Significance
Merkel cell carcinoma (MCC) is an aggressive skin cancer. While virus-negative [Merkel cell polyomavirus (MCV)] MCC contains inactivating mutations in RB and p53, MCV-positive MCC usually contains wild-type RB and p53. We demonstrate that MCV large T antigen binding to RB results in p53 activation, while MCV small T antigen reduces p53 activation by increasing levels of MDM2 and CK1, an activator of MDM4. Targeted degradation of CK1 by lenalidomide or a specific MDM4 inhibitor acts synergistically with MDM2 inhibitors to activate p53 and induce apoptosis. Our work uncovers the mechanism behind MCV control of p53 in MCC and demonstrates the utility of targeting MDM2 and MDM4 combinatorially in p53 wild-type tumors.
Keywords: p53, Merkel cell carcinoma, MDM2–MDM4, casein kinase 1 alpha, lenalidomide
Abstract
Merkel cell polyomavirus (MCV) contributes to approximately 80% of all Merkel cell carcinomas (MCCs), a highly aggressive neuroendocrine carcinoma of the skin. MCV-positive MCC expresses small T antigen (ST) and a truncated form of large T antigen (LT) and usually contains wild-type p53 (TP53) and RB (RB1). In contrast, virus-negative MCC contains inactivating mutations in TP53 and RB1. While the MCV-truncated LT can bind and inhibit RB, it does not bind p53. We report here that MCV LT binds to RB, leading to increased levels of ARF, an inhibitor of MDM2, and activation of p53. However, coexpression of ST reduced p53 activation. MCV ST recruits the MYC homologue MYCL (L-Myc) to the EP400 chromatin remodeler complex and transactivates specific target genes. We observed that depletion of EP400 in MCV-positive MCC cell lines led to increased p53 target gene expression. We suspected that the MCV ST–MYCL–EP400 complex could functionally inactivate p53, but the underlying mechanism was not known. Integrated ChIP and RNA-sequencing analysis following EP400 depletion identified MDM2 as well as CK1, an activator of MDM4, as target genes of the ST–MYCL–EP400 complex. In addition, MCV-positive MCC cells expressed high levels of MDM4. Combining MDM2 inhibitors with lenalidomide targeting CK1 or an MDM4 inhibitor caused synergistic activation of p53, leading to an apoptotic response in MCV-positive MCC cells and MCC-derived xenografts in mice. These results support dual targeting of MDM2 and MDM4 in virus-positive MCC and other p53 wild-type tumors.
Merkel cell carcinoma (MCC) is an aggressive neuroendocrine carcinoma of the skin with an incidence in the United States that has tripled in the last two decades (1, 2). In 2008, Feng et al. (3) discovered Merkel cell polyomavirus (MCV; MCPyV) clonally integrated in 8 of 10 MCC tumors. MCV-positive MCC contains integrated copies of the MCV genome and expresses small T antigen (ST) and a truncated form of large T antigen (LT) (4). MCC tumor-associated truncated LT retains the N-terminal LXCXE, RB-binding motif, but deletes the C-terminal DNA-binding and helicase domains required for viral replication (3). Expression of MCV ST and truncated LT can promote proliferation and transformation in several cell types, consistent with their oncogenic roles in MCC (5).
The prototypic polyomavirus Simian vacuolating virus 40 (SV40) LT binds to the retinoblastoma-associated protein RB (RB1) and the cellular tumor antigen p53 (TP53) and inactivates their tumor-suppressive functions (6). In contrast, MCV LT binds to RB, but not p53 (6). Next-generation sequencing of MCC reveals that virus-negative MCC typically harbors p53 and RB mutations along with a UV mutational signature (7, 8). In contrast, virus-positive MCC usually contains wild-type RB and p53 and no evidence for UV-induced mutations (7, 8). Given the presence of wild-type p53 in virus-positive MCC, we suspected that MCV T antigens could functionally inactivate p53 activity.
p53 is mutated in a wide variety of cancers. Alternatively, wild-type p53 can be functionally inactivated by overexpression of MDM2, a ubiquitin ligase targeting p53, or MDM4 (MDMX) (9, 10). MDM2 and MDM4 both have similar structures with N-terminal p53 binding and C-terminal RING domains (11). Although MDM4 does not directly ubiquitinate p53, its RING domain facilitates the recruitment of ubiquitin to MDM2 (11). MDM4 also has an autoinhibitory domain that reduces binding to p53 (12). The MDM4 autoinhibitory interaction can be relieved by casein kinase 1 alpha (CK1, CSNK1A1) (13).
Here, we demonstrate that MCV ST functions as a transcriptional activator to increase levels of MDM2 and CK1 that, in turn, cooperate with MDM4 to inhibit p53 function in MCC. We demonstrate the synergistic efficacy of targeting both MDM2 and MDM4 in MCC.
Results and Discussion
LT Activates and ST Dampens the p53 Response.
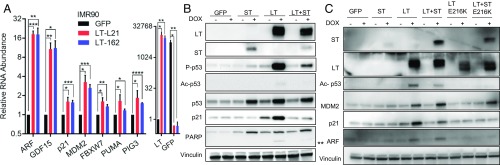
To study the effect of MCV T antigens on p53 in normal cells, a doxycycline-inducible vector expressing GFP or tumor-derived truncated or full-length forms of LT was introduced into IMR90 diploid lung fibroblasts (Fig. 1A and SI Appendix, Fig. S1A). The truncated forms of LT include L21 (encoding residues 1–292), 162 (residues 1–320), and 168 (residues 1–275), each containing an intact LXCXE motif (6). LT-L21 or -162 expression in IMR90 significantly increased levels of ARF and several p53 target genes, including GDF15 and p21 (CDKN1A), as assessed by quantitative RT-PCR (RT-qPCR) (Fig. 1A). Inhibition of RB activates the E2F transcription factors, leading to increased levels of ARF (10). ARF is a potent inhibitor of the major p53-degrading E3 ligase MDM2 (14). LT expression increased protein levels of p53 as well as phospho-serine 15 p53 (P-p53) and p21, indicative of p53 activation. Expression of LT-162 also led to increased levels of cleaved PARP(**), indicating an apoptotic response.
Fig. 1.
MCPyV LT activates and ST dampens the p53 response. (A) Inducible expression of truncated tumor isoforms of MCV LT increases ARF and p53 target genes in IMR90 cells. Expression of GFP or LT-L21 and -162 truncated LT was induced with doxycycline (DOX) treatment for 24 h. The LT, ARF, and p53 target gene RNA levels were normalized to those of the GFP-induced cells, whereas the GFP levels were normalized to the LT-L21 samples. Data are shown as mean ± SD. *P 0.05; **P 0.005; ***P 0.0005; ****P 0.00005 (Student t test). (B) IMR90 cells were induced to express GFP, ST, LT-L21, or LT-L21 with ST for 40 h. Lysates were prepared before (−) or after (+) DOX. Activation of p53 response is reflected by increased levels of p53, P-p53, acetyl-lysine 382 p53 (Ac-p53), p21, and cleaved PARP (**). (C) Expression of L21, but not an LT mutant in the LXCXE motif (E216K), activates p53 through inhibiting RB and inducing ARF.
LT and ST are coexpressed as splice variants from the integrated MCPyV viral DNA in MCC tumors. To mimic this in IMR90 cell lines, we introduced a genomic version of LT-L21 that coexpresses ST. When ST was coexpressed with LT-L21, lower levels of p53 activation (p21, P-p53, and Ac-p53) were observed compared with the response to LT-L21 only (Fig. 1B and SI Appendix, Fig. S1B). The results indicate that truncated LT can activate p53, while ST can reduce this response when coexpressed.
To determine whether increased levels of ARF required LT binding to RB, we introduced a point substitution mutation in the LXCXE motif (E216K) of LT-L21. When stably expressed in HCT116 cells, L21-E216K was unable to coprecipitate RB, but retained binding to VPS39 that binds to a different region of LT (SI Appendix, Fig. S1C) (5). When expressed in IMR90 cells, L21-E216K did not increase levels of ARF, p21, p53, or Ac-p53 (Fig. 1C). These results indicate that LT binding to RB contributes to increased levels of ARF and activation of the p53 response.
MDM2 and CK1 Are Transcriptional Targets of the ST–MYCL–EP400 Complex.
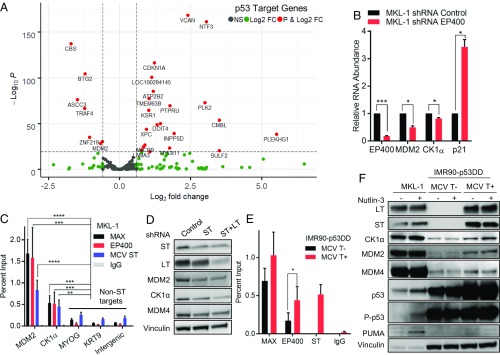
We recently reported that MCV ST recruits the MYC homologue MYCL (L-Myc) to the EP400 chromatin remodeler complex to bind specific gene promoters and activate their expression (15). To identify genes regulated by the ST–MYCL–EP400 complex in MKL-1 cells, RNA-sequencing (RNA-seq) was performed after depleting EP400 by using three different shRNAs (15). Using the reported RNA-seq results, we assessed changes in gene expression of known p53 target genes (9). EP400 depletion led to increased levels of many p53 target genes, including p21 (CDKN1A) (Fig. 2A). By using the fold change cutoff of 1.5, a total of 59 genes were up-regulated and 17 were down-regulated of 198 total p53 target genes (SI Appendix, Table S1) (9). EP400 depletion also led to a decrease in MDM2 E3 ligase and CK1 levels (Fig. 2B and SI Appendix, Fig. S2 A–C). MDM2 and MDM4 contain an N-terminal p53-binding domain that binds directly to the transactivation domain of p53 to block p53 activation (11). MDM4 can regulate its own activity toward p53. The p53-binding domain of MDM4 forms an intramolecular interaction with its central W motif region and thereby reduces binding to p53 (12). CK1 (CSNK1A) is a serine/threonine kinase that binds and phosphorylates MDM4, which in turn prevents this autoinhibitory interaction and activates MDM4 (13). RT-qPCR and Western blotting confirmed that p21 levels increased and MDM2 and CK1 levels decreased upon EP400 knockdown in MKL-1 cells (Fig. 2B and SI Appendix, Fig. S2C).
Fig. 2.
MDM2 and CK1 are transcriptional targets of the ST–MYCL–EP400 complex. (A) Volcano plot illustrating differentially expressed p53 target genes in MKL-1 MCC cell lines after depletion of EP400 with inducible shRNA relative to control shRNA. Each gene log2 fold change (FC) was plotted against the −log10 P value for statistical significance. Green dots indicate genes that meet the twofold change cutoff, and red dots signify adjusted P < 0.1. (B) RT-qPCR was performed with MKL-1 cells after shRNA was induced for 8 d. Reads were normalized to RPLP0 and uninduced samples. The experiment was performed three times and averaged. Data are shown as mean ± SD. *P 0.05; **P 0.005; ***P 0.0005; ****P 0.00005 (Student t test). (C) ChIP with MAX, EP400, ST, and IgG antibodies followed by quantitative PCR (qPCR) of indicated promoters in MKL-1 cells. ChIP-qPCR was performed three times with average percent input shown. (D) Depletion of MCV T antigens causes a reduction in the MDM2, CK1, and MDM4 levels. MKL-1 cells were transduced with shRNAs targeting ST or ST and LT. (E) ChIP-qPCR for the MDM2 promoter in IMR90 cells in the presence (+) or absence (−) of MCV T antigens. ChIP was performed five times. (F) Nutlin-3 treatment does not elicit p53 response, but MCV T antigens increase levels of MDM2, MDM4, and CK1 in IMR90 cells expressing p53DD. MKL-1 and IMR90-p53DD were treated with nutlin-3 (1 M) for 24 h.
ChIP-sequencing (ChIP-seq) with antibodies to ST, MAX (which dimerizes with MYCL), and EP400 revealed enrichment for the MDM2 and CK1 promoters (SI Appendix, Fig. S2E) (15). We performed ChIP-qPCR for MAX, ST, and EP400 and observed specific enrichment for these promoters (Fig. 2C). We depleted ST or ST and LT using specific shRNAs in MKL-1 cells and found that the levels of MDM2 and CK1 decreased (Fig. 2D) (16). This result together with the RT-qPCR and ChIP data indicates that MDM2 and CK1 are direct transcriptional targets of the ST–MYCL–EP400 complex. Of note, MDM4 levels decreased upon depletion of ST, although we did not find evidence for direct activation of MDM4 by ST. ST increases levels of CK1 that could serve to activate MDM4 activity toward p53.
Since MDM2 is a p53 target gene, it is possible that the MCV T antigens indirectly increase MDM2 levels by activating p53 (9). To exclude this possibility, we introduced a dominant-negative p53 (p53DD) that binds and inactivates the endogenous p53 into IMR90 cells (17). The IMR90-p53DD cells were further transduced with MYCL and MCV LT-L21 with ST (17). We detected ST binding to the MDM2 and CK1 promoters by ChIP-qPCR and observed that EP400 enrichment to the MDM2 promoter increased in the presence of MCV T antigens (Fig. 2E and SI Appendix, Fig. S2F). However, MDM2 inhibitor treatment with nutlin-3 did not increase p53, P-p53, and PUMA levels, indicating that p53 failed to become activated in the presence of p53DD (Fig. 2F). To further determine whether the ST–MYCL–EP400 complex binds to these promoters independently of p53, we depleted p53 in MKL-1 with shRNA and performed ChIP-qPCR of EP400 and ST (SI Appendix, Fig. S2G). We found that p53 depletion did not affect EP400 and ST enrichment to the MDM2 and CK1 promoters.
MDM4 Is Overexpressed in Virus-Positive MCC.
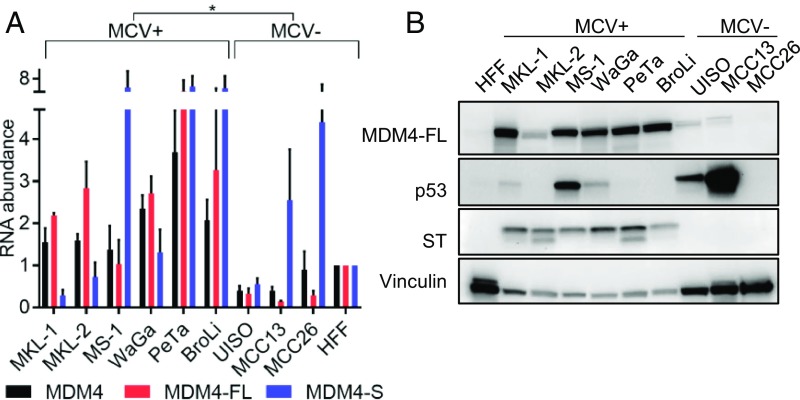
Overexpression of MDM4 can be found in some cancers with wild-type p53 (18). We used three sets of MDM4 primers to assess total MDM4 (all splice variants), MDM4-FL (full-length), and MDM4-S (short variant missing the RING domain) levels in MCC cell lines (18). Virus-positive (MKL-1, -2, MS-1, WaGa, PeTa, and BroLi) MCC cell lines had significantly higher levels of total MDM4 and MDM4-FL relative to the virus-negative (UISO, MCC13, and MCC26) MCC lines (Fig. 3A). Furthermore, the MDM4-FL to -S ratio was higher in virus-positive lines (SI Appendix, Fig. S3A). We assessed MDM4-FL protein levels in these MCC cell lines. Virus-positive MCC cell lines expressed high levels of MDM4-FL compared with virus-negative MCC cell lines (Fig. 3B). Of note, the virus-negative UISO cell line had a high MDM4-FL to -S ratio, but did not express abundant levels of MDM4 protein. LT’s activation of p53 in virus-positive MCC may create dependency on MDM2 and MDM4 expression. Targeted next-generation sequencing (Oncopanel) of 101 MCC tumors revealed frequent low-copy gains (three to eight copies) of MDM4 regardless of the presence of MCV (SI Appendix, Fig. S3B) (19).
Fig. 3.
MDM4 is overexpressed in MCV-positive MCC. (A) RNA from MCC cell lines and human foreskin fibroblasts (HFF) was harvested for RT-qPCR for MDM4 (total), MDM4-FL (full-length variant), and MDM4-S (short splice variant). MDM4 levels were normalized with the geomean of RPLP0, 18s rRNA, and beta-actin RNA controls. Data are shown as mean ± SD. *P 0.05 for MDM4-FL (Student t test). (B) Western blot of MCC cell lines and HFF with indicated antibodies. MS-1 and MCC13 overexpress p53 due to inactivating mutations, and MKL-2 and MCC26 do not express detectable levels of p53.
Inhibition of MDM2 and MDM4 Activates p53 in MCC.
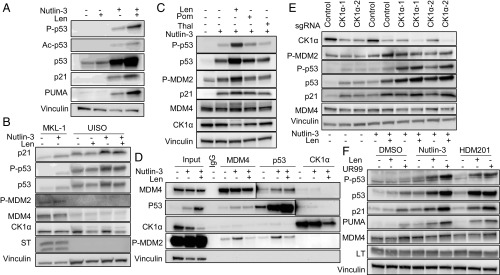
We observed that nutlin-3 MDM2 inhibitor treatment activated p53 as shown by increased levels of total p53, P-p53, Ac-p53, p21, and PUMA in MKL-1, WaGa, PeTa, and BroLi MCC cell lines containing wild-type p53, but not in MS-1 cells harboring an inactivating p53 mutation (Figs. 2F and 4A and SI Appendix, Fig. S2B) (20). Recent reports demonstrated that the CRBN (Cereblon) E3 ligase can specifically target CK1 for ubiquitination in the presence of lenalidomide (21, 22). We assessed whether lenalidomide could decrease CK1 levels and activate p53 in MCC cells. MKL-1 cells were treated with lenalidomide with or without cycloheximide to block protein synthesis. Although CK1 levels did not change appreciably with cycloheximide treatment alone for 6 h, levels rapidly decreased following the addition of lenalidomide (SI Appendix, Fig. S4A).
Fig. 4.
Inhibition of MDM2 and MDM4 enhances p53 activation in MCC cell lines. (A) Lenalidomide enhances p53 activation by nutlin-3 in MKL-1. MKL-1 cells were treated with nutlin-3 (5 M), lenalidomide (Len; 10 M), or both for 40 h. (B) MKL-1 (MCV+ MCC) or UISO (MCV− MCC) cells were treated with nutlin-3 (1 M) with or without lenalidomide (10 M) for 24 h. Of note, UISO has less MDM2 and MDM4 proteins than MKL-1. (C) Lenalidomide and, to a lesser degree, pomalidomide, but not thalidomide, cooperated with nutlin-3 to activate p53. MKL-1 cells were treated with nutlin-3 with lenalidomide, pomalidomide (10 M), or thalidomide (10 M) for 24 h. (D) Lenalidomide treatment reduced MDM4 binding to p53 and activated MDM2. MKL-1 cells were treated with nutlin-3 (5 M), lenalidomide (10 M), or both for 40 h and harvested for immunoprecipitation (IP) with antibodies to MDM4, p53, and CK1 followed by Western blotting. (E) Depletion of CK1 by CRISPR transduction enhanced p53 activation by nutlin-3. MKL-1 cells stably expressing each of two CK1 single-guide RNAs (sgRNAs) were treated with nutlin-3 (1 M) with or without lenalidomide for 24 h. Lenalidomide further decreased CK1 that sgRNAs did not completely deplete. (F) MDM4 inhibitor SC-24-UR99 (UR99) cooperated with MDM2 inhibitors in activating p53. MKL-1 cells were treated with nutlin-3 (1 M) or HDM201 (0.1 M) with or without lenalidomide (1 M) or UR99 (0.1 M).
Lenalidomide treatment alone led to a modest increase in p53 levels. However, when nutlin-3 and lenalidomide were combined, larger increases in p53 and p53 target genes were observed in the cell lines with wild-type p53 (Fig. 4A and SI Appendix, Fig. S4B). We assessed p53 stability in MKL-1 cells treated with nutlin-3, lenalidomide, or both in the presence of cycloheximide by quantitative Western blotting (SI Appendix, Fig. S4D). Lenalidomide significantly increased the stability of p53 in the presence of nutlin-3 (23). Depletion of CK1 by lenalidomide may decrease MDM4’s activities toward p53. We tested whether lenalidomide enhances p53 activation in virus-negative and p53 wild-type UISO cells, which express little MDM2 and MDM4 (Fig. 3 A and B). We observed that lenalidomide decreased the CK1 levels but failed to enhance p53 activation by nutlin-3 in UISO cells (Fig. 4B).
Structural analysis of CRBN revealed that lenalidomide could strongly promote the interaction with CK1, while the related compounds thalidomide and pomalidomide were much less capable of recruiting CK1 to CRBN (22). Compared with lenalidomide, pomalidomide and thalidomide did not reduce CK1 protein levels in MKL-1 cells (SI Appendix, Fig. S4C). Furthermore, pomalidomide or thalidomide did not enhance the p53 response to nutlin-3 in MKL-1 cells (Fig. 4C).
To test the contribution of CK1 to MDM4 binding to p53, we performed IP for MDM4, p53, and CK1 with lysates prepared from MKL-1 cells treated with nutlin-3 and lenalidomide for 40 h (Fig. 4D and SI Appendix, Fig. S4E). Lenalidomide and nutlin-3 reduced CK1 levels and increased p53 levels relative to nutlin-3 only. Despite the increased levels of p53, lenalidomide cotreatment with nutlin-3 did not increase coprecipitation of p53 by MDM4, CK1, or activated MDM2 compared with untreated or nutlin-3–treated controls. This suggests that reduced levels of CK1 decreased MDM4 binding to p53.
If CK1 enables MDM4 binding to p53, we expected that loss of CK1 would reduce MDM4 binding to p53 and enhance p53 activation (13). To test this, we depleted CK1 in MKL-1 by two independent CRISPR sgRNAs and assessed p53 activation following nutlin-3 treatment (Fig. 4E) (24). CRISPR knockout of CK1 led to increased p53 activity assessed by increased levels of activated MDM2 (phospho-serine 166; P-MDM2), P-p53, p53, and p21, indicating that a reduction of CK1, either by lenalidomide or sgRNAs, enhances p53 activation (25). Notably, addition of lenalidomide reduced CK1 protein that was not completely depleted by CRISPR and further increased the marks of p53 activation. We tested whether a specific MDM4 inhibitor (SC-24-UR99, UR99) can enhance the activities of MDM2 inhibitors nutlin-3 or HDM201 (Fig. 4F). UR99 specifically blocks MDM4 binding to p53. In a biochemical time-resolved FRET assay measuring the interaction between MDM4 and p53, the potency of UR99 was 1.4 nM (IC50), while its potency in the MDM2-p53 FRET was >10,000 nM. In a cellular bioluminescence resonance energy transfer (BRET) assay measuring the interaction between MDM4 and p53, its potency was 13 nM (IC50), and it was 10,000 nM in the MDM2-p53 BRET. In OCI-AML2 cells, which express high levels of MDM4, it can inhibit proliferation with a potency of 21 nM (50% growth inhibition). We found that inhibition of MDM4 by UR99 increased p53 activation by the MDM2 inhibitors.
Lenalidomide Synergizes with MDM2 Inhibitors to Induce Apoptosis in MCC Cell Lines.
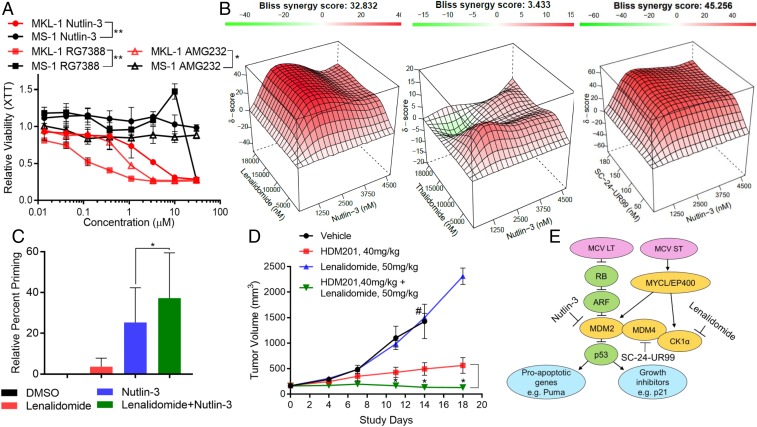
To test the effect of MDM2 inhibition on MCC cell viability, we treated MKL-1 (p53 wild type) and MS-1 (p53 mutant) cells with the MDM2 inhibitors nutlin-3, RG7388, or AMG232 for 96 h and performed a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) viability assay (26). MKL-1, but not MS-1, cells were sensitive to all three MDM2 inhibitors (Fig. 5A). We tested the effect of combining lenalidomide with nutlin-3, RG7388, and AMG232 and observed significantly improved cytotoxicity of all three MDM2 inhibitors when used with lenalidomide (SI Appendix, Fig. S5 A–D). Synergy testing using the Compusyn (SI Appendix, Fig. S6A) and Bliss (Fig. 5B) methodologies revealed synergistic activity for the combination of nutlin-3 or RG7388 with lenalidomide or UR99 (SI Appendix, Fig. S6 B–F) (27–30). In contrast, thalidomide had no evidence for synergy when used in combination with nutlin-3, consistent with its relatively reduced effects on CK1 levels (Fig. 5B).
Fig. 5.
Inhibition of MDM2 and CK1-MDM4 synergistically induces cell death by apoptosis. (A) MKL-1 and MS-1 cells were treated with MDM2 inhibitors, nutlin-3, RG7388, or AMG232, and XTT assay was performed after 96 h of treatment. **P 0.005 (multiple t test). (B) Bliss synergy test displays a strong synergy between nutlin-3 and lenalidomide or SC-24–UR99, but not with thalidomide. (C) BH3 profiling was performed with MKL-1 cells treated with lenalidomide, nutlin-3, or both drugs for 16 h. The experiment was performed three times. Data are shown as mean ± SD. *P 0.05 (Student t test). (D) MKL-1 MCC xenografts in SCID mice respond to the combinational treatment of HDM201 and lenalidomide. HDM201 (40 mg/kg), lenalidomide (50 mg/kg), or both drugs were administered orally daily, starting when xenograft tumors were . Data are shown as mean ± SEM. *P 0.05 (multiple t test between HDM201 and combination treatments). #The study was terminated because the tumor volume reached maximum permissible size. (E) MCPyV T antigens sensitize MCC for targeting the p53–MDM2–MDM4 pathway.
To determine whether lenalidomide affected the ability of nutlin-3 to cause cell death by apoptosis in MCC cells, we used dynamic BH3 profiling to measure changes in apoptotic priming upon treatment (31). Addition of lenalidomide to nutlin-3 enhanced the priming effect for apoptosis (Fig. 5C). To determine the efficacy of dual inhibition of MDM2 and MDM4 in vivo, we treated MKL-1 MCC xenografts with HDM201 (suitable for in vivo efficacy studies) with or without lenalidomide (Fig. 5D and SI Appendix, Fig. S5F and Table S3). We found that the addition of lenalidomide greatly enhanced the efficacy of HDM201, suggesting a potential for clinical utility of the combinational therapy for p53 wild-type tumors expressing MDM2 and MDM4. We propose a model where MCV T antigens increase the dependence on MDM2, MDM4, and CK1 to suppress p53 activity with therapeutic potential in virus-positive MCC (Fig. 5E).
Lenalidomide has been used to treat the hematologic malignancies, myelodysplastic syndrome (MDS), and multiple myeloma (21). It was reported that mutant CSNK1A1 predicts for a poor prognosis in lenalidomide-treated MDS (32). Furthermore, MDS with mutant p53 responds less well to lenalidomide and is more likely to progress to acute myeloid leukemia compared with MDS with wild-type p53, suggesting that lenalidomide’s effects may be partially dependent on inactivating MDM4 (33). Our work provides the rationale for the combination of lenalidomide with MDM2 inhibitors in MCC and perhaps in other solid tumors and hematologic malignancies containing wild-type p53.
Materials and Methods
Gene Expression Analysis.
The p53 targetome analysis is described in Allen et al. (9). RNA-seq data reported in Cheng et al. (15) were reanalyzed by using the DESeq2 and EnhancedVolcano R packages (34, 35).
Plasmids.
The GFP and T antigen cDNAs were Gateway (Invitrogen) cloned into pLIX_402 inducible empty or pLenti CMV Blast empty vector (w263-1), gifts from David Root, Broad Institute of MIT and Harvard, Cambridge, MA (Addgene 41394) and Eric Campeau, Zenith Epigenetics, Calgary, Canada (Addgene 17486), respectively. The sgRNA clones for CK1 (BRDN0001149315, BRDN0001145680) were gifts from John Doench and David Root (Addgene plasmid 76188, 76189), both at Broad Institute of MIT and Harvard, Cambridge, MA). pLKO-p53-shRNA-941 was a gift from Todd Waldman, Georgetown University School of Medicine, Washington, DC (Addgene 25637).
Supplementary Material
Acknowledgments
We thank Amy E. Shade (Harvard University); Anthony G. Letai [Dana-Farber Cancer Institute (DFCI)]; and Ensar Halilovic, Jutta Beyer, Bing Yu, and Claire Fabre (Novartis) for reagents and advice; Peter Howley (Harvard Medical School); Adam Bass and Thomas M. Roberts (DFCI); and Mitch Biermann (University of Wisconsin–Madison) for scientific advice.
Footnotes
Conflict of interest statement: M.C.-C. and S.F. are employed by Novartis Institute for Biomedical Research. R.A. was supported by Wellcome Trust DBT India Alliance Early Career Fellowship IA/E/14/1/501773. A patent was filed with the related work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818798116/-/DCSupplemental.
References
- 1.Becker JC, et al. Merkel cell carcinoma. Nat Rev Dis Primers. 2017;3:17077. doi: 10.1038/nrdp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson KG, et al. Merkel cell carcinoma: Current US incidence and projected increases based on changing demographics. J Am Acad Dermatol. 2017;78:457–463.e2. doi: 10.1016/j.jaad.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodig SJ, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest. 2012;122:4645–4653. doi: 10.1172/JCI64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol. 2013;87:6118–6126. doi: 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starrett GJ, et al. Merkel cell polyomavirus exhibits dominant control of the tumor genome and transcriptome in virus-associated Merkel cell carcinoma. MBio. 2017;8:e02079-16. doi: 10.1128/mBio.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harms PW, et al. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75:3720–3727. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen MA, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates S, et al. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 11.Nomura K, et al. Structural analysis of MDM2 RING separates degradation from regulation of p53 transcription activity. Nat Struct Mol Biol. 2017;24:578–587. doi: 10.1038/nsmb.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bista M, Petrovich M, Fersht AR. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci USA. 2013;110:17814–17819. doi: 10.1073/pnas.1317398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, et al. Autoinhibition of MDMX by intramolecular p53 mimicry. Proc Natl Acad Sci USA. 2015;112:4624–4629. doi: 10.1073/pnas.1420833112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beliveau A, Yaswen P. Soothing the watchman: Telomerase reduces the p53-dependent cellular stress response. Cell Cycle. 2007;6:1284–1287. doi: 10.4161/cc.6.11.4298. [DOI] [PubMed] [Google Scholar]
- 15.Cheng J, et al. Merkel cell polyomavirus recruits MYCL to the EP400 complex to promote oncogenesis. PLoS Pathog. 2017;13:1–31. doi: 10.1371/journal.ppat.1006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arora R, et al. Survivin is a therapeutic target in Merkel cell carcinoma. Sci Transl Med. 2012;4:133ra56. doi: 10.1126/scitranslmed.3003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berrios C, et al. Merkel cell polyomavirus small T antigen promotes pro-glycolytic metabolic perturbations required for transformation. PLoS Pathog. 2016;12:e1006020. doi: 10.1371/journal.ppat.1006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenos K, et al. Alternate splicing of the p53 inhibitor HDMX offers a superior prognostic biomarker than p53 mutation in human cancer. Cancer Res. 2012;72:4074–4084. doi: 10.1158/0008-5472.CAN-12-0215. [DOI] [PubMed] [Google Scholar]
- 19.Sholl LM, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houben R, et al. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J Invest Dermatol. 2013;133:2453–2460. doi: 10.1038/jid.2013.169. [DOI] [PubMed] [Google Scholar]
- 21.Krönke J, et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petzold G, Fischer ES, Thomä NH. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- 23.Spiotto MT, et al. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 2010;70:78–88. doi: 10.1158/0008-5472.CAN-09-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou B, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 26.Scudiero D, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- 27.Chou T, Motzer R, Tong Y, Bosl G. Computerized quantitation of synergism and antagonism of taxol, topotecan, and cisplatin against human teratocarcinoma cell growth: A rational approach to clinical protocol design. J Natl Cancer Inst. 1994;86:1517–1524. doi: 10.1093/jnci/86.20.1517. [DOI] [PubMed] [Google Scholar]
- 28.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: A web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33:2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding Q, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem. 2013;56:5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 30.Sun D, et al. Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2-p53 inhibitor in clinical development. J Med Chem. 2014;57:1454–1472. doi: 10.1021/jm401753e. [DOI] [PubMed] [Google Scholar]
- 31.Montero J, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160:977–989. doi: 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AE, et al. CSNK1A1 mutations and isolated del(5q) abnormality in myelodysplastic syndrome: A retrospective mutational analysis. Lancet Haematol. 2015;2:e212–e221. doi: 10.1016/S2352-3026(15)00050-2. [DOI] [PubMed] [Google Scholar]
- 33.Saft L, et al. p53 protein expression independently predicts outcome in patients with lower-risk myelodysplastic syndromes with del(5q) Haematologica. 2014;99:1041–1049. doi: 10.3324/haematol.2013.098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blighe K. 2018 Enhancedvolcano: Publication-ready volcano plots with enhanced colouring and labeling. Available at https://github.com/kevinblighe. Accessed November 23, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.