Significance
How transcription factors (TFs) activate transcription is a long-standing but still unsolved question. We analyzed serum response factor (SRF), a stimulus-responsive TF mediating immediate early gene (IEG) and cytoskeletal gene expression at single-molecule resolution. Cell stimulation enhanced SRF activity by increasing the number of long chromatin-associated SRF molecules in an oscillating pattern. Further, stimulation enhanced the SRF chromatin residence time, and SRF binding events segregated into three distinct residence time regimes (short, intermediate, and long bound). In summary, our single-molecule imaging study reveals highly dynamic and diverse SRF interactions with DNA. Thus, cell stimulation regulates TF activity by several interconnected mechanisms including nucleus−cytoplasm shuttling, TF phosphorylation, cofactor recruitment, and extension of chromatin residence time and enhancing chromatin-bound TF numbers.
Keywords: SRF, HaloTag, single molecule, neuron, transcription
Abstract
Serum response factor (SRF) mediates immediate early gene (IEG) and cytoskeletal gene expression programs in almost any cell type. So far, SRF transcriptional dynamics have not been investigated at single-molecule resolution. We provide a study of single Halo-tagged SRF molecules in fibroblasts and primary neurons. In both cell types, individual binding events of SRF molecules segregated into three chromatin residence time regimes, short, intermediate, and long binding, indicating a cell type-independent SRF property. The chromatin residence time of the long bound fraction was up to 1 min in quiescent cells and significantly increased upon stimulation. Stimulation also enhanced the long bound SRF fraction at specific timepoints (20 and 60 min) in both cell types. These peaks correlated with activation of the SRF cofactors MRTF-A and MRTF-B (myocardin-related transcription factors). Interference with signaling pathways and cofactors demonstrated modulation of SRF chromatin occupancy by actin signaling, MAP kinases, and MRTFs.
The transcription factor (TF) serum response factor (SRF) governs essential gene expression programs crucial to almost any cell type (1–3). This was revealed by Srf-directed mutagenesis in mice uncovering important SRF functions during embryogenesis, skin, liver, heart, muscle, and neuronal function (2, 4–6). One underlying SRF function in these organs is regulation of cell contractility and actin-based motility processes such as adhesion, migration, and cell growth (2). In addition, SRF conveys stimulus-dependent gene expression induced by growth factors, serum, and synaptic activity (3). SRF-directed genes fall into two major programs: cytoskeletal genes and immediate early genes (IEGs) (2, 3). SRF is the prototypical TF to connect Rho-actin cytoskeletal dynamics with a transcriptional response. Here, SRF interacts with MRTF (myocardin-related TFs, MRTF-A and MRTF-B) cofactors to govern expression of genes encoding for actin cytoskeletal proteins such as actin isoforms (Acta1, Acta2) and cytoskeleton-associated proteins (2). Further, SRF mediates the cellular IEG response, resulting in transient up-regulation of, e.g., c-Fos, Egr1, and Arc already several minutes after cell stimulation (4, 7). SRF’s pivotal role in IEG induction was demonstrated in SRF-deficient neurons that failed to induce many IEGs upon neuronal activation stimuli such as epileptic seizures or acute stress (8–11). Besides MRTF cofactors, SRF is coupled to TCF (ternary complex factors) partner proteins through MAP kinase signaling (2, 3). Both MRTFs and TCFs substantially contribute to serum-induced IEG induction, although both compete with each other for SRF binding (12, 13).
As with many TFs, including SRF, traditional models of TF function considered a rather static mechanism of TF−DNA interaction. This invokes stable TF binding to promoters already before and also after cell stimulation. For instance, classic genomic footprinting demonstrated constitutive SRF promoter occupancy at the c-Fos gene independent of the activation status (14). In contrast, chromatin immunoprecipitation (ChIP) data revealed inducible SRF binding at a majority of target genes upon serum (15) or neuronal stimulation (8). However, global methods such as ChIP might produce false-positive interactions (16, 17) and are still constrained by averaging over a multitude of cells and thereby not being able to resolve subpopulation TF binding events with different dynamics. Several techniques, including FRAP (Fluorescence Recovery After Photobleaching) and FCS (fluorescence correlation spectroscopy), were employed to investigate dynamic TF properties of individual populations (18). Another powerful technique for investigating TF binding dynamics is single-molecule tracking (SMT), bearing the advantage of measuring TF binding dynamics one molecule at a time (19–21). By applying these techniques in living cells, it was found that observed binding events of many TFs do not show a uniform behavior but segregate into different binding time regimes. To study TFs at single-molecule resolution, fusion proteins with specific tags, such as the HaloTag, that can be labeled with photostable organic dyes are analyzed in living cells. Such fusion proteins are monitored using light-sheet microscopy such as Highly Inclined and Laminated Optical sheet (HILO) microscopy (22). Here, molecules are selectively excited in a thin optical section, thereby increasing the signal-to-noise ratio. Up until now, live cell SMT studies have been performed with a few different TFs, including p53, CREB, Sox2, Oct4, c-Myc, STATs, and steroid receptors (23–32). These studies determined important parameters of TF dynamics, including chromatin residence times and chromatin-bound fractions. So far, most SMT studies identified two distinct residence time regimes of TFs, namely a short and a long binding fraction. Depending on the respective binding position on chromatin, TF binding events either lasted for several hundred microseconds (short binding fraction) or for several seconds (long binding fraction). It is important to note that TFs are not constitutively restricted to one binding regime but switch between, e.g., short and long binding states. Residence time of the long binding fraction varied depending on TF, cell type, and SMT experimental setup; however, the average residence time for the long binding fraction reported so far typically lasted a few seconds (e.g., 10 s to 15 s for p53 or Sox2; refs. 28 and 33). This TF fraction corresponds with transcriptionally active subnuclear domains (34, 35) and—for Sox2—predicted cell location within the four-cell embryo (36), thereby pointing at a functional relevance of this population. Besides residence time, a second parameter of transcriptional dynamics analyzed by SMT is the fraction of chromatin-bound molecules. Typically, the bound fraction of a TF population ranges between 10% and 40% of all molecules (28, 31). So far, most TF parameters were determined in basal conditions, and the impact of cell stimulation on single-molecule TF dynamics was not studied intensively. Single reports available showed little impact of neuronal stimulation on CREB residence time (27) whereas irradiation and hormones prolonged p53 (28) and GR/ER (24, 25, 30) residence times, respectively.
In this study, we provide a first SMT analysis of SRF employing two different cell types: fibroblasts and primary hippocampal neurons of mice. We investigated the impact of cell stimulation, providing detailed temporal resolution profiles of the long bound SRF fraction for two stimuli. We used serum and the growth factor BDNF (brain-derived neurotrophic factor), both established stimuli enhancing SRF activity in fibroblasts and neurons, respectively (15, 37). Our data for SRF resolved a surprisingly long average residence time, in the seconds to minute range, that was even further extended upon stimulation. The fraction of long bound molecules was rapidly augmented by stimulation within 20 min and reached a further peak at around 60 min after stimulation. In addition, our SMT data in fibroblasts and neurons argue for the presence of three distinct SRF residence time regimes, including short, intermediate, and long bound SRF molecules.
In summary, we provide further support for highly dynamic properties of TFs revealing alterations of residence times and binding fractions of SRF upon stimulation.
Materials and Methods
Cloning of Halo-SRF and Stable Cell Line Generation.
To observe SRF chromatin binding events in living cells, we cloned the WT murine Srf sequence (UniProtKB Q9JM73) or the murine Srf αI helix mutant (38) with four amino acid substitutions (corresponding to the human amino acid exchanges at positions L155T, Y158H, T159V, and T166H; synthetized by Genecust) into a lentiviral expression vector (24). The pLV-TetO construct contained the Halo sequence under the control of a Tet-inducible minimal CMV promoter. After inserting the Srf sequence, the vector was used to generate an NIH 3T3 cell line stably expressing N-terminally tagged Halo-SRF fusion proteins via lentiviral transduction (3T3 Halo-SRF cells). The N terminus has no obvious functional domain and was chosen since the C terminus harbors the transactivation domain (TAD) and we wanted to avoid any interference of TAD function by the HaloTag. Leaky expression of the noninduced construct was sufficient for all experiments, since low expression levels were required for resolving single molecules.
Quantitative Real-Time PCR.
We isolated total RNA with the ISOLATE II RNA/DNA/Protein kit (Bioline) according to manufacturer’s instructions. Reverse transcription was performed with 1 μg of RNA (NIH 3T3) or 0.5 µg of RNA (neurons) using random hexamers and reverse transcriptase (Promega). We performed quantitative real-time PCR (qPCR) on a Light Cycler 480II (Roche) with the Power SYBR green PCR master mix (Takara). Each sample was pipetted in doublets, and threshold cycle (Ct) values were calculated by the LC480 II Software. Expression was determined in relation to Gapdh RNA levels. Primer sequences are provided in SI Appendix, Table S1.
ChIP.
ChIP was performed according to the protocol of Nelson et al. (39) using 10-cm dishes of NIH 3T3 Halo-SRF cells; 2 μg/mL of anti-HALO (rabbit polyclonal; Promega) or IgG antibody (rabbit polyconal; Santa Cruz) were used for each IP. After purification of DNA (PCR purification kit; Qiagen), 2 μL of each IP and input were subjected to qPCR. Ct values obtained from HALO or IgG ChIP were normalized to the respective input values. Primer sequences are provided in SI Appendix, Table S1.
Single-Molecule Live Cell Imaging.
HaloTag labeling.
NIH 3T3 Halo-SRF cells were plated on heatable ∆T culture dishes with 0.17-mm glass bottoms (Bioptechs) the day before imaging. After attachment, we labeled the cells with 25 pM tetramethylrhodamine (TMR) ligand (Promega) or 0.8 pM to 1.0 pM silicone rhodamine (SiR) ligand (kindly provided by Kai Johnsson, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) to ensure single-molecule resolution by keeping the overall labeling density low. After labeling, cells were starved overnight in DMEM/0.05% FCS at 37 °C/5% CO2. Immediately before imaging, the starving medium was exchanged by phenol red-free OptiMEM/10% FCS to stimulate cells or by phenol red-free OptiMEM/0.05% FCS to investigate the binding behavior in starved cells. Cells were measured for a maximum of 120 min, and temperature was controlled by the Delta T5 µ-Environmental Culture Dish Controller (Bioptechs). Primary hippocampal neurons were prepared as described above and cultured for 3 days in vitro. Right before imaging, cells were labeled with 3.1 pM SiR ligand in NMEM/B27 for 15 min at 37 °C/5% CO2 and allowed to recover in NMEM/B27 for 30 min at 37 °C/5% CO2. Neurons were imaged in phenol red-free NMEM/B27 + 5 µg/mL of gentamycin. To stimulate cells, 10 ng/mL of BDNF was added to the medium before imaging.
SMT.
For single-molecule imaging, we used a custom-built fluorescence microscope built around a commercial microscope body (TiE; Nikon) as described in ref. 40. In brief, a 638-nm laser (IBEAM-SMART-640-S, 150 mW; Toptica) beam for SiR-labeled cells and a 515-nm laser (Cobolt Jive, 300 mW; Cobolt) beam for TMR-labeled cells was adjusted in size with a pinhole and focused on the back focal plane of a high-NA objective (100× 1.45 Plan Apo; Nikon) to achieve HILO. The fluorescence light was filtered and subsequently detected by an electron multiplying charge-coupled device (EMCCD) camera (iXon Ultra DU 897U; Andor). The setup was controlled with the NIS Elements software (Nikon) and a NIDAQ data acquisition card (National Instruments).
We used different camera integration times (τint) and varied the dark time between two consecutive frames to address different aspects of SRF binding kinetics. To track diffusing and bound molecules (see Fig. 2), we fixed τint to 10 ms and measured the displacement between two consecutive frames. Two consecutive frames were followed by a dark time of 5 s to allow for equilibration between diffusing and bound molecules. For measuring DNA residence times (see Figs. 3 and 6 and SI Appendix, Fig. S3), we used τint of 50 ms and inserted dark times between two consecutive frames varying from 100 ms to 60 s as described in ref. 24. The dark time sums up with the integration time to the time-lapse time (τtl). Additionally, continuous movies were acquired to resolve short binding events. To determine the proportion of long to all binding events during a short time interval, we made use of a recently developed illumination scheme called interlaced time-lapse microscopy (41). In short, two consecutive frames with τint = 50 ms were followed by a dark time of 2 s (see Figs. 4–6). Molecules surviving at least one dark period were classified as long binding events, whereas molecules appearing in at least two consecutive frames without or with an interspersed dark time were classified as all binding events (long or short). Ratios of long bound to all bound molecules were calculated for each cell. Only interlaced time-lapse microscopy (ITM) movies with a total molecule number (diffusing and binding) ranging between 200 and 1,000 were analyzed.
Results
Characterization of Functional Properties of Halo-Tagged SRF Molecules.
To analyze SRF at single-molecule resolution, we started with an established fibroblast cell line, i.e., murine NIH 3T3 cells stably expressing fusion proteins of the HaloTag connected to the N terminus of murine SRF (Halo-SRF; see Materials and Methods). The HaloTag system consists of the modified bacterial enzyme haloalkane dehalogenase that covalently binds cell-permeable ligands coupled to fluorophores such as TMR or SiR in living cells (20).
In the first set of experiments, we investigated the functionality of Halo-SRF in NIH 3T3 fibroblasts (Fig. 1). Compared with endogenous SRF levels, a 2.5-fold (2.5 ± 0.4; n = 4) overexpression of Halo-SRF was observed in immunoblots (Fig. 1B). The HaloTag has a molecular weight of ∼30 kDa, resulting in a Halo-SRF fusion protein of roughly 100 kDa. Halo-SRF molecules were constitutively localized in the nucleus (Fig. 1 C–E), as known for endogenous SRF. So far, SRF subcellular localization was not analyzed at superresolution. We employed superresolution microscopy technique dSTORM [direct stochastic optical reconstruction microscopy (42)] to image Halo-SRF molecules in nuclei (Fig. 1 D and E). In dSTORM, stochastic photoswitching of single fluorescent molecules between bright and dark states allows for localization of individual fluorophores beyond the diffraction limit, thereby achieving resolution of up to 20 nm. Our dSTORM data suggest SRF distribution throughout the entire nucleoplasm but sparing nucleoli (Fig. 1D). Halo-SRF molecules accumulated in clusters (arrows in Fig. 1E).
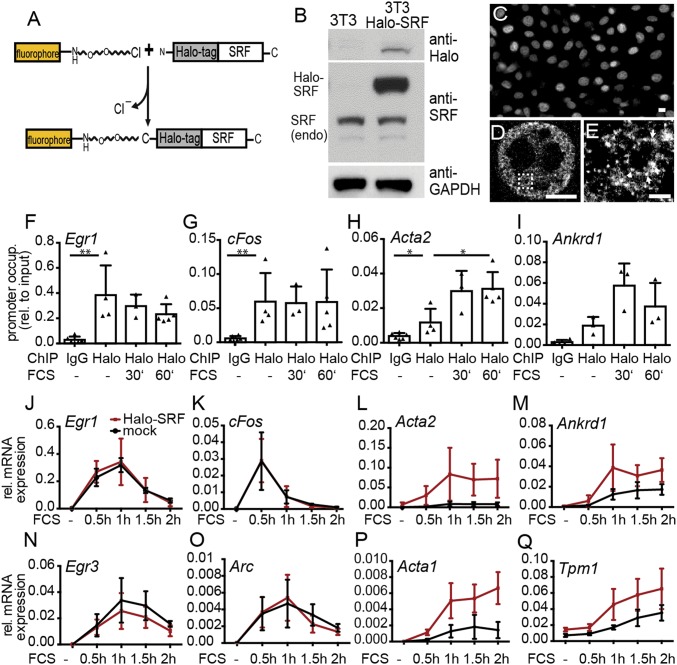
Fig. 1.
Characterization of Halo-SRF localization and function. (A) Scheme of SRF fused to the HaloTag at the N terminus. The HaloTag is an enzyme reacting with fluorophores such as TMR or SiR if modified by a chloroalkane linker. (B) Characterization of Halo-SRF overexpression in stably transfected NIH 3T3 cells (NIH 3T3 Halo-SRF) using immunoblotting. Halo-SRF was recognized by anti-Halo and anti-SRF directed antibodies. Endogenous SRF (endo) was detected with anti-SRF antibodies. (C) NIH 3T3 Halo-SRF cells stained with TMR revealed constitutive nuclear localization of Halo-SRF in conventional fluorescence microscopy. (D) The dSTORM microscopy of Halo-SRF molecules labeled with anti-Halo directed antibodies show individual SRF clusters distributed throughout the entire nucleoplasm. (E) A close-up view of the boxed area in D reveals Halo-SRF localization in clusters (arrows). (Scale bars: C, 10 μm; D, 5 μm; E, 1 μm.) (F–I) ChIP demonstrating occupancy of Halo-SRF at several SRF target genes. FCS stimulation did not alter Halo-SRF occupancy at IEG promoters of (F) Egr1 or (G) cFos but enhanced occupancy at cytoskeletal promoters such as (H) Acta2 and (I) Ankrd1. Each triangle represents an independent culture. Data are depicted as mean ± SD (*P ≤ 0.05; **P ≤ 0.01; Mann−Whitney u test). (J, K, N, and O) Halo-SRF overexpression did not alter gene expression profiles of IEGs in comparison with mock NIH 3T3 cells expressing endogenous SRF only. (L, M, P, and Q) Halo-SRF enhanced mRNA abundance of cytoskeletal genes in relation to mock transfected cells. Values are calculated from at least three independent cultures. Data are depicted as mean ± SD.
To further investigate Halo-SRF function, we characterized promoter binding (Fig. 1 F–I) and gene induction (Fig. 1 J–Q). In ChIP assays, Halo-SRF was bound to promoter elements of the IEGs Egr1 (Fig. 1F) and cFos (Fig. 1G) as well as actin cytoskeletal genes Acta2 (Fig. 1H) and Ankrd1 (Fig. 1I). Stimulation with serum (10% FCS) did not overtly alter Halo-SRF occupancy at IEGs but enhanced Halo-SRF promoter binding at both cytoskeleton-associated genes (Fig. 1 F–I), in line with previous ChIP data (15).
Next, gene induction of NIH 3T3 cells expressing endogenous SRF was compared with fibroblasts additionally expressing Halo-SRF (Fig. 1 J–Q). The genes Egr1 (Fig. 1J), cFos (Fig. 1K), Egr3 (Fig. 1N), and Arc (Fig. 1O) were strongly induced by serum administration 30 min to 1 h after stimulation. Expression profiles were almost identical for cells expressing endogenous SRF only and Halo-SRF, suggesting a physiological gene response not altered by Halo-SRF. Genes encoding for components of the actin cytoskeleton were also up-regulated by FCS; however, maximum induction was reached at 1 h or later (Fig. 1 L, M, P, and Q). For these genes, we observed further augmentation of mRNA levels by Halo-SRF (Fig. 1 L, M, P, and Q).
Overall, Halo-SRF expression showed comparable localization, promoter occupancy, and gene expression profiles as known for endogenous SRF.
Determination of Mobile and Chromatin-Associated Halo-SRF Molecules.
To visualize single Halo-SRF molecules in living cells, NIH 3T3 cells were stained with SIR ligand before imaging. Subsequently, we used HILO microscopy, exciting single Halo-SRF molecules in a thin light sheet of ∼2 μm. To identify the percentage of chromatin-bound molecules compared with diffusing molecules, we employed an illumination scheme consisting of two consecutive frames (10 ms of illumination each) interspersed with 5 s of dark time to allow for equilibration between diffusing and bound molecules (Fig. 2A). Single molecules in movies were tracked with previously described tracking software (see Materials and Methods).
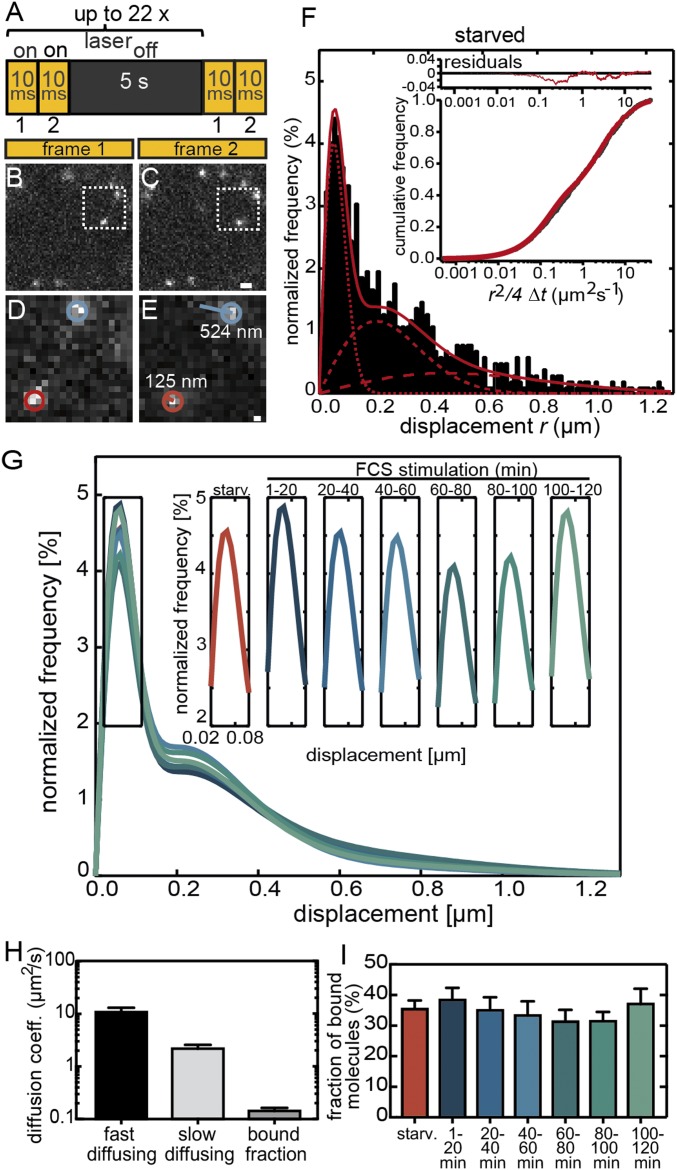
Fig. 2.
Analysis of Halo-SRF binding fractions in unstimulated and stimulated fibroblasts. (A) Illumination scheme indicating on and off times of laser illumination. (B and C) Typical jump distances of individual molecules between two frames are illustrated by two exemplary molecules. (D and E) Magnified areas of the regions marked by a dashed boxes in B and C. The molecule labeled in blue was mobile and moved by 524 nm, whereas the red molecule was rather immobile and had a jump distance of only 125 nm. (Scale bar: B and C, 1 μm; D and E, 160 nm.) (F) Distribution of single-molecule displacements in 10-ms integration time (n = 1,018 molecules, n = 21 cells) normalized to all molecules detected in serum-starved NIH 3T3 cells (black bars). The distributions were fitted with a three-component diffusion model (red line; single components in dotted red lines; SI Appendix, Eqs. I and II) where the lowest jump distances are representative of chromatin-bound molecules. (Inset) The residuals (red line in upper part) from the three-component fit (red line in lower part) to the cumulative distribution of all calculated squared displacements per 10-ms integration time for a 2D diffusion (in black). (G) Three-component fits including three diffusion coefficients to the measured displacement histograms for starved NIH 3T3 cells and consecutive 20-min time intervals of serum stimulation (for N numbers, see SI Appendix, Fig. S1). (Inset) Changes in the amplitude of the bound fraction only for all 20-min time intervals. (H) Average diffusion coefficients (mean ± SEM) for all three Halo-SRF fractions determined from a global fit to cumulative histograms of unstimulated and stimulated cells computed from all timepoints of serum stimulation. Values were calculated by bootstrapping. (I) The ratio of bound molecules to all molecules is depicted in starved cells and along the several timepoints of FCS application (mean ± SEM). Between 1 min to 20 min and 100 min to 120 min, FCS slightly enhanced the fraction of bound molecules. Values were calculated by bootstrapping.
We measured the displacement (“jump distances”) of individual Halo-SRF molecules between two 10-ms frames in starved as well as stimulated cells (Fig. 2 A, B, and E) as reported previously (40). Since SRF regulates IEG and actin cytoskeletal genes with different temporal profiles (Fig. 1), we analyzed the fraction of chromatin-bound SRF molecules in 20-min time intervals after serum addition for up to 2 h (Fig. 2 G and I).
SRF molecules showed a distribution of jump distances within the 10-ms interval ranging between few nanometers (value indicative of localization precision) up to 1.2 μm, corresponding to immobilized and highly mobile Halo-SRF molecules, respectively (Fig. 2 B–F and Movie S1). The measured jump distances of all recorded molecules for each condition were plotted in a histogram (starved condition in Fig. 2F; other conditions in SI Appendix, Fig. S1). Since the nucleoplasm is packed with chromatin and proteins, we would not expect free Brownian but rather anomalous diffusion (43). Indeed, for all conditions, the distribution of single-molecule displacements was fitted best with a three-component diffusion model (Fig. 2H) as previously seen for STAT1, p53, and c-Myc (26, 28, 31). We approximated the diffusive behavior with a model of two diffusion components in addition to a component resulting from the bound population, as reported for other TFs previously (24, 26, 28, 31). Here, the slowest diffusion component is representative for chromatin-bound molecules. Since we do not expect cell stimulation to change diffusion coefficients, we globally fitted the diffusion coefficients for all conditions, whereas fractions were fitted individually for each condition (see Materials and Methods).
For the immobile chromatin-bound fraction, we determined an average diffusion coefficient of D1 = 0.14 ± 0.02 μm2/s (Fig. 2H) corresponding to a localization precision of the microscopic setup of 75 nm in line with previous reports for STAT1 and Sox2 (23, 31). Additionally, we observed a slow diffusing component with D2 = 2.18 ± 0.40 μm2/s and a fast diffusing component with D3 = 10.82 ± 2.23 μm2/s (Fig. 2H). Already, in starved cells, 35% of all detected Halo-SRF molecules are bound to DNA, whereas 65% are either slowly or quickly diffusing (Fig. 2I). This is a relatively high percentage compared with previous reports on p53 having solely 10% of chromatin-bound molecules in the unstimulated state (28). Serum stimulation slightly elevated the bound fraction by 2 to 3% at two time intervals (Fig. 2I), 1 min to 20 min (38.4 ± 4.0%) and 100 min to 120 min (37.0 ± 5.0%) compared with starved fibroblasts (35.4 ± 2.8%).
In summary, the mobility behavior of single Halo-SRF molecules was characterized with a three-component diffusion model. Roughly one-third of all molecules were chromatin-associated, a fraction slightly elevated upon stimulation (SI Appendix, Table S2).
SRF Residence Time Is Prolonged by Cell Stimulation.
In the previous experiment, we determined an immobilized fraction of ∼1/3 of all molecules potentially bound to chromatin (Fig. 2). Those bound molecules typically segregate into two categories, short and long bound TF molecules, as reported in the literature (20). Short binding (<1 s) molecules are considered to bind unspecifically to chromatin for target search. In contrast, longer binding molecules (>1 s) are considered to bind to specific promoter sequences in transcriptionally active regions (35). In the next step, we measured SRF residence times with and without cell stimulation for such fractions of chromatin-bound SRF molecules (Fig. 3).
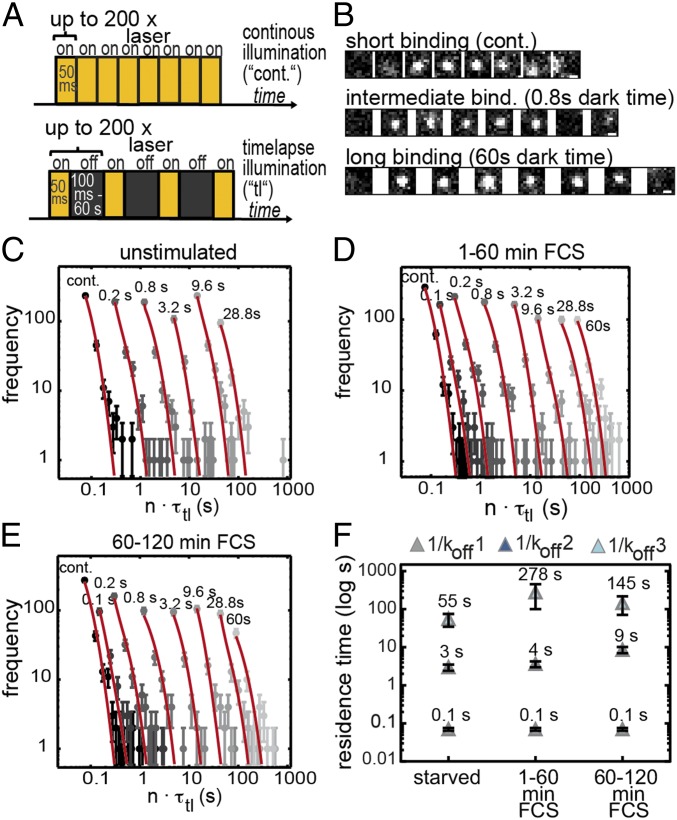
Fig. 3.
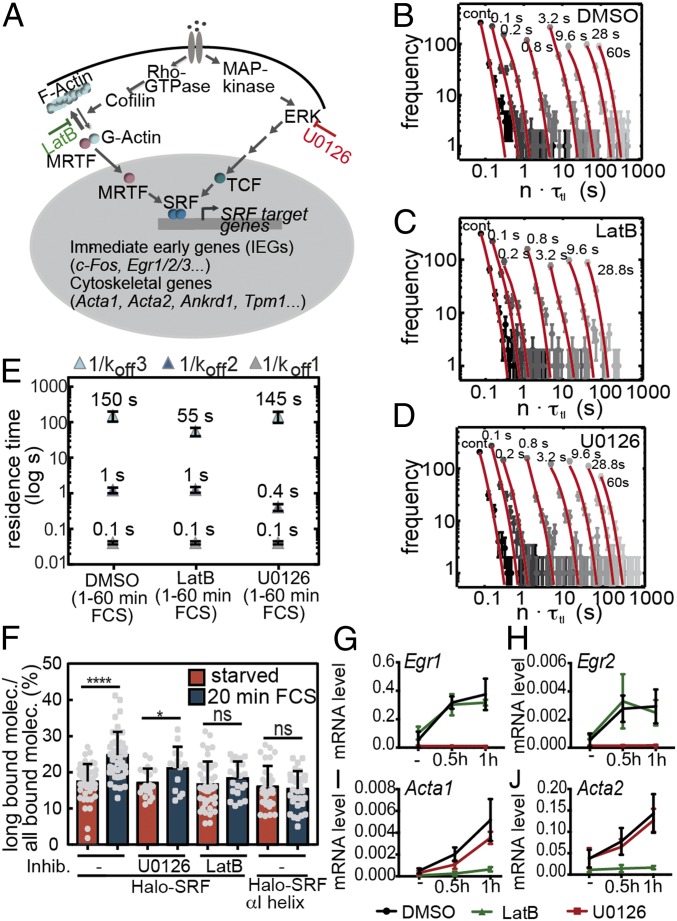
Halo-SRF residence time increases with cell stimulation in fibroblasts. (A) Illumination schemes for measuring residence times. Continuous illumination is depicted (Top). In addition, different dark times (black bars) ranging from 100 ms to 60 s were included between two 50-ms illuminations (Bottom). (B) Representative images showing single-molecule binding events for a short, intermediate, and long bound molecule. Dark times between two illuminated frames are indicated. (Scale bar: 500 nm.) (C–E) Histograms with fluorescent “on” times (gray circles) of (C) serum-starved cells (n = 1,409 bound molecules, n = 26 cells) or (D and E) FCS-stimulated cells for (D) 1 min to 60 min (n = 1,811 bound molecules, n = 61 cells) or (E) 60 min to 120 min (n = 1,308 bound molecules, n = 46 cells). The histograms were fitted globally with a three-component decay model to calculate average residence times of SRF (red; SI Appendix, Eq. III). Time-lapse times are indicated above the respective fit to the data points. Error bars indicate SD. (F) Computed average chromatin residence times for short (1/koff1, gray), intermediate (1/koff2, dark blue), and long bound (1/koff3, light blue) SRF molecules. Within 1 min to 60 min of serum stimulation (FCS), the average long residence time (dark-blue rectangle) increased fivefold, from 55 s to 278 s, and decreased to ∼145 s in the second hour of serum application. The average residence time for the intermediate fraction was unchanged at 1 min to 60 min of FCS stimulation, whereas it increased from 60 min to 120 min. Error bars indicate SD.
To determine chromatin residence times, we made use of a published time-lapse illumination scheme (24) having 50-ms laser exposure times interspersed with varying dark times ranging from 0.1 s to 60 s (Fig. 3A). Additionally, continuous illumination movies (“cont.”) were acquired to resolve short binding events (Fig. 3 A, B, and E). TFs “surviving” longest dark times between two illuminations have highest residence times. Different dark times are necessary to accurately determine residence times, since, in movies recording short dark times, long binding events are not fully covered due to photobleaching, and residence times are underestimated, whereas long dark times fail to track faster transient binding events (24, 30).
We analyzed serum-starved NIH 3T3 cells along with cells stimulated with FCS for 1 min to 60 min or 60 min to 120 min (Movie S2). Since some movies with longest dark times took up to 40 min, analysis of 20-min time intervals as done before (Fig. 2) was precluded. For each condition, we recorded the time a bound molecule was visible (Fig. 3B) and collected these times in residence time histograms for starved cells (Fig. 3C) and cells stimulated for the first (Fig. 3D) or second (Fig. 3E) hour with FCS. SRF molecules remaining within an area of up to 0.32 μm2 for at least two frames were considered bound (Fig. 3B; see Materials and Methods). To determine a fitting model characterizing our data best, we calculated the reduced χ2 (χ2/ν) for a one-, two-, and three-rate decay model. The three-rate decay model including three dissociation rate constants was the most appropriate model, with χ2/ν = 1.63 compared with the two-rate model (χ2/ν = 1.81) and the one-rate model (χ2/ν = 1.83). The three dissociation rate constants (koff1, koff2, and koff3) correspond to three residence time regimes characterized by the corresponding average residence time (short, intermediate, or long; Fig. 3 C–E and F). The interaction of an SRF molecule at a particular chromatin position belongs to one of these three residence time regimes, but the same SRF molecule might switch to a different fraction at any later timepoint, depending on the local binding environment.
From the three off-rate constants (koff1, koff2, and koff3) obtained from the fit, we calculated the average residence time for all three residence time regimes with and without stimulation (Fig. 3F). In starved cells, the longest bound fraction of SRF molecules persisted on chromatin already for an average of approximately 1 min (55 ± 20 s). Interestingly, FCS stimulation increased the duration of the average binding time of longest bound SRF molecules fivefold, now resulting in more than 4 min (278 ± 178 s). This elevation was transient, since, in the second hour of stimulation, the average binding time decreased again (145 ± 74 s). For the intermediate bound fraction, the average residence time in starved cells was approximately 3 s (3 ± 1 s; Fig. 3F). Upon FCS stimulation, the residence time of the intermediate fraction almost remained constant within the first hour (4 ± 1 s) and more than doubled to 9 s (9 ± 2 s) in the second hour of stimulation. The average residence time of the short bound fraction was 0.1 s (0.1 ± 0.01 s; globally fitted for all conditions; Fig. 3F).
Taken together, we observed three residence time regimes for the binding of SRF molecules with short, intermediate, or long residence times on chromatin (summarized in SI Appendix, Table S2). The long residence time was increased in the first and, to a somewhat lower extent, in the second hour after FCS stimulation.
Cell Stimulation Enhances the Long Chromatin-Bound SRF Fraction at Discrete Time Intervals.
SRF target genes were regulated within minutes after cell stimulation (Fig. 1). In the previous experiment, we determined accurate residence times in 60-min time frames (Fig. 3). However, this temporal resolution was not suited to resolve more rapid changes in the SRF-mediated gene expression profile. To investigate long binding events during short time intervals (i.e., 20 min) after stimulation, we employed a new illumination protocol, termed “interlaced time-lapse microscopy” (41). ITM allows for calculation of fractions of long bound and short bound Halo-SRF molecules. Of note, the ITM ratio we measured likely involves alterations in the fraction of long bound molecules but also changes of the residence time, since the probability for a molecule to survive a 2-s dark time is higher the longer the average residence time is. In this illumination scheme, two subsequent image acquisitions (each 50-ms exposure times) are followed by a rather long dark time of 2 s (Fig. 4A). Bound molecules “surviving” at least one dark time in an area of up to 0.08 µm2 were classified as long binding events, whereas molecules appearing in the same spot in at least two consecutive frames without or with an interspersed dark time were classified as all binding events (Fig. 4A). The ratio of long bound to all bound molecules was calculated within one movie for each cell. We chose a dark time of 2 s to cover especially the percentage of intermediate (∼3 s to 9 s; Fig. 3F) plus long (∼2 min to 5 min; Fig. 3F) binding molecules, since binding events of >1 s are presumed to be specific (35). For unspecific short binding events (∼0.07 s), it is highly unlikely to “survive” a dark time of 2 s or more.
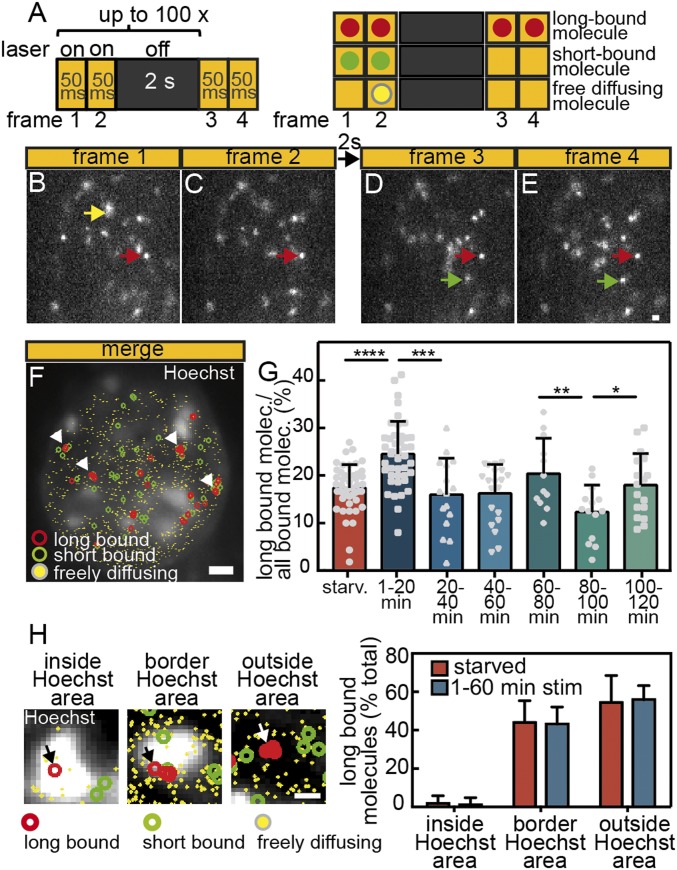
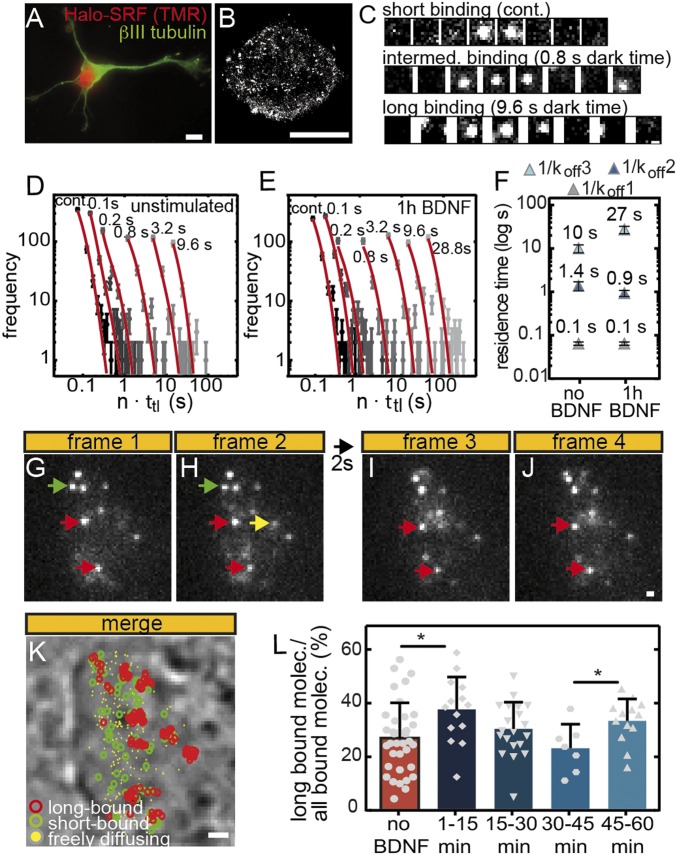
Fig. 4.
The fraction of long bound Halo-SRF molecules is enhanced by serum stimulation. (A) ITM illumination scheme. Molecules are tracked over repeated cycles of 2 × 50-ms exposures interspersed by 2 s of dark time (Left). Scheme illustrating different types of single-molecule events (Right). Long bound molecules (red dot) are present over ≥1 dark times, whereas short bound molecules (green dots) are only detectable over 2 × 50-ms illuminations. Freely diffusing molecules are detected in one frame only (yellow dot). (B–E) Representative examples of molecules imaged over four frames and one dark time interval. A freely diffusing molecule is present in frame 1 only (yellow arrow). A long binding molecule is present in all four frames (red arrows), whereas the short bound molecule (green) is present in frames 3 and 4 (green arrows). (Scale bar: 1 μm.) (F) All frames of an ITM movie were merged, and long bound, short bound, and freely diffusing molecules are highlighted by colors. The nucleus was counterstained with Hoechst to outline nuclear territories and borders, and binding events were overlaid. Arrows point at accumulations of long binding events. (Scale bar, 2 μm.) (G) NIH 3T3 cells expressing Halo-SRF were starved or stimulated for indicated time intervals with 10% FCS. During stimulation, the fraction of long bound molecules showed a periodic behavior peaking at 1 min to 20 min, 60 min to 80 min, and 100 min to 120 min after FCS addition (mean ± SD; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; t test). Each gray circle, square, or triangle indicates one cell analyzed (starved: n = 47 cells; 1 min to 20 min: n = 43 cells; 20 min to 40 min: n = 16 cells; 40 min to 60 min: n = 20 cells; 60 min to 80 min: n = 12 cells; 80 min to 100 min: n = 14 cells; 100 min to 120 min: n = 17 cells). (H) Costaining of nuclei for Halo-tagged SRF molecules (red circles) and Hoechst (white signals). Example of long bound SRF molecule (arrow) present inside the white Hoechst-positive area (Left), at the border (Center Left) or outside the Hoechst signal (Center Right). (Right) Quantification shows localization of SRF molecules almost exclusively outside the Hoechst area or at the border but rarely inside the Hoechst-positive area. No differences were observed in starved or stimulated cells (starved: n = 5 cells; 1 min to 60 min FCS: n = 12 cells). (Scale bar, 400 nm.)
ITM movies of NIH 3T3 cells expressing Halo-SRF revealed a high percentage of long binding events surviving more than 2 s (examples labeled with red arrows in Fig. 4 B–E). Furthermore, we observed short bound (green arrows) and freely diffusing (yellow arrow) molecules (Fig. 4 B–E). Merging all single-molecule events within a 4-min ITM movie allows for mapping localization of long plus short binding events and diffusing molecules within the nucleoplasm (Fig. 4F). Interestingly, several long binding events appear to accumulate in clusters at various positions throughout the nucleus (arrows in Fig. 4F). Closer inspection revealed preferential localization of such long SRF binding events outside (55% of all long binding events) but rarely (<1%) inside Hoechst-positive areas, indicative of heterochromatin regions, in the nucleus (Fig. 4 F and H). In addition, we observed an abundance of ∼45% of all long binding events at the border between Hoechst-positive and Hoechst-negative areas (arrow in Fig. 4H, Center Left). This localization of SRF molecules was comparable in starved and FCS stimulated cells (Fig. 4H).
As mentioned above, ITM allows classifying of molecules into binding time populations during short measurement intervals, thus enabling high time resolution of changes in binding behavior. We analyzed changes in the long bound proportion of Halo-SRF molecules upon FCS stimulation (Fig. 4G). As before (Fig. 2), we analyzed 20-min time bins after FCS administration. In our initial results, analyzing all binding events regardless of the residence duration, we already noted a small increase in chromatin association upon stimulation (Fig. 2). With ITM, we focused on the fraction of long binding events only and observed a specific increase of this fraction in the first 20 min after FCS application (Fig. 4G). In starved fibroblasts, 17.4 ± 4.9% of all Halo-SRF molecules were long bound, i.e., chromatin-associated for 2 s or more. In the first 20 min after FCS stimulation, we observed 8% more long bound molecules (24.8 ± 6.4%) compared with starved cells (SI Appendix, Table S2). In fact, there was a considerable cell-to-cell variability, with some cells reaching 40% or more of long bound SRF molecules (gray symbols in Fig. 4G). Interestingly, this elevation in the long bound chromatin-associated fraction was only transient and declined to the level observed in starved cells until 60 min of stimulation. After 60 min, a second rise in the long bound SRF fraction was observed, now reaching 20% (20.4 ± 7.5%). Thereafter, we again observed a drop in the long bound fraction, slightly increasing only after 100 min (Fig. 4G). Thus, we obtained a quasi periodic pattern of increased SRF binding activity approximately every 40 min.
Data above suggest a rhythmic pattern of long-term SRF association with chromatin over the 2-h stimulation period. SRF-mediated transcription strongly depends on interaction with partner proteins of the MRTF family (13, 15, 44). Therefore, we analyzed whether long SRF associations correlated with activation of MRTF-A and MRTF-B. Both SRF partner proteins are activated by shuttling to the nucleus upon cell stimulation (SI Appendix, Fig. S2) (44). We noted two peaks of nuclear MRTF-A and MRTF-B entry, with MRTF-A reaching maximum nuclear levels at 20 min and MRTF-B at 60 and 100 min after stimulation (SI Appendix, Fig. S2). These peaks matched time intervals of prolonged SRF association with chromatin uncovered above in ITM experiments (Fig. 4G).
In summary, we observed distinct peaks of long bound SRF association with chromatin at 20, 60, and 100 min of cell stimulation.
Interference with MAP Kinase Signaling, Actin Polymerization, and MRTF Cofactors Modulates SRF Residence Time.
SRF activity is regulated by two classes of partner proteins: MRTFs whose nuclear availability is controlled by Rho/actin signaling and TCFs associated with MAP kinase signaling (Fig. 5A). Since we saw a correlation of the percentage of long binding events with MRTF-A and MRTF-B activity (Fig. 4G and SI Appendix, Fig. S2), we tested whether such signaling partners affect SRF residence time in fibroblasts (Fig. 5E).
Fig. 5.
Actin and MAP kinase signaling modulate SRF residence time. (A) Scheme of RhoA-actin-MRTF and MAP kinase-TCF signaling pathways resulting in SRF activation. LatB and U0126 were used to block actin polymerization and MAP kinase signaling, respectively. (B–D) Residence time histograms of (B) DMSO (n = 1,838 bound molecules, n = 65 cells), (C) LatB (n = 1,522 bound molecules, n = 58 cells), and (D) U0126 (n = 1,795 bound molecules, n = 66 cells) treated cells. The histograms were fitted globally with a three-component decay model (red; SI Appendix, Eq. III). Time-lapse times are indicated above the data points. Error bars indicate SD. (E) NIH 3T3 cells expressing Halo-SRF were preincubated with DMSO or the inhibitors LatB or U0126 and stimulated for 60 min with FCS. The distribution of residence times followed a three-rate global fit with a long (light blue), intermediate (dark blue), and short (gray) bound fraction of Halo-SRF molecules. LatB incubation reduced the residence time of the long bound fraction, whereas this was unaffected by MAP kinase signaling. Residence time of the intermediate bound fraction was reduced by U0126 but not LatB treatment. (F) ITM measurement of starved or 20-min FCS-stimulated WT Halo-SRF cells untreated or pretreated with LatB or U0126 and of cells expressing the SRF mutant protein Halo-SRF αI helix. Without inhibitors, FCS increased the long bound Halo-SRF fraction (as shown before; see Fig. 4G), whereas LatB but not U0126 inhibited this response. In Halo-SRF αI helix-expressing cells, FCS did not induce an increase in the long bound fraction of the SRF mutant protein (mean ± SD; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; t test). Each gray circle or square indicates one cell analyzed (Halo-SRF starved: n = 47 cells; Halo-SRF 1 min to 20 min: n = 43 cells; Halo-SRF starved/U0126: n = 19 cells; Halo-SRF 1 min to 20 min/U0126: 14 cells; Halo-SRF starved/LatB: n = 43 cells; Halo-SRF 1 min to 20 min/LatB: 21 cells; Halo-SRF αI helix starved: n = 31 cells; Halo-SRF αI helix 1 min to 20 min: 34 cells). (G−J) The qPCR measuring endogenous mRNA levels of SRF target genes (n ≥ 3 independent cultures/each timepoint). U0126 interfered with IEG induction of (G) Egr1 and (H) Egr2, whereas LatB reduced expression of actin isoforms (I) Acta1 and (J) Acta2. Data are depicted as mean ± SD.
For this, NIH 3T3 were incubated with Latrunculin B (LatB), preventing actin polymerization and inhibiting nuclear MRTF entry (45, 46). To interfere with MAP kinase signaling, the established ERK inhibitor U0126 was employed (37). Either inhibitor or DMSO as control was applied to cells before stimulation with FCS for 1 h. Again we measured SRF binding times for different time-lapse conditions using the same illumination regime as before (Fig. 3A).
As for Fig. 3, we tested a one-, two-, or three-rate decay model for fitting the residence time histograms of all conditions. The three-rate decay model with χ2/ν = 1.38 was the best-fitting model for our data compared with the two-rate model (χ2/ν = 1.52) and the one-rate model (χ2/ν = 1.67), corroborating our previous findings (Fig. 3). Thus, cells stimulated with FCS in the presence of DMSO (control) or inhibitors revealed segregation of all bound molecules into three fractions: short, intermediate, and long bound molecules (Fig. 5E). Interference with Rho/actin signaling by LatB specifically decreased the residence time of the long binding SRF fraction in the first hour of stimulation (151 ± 47 s vs. 55 ± 14 s; Fig. 5 B–E), reaching a time comparable to starved cells (55 ± 21 s). In contrast, MAP kinase inhibition did not alter the long bound residence time (Fig. 5 D and E). Instead, the residence time of the intermediate bound fraction was decreased by MAP kinase inhibition (DMSO: 1.2 ± 0.2 s vs. U0126: 0.4 ± 0.1 s), whereas this was unaffected by LatB (Fig. 5 C and E). Besides residence times (Fig. 5 B–E), we also analyzed LatB and U0126 in ITM experiments focusing on starved cells and cells stimulated with FCS for the first 20 min (Fig. 5F). LatB application prevented formation of an increased long bound SRF fraction after cell stimulation, whereas U0126 did not show this effect (Fig. 5F). These ITM results corroborate previous findings on the impact of both inhibitors on the long SRF residence time (Fig. 5E).
We next analyzed the impact of both inhibitors on IEGs and cytoskeletal gene expression using qPCR (Fig. 5 G–J). In line with previous reports, we noted a strong dependence of IEG induction on MAP kinases (Fig. 5 G and H). In contrast, expression of several actin genes was predominantly inhibited by LatB (Fig. 5 I and J).
Since LatB-mediated interference of actin/MRTF signaling reduced the long SRF residence time, we more directly addressed the question of whether MRTFs modulate SRF residence time (SI Appendix, Fig. S3). For this, NIH 3T3 cells were transfected with siRNAs targeting both Mrtfa and Mrtfb. Indeed, mRNA abundance of both genes was reduced by Mrtfa/b-directed siRNAs in comparison with control siRNAs (SI Appendix, Fig. S3 A and B). This resulted in down-regulation of mRNA levels for several SRF/MRTF target genes including Arc, Egr2, Acta1, and Acta2 (SI Appendix, Fig. S3 C–F), although at lower efficacy than LatB treatment (Fig. 5 G–J).
Subsequently, SRF residence times were measured within the first hour of FCS stimulation in cells transfected either with control siRNA or Mrtfa/Mrtfb-directed siRNA. As before (Figs. 3 and 5), we used a three-rate decay model to characterize the SRF residence time regimes. Upon Mrtfa and Mrtfb depletion, we observed a decrease in the average residence time for both the intermediate and long bound SRF fraction (SI Appendix, Fig. S3G), although to a lower extent than with LatB inhibition (Fig. 5 E and F).
In addition to siRNA-mediated down-regulation of endogenous MRTF levels (SI Appendix, Fig. S3), we took advantage of an SRF mutant (SRF αI helix) that harbors point mutations in the alpha helix, thereby impairing SRF interaction with MRTFs (38). An NIH 3T3 cell line stably expressing the SRF αI helix mutant protein was left unstimulated or FCS-stimulated for 20 min, followed by imaging with the ITM protocol (Fig. 5F). In contrast to an enhanced fraction of chromatin-associated WT SRF proteins (Figs. 4G and 5F), FCS failed to enhance the fraction of long bound SRF molecules if MRTF interaction was precluded in the SRF αI helix mutant protein expressing NIH 3T3 cell line (Fig. 5F). This suggests that, in WT fibroblasts, SRF−MRTF cooperativity can enhance the fraction of long SRF binding events to chromatin.
Taken together, we report a function for MAP kinases, actin polymerization, and MRTFs in adjusting SRF residence times and long bound fractions.
SRF Residence Time Is Enhanced Upon Growth Factor Stimulation in Primary Neurons.
In experiments described until now, we uncovered changes in SRF binding behavior in highly proliferative fibroblasts upon stimulation (Figs. 1–5). To test whether SRF binding dynamics are conserved among different cell types, we turned toward mouse primary hippocampal neurons, a postmitotic cell type highly dependent on SRF function (4, 47). Since SRF is activated by growth factors, we tested whether growth factor stimulation affected the residence time of SRF in neurons. Here, the neurotrophic growth factor BDNF was employed to measure SRF residence times after stimulation. BDNF engages with TrkB receptor tyrosine kinases and both Rho/actin and MAP kinase signaling to activate SRF (37, 48). To analyze cell function as closely as possible to in vivo conditions, we used primary postmitotic mouse hippocampal neurons recapitulating several aspects of neuronal differentiation in culture (Fig. 6).
Fig. 6.
BDNF enhances SRF residence time and fraction of long bound SRF molecules in primary neurons. (A) Primary hippocampal neuron stained for neuron-specific βIII tubulin (green) and Halo-SRF with TMR (red). (Scale bar, 5 μm.) (B) A dSTORM picture of neuronal nucleus stained with anti-Halo antibodies to label Halo-SRF. (Scale bar: 5 μm.) (C) Representative traces for a short, intermediate, and long bound molecule. Time-lapse times between two illuminated frames are indicated. (Scale bar: 500 nm.) (D and E) Residence time histograms of (D) unstimulated neurons (n = 1,559 bound molecules, n = 78 cells) and (E) neurons stimulated for 1 h with BDNF (n = 1,530 bound molecules, n = 107 cells). The histograms were fitted globally with a three-component decay model (red). Dark times are indicated above the data points. Error bars indicate SD. (F) The 1/koff1, 1/koff2, and 1/koff3 represent average chromatin residence times for short, intermediate, and long bound SRF molecules, respectively. Within 1 h of BDNF stimulation, the long binding fraction (light blue) increased threefold, to 27 s. (G–J) Representative ITM examples of molecules imaged over four frames and one dark time interval in one neuron. A freely diffusing molecule is present in one frame only (yellow arrow). Two long binding molecules present in all four frames are labeled with red arrows. A short bound molecule is present in frames 1 and 2 (green arrows). (Scale bar: 500 nm.) (K) All frames of an ITM movie were merged, and long bound, short bound, and freely diffusing molecules are highlighted by colors. The nucleus is visible under phase contrast. (Scale bar, 2 μm.) (L) Stimulation rapidly enhanced the long bound fraction in the first 15 min after BDNF addition in relation to unstimulated neurons. At 45 min to 60 min of stimulation, the fraction of long bound molecules was peaking again (mean ± SD; *P ≤ 0.05; t test). Each gray symbol indicates one cell analyzed (unstimulated: n = 36 cells; 1 min to 15 min: n = 14 cells; 15 min to 30 min: n = 20 cells; 30 min to 45 min: n = 7 cells; 45 min to 60 min: n = 13 cells).
Neurons with Halo-SRF expression revealed robust nuclear Halo-SRF localization (Fig. 6A). As seen for fibroblasts (Fig. 1), dSTORM imaging revealed localization of individual SRF clusters throughout the nucleus (Fig. 6B). We employed immunocytochemistry to monitor Halo-SRF expression in fixed neurons, to exclude nonneuronal cells also present in the culture. In neurons, a twofold Halo-SRF overexpression (195.3 ± 9.3 rel. units; n = 34 neurons) compared with endogenous SRF levels only (114.5 ± 6.5 rel. units; n = 21 neurons) was observed. Halo-SRF overexpression did not interfere with BDNF-mediated gene induction of endogenous mRNA levels of IEGs (SI Appendix, Fig. S4). In contrast to fibroblasts, actin cytoskeletal genes analyzed in this study were not induced by BDNF in neurons (SI Appendix, Fig. S4), in line with a previous report (37).
Next, we determined the residence time of Halo-SRF in unstimulated neurons and neurons stimulated for 1 h with BDNF (Fig. 6 C–F and Movie S4). The imaging scheme was identical to Fig. 3A. As for the case of fibroblasts (Figs. 3 and 5 and SI Appendix, Fig. S3), a three-rate decay model was best at characterizing our data, with χ2/ν = 1.68, compared with a two-rate model (χ2/ν = 1.89) and a one-rate model (χ2/ν = 2.58). Thus, also in postmitotic neuronal cells, Halo-SRF molecules segregated into three populations: short, intermediate, and long bound molecules (Fig. 6 D–F). The average residence time of the short bound fraction in unstimulated and BDNF-stimulated neurons (0.1 ± 0.01 s; Fig. 6F) was similar to the one observed in fibroblasts (Fig. 3). The residence time of the intermediate bound fraction was calculated with 1 ± 0.3 s (Fig. 6F), thus being in a similar range as observed in fibroblasts (Figs. 3 and 5 and SI Appendix, Table S2). However, the average binding time of the long bound fraction in unstimulated neurons was shorter, 10 ± 2 s (Fig. 6F), compared with fibroblast, 55 s. Notably, similar to what was observed in fibroblasts, BDNF stimulation enhanced the residence time of the long bound fraction threefold, now averaging 27 ± 5 s (Fig. 6F). Thus, similar to serum stimulation in fibroblasts (Fig. 3), growth factor signaling in neurons enhanced SRF residence time of the long bound fraction during the first hour of stimulation (Fig. 6).
To compare stimulation in time intervals as applied to fibroblasts (Figs. 2 and 4), we also used ITM in neurons (Fig. 6 G–L and Movie S5). Similar to fibroblasts (Fig. 4), we observed short bound (green arrows), long bound (red arrows), and freely diffusing molecules (yellow arrow) also in primary neurons (Fig. 6 G–J). In unstimulated cells, the average percentage of long binding events was 27.1 ± 2.1% (Fig. 6L). Similar to fibroblasts (Fig. 4), the fraction of long binding SRF molecules was increased by BDNF in the first 15 min by 10%, to 36.3 ± 2.7% (Fig. 6L). After 15 min, this fraction decreased until 45 min after BDNF administration (Fig. 6L). Thereafter, as seen for fibroblasts (Fig. 4), a significant second increase in the percentage of long binding events was detected between 45 min and 60 min of BDNF stimulation (Fig. 6L).
In summary, in hippocampal neurons, we showed prolonged SRF residence time and an increased fraction of long binding SRF molecules by the growth factor BDNF.
Discussion
ITM Revealed an Increased Long Bound SRF Fraction After Cell Stimulation.
So far, the impact of cell stimulation particularly of growth factors has not been investigated with many TFs in SMT studies available. We employed ITM to focus on long bound chromatin-associated SRF fractions. Since, for this fraction, SRF binds for 2 s or more, we assume occupancy of these molecules at specific SRF promoters (see red lines in Fig. 7B). In fibroblasts, we observed an enhanced fraction of such long bound SRF binding events to chromatin at certain time intervals after stimulation, in a periodic pattern (Figs. 4 and 7B). Here, more long bound SRF molecules were present at around 20 and 60 min after stimulation. This temporal profile of the long bound SRF fraction was conserved in two cell types: proliferative fibroblasts and postmitotic neurons (Figs. 4 and 6). The wave-like pattern we obtained is comparable to the behavior of other TFs, including ER (18). For instance, dynamic ER oscillations binding to specific promoters were observed in a 40-min pattern, similarly to our findings (Figs. 4, 6, and 7B). In our study, this rhythmic pattern correlated with the nuclear abundance of the two SRF cofactors, MRTF-A and MRTF-B, that accumulated at earlier and later timepoints, respectively, after stimulation in the nucleus (SI Appendix, Fig. S2). MRTF−SRF interactions exclude nucleosomes from SRF-directed promoters, resulting in enhanced RNA polymerase II recruitment (15). Thus, nuclear MRTF abundance might contribute to the enhanced fraction of long SRF binding events at serum-inducible target genes. We could further support this finding by ITM experiments with cells pretreated with the actin polymerization inhibitor LatB and cells expressing an SRF mutant protein (SRF αI helix) incapable of interacting with MRTFs (Fig. 5). Both inhibition of MRTF nuclear entry (with LatB) and direct prevention of SRF−MRTF interaction reduced the increase of long binding events as seen for untreated WT cells in the first 20 min of serum stimulation (Fig. 5).
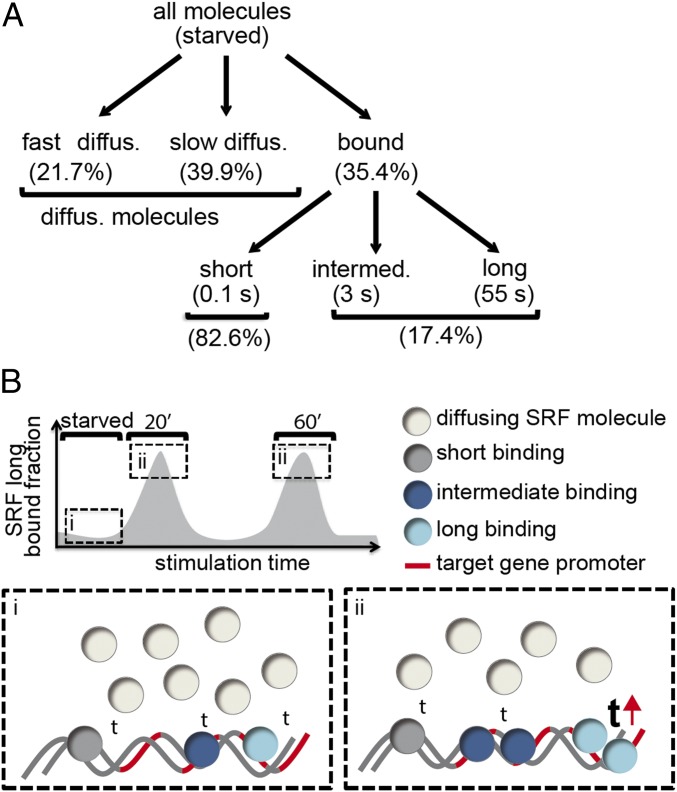
Fig. 7.
Summary scheme. (A) Scheme depicting segregation of all SRF fractions in starved fibroblast cells. (B) Scheme depicting changes in residence time (“t”) and long bound fraction of SRF molecules in unstimulated (i) and stimulated (ii) cells. After stimulation, the residence time of the long bound fraction increased (see upward-directed red arrow).
Of note is that long binding SRF events appeared to cluster at several positions in the nucleus (Figs. 4F and 6K). Such TF accumulations are reminiscent of so-called transcriptional hot spots or hubs enriched with activated RNA Polymerase II resulting in enhanced mRNA production (49). In agreement, long binding SRF events almost exclusively took place outside chromatin-dense areas (stained with Hoechst) or at the border, but rarely inside these bona fide heterochromatic areas (Fig. 4H). This suggests abundance of these long bound SRF molecules in transcriptionally active euchromatin areas. Thus, similar to TFs such as CREB (27), transcription of SRF-dependent genes might also take place in such transcriptional hubs.
Taken together, the enhanced proportion of long bound SRF molecules correlated well with MRTF−SRF cooperativity, indicating a role for MRTFs in mediating long SRF binding events.
Cell Stimulation Prolongs the SRF Residence Time After Stimulation.
ITM cannot determine absolute SRF residence times. We showed, in independent residence time measurements, an average chromatin association of long bound SRF molecules in starved fibroblasts for ∼60 s (Fig. 3). Thus, already in unstimulated cells, we observed relatively long residence times for SRF compared with other prototypical TFs such as p53, CREB, SOX2, GR, or ER (23, 24, 27, 28, 30). Similar long residence times in the minute range as seen for SRF have been reported only for general transcription regulators, including CTCF (40) and TBP (50). For quiescent neurons, the long residence time was approximately sixfold lower than in fibroblasts, averaging 10 s (Fig. 6). This suggests cell type-specific differences in residence times for SRF. We noted IEG induction in both cell types, whereas there were differences in cytoskeletal gene induction between fibroblasts and neurons (Fig. 1 and SI Appendix, Fig. S4). In fibroblasts, cytoskeletal genes selected in this study were up-regulated by stimulation, but not in neurons (Fig. 1 and SI Appendix, Fig. S4). However, neurons also induce further cytoskeletal genes, as shown before (51, 52). In general, such cell type-specific gene expression programs might contribute to differences in residence times observed. In addition, cell proliferation signals, continuously present in fibroblasts but not in postmitotic neurons, might account for these differences in residence time.
Besides unstimulated conditions, we provided a detailed SMT analysis under cell stimulating conditions. BDNF or FCS prolonged SRF residence times threefold to fivefold within the first hour in neurons or fibroblasts (Figs. 3 and 6). This points to a conserved mechanism of regulating SRF activity through enhancing the length of SRF interaction with chromatin independent of cell type (Fig. 7B, “t”). Besides SRF, other TFs were also shown to have longer duration of chromatin binding upon cell activation, including p53, ER, and, e.g., GR (24, 25, 28, 30). In contrast, CREB, a TF related to SRF, did only weakly respond with an increased residence time but rather showed an enhanced number of transcriptional hot spots (27). This points to TF-specific regulation by several available mechanisms, including modulation of residence times, enhancing TF numbers bound to chromatin, or recruitment to specific gene loci (Fig. 7B).
Population Dynamics of TF Molecules Includes Three Residence Time Regimes.
SMT together with FRAP and FCS experiments provided insight into dynamic TF behavior (18). This included separation of a TF population into different binding time categories with distinct biophysical properties (Fig. 7A), rather than assuming a homogenous TF population exerting static and stable TF−chromatin interactions (18, 20). Previous SMT experiments identified two TF subpopulations, distinguishable by short and long residence times (23, 24, 27, 28, 30). In this study, a three-rate model was best suited to characterize SRF dynamics (Fig. 7). This observation was made in fibroblasts and neurons, suggesting conserved SRF residence time regimes in diverse cell types (SI Appendix, Table S2). So far, molecules with a residence time of >1 s are considered to result in robust mRNA transcription at specific promoters (35). Therefore, we assume unspecific chromatin binding of the short bound fraction, whereas both intermediate and long bound fractions are involved in specific chromatin association at SRF target genes (see red lines in Fig. 7B). The residence time of the intermediate fraction was not obviously altered within the first hour of stimulation (see also Fig. 7B). Notably, in fibroblasts, the intermediate bound fraction was affected by MAP kinase signaling (Fig. 5), typically associated with SRF-mediated IEG expression. This might indicate a functional role of the intermediate bound SRF fraction in IEG regulation, allowing for more rapid exchanges of SRF at IEG promoters. In opposite to the intermediate bound fraction, we observed an increase in the residence time for long bound SRF molecules in both cell types after stimulation. This long bound SRF fraction was sensitive toward inhibition of Rho-actin-MRTF signaling in fibroblasts (Fig. 5 and SI Appendix, Fig. S3). This points to a function of the Rho-actin-MRTF axis, usually associated with cytoskeletal gene regulation, in prolonging the residence time and enhancing the fraction of long bound SRF molecules in fibroblasts.
In summary, our data for SRF describe the dynamic potential of TFs to stratify into more than two different residence time regimes with distinct residence times and sensitivity toward signaling inhibitors.
Supplementary Material
Acknowledgments
We are grateful to Lena Schulze and Anja Palmer for help with the HILO microscopy. The work has been supported by the Deutsche Forschungsgemeinschaft through the Collaborative Research Center 1149 “Danger Response, Disturbance Factors and Regenerative Potential after Acute Trauma” (B.K.), Grant KN543/6-1 (to B.K.), Grant MI749/7-1 (to J.M.), and Grant GE 2631/1-1 (to J.C.M.G.). This project is funded by a joint (B.K. and J.M.) PhD stipend (to L.H.) by the International Graduate School of Molecular Medicine at Ulm University. B.K. is supported by the Paul und Marlene Hepp-Stiftung. The work was further supported by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program No. 637987 ChromArch (to J.C.M.G.) and the German Academic Scholarship Foundation (M.R.). The authors thank the Ulm University Center for Translational Imaging for its support. J.M. and J.C.M.G. acknowledge support by the Collaborative Research Centre 1279.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812734116/-/DCSupplemental.
References
- 1.Miano JM. Serum response factor: Toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 2.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posern G, Treisman R. Actin’ together: Serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Knöll B, Nordheim A. Functional versatility of transcription factors in the nervous system: The SRF paradigm. Trends Neurosci. 2009;32:432–442. doi: 10.1016/j.tins.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Miano JM. Role of serum response factor in the pathogenesis of disease. Lab Invest. 2010;90:1274–1284. doi: 10.1038/labinvest.2010.104. [DOI] [PubMed] [Google Scholar]
- 6.Small EM. The actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J Cardiovasc Transl Res. 2012;5:794–804. doi: 10.1007/s12265-012-9397-0. [DOI] [PubMed] [Google Scholar]
- 7.Benito E, Barco A. The neuronal activity-driven transcriptome. Mol Neurobiol. 2015;51:1071–1088. doi: 10.1007/s12035-014-8772-z. [DOI] [PubMed] [Google Scholar]
- 8.Kuzniewska B, Nader K, Dabrowski M, Kaczmarek L, Kalita K. Adult deletion of SRF increases epileptogenesis and decreases activity-induced gene expression. Mol Neurobiol. 2016;53:1478–1493. doi: 10.1007/s12035-014-9089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lösing P, et al. SRF modulates seizure occurrence, activity induced gene transcription and hippocampal circuit reorganization in the mouse pilocarpine epilepsy model. Mol Brain. 2017;10:30. doi: 10.1186/s13041-017-0310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramanan N, et al. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 11.Zimprich A, et al. A robust and reliable non-invasive test for stress responsivity in mice. Front Behav Neurosci. 2014;8:125. doi: 10.3389/fnbeh.2014.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gualdrini F, et al. SRF co-factors control the balance between cell proliferation and contractility. Mol Cell. 2016;64:1048–1061. doi: 10.1016/j.molcel.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, et al. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 14.Herrera RE, Shaw PE, Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- 15.Esnault C, et al. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teves SS, et al. A dynamic mode of mitotic bookmarking by transcription factors. eLife. 2016;5:e22280. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Tjian R. Visualizing transcription factor dynamics in living cells. J Cell Biol. 2018;217:1181–1191. doi: 10.1083/jcb.201710038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao S, Xue B, Sun Y. Intranucleus single-molecule imaging in living cells. Biophys J. 2018;115:181–189. doi: 10.1016/j.bpj.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gebhardt JC, et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groeneweg FL, et al. Quantitation of glucocorticoid receptor DNA-binding dynamics by single-molecule microscopy and FRAP. PLoS One. 2014;9:e90532. doi: 10.1371/journal.pone.0090532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izeddin I, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife. 2014;3:02230. doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitagawa H, et al. Activity-dependent dynamics of the transcription factor of cAMP-response element binding protein in cortical neurons revealed by single-molecule imaging. J Neurosci. 2017;37:1–10. doi: 10.1523/JNEUROSCI.0943-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loffreda A, et al. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat Commun. 2017;8:313. doi: 10.1038/s41467-017-00398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40:e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paakinaho V, et al. Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat Commun. 2017;8:15896. doi: 10.1038/ncomms15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speil J, et al. Activated STAT1 transcription factors conduct distinct saltatory movements in the cell nucleus. Biophys J. 2011;101:2592–2600. doi: 10.1016/j.bpj.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie L, et al. A dynamic interplay of enhancer elements regulates Klf4 expression in naïve pluripotency. Genes Dev. 2017;31:1795–1808. doi: 10.1101/gad.303321.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clauß K, et al. DNA residence time is a regulatory factor of transcription repression. Nucleic Acids Res. 2017;45:11121–11130. doi: 10.1093/nar/gkx728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morisaki T, Müller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun. 2014;5:4456. doi: 10.1038/ncomms5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White MD, et al. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell. 2016;165:75–87. doi: 10.1016/j.cell.2016.02.032. [DOI] [PubMed] [Google Scholar]
- 37.Meier C, Anastasiadou S, Knöll B. Ephrin-A5 suppresses neurotrophin evoked neuronal motility, ERK activation and gene expression. PLoS One. 2011;6:e26089. doi: 10.1371/journal.pone.0026089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaromytidou AI, Miralles F, Treisman R. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol Cell Biol. 2006;26:4134–4148. doi: 10.1128/MCB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 40.Agarwal H, Reisser M, Wortmann C, Gebhardt JCM. Direct observation of cell-cycle-dependent interactions between CTCF and chromatin. Biophys J. 2017;112:2051–2055. doi: 10.1016/j.bpj.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisser M, et al. Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development. Nat Commun. 2018;9:5218. doi: 10.1038/s41467-018-07731-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 43.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: Balancing stability and plasticity. Biochim Biophys Acta. 2008;1783:2061–2079. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 44.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 45.Posern G, Miralles F, Guettler S, Treisman R. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 2004;23:3973–3983. doi: 10.1038/sj.emboj.7600404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knöll B, et al. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat Neurosci. 2006;9:195–204. doi: 10.1038/nn1627. [DOI] [PubMed] [Google Scholar]
- 48.Kalita K, Kharebava G, Zheng JJ, Hetman M. Role of megakaryoblastic acute leukemia-1 in ERK1/2-dependent stimulation of serum response factor-driven transcription by BDNF or increased synaptic activity. J Neurosci. 2006;26:10020–10032. doi: 10.1523/JNEUROSCI.2644-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho WK, et al. RNA polymerase II cluster dynamics predict mRNA output in living cells. eLife. 2016;5:e13617. doi: 10.7554/eLife.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: Constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li CL, Sathyamurthy A, Oldenborg A, Tank D, Ramanan N. SRF phosphorylation by glycogen synthase kinase-3 promotes axon growth in hippocampal neurons. J Neurosci. 2014;34:4027–4042. doi: 10.1523/JNEUROSCI.4677-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mokalled MH, Johnson A, Kim Y, Oh J, Olson EN. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development. 2010;137:2365–2374. doi: 10.1242/dev.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.