. [DOI] [PubMed] [Google Scholar]
- 12.Dombrowski C, Cisneros L, Chatkaew S, Goldstein RE, Kessler JO. Self-concentration and large-scale coherence in bacterial dynamics. Phys Rev Lett. 2004;93:098103. doi: 10.1103/PhysRevLett.93.098103. [DOI] [PubMed] [Google Scholar]
- 13.Sokolov A, Aranson IS, Kessler JO, Goldstein RE. Concentration dependence of the collective dynamics of swimming bacteria. Phys Rev Lett. 2007;98:158102. doi: 10.1103/PhysRevLett.98.158102. [DOI] [PubMed] [Google Scholar]
- 14.Zhang HP, Be’er A, Florin EL, Swinney HL. Collective motion and density fluctuations in bacterial colonies. Proc Natl Acad Sci USA. 2010;107:13626–13630. doi: 10.1073/pnas.1001651107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiguchi D, Nagai KH, Chaté H, Sano M. Long-range nematic order and anomalous fluctuations in suspensions of swimming filamentous bacteria. Phys Rev E. 2017;95:020601. doi: 10.1103/PhysRevE.95.020601. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Liu S, Shi Xq, Chate H, Wu Y. Weak synchronization and large-scale collective oscillation in dense bacterial suspensions. Nature. 2017;542:210–214. doi: 10.1038/nature20817. [DOI] [PubMed] [Google Scholar]
- 17.Bi D, Yang X, Marchetti MC, Manning ML. Motility-driven glass and jamming transitions in biological tissues. Phys Rev X. 2016;6:021011. doi: 10.1103/PhysRevX.6.021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, et al. Correlating cell shape and cellular stress in motile confluent tissues. Proc Natl Acad Sci USA. 2017;114:12663–12668. doi: 10.1073/pnas.1705921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaller V, Weber C, Semmrich C, Frey E, Bausch AR. Polar patterns of driven filaments. Nature. 2010;467:73–77. doi: 10.1038/nature09312. [DOI] [PubMed] [Google Scholar]
- 20.Sumino Y, et al. Large-scale vortex lattice emerging from collectively moving microtubules. Nature. 2012;483:448–452. doi: 10.1038/nature10874. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez T, Chen DTN, DeCamp SJ, Heymann M, Dogic Z. Spontaneous motion in hierarchically assembled active matter. Nature. 2012;491:431–434. doi: 10.1038/nature11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keber FC, et al. Topology and dynamics of active nematic vesicles. Science. 2014;345:1135–1139. doi: 10.1126/science.1254784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillamat P, Ignes-Mullol J, Sagues F. Control of active liquid crystals with a magnetic field. Proc Natl Acad Sci USA. 2016;113:5498–5502. doi: 10.1073/pnas.1600339113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu KT, et al. Active matter transition from turbulent to coherent flows in confined three-dimensional active fluids. Science. 2017;355:eaal1979. doi: 10.1126/science.aal1979. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki K, Miyazaki M, Takagi J, Itabashi T, Ishiwata S. Spatial confinement of active microtubule networks induces large-scale rotational cytoplasmic flow. Proc Natl Acad Sci USA. 2017;114:2922–2927. doi: 10.1073/pnas.1616001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellis PW, et al. Curvature-induced defect unbinding and dynamics in active nematic toroids. Nat Phys. 2017;14:85–90. [Google Scholar]
- 27.Palacci J, Sacanna S, Steinberg AP, Pine DJ, Chaikin PM. Living crystals of light-activated colloidal surfers. Science. 2013;339:936–940. doi: 10.1126/science.1230020. [DOI] [PubMed] [Google Scholar]
- 28.Yan J, et al. Reconfiguring active particles by electrostatic imbalance. Nat Mater. 2016;15:1095–1099. doi: 10.1038/nmat4696. [DOI] [PubMed] [Google Scholar]
- 29.Bricard A, Caussin JB, Desreumaux N, Dauchot O, Bartolo D. Emergence of macroscopic directed motion in populations of motile colloids. Nature. 2013;503:95–98. doi: 10.1038/nature12673. [DOI] [PubMed] [Google Scholar]
- 30.Theurkauff I, Cottin-Bizonne C, Palacci J, Ybert C, Bocquet L. Dynamic clustering in active colloidal suspensions with chemical signaling. Phys Rev Lett. 2012;108:268303. doi: 10.1103/PhysRevLett.108.268303. [DOI] [PubMed] [Google Scholar]
- 31.Jiang HR, Yoshinaga N, Sano M. Active motion of a janus particle by self-thermophoresis in a defocused laser beam. Phys Rev Lett. 2010;105:268302. doi: 10.1103/PhysRevLett.105.268302. [DOI] [PubMed] [Google Scholar]
- 32.Buttinoni I, et al. Dynamical clustering and phase separation in suspensions of self-propelled colloidal particles. Phys Rev Lett. 2013;110:238301. doi: 10.1103/PhysRevLett.110.238301. [DOI] [PubMed] [Google Scholar]
- 33.Deseigne J, Dauchot O, Chate H. Collective motion of vibrated polar disks. Phys Rev Lett. 2010;105:098001. doi: 10.1103/PhysRevLett.105.098001. [DOI] [PubMed] [Google Scholar]
- 34.Kumar N, Soni H, Ramaswamy S, Sood AK. Flocking at a distance in active granular matter. Nat Commun. 2014;5:4688. doi: 10.1038/ncomms5688. [DOI] [PubMed] [Google Scholar]
- 35.Weber CA, et al. Long-range ordering of vibrated polar disks. Phys Rev Lett. 2013;110:208001. doi: 10.1103/PhysRevLett.110.208001. [DOI] [PubMed] [Google Scholar]
- 36.Narayan V, Ramaswamy S, Menon N. Long-lived giant number fluctuations in a swarming granular nematic. Science. 2007;317:105–108. doi: 10.1126/science.1140414. [DOI] [PubMed] [Google Scholar]
- 37.Lushi E, Wioland H, Goldstein RE. Fluid flows created by swimming bacteria drive self-organization in confined suspensions. Proc Natl Acad Sci USA. 2014;111:9733–9738. doi: 10.1073/pnas.1405698111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wioland H, Woodhouse FG, Dunkel J, Goldstein RE. Ferromagnetic and antiferromagnetic order in bacterial vortex lattices. Nat Phys. 2016;12:341–345. doi: 10.1038/nphys3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duclos G, Erlenkämper C, Joanny JF, Silberzan P. Topological defects in confined populations of spindle-shaped cells. Nat Phys. 2016;13:58–62. [Google Scholar]
- 40.Saw TB, et al. Topological defects in epithelia govern cell death and extrusion. Nature. 2017;544:212–216. doi: 10.1038/nature21718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawaguchi K, Kageyama R, Sano M. Topological defects control collective dynamics in neural progenitor cell cultures. Nature. 2017;545:327–331. doi: 10.1038/nature22321. [DOI] [PubMed] [Google Scholar]
- 42.Creppy A, Praud O, Druart X, Kohnke PL, Plouraboue F. Turbulence of swarming sperm. Phys Rev E. 2015;92:2722–2722. doi: 10.1103/PhysRevE.92.032722. [DOI] [PubMed] [Google Scholar]
- 43.DeCamp SJ, Redner GS, Baskaran A, Hagan MF, Dogic Z. Orientational order of motile defects in active nematics. Nat Mater. 2015;14:1110–1115. doi: 10.1038/nmat4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Xq, Ma Yq. Topological structure dynamics revealing collective evolution in active nematics. Nat Commun. 2013;4:3013. doi: 10.1038/ncomms4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao T, Blackwell R, Glaser MA, Betterton MD, Shelley MJ. Multiscale polar theory of microtubule and motor-protein assemblies. Phys Rev Lett. 2015;114:048101. doi: 10.1103/PhysRevLett.114.048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramaswamy S, Simha RA, Toner J. Active nematics on a substrate: Giant number fluctuations and long-time tails. Europhys Lett. 2003;62:196–202. [Google Scholar]
- 47.Baskaran A, Marchetti MC. Enhanced diffusion and ordering of self-propelled rods. Phys Rev Lett. 2008;101:268101. doi: 10.1103/PhysRevLett.101.268101. [DOI] [PubMed] [Google Scholar]
- 48.Pismen LM. Dynamics of defects in an active nematic layer. Phys Rev E. 2013;88:050502, and erratum (2014) 89:069901. doi: 10.1103/PhysRevE.88.050502. [DOI] [PubMed] [Google Scholar]
- 49.Ngo S, et al. Large-scale chaos and fluctuations in active nematics. Phys Rev Lett. 2014;113:038302. doi: 10.1103/PhysRevLett.113.038302. [DOI] [PubMed] [Google Scholar]
- 50.Giomi L, Bowick MJ, Ma X, Marchetti MC. Defect annihilation and proliferation in active nematics. Phys Rev Lett. 2013;110:228101. doi: 10.1103/PhysRevLett.110.228101. [DOI] [PubMed] [Google Scholar]
- 51.Thampi SP, Golestanian R, Yeomans JM. Velocity correlations in an active nematic. Phys Rev Lett. 2013;111:118101. doi: 10.1103/PhysRevLett.111.118101. [DOI] [PubMed] [Google Scholar]
- 52.Blow ML, Thampi SP, Yeomans JM. Biphasic, lyotropic, active nematics. Phys Rev Lett. 2014;113:248303. doi: 10.1103/PhysRevLett.113.248303. [DOI] [PubMed] [Google Scholar]
- 53.Giomi L. Geometry and topology of turbulence in active nematics. Phys Rev X. 2015;5:031003. [Google Scholar]
- 54.Zhou S, Sokolov A, Lavrentovich OD, Aranson IS. Living liquid crystals. Proc Natl Acad Sci USA. 2014;111:1265–1270. doi: 10.1073/pnas.1321926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Genkin MM, Sokolov A, Lavrentovich OD, S. Topological defects in a living nematic ensnare swimming bacteria. Phys Rev X. 2017;7:011029. [Google Scholar]
- 56.Doostmohammadi A, Thampi SP, Yeomans JM. Defect-mediated morphologies in growing cell colonies. Phys Rev Lett. 2016;117:048102. doi: 10.1103/PhysRevLett.117.048102. [DOI] [PubMed] [Google Scholar]
- 57.Doostmohammadi A, et al. Celebrating soft matter’s 10th anniversary: Cell division: A source of active stress in cellular monolayers. Soft Matter. 2015;11:7328–7336. doi: 10.1039/c5sm01382h. [DOI] [PubMed] [Google Scholar]
- 58.Zhang HP, Be’er A, Smith RS, Florin EL, Swinney HL. Swarming dynamics in bacterial colonies. Europhys Lett. 2009;87:48011. [Google Scholar]
- 59.Sokolov A, Aranson IS. Physical properties of collective motion in suspensions of bacteria. Phys Rev Lett. 2012;109:248109. doi: 10.1103/PhysRevLett.109.248109. [DOI] [PubMed] [Google Scholar]
- 60.Giomi L, Bowick MJ, Mishra P, Sknepnek R, Cristina Marchetti M. Defect dynamics in active nematics. Philos Trans A Math Phys Eng Sci. 2014;372:20130365. doi: 10.1098/rsta.2013.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang R, Kumar N, Ross JL, Gardel ML, de Pablo JJ. Interplay of structure, elasticity, and dynamics in actin-based nematic materials. Proc Natl Acad Sci USA. 2017;115:E124–E133. doi: 10.1073/pnas.1713832115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hemingway EJ, Mishra P, Marchetti MC, Fielding SM. Correlation lengths in hydrodynamic models of active nematics. Soft Matter. 2016;12:7943–7952. doi: 10.1039/c6sm00812g. [DOI] [PubMed] [Google Scholar]
- 63.Guillamat P, Ignes-Mullol J, Sagues F. Taming active turbulence with patterned soft interfaces. Nat Commun. 2017;8:564. doi: 10.1038/s41467-017-00617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doostmohammadi A, Adamer MF, Thampi SP, Yeomans JM. Stabilization of active matter by flow-vortex lattices and defect ordering. Nat Commun. 2016;7:10557. doi: 10.1038/ncomms10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doostmohammadi A, Ignés-Mullol J, Yeomans JM, Sagués F. Active nematics. Nat Commun. 2018;9:3246. doi: 10.1038/s41467-018-05666-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thampi SP, Golestanian R, Yeomans JM. Active nematic materials with substrate friction. Phys Rev E. 2014;90:062307. doi: 10.1103/PhysRevE.90.062307. [DOI] [PubMed] [Google Scholar]
- 67.Putzig E, Redner GS, Baskaran A, Baskaran A. Instabilities, defects, and defect ordering in an overdamped active nematic. Soft Matter. 2016;12:3854–3859. doi: 10.1039/c6sm00268d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srivastava P, Mishra P, Marchetti MC. Negative stiffness and modulated states in active nematics. Soft Matter. 2016;12:8214–8225. doi: 10.1039/c6sm01493c. [DOI] [PubMed] [Google Scholar]
- 69.Guillamat P, Ignes-Mullol J, Shankar S, Marchetti MC, Sagues F. Probing the shear viscosity of an active nematic film. Phys Rev E. 2016;94:060602. doi: 10.1103/PhysRevE.94.060602. [DOI] [PubMed] [Google Scholar]
- 70.Jeffery GB. The motion of ellipsoidal particles immersed in a viscous fluid. Proc R Soc A Math Phys Eng Sci. 1922;102:161–179. [Google Scholar]
- 71.Vicsek T, Czirok A, Benjacob E, Cohen I, Shochet O. Novel type of phase-transition in a system of self-driven particles. Phys Rev Lett. 1995;75:1226–1229. doi: 10.1103/PhysRevLett.75.1226. [DOI] [PubMed] [Google Scholar]
- 72.Chate H, Ginelli F, Montagne R. Simple model for active nematics: Quasi-long-range order and giant fluctuations. Phys Rev Lett. 2006;96:180602. doi: 10.1103/PhysRevLett.96.180602. [DOI] [PubMed] [Google Scholar]
- 73.Kumar N, Zhang R, de Pablo JJ, Gardel ML. Tunable structure and dynamics of active liquid crystals. Sci Adv. 2018;4:eaat7779. doi: 10.1126/sciadv.aat7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.