Significance
AML1-ETO–containing transcription factor complex (AETFC) is a recently identified unique multiprotein complex through which AML1-ETO controls leukemogenesis. In this study, we found a functionally important AETFC heterogeneity defined by the three E proteins: while HEB and E2A facilitate leukemogenesis, E2-2 functions as a negative regulator. The underlying mechanism involves E2-2–mediated redistribution of AETFC to specific target genes, enabling a dendritic differentiation of the leukemic cells. Consistent with the fact that E2-2 is absent in AML1-ETO–expressing leukemic cell lines, E2-2 silencing/repression may occur as a cooperative event in some t(8;21) AML patients. An E2-2 target gene is identified as a predictor of relapse in the patients. This study thus provides insights into the mechanism of AML1-ETO–associated leukemogenesis and carries prognostic/therapeutic implications.
Keywords: AML1-ETO, E protein, AETFC, acute myeloid leukemia, dendritic cell
Abstract
The AML1-ETO fusion protein, generated by the t(8;21) chromosomal translocation, is causally involved in nearly 20% of acute myeloid leukemia (AML) cases. In leukemic cells, AML1-ETO resides in and functions through a stable protein complex, AML1-ETO–containing transcription factor complex (AETFC), that contains multiple transcription (co)factors. Among these AETFC components, HEB and E2A, two members of the ubiquitously expressed E proteins, directly interact with AML1-ETO, confer new DNA-binding capacity to AETFC, and are essential for leukemogenesis. However, the third E protein, E2-2, is specifically silenced in AML1-ETO–expressing leukemic cells, suggesting E2-2 as a negative factor of leukemogenesis. Indeed, ectopic expression of E2-2 selectively inhibits the growth of AML1-ETO–expressing leukemic cells, and this inhibition requires the bHLH DNA-binding domain. RNA-seq and ChIP-seq analyses reveal that, despite some overlap, the three E proteins differentially regulate many target genes. In particular, studies show that E2-2 both redistributes AETFC to, and activates, some genes associated with dendritic cell differentiation and represses MYC target genes. In AML patients, the expression of E2-2 is relatively lower in the t(8;21) subtype, and an E2-2 target gene, THPO, is identified as a potential predictor of relapse. In a mouse model of human t(8;21) leukemia, E2-2 suppression accelerates leukemogenesis. Taken together, these results reveal that, in contrast to HEB and E2A, which facilitate AML1-ETO–mediated leukemogenesis, E2-2 compromises the function of AETFC and negatively regulates leukemogenesis. The three E proteins thus define a heterogeneity of AETFC, which improves our understanding of the precise mechanism of leukemogenesis and assists development of diagnostic/therapeutic strategies.
Hematopoiesis is governed by a number of critical transcription factors that cooperatively regulate gene expression and intrinsically determine cell fate (1). Recent genome-wide studies further highlight the role of combinatorial and dynamic interactions of these transcription factors in regulation of hematopoietic stem cell (HSC) self-renewal and differentiation (2, 3). In leukemia, genes encoding transcription factors are frequently involved in chromosomal translocation and mutation (4), and the resulting fusion/mutant proteins often acquire new interactions with other transcription factors and cofactors, thereby altering transcriptional networks and blocking hematopoietic differentiation (5). Targeting the leukemogenic transcription factors and restoring differentiation thus represents an attractive strategy for leukemia therapy (6, 7).
The AML1-ETO fusion protein, generated by the t(8;21) translocation, is causally involved in nearly 20% of acute myeloid leukemia (AML) cases (8). AML1-ETO is mainly associated with the French–American–British (FAB) classified M2 AML, but it is also observed in ∼6% of M1 and in lower percentages of M0, M4, and M5 AML and other myeloproliferative syndromes (9). Although patients with t(8;21) AML generally respond well to chemotherapy, and thus have a relatively favorable prognosis, a considerable number of patients relapse and suffer poor clinical outcome (9). These observations together indicate a heterogeneity within t(8;21) AML and emphasize the necessity for understanding the precise mechanisms and for developing personalized therapeutics. The t(8;21) most likely occurs in human HSCs (10), in which the expression of AML1-ETO enhances HSC self-renewal (11). AML1-ETO has also been shown to block differentiation of multiple hematopoietic lineages, including granulocytes (12–14), erythrocytes (15), and lymphocytes (16). These differentiation blockages are mainly attributed to the capacities of AML1-ETO to directly interact and interfere with key transcription factors that regulate each hematopoietic lineage (17). Strikingly, however, it has been noted that the AML1-ETO–mediated differentiation blockages are likely incomplete (18, 19). Although the underlying mechanism remains unclear, this notion explains the consistently observed presence of AML1-ETO transcripts in t(8;21) AML patients with long-term remission (20, 21), as well as in some differentiated cells in primary leukemia samples (10).
In contrast to the dynamic interactions of AML1-ETO with many transcription factors (e.g., ETS family proteins, C/EBPα and GATA1) (13, 22, 23) and cofactors (e.g., N-CoR/SMRT, HDACs, p300, PRMT1, and JMJD1C) (24–29), we recently found that the majority, if not all, of AML1-ETO actually resides in a stable AML1-ETO–containing transcription factor complex (AETFC) (30). In leukemic cells, the multivalently interacting AETFC components stabilize each other, colocalize, and cooperatively regulate target genes; and the integrity and proper conformation of AETFC are critically important for AML1-ETO–mediated leukemogenesis. Of note, AETFC can recognize target genes not only through the RUNT domain of AML1-ETO, but also through other DNA-binding transcription factors within the complex, particularly the E-box binding factors—thus explaining the mechanistic complexity for AML1-ETO regulation of a wide variety of target genes (30).
Among the AETFC components, E2A (also known as TCF3) and HEB (also known as TCF12) are two members of the E protein subfamily (class I) of basic helix–loop–helix (bHLH) transcription factors. Surprisingly, the third E protein E2-2 (also known as TCF4) is absent from AETFC (30). Originally isolated as transcription factors binding the E-box DNA motifs in Ig enhancers (31, 32), the three E proteins are ubiquitously expressed and mainly function through heterodimerization with the tissue-specific class II bHLH transcription factors, thereby synergistically recognizing E-boxes (33). On the other hand, E-protein activities can be inhibited by the ID subfamily (class V) bHLH transcription factors, which lack the basic region for DNA binding but heterodimerize with E proteins and prevent them from binding to DNA (34). These functionally synergistic and antagonistic cross-interactions of E proteins with other bHLH subfamilies form a transcriptional feedback loop, which represents an important general mechanism regulating cell fate decision in various developmental systems (35). Indeed, during hematopoiesis, E proteins are broadly expressed and play important roles in multiple steps of lineage commitment (36). In particular, E2A is essential for B-cell development (37), in which E2-2 and HEB also play a minor role (38). HEB and E2A, probably functioning as a heterodimer, are involved in T-cell development (39). E2-2 recently has been identified as an important regulator of dendritic cell development (40). Furthermore, the E-protein/ID regulatory axis has been implicated in mixed lineage leukemia (MLL)-associated AML and may serve as a therapeutic target (41–43). Given the strong similarities among the E proteins in sequence and structure, it is important to clarify whether their different functions in developmental regulation rely on their intrinsic structural divergences or subtly different expression patterns.
In this study, we investigated the different roles of the three E proteins in AML1-ETO–mediated leukemogenesis, which is potentially related to an AETFC heterogeneity defined by the different E proteins. Interestingly, despite their broad expression in normal hematopoietic stem and progenitor cells (HSPCs), E2-2, but not E2A and HEB, is silenced in the cell lines derived from t(8;21) AML patients. We hypothesize that suppression of E2-2 may confer an advantage for AML1-ETO–mediated leukemogenesis. Given the specific function of E2-2 in dendritic cell development, it is possible that, relative to E2A and HEB, the E2-2–driven dendritic cell differentiation is less efficiently blocked by AML1-ETO. Therefore, HSPCs with a lower E2-2 expression/activity may have a better chance to be blocked from differentiation and thereafter to be transformed by AML1-ETO. We performed functional and mechanistic studies to examine these hypotheses. Our results also suggest that the dendritic differentiation of leukemic cells could be achieved in the presence of AML1-ETO, and thus provide a valuable clue for development of targeted and immune-based therapeutics.
Results
The Absence of E2-2 from AETFC Is Due to Its Loss of Expression in t(8;21) AML Cell Lines.
In our unbiased biochemical purification and characterization of AETFC from Kasumi-1 cells, large amounts of E2A and HEB proteins were consistently detected, while E2-2 was completely undetectable (30). This surprising observation could be explained by two possibilities: (i) a structural difference of E2-2 from E2A and HEB that precludes interactions with AML1-ETO and, consequently, integration into AETFC; or (ii) lack of E2-2 expression in Kasumi-1 cells. In an examination of the first possibility, we compared the ability of the three E proteins (ectopically expressed with AML1-ETO) to interact with AML1-ETO by coimmunoprecipitation (co-IP) assay. The results show that E2-2 and HEB bind AML1-ETO with a comparable affinity, while E2A exhibits a slightly weaker interaction (Fig. 1A). In support of this result, the E2-2 p300/CBP and ETO target (PCET) and NHR2-binding (N2B) motifs, which bind the NHR1 and NHR2 domains of AML1-ETO, respectively (30, 44), are almost identical to those of HEB (SI Appendix, Fig. S1 A–C), whereas the E2A PCET and N2B motifs are relatively divergent (SI Appendix, Fig. S1 B and C). Indeed, the E2A PCET has been shown to mediate a weaker interaction with NHR1 of AML1-ETO (45). Notably, the HLH domains of the three E proteins show a different phylogenic relationship—the E2-2 HLH is more divergent than E2A and HEB, implying a potentially distinct DNA-binding specificity of E2-2 (SI Appendix, Fig. S1D). Thus, these analyses suggest that a structural difference may not be an adequate explanation for the absence of E2-2 from AETFC.
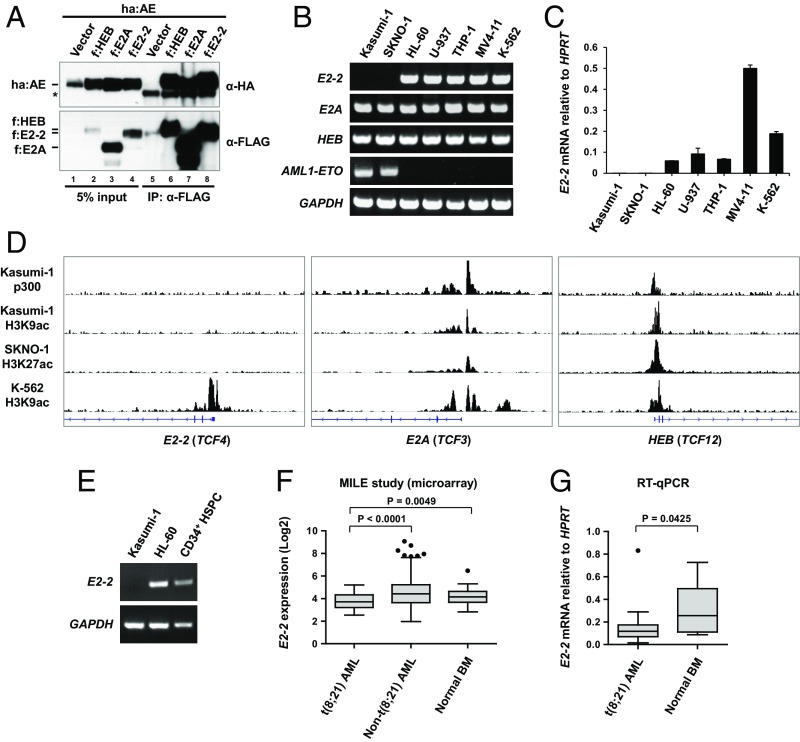
Fig. 1.
The absence of E2-2 from AETFC is due to its loss of expression in t(8;21) AML cells. (A) Co-IP experiment showing that AML1-ETO can be copurified with E2-2, E2A, and HEB. Each of the FLAG-tagged (f) E proteins was independently coexpressed with HA-tagged AML1-ETO (ha:AE) in 293T cells, followed by co-IP with anti-FLAG antibody and Western blot with indicated antibodies. The asterisk denotes IgG signals. (B) RT-PCR analysis of indicated genes in different leukemic cell lines. Note that E2-2 is undetectable in the two t(8;21) cell lines (Kasumi-1 and SKNO-1) but expressed well in other cells, whereas E2A and HEB are broadly expressed. (C) RT-qPCR analysis of E2-2 mRNA levels relative to the housekeeping gene HPRT in leukemic cells. Data are presented as mean ± SD of triplicate reactions; the E2-2 mRNA levels in Kasumi-1 and SKNO-1 cells are very low so that only one of the triplicate reactions was produced. (D) ChIP-seq data of p300 and histone H3 acetylation at the genomic loci of the three E protein genes. Of note is the specific absence of these transcriptional activation marks at the E2-2 promoter in Kasumi-1 and SKNO-1 cells. (E) E2-2 is normally expressed in primary human HSPCs, as indicated by the RT-PCR result. (F) Expression levels of E2-2 in t(8;21) AML patient samples (n = 40) are statistically lower than non-t(8;21) AML (n = 502) and normal bone marrow (BM) samples (n = 73). Data were obtained from the International Microarray Innovations in Leukemia (MILE) Study. (G) RT-qPCR analysis of E2-2 expression in t(8;21) AML (n = 23) and normal BM samples (n = 11).
To test the second possibility, we determined E2-2, E2A and HEB mRNA levels by RT-PCR analysis of Kasumi-1 and SKNO-1 [also t(8;21) patient-derived] cells, as well as several other hematopoietic/leukemic cell lines that included HL-60 (M2 AML; AML1-ETO–negative), U-937 (monocyte derived from histiocytic lymphoma), THP-1 (M5 AML), MV4-11 (biphenotypic B-myelomonocytic leukemia), K-562 (chronic myelogenous leukemia), and Namalwa (Burkitt’s lymphoma). The results showed that, while E2A and HEB are broadly expressed in all cell lines, E2-2 mRNA is not detectable in Kasumi-1 and SKNO-1 cells but is normally expressed in other cells (Fig. 1B and SI Appendix, Fig. S2A). This result was further confirmed by quantitative RT-PCR (RT-qPCR) analysis, in which E2-2 was barely detected in Kasumi-1 and SKNO-1 cells (Fig. 1C). Consistent with the gene expression pattern, chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-seq) analysis of p300 binding (27) and histone acetylation (46–48) revealed that these epigenetic marks for transcriptional activation are present at the E2-2 promoter in K562 cells but not in Kasumi-1 and SKNO-1 cells (Fig. 1D, Left). In contrast, the E2A and HEB promoters show activation marks in all cell types (Fig. 1D, Middle and Right). These results indicate that, despite being considered as a ubiquitously expressed gene, E2-2 is specifically silenced in the t(8;21) cell lines, which explains why the E2-2 protein is absent from AETFC.
Expression of E2-2 in Primary HSPCs and Leukemic Samples.
To determine the expression of E2-2 in primary human HSPCs, from which the t(8;21) leukemias originate, we purified CD34+ HSPCs from umbilical cord blood and analyzed E2-2 expression by RT-PCR. The result showed that E2-2 is normally expressed in these cells (Fig. 1E). Other studies of gene expression profiling of different hematopoietic lineages in human (3, 49) and mouse (50, 51) also showed relatively high E2-2 expression in HSPCs. We then assessed the expression of E2-2 in primary leukemia and normal bone marrow (BM) samples. Analysis of a microarray-based gene expression profiling of leukemia performed by the International Microarray Innovations in Leukemia (MILE) Study Group (52) demonstrated that the E2-2 expression is significantly lower in t(8;21) leukemia compared with non-t(8;21) AML and normal BM samples (Fig. 1F). Indeed, our own RT-qPCR analysis of t(8;21) AML patient (n = 11) versus normal BM (n = 23) samples also indicated a significant decrease of E2-2 expression in the t(8;21) AML patient samples, even though a considerably diverse expression of E2-2 exists in the t(8;21) AML samples (Fig. 1G).
E2-2 Is Not a Direct Target Gene of AML1-ETO.
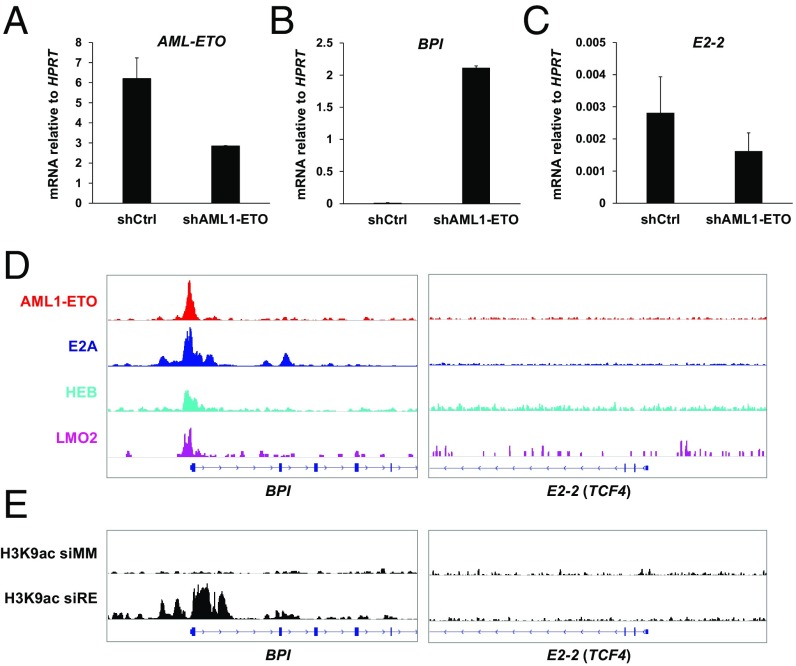
Both the complete silence of E2-2 in t(8;21) leukemic cell lines and the relatively lower expression in primary t(8;21) leukemia samples imply that E2-2 could be directly repressed by AML1-ETO. We examined this possibility by determining whether E2-2 can be up-regulated by knockdown of AML1-ETO in Kasumi-1 cells (Fig. 2A). The BPI gene, which has been known to be directly repressed by AML1-ETO (53), was used as a positive control. The results showed that, while the AML1-ETO knockdown can dramatically activate BPI (Fig. 2B), it cannot up-regulate E2-2 (Fig. 2C). To investigate whether AML1-ETO directly binds to the E2-2 gene, we analyzed our ChIP-seq data for AETFC in Kasumi-1 cells. The results showed that none of the tested AETFC components bind to E2-2 but that they all bind to the BPI promoter (Fig. 2D). Furthermore, knockdown of AML1-ETO led to a gain of histone H3 lysine 9 acetylation (H3K9ac) at the BPI promoter but not the E2-2 locus (Fig. 2E), reflecting a consistent silencing of E2-2 at the chromatin level. Ectopic expression of AML1-ETO in human HSPCs also failed to significantly down-regulate E2-2 (SI Appendix, Fig. S2B). Thus, these results clearly indicate that E2-2 is not a direct target gene of AML1-ETO. To test whether reversal of epigenetic gene silencing mechanism(s) may release the repression of E2-2, we analyzed previously published (54) gene expression profiles of Kasumi-1 cells treated with the DNA methyltransferase inhibitor Decitabine (DAC), the histone deacetylase inhibitor Trichostatin A (TSA), and the chemotherapeutic agent cytarabine (Ara-C). The results showed that the Kasumi-1 cells treated with DAC (500 nM) and TSA (300 nM) have higher E2-2 expression levels relative to those treated with Ara-C (100 nM) or a low dosage of DAC (10 nM) (SI Appendix, Fig. S3), supporting the possibility that E2-2 is silenced by an epigenetic mechanism that is associated with DNA methylation and histone modification.
Fig. 2.
E2-2 is not a direct target gene of AML1-ETO in Kasumi-1 cells. (A) RT-qPCR showing knockdown of AML1-ETO by shRNA in Kasumi-1 cells. Data are presented as mean ± SD of triplicate reactions. (B) RT-qPCR showing that BPI, which is known to be directly repressed by AML1-ETO, is up-regulated by AML1-ETO knockdown. (C) RT-qPCR showing that E2-2 remains silenced upon AML1-ETO knockdown. (D) ChIP-seq showing that BPI, but not E2-2, is directly bound by AETFC components. (E) ChIP-seq showing that AML1-ETO knockdown leads to a gain of histone H3 acetylation at the BPI promoter, but not at the E2-2 promoter. The data are consistent with an inactive state of E2-2. ChIP-seq data for Kasumi-1 cells treated with mismatch siRNA (siMM) and AML1-ETO siRNA (siRE) are from Ptasinska et al. (46).
Restoration of E2-2 Expression Selectively Inhibits the Growth of AML1-ETO–Expressing Leukemic Cells.
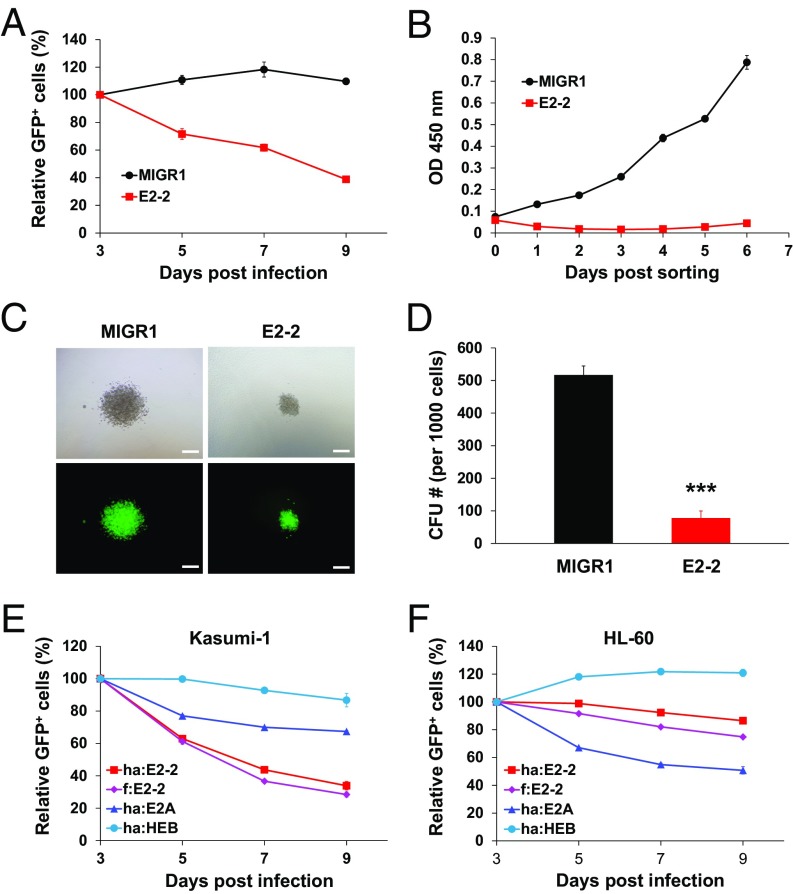
To define the function of E2-2 in AML1-ETO–expressing leukemic cells, we transduced Kasumi-1 cells with an E2-2–expressing retrovirus in which a green fluorescent protein (GFP) reporter was linked downstream of E2-2 in the same mRNA by an internal ribosome entry site (IRES). The vector containing GFP alone (MIGR1) was used as a control. We cocultured the GFP+ and GFP− cells and measured the percentage of GFP+ cells to indicate their competitive growth ability. The results showed that the percentage of E2-2–expressing cells significantly decreased over time, whereas the MIGR1-transduced cells remained unchanged, indicating that the restoration of E2-2 expression inhibits cell growth (Fig. 3A). We then sorted the GFP+ cells using flow cytometry and assessed cell growth with the Cell Counting Kit-8 (CCK-8) assay. The results showed that the E2-2 expression significantly inhibits the growth/viability of Kasumi-1 cells (Fig. 3B). We also performed a colony formation assay with methylcellulose medium supporting myeloid cell growth, and the results showed that the E2-2–expressing cells generate much smaller and fewer numbers of colonies than do control (MIGR1) cells (Fig. 3 C and D).
Fig. 3.
Restoration of E2-2 expression selectively inhibits the growth of AML1-ETO–expressing leukemic cells. (A) Competitive coculture assay showing a growth disadvantage of the E2-2–transduced Kasumi-1 cells. Kasumi-1 cells were infected with retroviruses that contain E2-2 or control vector (MIGR1), and their growth was interpreted by the percentage of the infected (GFP+) cells within total cells over time. As it takes ∼3 d for the GFP expression to reach its peak, the percentages were normalized to the values at day 3 postinfection. (B) Growth curves of E2-2–transduced and control cells, showing that E2-2 expression inhibits the growth/viability of Kasumi-1 cells. The GFP+ cells were sorted by flow cytometry at day 3, and the viable cell numbers over time in the culture were assessed by measuring optical density (OD) at wavelength of 450 nm by CCK-8 assay. (C) Colony formation assay showing that the Kasumi-1 cells expressing E2-2 form smaller colonies compared with the control (MIGR1) cells. Upper and Lower show the same representative colonies in bright-field and fluorescent views. (Scale bar, 200 μm.) (D) Quantification of the colony formation units (CFU) of the cells analyzed in C, showing that the number of CFUs formed by the E2-2–expressing Kasumi-1 cells is much lower compared with the control cells. n = 3; mean ± SD; ***P < 0.001. (E and F) Different effects of overexpression of E2-2, E2A, and HEB in Kasumi-1 and HL-60 cells. While the growth-inhibitory effect of E2-2 is evident in Kasumi-1 cells (E), this E2-2 effect is very modest, and virtually ranked between E2A and HEB, in HL-60 cells (F). The growth curves (A, B, E, and F) are representatives of at least two independent experiments with triplicate wells each time and are presented as mean ± SD.
To determine whether the growth inhibition is specific for E2-2 relative to other E proteins, we transduced Kasumi-1 cells with E2-2 (tagged by either FLAG or HA), E2A, and HEB, and found that only E2-2 dramatically inhibits cell growth, whereas E2A and HEB exert little effect (Fig. 3E). We then determined whether this effect is specific for AML1-ETO–expressing leukemic cells by a similar analysis in a cell line (HL-60) derived from a non-t(8;21) M2 AML patient (55). The results indicate that E2-2 exerts only a very modest effect that lies between those shown by E2A and HEB (Fig. 3F). These results thus strongly suggest that E2-2 specifically inhibits the growth of t(8;21) leukemic cells.
The bHLH DNA-Binding Domain Is Important for the Activity of E2-2.
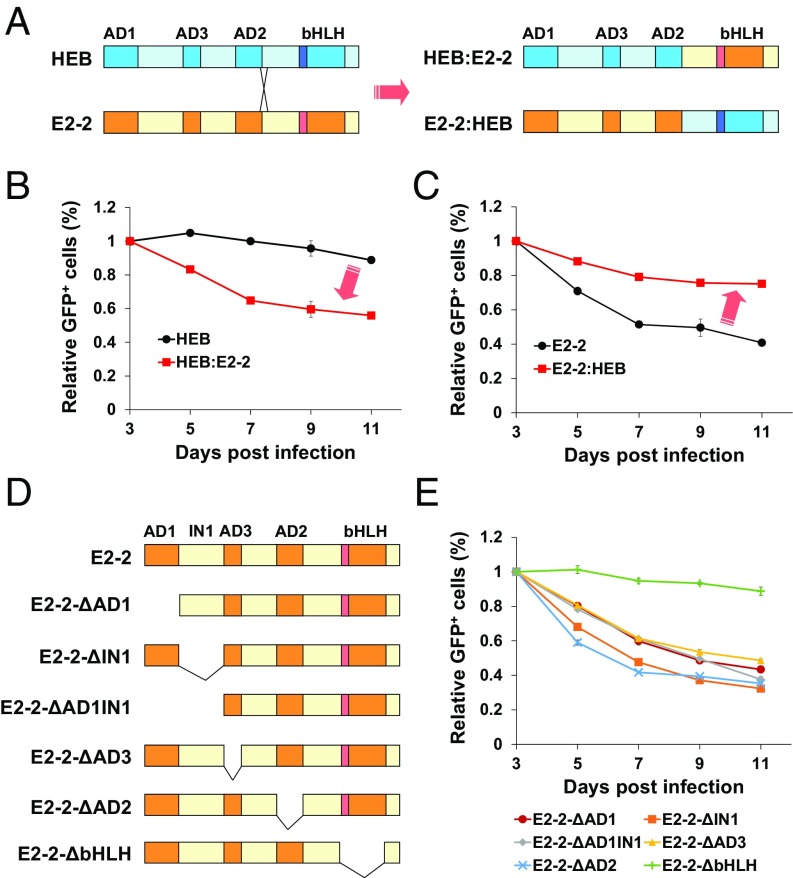
As classical transcription factors, E2-2, E2A, and HEB similarly exert their DNA-binding and transcriptional activation functions through their separable C-terminal bHLH domain and N-terminal activation domains (AD1–3), respectively (56). To determine whether the inhibitory effect of E2-2 in AML1-ETO–expressing leukemic cells can be attributed to either the DNA-binding domain (DBD) or the activation domain (AD), we swapped the DBD and AD parts between E2-2 and HEB (Fig. 4A) and assessed the chimeric proteins for their ability to inhibit the growth of Kasumi-1 cells. Interestingly, the results showed that the HEB derivative carrying the E2-2 DBD (designated as HEB:E2-2) acquires the inhibitory activity (Fig. 4B), whereas the E2-2 derivative carrying HEB DBD (designated as E2-2:HEB) loses this activity (Fig. 4C). These results suggest that the specific role of E2-2 requires a distinct DNA-binding specificity that can possibly target E2-2 to specific target genes. This notion is also consistent with the fact that the E2-2 HLH domain is structurally divergent from the E2A and HEB HLH domains (SI Appendix, Fig. S1D).
Fig. 4.
The bHLH DNA-binding domain is important for the ability of E2-2 to inhibit the growth of Kasumi-1 cells. (A) A schematic representation of the strategy for swapping the DNA-binding and the transcriptional activation domains between E2-2 and HEB. (B) Gain of the growth-inhibitory function by the chimeric HEB protein containing the E2-2 DNA-binding domain (HEB:E2-2), as indicated by the experiment determining the competitive growth of the Kasumi-1 cells expressing the indicated proteins (GFP+) versus control (GFP−) cells. (C) Loss of the growth-inhibitory function by the chimeric E2-2 protein containing the HEB DNA-binding domain (E2-2:HEB). (D) A schematic representation of the deletion of individual domains of E2-2. (E) Elimination of the growth-inhibitory function of E2-2 in Kasumi-1 cells by deletion of the bHLH DNA-binding domain but not other domains. The growth curves (B, C, and E) are representatives of at least two independent experiments with triplicate wells each time and are presented as mean ± SD.
To further define the role of the different E2-2 domains, we made a serial deletion of each domain and assessed the functions of these deletion mutants (Fig. 4D). The results show that only deletion of the bHLH domain abolishes the E2-2 inhibition of Kasumi-1 cell growth, whereas deletions of other individual domains show little effect (Fig. 4E). A Western blot showed that all these mutants were expressed properly (SI Appendix, Fig. S4). Taken together, these results suggest that the E2-2 bHLH DNA-binding domain is important for the activity of E2-2 in inhibiting the growth of AML1-ETO–expressing leukemic cells.
E2-2 Overexpression Leads to Repression of MYC Target Genes.
To explore the mechanisms underlying the different roles of E2-2, E2A, and HEB in AML1-ETO–associated leukemic cells, we performed RNA massively parallel sequencing (RNA-seq) and ChIP-seq analyses of Kasumi-1 cells transduced with MIGR1-based retroviruses expressing E2-2, E2A, or HEB. As each E protein can be efficiently integrated into AETFC, we observed considerable overlapping patterns among the three E proteins both in their binding to genomic sites/genes and in their regulation of gene expression, which is consistent with their structural similarity. For example, despite possible competitions between the three E proteins, alignment of their binding sites indicated a genome-wide colocalization (SI Appendix, Fig. S5A); sequence analysis of these regions revealed similar overrepresented transcription factor binding sites (SI Appendix, Fig. S5B); and gene expression analysis demonstrated significant overall correlations of the genes up- and down-regulated by E2-2, E2A, and HEB (SI Appendix, Fig. S5 C and D).
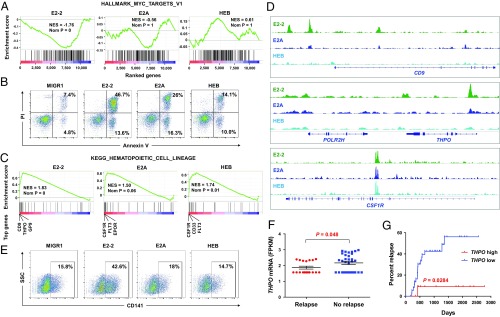
We next performed gene set enrichment analysis (GSEA) of the gene expression data to identify biological features and pathways underlying the different roles of the E proteins in Kasumi-1 cells. One of the striking findings was that the MYC target genes are significantly repressed by E2-2, but not by E2A or HEB (Fig. 5A). Notably, MYC recently has been identified as a critical player in preventing cellular apoptosis and differentiation, and this function is highly specific for t(8;21) leukemia relative to other leukemic cells (57). Indeed, a gene ontology (GO) analysis of genes differentially expressed in E2-2–overexpressing Kasumi-1 cells indicated that the genes related to hematopoietic cell lineage and apoptotic processes are most significantly affected (SI Appendix, Fig. S6). We therefore analyzed apoptosis of the cells by double staining with Annexin V-FITC and propidium iodide (PI). The results show that the percentage of apoptotic cells in the E2-2–expressing Kasumi-1 cells is dramatically higher than the percentages of E2A- and HEB-expressing cells (Fig. 5B and SI Appendix, Fig. S7 A and B).
Fig. 5.
E2-2 represses MYC target genes and activates dendritic genes to promote dendritic differentiation, which is shown to be relevant to clinical outcome of t(8;21) AML patients. (A) Gene set enrichment analysis (GSEA) of the RNA-seq data indicating that the MYC target genes are negatively correlated with the genes regulated by E2-2, but not E2A or HEB, in Kasumi-1 cells. (B) Flow cytometry analysis of apoptosis of the Kasumi-1 cells transduced with indicated E proteins. Note that more significant apoptosis is induced by overexpression of E2-2 relative to E2A and HEB in Kasumi-1 cells. Combination of Annexin V and propidium iodide (PI) staining allows detection of early (Annexin V+/PI−) and late apoptotic (Annexin V+/PI+) cells. See more detailed analysis in SI Appendix, Fig. S7. (C) GSEA revealing that the gene set of hematopoietic lineages is significantly enriched in the up-regulated genes by E2-2, E2A, and HEB. However, the rank of genes up-regulated by E2-2 is distinguishable by several top-ranked dendritic genes. (D) ChIP-seq data showing that some dendritic genes are specifically bound by E2-2, whereas many commonly regulated genes are cobound by the three E proteins. (E) Flow cytometry analysis of the presence of the dendritic cell surface marker CD141 on the Kasumi-1 cells transduced with indicated E proteins, showing that overexpression of E2-2 enhances dendritic differentiation. The cells were cultured in a dendritic cell medium. (F) Expression of THPO in t(8;21) AML patient samples collected at diagnosis. Note that the THPO mRNA level is statistically lower in the patients who later suffered relapse (n = 18) than those who did not (n = 32), and that the patients with highest TPHO expression tend not to relapse. (G) Relapse curves of patients with high and low THPO expression. Cutoff point was set as 25% so that 13 patients with high THPO and 37 patients with low THPO were analyzed.
E2-2 Targets Dendritic Genes and Promotes Dendritic Differentiation of AML1-ETO–Expressing Leukemic Cells.
Reflecting a potential for cellular differentiation induced by E proteins, a KEGG gene set containing markers for hematopoietic lineages was significantly enriched in the up-regulated genes by all three E proteins (Fig. 5C). However, it is notable that the top-ranked genes up-regulated by E2-2 are virtually distinct from those of E2A and HEB (Fig. 5C). Consistent with the relatively specific role of E2-2 in dendritic cell differentiation, the top-ranked hematopoietic lineage genes (e.g., CD9, THPO, and GP9) up-regulated by E2-2 are important markers and/or regulators of dendritic cells (58–60) (Fig. 5C). In contrast, E2A and HEB share common top-ranked genes (e.g., CSF1R and FLT3) that are relatively lower ranked in the E2-2–up-regulated genes. In strong support of this finding, our ChIP-seq data also showed that the CD9 and THPO gene loci are directly targeted by E2-2, but not E2A or HEB, whereas many other genes (e.g., CSF1R) are equally bound and regulated by all E proteins (Fig. 5D). These results suggest that, despite considerable similarity among the three E proteins, E2-2 can specifically target some genes related to dendritic cell differentiation.
To determine whether the up-regulation of dendritic genes by E2-2 really associates with a phenotypic dendritic differentiation of leukemic cells, we performed flow cytometry analysis of surface markers of the Kasumi-1 cells expressing ectopic E2-2, E2A, and HEB. Remarkably, we observed the emergence of a population of potential dendritic cells marked by CD141 (also known as thrombomodulin, or THBD) (61–63) from the E2-2–expressing Kasumi-1 cells (SI Appendix, Fig. S8A), and the percentage of these cells can be increased to over 40% in a dendritic cell culturing medium (Fig. 5E). In contrast, the granulocytic marker CD11b (also known as ITGAM) and the monocytic marker CD14 were not detected in these cells (SI Appendix, Fig. S8B). These results thus suggest that E2-2 can fulfill unique functions in regulating specific target genes and promoting dendritic differentiation of the AML1-ETO–associated leukemic cells.
High THPO Expression Is Associated with a Low Relapse Rate of t(8;21) AML Patients.
To test whether the molecular features could predict clinical outcomes of the patients, we analyzed 50 patients with t(8;21) AML by performing RNA-seq of their leukemic cells collected at diagnosis. These patients all achieved complete remission upon chemotherapy, but 18 of them suffered relapse. A comparison of differentially expressed genes between the relapsed and nonrelapsed patients revealed that THPO expression is statistically higher in the nonrelapsed patients (Fig. 5F), and this finding was supported by a GSEA result showing that a set of THPO neighborhood genes (MORF_THPO) (64) is also significantly enriched in the nonrelapsed patient samples (SI Appendix, Fig. S9A). Although the THPO signaling pathway that is mediated by the THPO receptor (i.e., the proto-oncoprotein MPL) previously has been implicated in regulation of AML1-ETO leukemogenesis (65, 66), the leukemia cellular autonomous expression of THPO has not been studied. Of note, while being highly expressed in dendritic cells (3), THPO is not expressed in normal HSPCs (SI Appendix, Fig. S9B) (3, 67). Thus, elevated THPO mRNA levels in the leukemic cells of some t(8;21) AML patients may reflect a partial dendritic differentiation of the cells. In contrast, E2-2 is ubiquitously expressed in all cell types, and therefore it is difficult to detect the cell population-specific alteration of E2-2 expression that may contribute to leukemia progression, explaining why we did not observe a correlation between E2-2 expression and patient relapse or a coexpression pattern between E2-2 and THPO in the herein analyzed total BM samples. Nonetheless, a gene expression analysis (SI Appendix, Fig. S9B) of purified populations of hematopoietic cells (3) demonstrated a closely correlated expression of E2-2 and THPO mRNAs during differentiation of HSCs to plasmacytoid dendritic cells, supportive of our functional studies. Interestingly, by comparison of the t(8;21) AML patients’ THPO mRNA levels, the patients with the highest THPO expression appear separable from other patients and virtually all reside in the nonrelapse group (Fig. 5F), implying that a stratification of patients according to their THPO mRNA levels at a defined cutoff point may help predict relapse risk. Indeed, when we choose a cutoff value of 25% to stratify these patients into THPO-higher (n = 13) and -lower (n = 37) groups, the Kaplan–Meier curves show that the patients with high THPO expression have a significantly lower chance of relapse (P = 0.0284; Fig. 5G). Thus, these results suggest that the elevated THPO expression in patient leukemia samples can be considered as a potential predictor of nonrelapse in t(8;21) AML patients.
E2-2 Suppression Accelerates Leukemogenesis in Vivo.
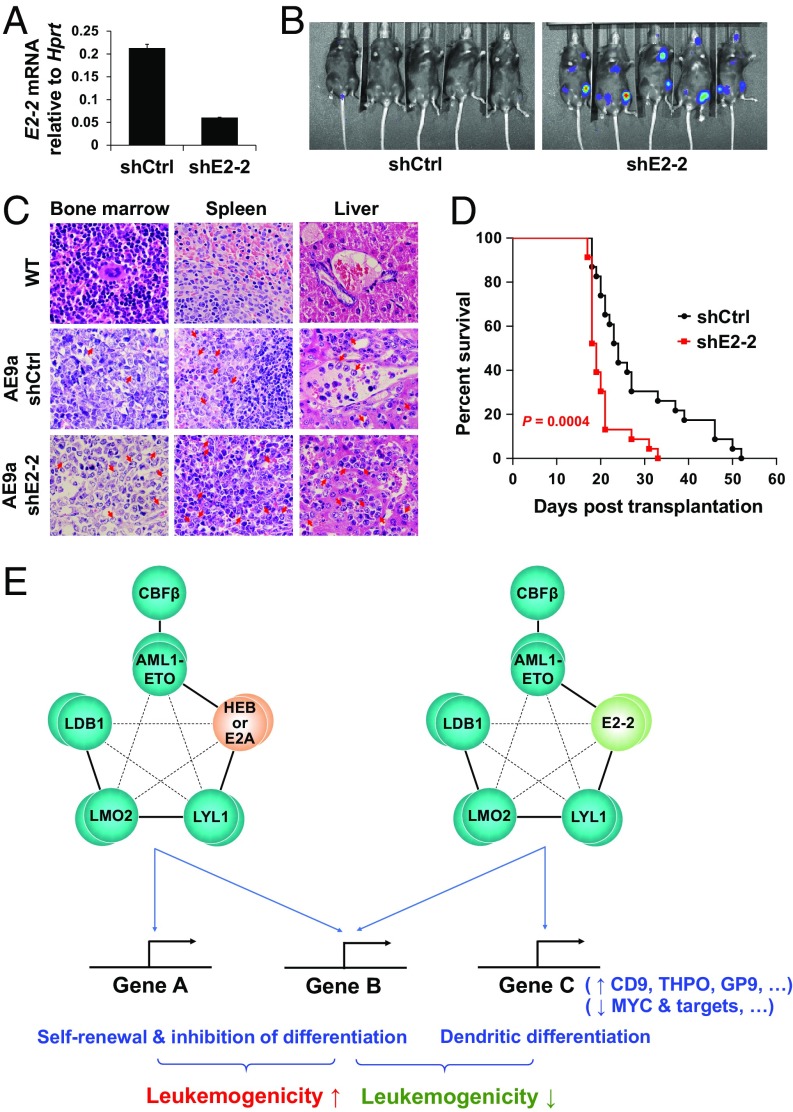
We previously found that E2A and HEB are important for AML1-ETO–mediated leukemogenesis. Thus, in a mouse model for human t(8;21) leukemia that was generated by transducing mouse HSPCs with AML1-ETO9a (a highly leukemogenic short isoform of AML1-ETO), knockdown of E2A and HEB significantly delayed leukemogenesis (30). Equivalent to human HSPCs, the mouse HSPCs express all three E proteins (Fig. 6A), allowing investigation of the role of E2-2 in AML1-ETO9a–driven leukemogenesis. To this end, we performed shRNA-mediated knockdown of E2-2 in the cells (Fig. 6A) and transplanted them into sublethally irradiated recipient mice. Since a luciferase reporter gene had been integrated into these cells (30, 68), bioluminescence imaging was used to monitor the growth of leukemic cells in vivo. At 20 d posttransplantation, the bioluminescent signal was readily detected in the mice carrying E2-2 knockdown cells, but it was very weak in the mice carrying control cells (Fig. 6B). Organ sections of the mice showed invasion of myeloid leukemia cells into their spleens and livers (Fig. 6C). Furthermore, the mice carrying E2-2 knockdown cells showed comparable levels of AML-ETO9a in leukemic cells (SI Appendix, Fig. S10A) but more severe splenomegaly (SI Appendix, Fig. S10B) and significantly shorter survival time compared with the control mice (P = 0.0004; Fig. 6D). These results indicate that suppression of E2-2 accelerates AML1-ETO9a–driven leukemogenesis, which is opposite to the effect of E2A/HEB knockdown in the same mouse model. Notably, gene expression analysis of flow cytometry-purified leukemic cells of two mouse models, namely the AML1-ETO9a– and Tet2−/−;AML1-ETO–driven leukemic mouse models (69, 70), suggested that E2-2 could be suppressed in their leukemic cells in comparison with the related nonleukemic cells (SI Appendix, Fig. S11 A and B) or the control granulocyte macrophage progenitors (SI Appendix, Fig. S11C), respectively. This observation may also depict leukemia progression in the t(8;21) AML patients. To assess the effect of E2-2 suppression in human normal HSPCs, we performed shRNA-mediated knockdown of E2-2 in human CD34+ HSPCs (SI Appendix, Fig. S12A), and the results showed that E2-2 knockdown slightly increases the capacity of the cells to form myeloid colonies (SI Appendix, Fig. S12B), which may reflect a skewed balance of cell differentiation from a dendritic to a myeloid linage. Thus, although E2-2 is apparently dispensable for development and function of HSCs (36), its capacity in hematopoietic lineage commitment and differentiation could cast it in an important role in regulating leukemogenesis.
Fig. 6.
Suppression of E2-2 accelerates AML1-ETO–associated leukemogenesis in vivo. (A) shRNA-mediated knockdown of E2-2 in the AML1-ETO9a–induced, transplantable mouse leukemic cells. Data are presented as mean ± SD of triplicate reactions. (B) Representative bioluminescent imaging of the mice transplanted with leukemic cells expressing control and E2-2 shRNAs. Imaging was performed at 20 d posttransplantation. (C) Representative HE-stained organ sections from wild-type (WT) and leukemic mice showing the leukemic cells in BM and their invasion into spleen and liver. Arrowheads denote leukemic cells. (D) Kaplan–Meier survival curves of the mice, indicating that mice harboring E2-2 knockdown cells have a significantly shorter survival time (n = 23 in each group). (E) A working model for the different roles of E proteins in AML1-ETO–associated leukemia. E2-2, E2A, and HEB can be equally and competitively integrated into AETFC, but their subtly different capacities in recognizing target genes may define a heterogeneity of AETFC; some E2-2–specific target genes, including the dendritic and MYC target genes, underlie the negative role of E2-2 in AML1-ETO–associated leukemogenesis.
Discussion
In this study, we found that E2-2 is specifically silenced in t(8;21) patient-derived leukemic cell lines, thereby explaining why E2-2 is completely absent from AETFC. In confirmation of the hypothesis that E2-2 negatively regulates AML1-ETO–associated leukemogenesis, we discovered that E2-2 plays a distinct role relative to E2A and HEB, although the three E proteins, if present, can equally integrate into AETFC. Our functional and genome-wide studies suggest a model for the mechanism (Fig. 6E), in which the different E proteins and related heterogenous AETFC complexes may target specific genes that differentially alter cell fates and contribute to leukemogenesis. Restoring E2-2 expression fulfils a potential for dendritic differentiation of the cells in the presence of AML1-ETO, and suppression of E2-2 accelerates leukemogenesis in vivo. Thus, this study provides insights into the mechanism of AETFC regulation of gene expression and leukemogenesis and valuable information for identifying potential therapeutic targets.
AETFC is a stable complex containing multiple transcription factors and cofactors, including the oligomerized AML1-ETO, the AML1-binding partner CBFβ, E proteins E2A and HEB, the hematopoietic class II bHLH transcription factor LYL1, the LIM-domain protein LMO2, and its interacting partner LDB1 (30). The stability of AETFC is achieved by strong multivalent interactions among these components, which colocalize genome-wide and stabilize each other. However, this stability does not necessarily mean that AETFC is homogenous. In fact, the involvement of two E proteins (E2A and HEB) in AETFC already implies a potential heterogeneity. In support of this notion, we previously found that knockdown of either E2A or HEB in Kasumi-1 cells is inefficient in disrupting AETFC, whereas the joint knockdown of E2A and HEB dramatically decreases the protein levels of AETFC components (30). Notably, compared with E2A and HEB, both of which positively contribute to AML1-ETO–mediated leukemogenesis, we herein have found that E2-2 plays a negative role. Although the absence of E2-2 in the AML1-ETO–expressing leukemic cell lines makes it unfeasible to directly purify an E2-2–containing AETFC, our studies in primary cells and overexpression experiments suggest that E2-2 is expressed normally in the HSPCs that originally acquire AML1-ETO and can be integrated into AETFC in the same manner as E2A and HEB. Thus, E2-2, E2A, and HEB can define an AETFC heterogeneity in which these three E proteins can be integrated competitively or cooperatively, allowing them to play both overlapping and distinct functions.
In parallel with herein-identified distinct functions of E2-2 relative to E2A and HEB in AML1-ETO–mediated leukemogenesis, functional differences of the E proteins in normal development have been evidently observed in mouse genetic studies. In particular, knockout of E2-2, E2A, or HEB in mice predominantly affects the development of plasmacytoid dendritic cells (40), B cells (37), and T cells (38), respectively. The underlying mechanism can be explained in two different ways: expression levels versus intrinsic structural properties of the E proteins. First, although the E proteins are broadly expressed in different tissues, their expression levels are not equal and the functionally dominant E protein in a certain tissue could be relatively highly expressed or tightly regulated at the transcriptional level. In supportive of this possibility, it has been observed that a combined dosage of E2-2, E2A, and HEB regulates B-cell development (38) and, interestingly, that endogenous E2A can be functionally replaced by E2A-promoter–driven expression of HEB in supporting B-cell development (71). These observations suggest that the functional specificity of E proteins could be determined at least partially by their expression levels and independently of their structural divergence. Second, and in complete contrast, the intrinsic structural characteristics of E proteins can also contribute to their tissue-specific functions. For example, E2A has been found to form a homodimer that is restricted in B cells (72), and this homodimerization is determined either by its phosphorylation status (73) or by an intermolecular disulfide bound (74), neither of which is observed in E2-2 or HEB. In this study, using an approach of domain swapping between E2-2 and HEB in the same cellular context, we have unequivocally shown that the bHLH DNA-binding domain is critical for the functional specificity of E2-2, which is in agreement with the divergence of the E2-2 bHLH domain from the E2A and HEB bHLH domains. By ChIP-seq and RNA-seq analyses, we also confirmed the differential recognition of target genes by E2-2 relative to E2A and HEB. Thus, our results provide conclusive evidence for the above-described second mechanism, in which the intrinsic structural characteristics of the E2-2 bHLH domain define the functional specificity by targeting specific genes.
It is striking that E2-2 is completely silenced in both Kasumi-1 and SKNO-1 cell lines, and it remains a mystery as to when and how E2-2 is silenced during the progression of these leukemia cases. At least in these two cell lines, ChIP-seq data suggest that E2-2 is not directly repressed by AML1-ETO, and that it is likely silenced by a more stable epigenetic mechanism. Although we cannot retrace the clonal evolutionary history of the leukemia patients from whom the two cell lines were derived, it would be interesting to investigate this aspect in live patients and to determine when the E2-2 silencing/repression occurs. Given that t(8;21) is believed to be produced in HSPCs in which E2-2 is normally expressed, the E2-2 silencing/repression should emerge as a secondary event after the expression of AML1-ETO. Further pursuit of this issue with innovative single-cell approaches would be very helpful to understand whether and how E2-2 contributes to clonal evolution of leukemia cells.
Blockage of differentiation has been considered to be one of the major mechanisms by which AML1-ETO drives leukemogenesis (75, 76). Differentiation therapy, a concept already being exceptionally successful in treatment of acute promyelocytic leukemia (77), thus has been sought to treat AML1-ETO–associated leukemia. While remaining far from a success in the clinic, experiments with cultured cells and animal models have shown that differentiation of AML1-ETO–associated leukemia can be achieved by application of certain drugs (e.g., histone deacetylase inhibitors) (78–80) or by overexpression of transcription factors such as C/EBPα (81). Notably, differentiation of these cells is mostly accompanied by degradation of AML1-ETO. Indeed, siRNA-mediated AML1-ETO knockdown per se can increase the susceptibility of these cells to differentiation (82). In this study, however, we found that overexpression of E2-2 can induce a partial dendritic differentiation of the cells without degradation of AML1-ETO. As an explanation for this observation, it is possible that the ability of AML1-ETO to inhibit E2-2–mediated dendritic differentiation is not as strong as differentiation toward myeloid and erythroid lineages. Interestingly, the ETO family protein MTG16/ETO2 has been shown to cooperate with E2-2 to promote dendritic differentiation (83). Along this line, it is conceivable that, during leukemia progression, if AML1-ETO emerges in an HSPC that highly expresses E2-2 and is committed to the dendritic lineage, AML1-ETO may not be able to efficiently block differentiation to cause leukemogenesis; suppression/silencing of E2-2 thus offers this AML1-ETO–positive cell an advantage to propagate as leukemia. Last, as dendritic cells are antigen-presenting cells that stimulate immune responses, efforts have been made to convert leukemic cells directly into dendritic cells to present leukemia-specific antigen to immune effector cells (84, 85). To this end, our study provides a promising potential for an efficient dendritic differentiation of AML1-ETO–associated leukemia and for development of immune-based therapeutics for t(8;21) leukemia.
Materials and Methods
Cell Culture, Proliferation, Differentiation, and Colony Assays.
Kasumi-1 cells were cultured in RPMI medium 1640 (Gibco) supplemented with 10% FBS and penicillin/streptomycin. Cell proliferation and viability was determined by cell counting and CCK-8 (Bimake). Competitive growth (dis)advantage was evaluated by calculating the percentage of analyzed (e.g., GFP+) cells in total cells at different time points. Dendritic cell differentiation was induced by adding 20 ng/mL recombinant human stem cell factor (hSCF), 20 ng/mL recombinant human granulocyte–macrophage colony stimulating factor (hGM-CSF), 20 ng/mL recombinant human interleukin-4 (hIL-4), and 100 ng/mL recombinant human fms-like tyrosine kinase 3 ligand (hFLT3L), as previously reported (86). For colony assay (87), 1,000 cells were cultured in Methocult H4435 medium (StemCell Technologies), and the colony number was scored at day 7 or indicated time points under an inverted microscope.
Gene Overexpression and Knockdown.
Gene overexpression and knockdown were achieved by MIGR1-based retrovirus and pLKO.1-based lentivirus systems, respectively. For preparation of retroviruses, 293T cells were transfected with MIGR1-based plasmids and packaging plasmids VSVG and gag-pol by Lipofectamine 2000 (Invitrogen). For lentiviruses, 293T cells were transfected with pLKO.1-based plasmids and packaging plasmids pMD2G and PsPAx2. Viral supernatant was collected at 48 h after transfection and filtered with 0.45-μm Millex filters. Leukemic/hematopoietic cells were infected with 30% (vol/vol) filtered virus in the presence of 8 μg/mL Polybrene (Sigma) and centrifuged at 1,200 × g for 90 min at 37 °C. Medium was changed 12–20 h after infection. To improve efficiency, the cells could be infected twice. The AML1-ETO shRNA vector was reported previously (30), and the other shRNA vectors were purchased from Open Biosystems. The efficiency of knockdown was determined by RT-qPCR at 3 d after infection.
Co-IP Assay.
For coexpression of exogenous proteins, 293T cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen), and leukemic cells were infected with retroviruses expressing genes of interest. Cell lysis, protein binding, and immunoprecipitation were performed with T/G lysis buffer (20 mM Tris⋅HCl, pH 7.5, 300 mM NaCl, 50 mM NaF, 2 mM EDTA, 1% Triton X-100, and 20% glycerol), and proteins were analyzed by Western blot.
RT-PCR and RT-qPCR.
Total RNA was extracted with the TRIzol Reagent (Invitrogen), and cDNA was synthesized by the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). Standard PCR and qPCR were performed with the KOD Plus system (TOYOBO) and the SYBR Premix Ex Taq GC system (TaKaRa), respectively.
Flow Cytometry.
Flow cytometry analysis was performed on the BD LSRFortessa cell analyzer. An antibody for CD141 (BDCA-3) (MACS Miltenyi Biotec) was used to assess the CD141 expression and dendritic cell differentiation. Cell apoptosis analysis were performed by incubating 1 × 106 cells with APC Annexin V (BD Pharmingen) and PI at room temperature for 15 min, followed by flow cytometry analysis.
ChIP-Seq.
ChIP assay was performed as previously reported (88). In brief, GFP+ Kasumi-1 cells were sorted by flow cytometry at day 3 after retrovirus infection, cross-linked with 1% formaldehyde for 10 min, and stopped by 125 mM glycine. After lysis and sonication, 3 μg of HA-Tag (C29F4) rabbit monoclonal antibody (Cell Signaling Technology) was incubated with chromatin samples overnight at 4 °C, and 20 μL of rProtein A/G Beads 4FF (Smart Lifesciences) were used for immunoprecipitation. The DNA were purified with the MinElute PCR Purification Kit (QIAGEN) and subjected to library construction and next-generation sequencing with the Illumina systems. ChIP-seq reads were aligned to the hg19 version of human reference genome using Bowite2 (version 2.2.9). MACS2 (version 2.1.1) callpeak algorithm was used to identify regions of ChIP-seq enrichment over background. Bigwig files were then produced by MACS2 bdgcmp algorithm and visualized in IGV.
RNA-Seq.
GFP+ Kasumi-1 cells were harvested at day 4 after virus infection. RNA was extracted with the TRIzol Reagent (Invitrogen). Libraries were generated using the TruSeq RNA Sample Preparation Kit, version 2 (Illumina), and sequenced with Illumina HiSeq 2500. TopHat (version 2.0.9) was used to align the reads to the genome and HTseq was used to calculate fragments per kilobase of exon model per million mapped reads (FPKM) of the genes. GSEA and GO analyses were performed to interpret the function and mechanism of the differentially expressed genes. ChIP-seq and RNA-seq data were deposited in the GEO database (accession no. GSE114644) (89).
Mouse Leukemia Transplantation Assay.
The transplantable AML1-ETO9a–driven leukemic cells were infected with lentiviruses expressing control or E2-2 shRNAs, screened by puromycin for 3 d, and transplanted into sublethally irradiated recipient mice by tail vein injection. Leukemia development was determined by flow cytometry and morphological analyses of peripheral blood and BM cells. For bioluminescent imaging of leukemic cells in vivo, bioluminescent imaging was performed using an IVIS100 imaging system upon injection of the substrate of luciferase, d-Luciferin. Overall survival of the mice was analyzed by the Kaplan–Meier method, and the statistical significance was evaluated by the log rank test.
Patients and Samples.
Fifty children with t(8;21) AML were diagnosed and treated in the Shanghai Children’s Medical Center, and this study was approved by the Institutional Review Board of the center. Leukemic cells of these patients were subjected to RNA-seq following the protocol as described above. The patients were stratified into groups based on their gene expression patterns, and the Kaplan–Meier method and log rank test were used to analyze relapse rate of different groups of the patients.
Supplementary Material
Acknowledgments
We thank Y. Zhai, X. Miao, S. Yan, Y. Chen, K. Wang, and Y. Wang at the Shanghai Institute of Nutrition and Health core facilities for technical support. This work was supported by the National Key Research and Development Plan of China [Grants 2018YFA0107802 (to X.-J.S.) and 2018YFA0107202 (to L.W.)]; the Chinese Academy of Sciences (CAS) Bureau of Frontier Sciences and Education Program [Grant QYZDBSSW-SMC027 (to L.W.)]; the National Natural Science Foundation of China (NSFC) General Program [Grants 81470316 and 81670094 (to X.-J.S.), 81470334 and 81670122 (to L.W.), and 81270623 (to S.S.)]; the NSFC Excellent Young Scholar Program [Grant 81622003 (to L.W.)]; Chinese National Key Basic Research Project 2013CB966801 (to X.-J.S.); the National Key Research and Development Plan of China [Grant 2016YFC0902202 (to L.W.)]; NIH R01 Grants CA163086 and CA178765 (to R.G.R.) and CA166835 (to S.D.N.); Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant 20152506 (to X.-J.S.); CAS Bureau of Major R&D Program [Grant XDA12010310 (to L.W.)]; the Shanghai Pudong Innovation Fund [Grant PDJ2014-04 (to J. Song)]; the fourth round of Three-Year Public Health Action Plan Grant GWIV-25 (to S.S.); Shanghai Municipal Science and Technology Major Project 2017SHZDZX01 (to F.L.); 111 Project B17029 (to X.-J.S., R.G.R., and S.-J.C.); and the Samuel Waxman Cancer Research Foundation. X.-J.S. and L.W. were supported by the 1000 Talents Program for Young Scholars.
Footnotes
Conflict of interest statement: S.D.N. has coauthored papers with J.L. and O.A.-W. but has not collaborated directly with them. R.G.R. and J.L. are both investigators on a program grant but on separate projects.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE114644).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809327116/-/DCSupplemental.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: Genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 5.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat AS, Vakoc CR. Targeting transcription factors in cancer. Trends Cancer. 2015;1:53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Thé H. Differentiation therapy revisited. Nat Rev Cancer. 2018;18:117–127. doi: 10.1038/nrc.2017.103. [DOI] [PubMed] [Google Scholar]
- 8.Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–5679. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- 9.Marcucci G, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): A cancer and leukemia group B study. J Clin Oncol. 2005;23:5705–5717. doi: 10.1200/JCO.2005.15.610. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci USA. 2000;97:7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulloy JC, et al. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002;99:15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- 12.Ahn MY, et al. Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO. Proc Natl Acad Sci USA. 1998;95:1812–1817. doi: 10.1073/pnas.95.4.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westendorf JJ, et al. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonks A, et al. Expression of AML1-ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia. 2004;18:1238–1245. doi: 10.1038/sj.leu.2403396. [DOI] [PubMed] [Google Scholar]
- 15.Tonks A, et al. The AML1-ETO fusion gene promotes extensive self-renewal of human primary erythroid cells. Blood. 2003;101:624–632. doi: 10.1182/blood-2002-06-1732. [DOI] [PubMed] [Google Scholar]
- 16.Schwieger M, et al. AML1-ETO inhibits maturation of multiple lymphohematopoietic lineages and induces myeloblast transformation in synergy with ICSBP deficiency. J Exp Med. 2002;196:1227–1240. doi: 10.1084/jem.20020824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson LF, Zhang DE. The 8;21 translocation in leukemogenesis. Oncogene. 2004;23:4255–4262. doi: 10.1038/sj.onc.1207727. [DOI] [PubMed] [Google Scholar]
- 18.Nimer SD, Moore MA. Effects of the leukemia-associated AML1-ETO protein on hematopoietic stem and progenitor cells. Oncogene. 2004;23:4249–4254. doi: 10.1038/sj.onc.1207673. [DOI] [PubMed] [Google Scholar]
- 19.Elagib KE, Goldfarb AN. Oncogenic pathways of AML1-ETO in acute myeloid leukemia: Multifaceted manipulation of marrow maturation. Cancer Lett. 2007;251:179–186. doi: 10.1016/j.canlet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993;82:712–715. [PubMed] [Google Scholar]
- 21.Miyamoto T, et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87:4789–4796. [PubMed] [Google Scholar]
- 22.Mao S, Frank RC, Zhang J, Miyazaki Y, Nimer SD. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: Implications for the pathogenesis of t(8;21)-positive leukemias. Mol Cell Biol. 1999;19:3635–3644. doi: 10.1128/mcb.19.5.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elagib KE, et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 24.Gelmetti V, et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutterbach B, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science. 2011;333:765–769. doi: 10.1126/science.1201662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shia WJ, et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood. 2012;119:4953–4962. doi: 10.1182/blood-2011-04-347476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen M, et al. JMJD1C is required for the survival of acute myeloid leukemia by functioning as a coactivator for key transcription factors. Genes Dev. 2015;29:2123–2139. doi: 10.1101/gad.267278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun XJ, et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature. 2013;500:93–97. doi: 10.1038/nature12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 32.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 33.Massari ME, Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharya A, Baker NE. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell. 2011;147:881–892. doi: 10.1016/j.cell.2011.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 37.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.in ’t Hout FE, van der Reijden BA, Monteferrario D, Jansen JH, Huls G. High expression of transcription factor 4 (TCF4) is an independent adverse prognostic factor in acute myeloid leukemia that could guide treatment decisions. Haematologica. 2014;99:e257–e259. doi: 10.3324/haematol.2014.110437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghisi M, et al. Id2 and E proteins orchestrate the initiation and maintenance of MLL-rearranged acute myeloid leukemia. Cancer Cell. 2016;30:59–74. doi: 10.1016/j.ccell.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Man N, et al. Differential role of Id1 in MLL-AF9-driven leukemia based on cell of origin. Blood. 2016;127:2322–2326. doi: 10.1182/blood-2015-11-677708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 45.Gow CH, Guo C, Wang D, Hu Q, Zhang J. Differential involvement of E2A-corepressor interactions in distinct leukemogenic pathways. Nucleic Acids Res. 2014;42:137–152. doi: 10.1093/nar/gkt855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ptasinska A, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26:1829–1841. doi: 10.1038/leu.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Micol JB, et al. ASXL2 is essential for haematopoiesis and acts as a haploinsufficient tumour suppressor in leukemia. Nat Commun. 2017;8:15429. doi: 10.1038/ncomms15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rapin N, et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood. 2014;123:894–904. doi: 10.1182/blood-2013-02-485771. [DOI] [PubMed] [Google Scholar]
- 50.Chambers SM, et al. Hematopoietic fingerprints: An expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Tullio A, et al. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc Natl Acad Sci USA. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haferlach T, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunne J, et al. siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene. 2006;25:6067–6078. doi: 10.1038/sj.onc.1209638. [DOI] [PubMed] [Google Scholar]
- 54.Tsai HC, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dalton WT, Jr, et al. HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood. 1988;71:242–247. [PubMed] [Google Scholar]
- 56.Chen WY, et al. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev. 2013;27:1596–1609. doi: 10.1101/gad.216192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weng S, et al. Restoration of MYC-repressed targets mediates the negative effects of GM-CSF on RUNX1-ETO leukemogenicity. Leukemia. 2017;31:159–169. doi: 10.1038/leu.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unternaehrer JJ, Chow A, Pypaert M, Inaba K, Mellman I. The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc Natl Acad Sci USA. 2007;104:234–239. doi: 10.1073/pnas.0609665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W, et al. Thrombopoietin cooperates with FLT3-ligand in the generation of plasmacytoid dendritic cell precursors from human hematopoietic progenitors. Blood. 2004;103:2547–2553. doi: 10.1182/blood-2003-09-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verschoor A, et al. A platelet-mediated system for shuttling blood-borne bacteria to CD8α+ dendritic cells depends on glycoprotein GPIb and complement C3. Nat Immunol. 2011;12:1194–1201. doi: 10.1038/ni.2140. [DOI] [PubMed] [Google Scholar]
- 61.Bachem A, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jongbloed SL, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poulin LF, et al. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207:1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chou FS, et al. The thrombopoietin/MPL/Bcl-xL pathway is essential for survival and self-renewal in human preleukemia induced by AML1-ETO. Blood. 2012;120:709–719. doi: 10.1182/blood-2012-01-403212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pulikkan JA, et al. Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling. Blood. 2012;120:868–879. doi: 10.1182/blood-2012-03-414649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Decker M, Leslie J, Liu Q, Ding L. Hepatic thrombopoietin is required for bone marrow hematopoietic stem cell maintenance. Science. 2018;360:106–110. doi: 10.1126/science.aap8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, et al. Regulation of AKT signaling by Id1 controls t(8;21) leukemia initiation and progression. Blood. 2015;126:640–650. doi: 10.1182/blood-2015-03-635532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan M, et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 70.Rasmussen KD, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhuang Y, Barndt RJ, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen CP, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sloan SR, Shen CP, McCarrick-Walmsley R, Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 75.Hatlen MA, Wang L, Nimer SD. AML1-ETO driven acute leukemia: Insights into pathogenesis and potential therapeutic approaches. Front Med. 2012;6:248–262. doi: 10.1007/s11684-012-0206-6. [DOI] [PubMed] [Google Scholar]
- 76.Lin S, Mulloy JC, Goyama S. RUNX1-ETO leukemia. Adv Exp Med Biol. 2017;962:151–173. doi: 10.1007/978-981-10-3233-2_11. [DOI] [PubMed] [Google Scholar]
- 77.Wang ZY, Chen Z. Acute promyelocytic leukemia: From highly fatal to highly curable. Blood. 2008;111:2505–2515. doi: 10.1182/blood-2007-07-102798. [DOI] [PubMed] [Google Scholar]
- 78.Wang J, Saunthararajah Y, Redner RL, Liu JM. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1-ETO leukemia cells. Cancer Res. 1999;59:2766–2769. [PubMed] [Google Scholar]
- 79.Liu S, et al. Targeting AML1/ETO-histone deacetylase repressor complex: A novel mechanism for valproic acid-mediated gene expression and cellular differentiation in AML1/ETO-positive acute myeloid leukemia cells. J Pharmacol Exp Ther. 2007;321:953–960. doi: 10.1124/jpet.106.118406. [DOI] [PubMed] [Google Scholar]
- 80.Bots M, et al. Differentiation therapy for the treatment of t(8;21) acute myeloid leukemia using histone deacetylase inhibitors. Blood. 2014;123:1341–1352. doi: 10.1182/blood-2013-03-488114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pabst T, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 82.Heidenreich O, et al. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood. 2003;101:3157–3163. doi: 10.1182/blood-2002-05-1589. [DOI] [PubMed] [Google Scholar]
- 83.Ghosh HS, et al. ETO family protein Mtg16 regulates the balance of dendritic cell subsets by repressing Id2. J Exp Med. 2014;211:1623–1635. doi: 10.1084/jem.20132121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moldenhauer A, et al. Histone deacetylase inhibition improves dendritic cell differentiation of leukemic blasts with AML1-containing fusion proteins. J Leukoc Biol. 2004;76:623–633. doi: 10.1189/jlb.1103581. [DOI] [PubMed] [Google Scholar]
- 85.Anguille S, et al. Dendritic cells as pharmacological tools for cancer immunotherapy. Pharmacol Rev. 2015;67:731–753. doi: 10.1124/pr.114.009456. [DOI] [PubMed] [Google Scholar]
- 86.Helft J, et al. Dendritic cell lineage potential in human early hematopoietic progenitors. Cell Rep. 2017;20:529–537. doi: 10.1016/j.celrep.2017.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Man N, et al. Caspase-3 controls AML1-ETO-driven leukemogenesis via autophagy modulation in a ULK1-dependent manner. Blood. 2017;129:2782–2792. doi: 10.1182/blood-2016-10-745034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu N, et al. 2018 Different roles of E proteins in t(8;21) leukemia: E2-2 compromises the function of AETFC and negatively regulates leukemogenesis. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114644. Deposited May 18, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.