Significance
Pathogens are omnipresent and by definition detrimental to their hosts. Pathogens thus exert high selection on their hosts, which, if adapting, can exert similar levels of selection on the pathogen, resulting in ongoing cycles of reciprocal adaptation between the antagonists. Such coevolutionary interactions have a central influence on the evolution of organisms. Surprisingly, we still know little about the exact selection dynamics and the genome regions involved. Our study uses a controlled experimental approach with an animal host to dissect coevolutionary selection. We find that distinct selective processes underlie rapid coadaptation in the two antagonists, including antagonistic frequency-dependent selection on toxin gene copy number in the pathogen, while the host response is likely influenced by changes in multiple genome regions.
Keywords: host–pathogen coevolution, Red Queen hypothesis, population genomics, copy number variation, Caenorhabditis elegans
Abstract
Red Queen dynamics, involving coevolutionary interactions between species, are ubiquitous, shaping the evolution of diverse biological systems. To date, information on the underlying selection dynamics and the involved genome regions is mainly available for bacteria–phage systems or only one of the antagonists of a eukaryotic host–pathogen interaction. We add to our understanding of these important coevolutionary interactions using an experimental host–pathogen model, which includes the nematode Caenorhabditis elegans and its pathogen Bacillus thuringiensis. We combined experimental evolution with time-shift experiments, in which a focal host or pathogen is tested against a coevolved antagonist from the past, present, or future, followed by genomic analysis. We show that (i) coevolution occurs rapidly within few generations, (ii) temporal coadaptation at the phenotypic level is found in parallel across replicate populations, consistent with antagonistic frequency-dependent selection, (iii) genomic changes in the pathogen match the phenotypic pattern and include copy number variations of a toxin-encoding plasmid, and (iv) host genomic changes do not match the phenotypic pattern and likely involve selective responses at more than one locus. By exploring the dynamics of coevolution at the phenotypic and genomic level for both host and pathogen simultaneously, our findings demonstrate a more complex model of the Red Queen, consisting of distinct selective processes acting on the two antagonists during rapid and reciprocal coadaptation.
More than 40 y ago, Van Valen (1) proposed the Red Queen hypothesis stating that evolutionary lineages persist only if they continuously change and adapt to ongoing selective pressures. A later refinement of the hypothesis put the spotlight on host–pathogen interactions (2, 3): Because these interactions are antagonistic and many pathogens depend on their hosts for survival, they are likely shaped by repeated cycles of adaptation and counteradaptation from both partners. The consequences of these dynamics are potentially far-reaching. For example, they may account for the extremely high rates of molecular evolution in host immunity and pathogen virulence genes (4, 5), they might favor the evolution and maintenance of outcrossing and sexual reproduction (6, 7), they might generally promote genetic diversity (3), and they can also affect a diversity of life history traits (8). Despite the enormous number of studies on host–pathogen coevolution, two core aspects are as yet only poorly understood: the genome regions targeted by reciprocal selection and the resulting allele frequency dynamics in these regions.
Two main dynamics are generally proposed as alternative mechanisms underlying host–pathogen coevolution: recurrent selective sweeps (RSS) and antagonistic frequency-dependent selection (aFDS) (2). Under RSS, the two antagonists sequentially accumulate adaptive mutations, each one sweeping to fixation through the host or pathogen population. Under aFDS, genetic diversity is maintained, and adaptive alleles oscillate in frequency as a result of selection against the most common host or pathogen type. The relevance of these two selection mechanisms can be assessed through time-shift experiments, where fitness-related traits for one of the antagonists is evaluated in the presence of the other antagonist from the evolutionary past, present, and future (9, 10). The expectation for RSS is that the focal organism produces high fitness toward its past and low fitness toward its future antagonist. Under aFDS the faster-evolving partner (usually the pathogen) is expected to have higher fitness in the presence of the contemporaneous antagonist than those from the evolutionary past or future (11). To date, such time-shift experiments have primarily been applied to bacteria–phage models, revealing that coevolution leads to RSS dynamics initially (12, 13) and possibly aFDS dynamics at later time points depending on nutrient availability, spatial structure, and fluctuating environments (11, 14, 15). Time-shift experiments have occasionally been used for eukaryotic unicellular hosts, for example, unicellular algae and coevolving viruses, revealing transient RSS dynamics followed by the dominance of a generally resistant host (16). Time-shift experiments with multicellular hosts were, for example, performed for the water flea Daphnia magna and the flax Linum marginale host systems, in both cases suggesting aFDS as the main determinant of coevolution (17, 18).
Conclusive information on the involved selection dynamics further requires a combination of the time-shift experiments with data on allele frequency changes, using, for example, whole-genome sequence analysis. Several previous studies applied these two approaches (see summary in SI Appendix, Table S1). However, many of these studies had a different focus and thus did not quantify allele frequency changes across time. For example, previous genome analysis of host–phage coevolution focused on sequencing phage genomes only at the end point of the experiment (4) or only for a single phage gene across several time points (14). In general, longitudinal whole-genome analysis of coevolving populations remains rare and usually includes only one antagonist (SI Appendix, Table S1). In fact, whole-genome sequence analysis for both antagonists has been performed only for a bacteria–phage interaction, revealing RSS, as predicted by time-shift experiments, and an influence of phage diversity on selective sweeps dynamics (19, 20).
The objectives of the current study were to enhance our understanding of the selection dynamics and the genomic basis of host–pathogen coevolution by using (i) a multicellular animal host system, which can be easily manipulated and which allows cryopreservation for later time-shift experiments, (ii) laboratory-based coevolution experiments with control over interaction type (i.e., coevolution or not), population size, generation time (the latter only for the host), and high replication, (iii) time-shift experiments for the two antagonists, using cryopreserved material from the evolution experiment for both host and pathogen, and (iv) whole-genome sequence analysis for both antagonists, taking advantage of available high-quality reference genomes (Fig. 1). In detail, we used the nematode host Caenorhabditis elegans and its pathogen Bacillus thuringiensis as a model (21, 22). We implemented methods for nematode cultivation and worm–pathogen purification (Methods) to allow performance of evolution experiments at high throughput. To initiate coevolution in our experiments, we used genetically diverse host populations (with 3,000 individuals each, including >370,000 segregating sites per population) and infected them with a single B. thuringiensis strain, MYBT18247 (SI Appendix, Figs. S1–S3). In the coevolution treatment, host and pathogen populations were continuously paired to enforce reciprocal adaptation, while the control treatments were identical but without exposure to the respective antagonists (Methods, 16 biological replicates and 23 transfers per treatment). After the evolution experiment, we used the regularly frozen evolved hosts and pathogens for measuring changes in fitness by their exposure to the respective ancestral antagonists. We then characterized the selection dynamics using reciprocal time-shift experiments (all replicate populations, three time points) in combination with population genomic analysis of both coevolved hosts and pathogens (six replicate populations and three time points for each antagonist and treatment).
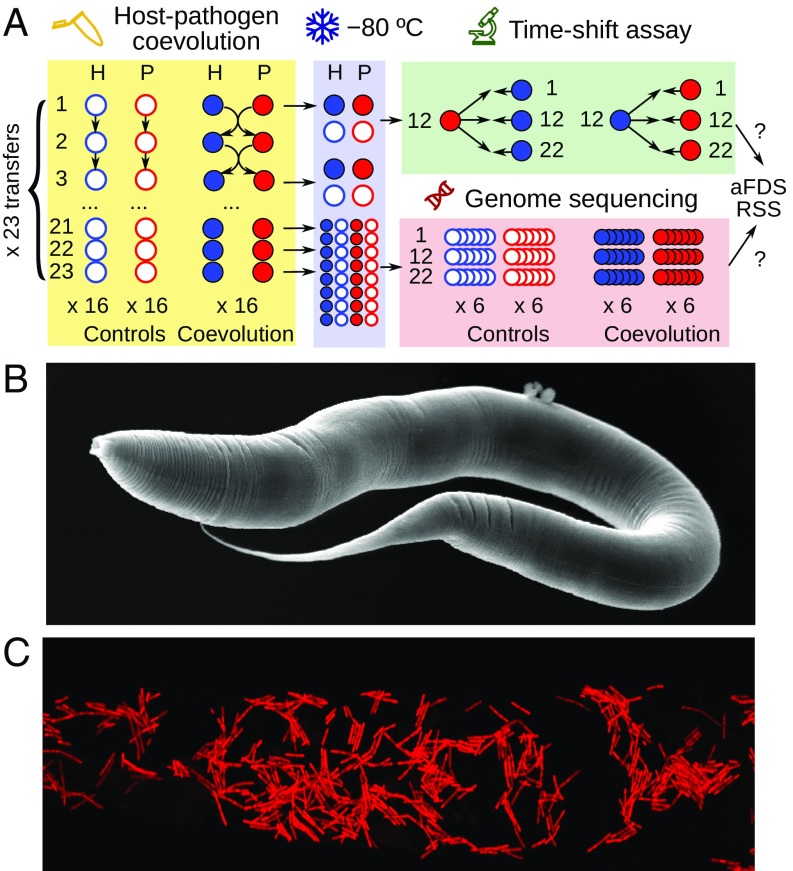
Fig. 1.
A Caenorhabditis elegans–Bacillus thuringiensis model for experimental host–pathogen coevolution. (A) Overview of the general design of the evolution experiment, the subsequent phenotyping and time-shift experiment, and focus of the genomic analysis. H, host; P, pathogen. (B) Scanning electron micrograph of a C. elegans hermaphrodite. (C) Illustration of B. thuringiensis, labeled by red fluorescence, inside an infected and killed C. elegans.
Results and Discussion
Coevolution Caused a General Increase in Host and Pathogen Fitness.
For our phenotypic analysis of the experimentally evolved hosts and pathogens, we focused on the fitness traits that were targeted by selection in our experimental protocol: host egg number and pathogen cell number inside a host body at exactly the transfer time point (Methods). We found that in the presence of the ancestral pathogen, the coevolved hosts had significantly more eggs than the control and ancestor, both at the end of the evolution experiment (Fig. 2A and SI Appendix, Table S2) and across the 23 transfers (SI Appendix, Fig. S4 and Tables S3 and S4). The host populations from the control treatment did not differ significantly from the ancestor (Fig. 2A and SI Appendix, Fig. S4 and Table S2). These changes occurred independently of changes in resistance (SI Appendix, Fig. S5A and Tables S5 and S6), a trait not under direct selection in our experiment. To estimate fitness of B. thuringiensis, the evolved pathogen lines were competed against a labeled ancestral clone for one infection cycle within the host. Bacterial fitness here is mainly determined by replication inside the host because the experimental medium did not support B. thuringiensis growth (SI Appendix, Fig. S7). In these competitions, coevolved pathogens produced significantly more cells than ancestral and control-evolved bacteria (Fig. 2B and SI Appendix, Tables S7 and S8). The control lines did not differ from the ancestral clone (however, see relative fitness in broth, SI Appendix, Fig. S7 and Tables S9 and S10). Pathogen fitness evolved independent of virulence (SI Appendix, Fig. S5 B and C and Tables S11 and S12), which, like host resistance, was not directly targeted by selection in our experiments. We conclude that both host and pathogen adapted to the presence of their coevolving antagonist.
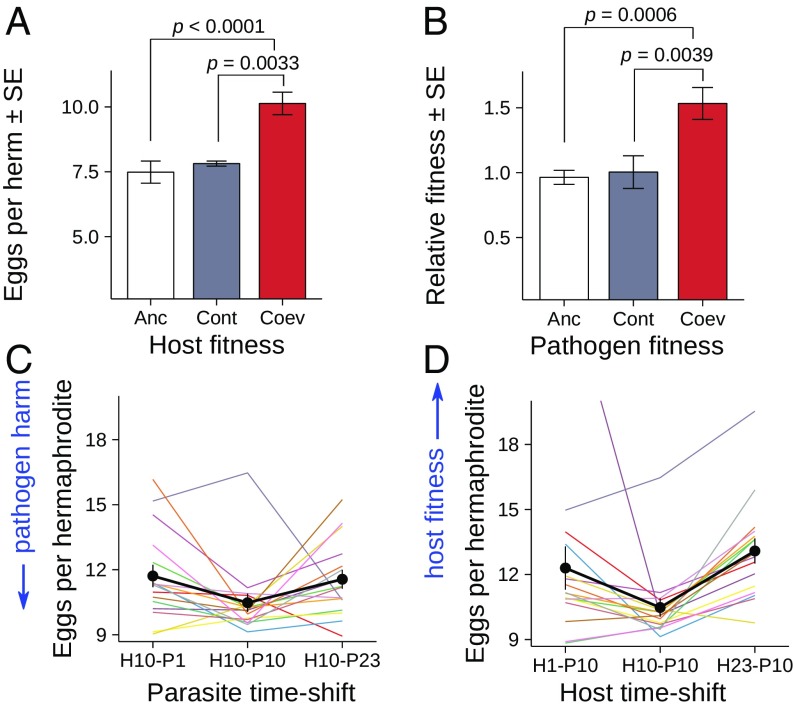
Fig. 2.
Adaptation in both host and pathogen during experimental coevolution. (A) Increased host fitness after coevolution, measured as average number of eggs per hermaphrodite upon exposure to the ancestral pathogen (eight populations per treatment, 30 hermaphrodites per population). (B) Increased pathogen fitness after coevolution, measured as the competitive ability of an evolved bacterium against a gfp-labeled ancestral strain during one infection cycle (16 lines per treatment, three technical replicates per line). Variation in A and B was assessed with the Wilcoxon rank sum test, adjusted for multiple comparisons (three nonindependent tests) with the Holm–Bonferroni method. Anc, ancestor; Coev, coevolution treatment; Cont, control treatment. (C) Time-shift experiments for focal hosts from transfer 10 (H10) exposed to past, contemporaneous, and future pathogens (from transfers 1, 10, and 23; P1, P10, and P23, respectively). (D) Similar time-shift experiments using focal pathogens from transfer 10 (P10). In C and D, the host and pathogen were always matched with the coevolved antagonist from the same replicate population. Each line represents an independent replicate; the black lines and bars show the average across replicates with SEs (16 lines, 30 hermaphrodites per line).
Time-Shift Experiments Indicated Antagonistic Frequency-Dependent Selection Dynamics for Both Antagonists.
We performed time-shift experiments to find out whether the antagonists adapted reciprocally to each other and, if so, whether adaptation was determined by RSS or aFDS. We ensured comparability of results for the two antagonists by assaying the same trait for both, namely, the effect on host egg production as one of the properties directly under coevolutionary selection. All 16 host populations from transfer 10 (H10, the focal time point) were infected with the corresponding pathogen populations from three different time points, including one from the past (P1, transfer 1), the present (P10, transfer 10), and the future (P23, transfer 23). In 10 out of 16 coevolved lines, pathogen P10 had the strongest negative effect on host fertility (Fig. 2C and SI Appendix, Table S13). This result is consistent with aFDS dynamics, where the pathogen is best adapted to the contemporaneous host. Similarly, when the pathogen from transfer 10 (P10) was used to infect the corresponding host populations from the past (H1), the present (H10), and the future (H23), the contemporaneous host H10 had the lowest fitness (Fig. 2D and SI Appendix, Table S13), again in line with aFDS.
The pattern of temporal genotype-specific coadaptation was additionally confirmed by considering all possible host–pathogen combinations from transfers 1, 10, and 23 (SI Appendix, Fig. S9), further supporting the observation that the P10 pathogen was specifically adapted to H10 but not the hosts from the past or future, while the H10 host changed in a specific way toward the P10 antagonist but not those from the past or future (in this case, indicating maladaptation). Moreover, to test the robustness of these results, we repeated the entire time-shift experiment using populations from other close time points (transfers 12 and 22 instead of transfers 10 and 23, respectively). We assayed 12 out of the 16 coevolved host–pathogen lines and included three technical replicates and similarly found that the contemporaneous combinations produced the lowest host reproduction measurements (SI Appendix, Fig. S8 and Table S14).
Based on these two independent time-shift experiments and always at least 12 independent biological replicates, we conclude that experimental coevolution between C. elegans and B. thuringiensis produces a highly robust phenotypic pattern that is most consistent with rapid reciprocal temporal coadaptation between the antagonists, a high level of parallel evolution across the independently evolving replicate populations, and also the involvement of aFDS dynamics. The latter finding concurs with similar aFDS patterns during host–pathogen coevolution in other multicellular hosts, such as the water flea D. magna or the flax L. marginale (17, 18), while unicellular host systems were most commonly found to be subject to RSS (16, 23), with aFDS observed only under certain conditions [e.g., changes in spatial structure or the presence of different phage types (11, 14, 15)]. The aFDS-like pattern arose in our experiments despite the experiment being initiated from a single clone of the pathogen (the host population had comprehensive genetic variation). This experimental design was chosen to reflect natural conditions, where the pathogen is commonly subjected to a severe bottleneck during infection (refs. 24 and 25; see also SI Appendix, Material and Methods). Nevertheless, the aFDS-like phenotypic response for both antagonists indicates that variation arose rapidly within the pathogen populations.
Host Coevolutionary Adaptation Is Associated with Large Genomic Regions on Chromosomes II, III, and X.
The results of the time-shift experiment implied that host and pathogen genotypes oscillated in frequency for at least one cycle. To test whether the genomic regions responsible for adaptation similarly follow the observed phenotypic pattern, we analyzed whole-genome sequences for six matching coevolved C. elegans and B. thuringiensis populations from three time points (transfers 1, 12, and 22). We specifically chose replicate populations that produced highly consistent patterns of temporal coadaptation (SI Appendix, Fig. S8), strongly suggesting parallel evolution. We also included six control populations from the same time points. We first focused on C. elegans populations. The ancestral population of the host had high standing genetic diversity, with more than 370,000 segregating biallelic sites (SI Appendix, Table S17). Based on this high variation level, we attempted to identify signatures of aFDS that matched the observed phenotypic pattern by estimating selection coefficients from allele frequency changes during two consecutive intervals: between transfers 1 and 12 and between transfers 12 and 22 (s1–12 and s12–22, respectively; Fig. 3 and Methods). Under aFDS, the direction of pathogen-mediated selection is expected to change, and therefore, the dynamics should be characterized by contrasting signs of s between the time periods (i.e., either positive–negative or negative–positive combinations; Fig. 3 C and E). We also considered RSS and incomplete selective sweeps, which would be evident with selection coefficients of similar signs across intervals (Fig. 3 C and E). To enhance the power of our statistical analysis, we focused on selection signatures that were consistent across the replicate populations, thus following the high parallelism observed at the phenotypic level.
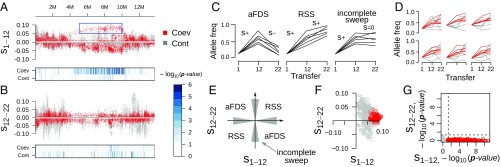
Fig. 3.
Selective sweep dynamics of specific genome regions in coevolving C. elegans populations. (A) The distribution of the selection coefficient s along chromosome III during the first half of the experiment (transfers 1–12) estimated for coevolving (Coev; red) and control (Cont; gray) populations. The estimation of s is based on allele frequency changes of biallelic SNPs in six replicate populations. The heat maps below the main panel indicate genome regions with a significant change in allele frequency (more intense color corresponds to higher significance). The blue box highlights the genome region supported by many significant SNPs and high values of s (>0.05). (B) The estimated selection coefficients along chromosome III for the second half of the experiment (transfers 12–22). (C) Hypothetical allele trajectories expected for aFDS (Left), RSS (Middle), and incomplete sweeps (Right). (D) The actual allele trajectories for six representative SNPs from the highlighted region on A. (E) Hypothetical change in selection for three types of coevolutionary dynamics. If the SNPs observed are in complete linkage with the loci under selection, then a change in sign of the selection coefficient is expected for aFDS but not RSS dynamics. The shaded areas close to the axes correspond to incomplete sweeps. (F) The change in s for SNPs within the candidate region on chromosome III from A (n = 489). (G) The corresponding P values for the same SNPs, showing that allele frequency changes were significant from transfer 1 to 12 but not from transfer 12 to 22.
We identified several regions with significant selection coefficients under coevolution but not control conditions, suggesting that these regions are under pathogen-mediated selection (SI Appendix, Fig. S9). The most striking pattern was found for a large region on chromosome III (III: in between megabase pairs 5–10) with hundreds of SNPs showing significant changes in allele frequencies during the first 12 transfers (Fig. 3 A and D and SI Appendix, Fig. S10). Such a uniform pattern in many SNPs illustrates how pathogen-mediated selection can cause dramatic changes in just a few generations. During the second half of coevolution (transfers 12–22), only a small subset of these SNPs continued to yield significant selection coefficients (Fig. 3 B, F, and G and SI Appendix, Fig. S12, III: megabase pairs 5–10), and these tended to have the same sign (Fig. 3F and SI Appendix, Fig. S12, III: megabase pairs 8–10), a result that is inconsistent with aFDS. Similar observations were made for the other identified candidate regions, including two on the X chromosome (X: in between megabase pairs 7–9 and X: in between megabase pairs 15.9–16.2; SI Appendix, Figs. S13 and S14).
In general, fewer significant s were identified for the second half of experimental coevolution (441 versus 9,397 significant SNPs for the second and first halves, respectively). It is possible that the increasing divergence among replicate populations may have made replicate-specific adaptations statistically indistinguishable from genetic drift. The few significant regions in the second half included, for example, one on chromosome II, in between megabase pairs 10–12 (SI Appendix, Fig. S15); chromosome III, megabase pairs 8–10 (SI Appendix, Fig. S12); and chromosome X, megabase pairs 9–10.5 (SI Appendix, Fig. S16). Again, contrary to expectations, we did not find changes in the direction of selection for these particular SNPs. In addition, almost all of the identified regions under selection were confirmed through an independent reanalysis of the data using FST statistics (SI Appendix, Fig. S17). The size of the affected region may not necessarily indicate that many loci are under selection but could result from high linkage due to the low recombination rates typical for the central regions of C. elegans chromosomes (26). These regions nevertheless include genes with functions related to the observed phenotypic variations, including fertility [e.g., a coevolution-specific duplication containing trd-1 (27); SI Appendix, Figs. S18 and S19] and immunity-related genes like C-type lectin-like genes (ref. 28; for a complete list of change-of-function mutations see Dataset S2).
We conclude that none of the identified genome regions with significant changes under coevolution showed allele trajectories that match the expected patterns of the time-shift experiments. The lack of support for aFDS in the genomic data are unlikely due to a lack of statistical power. For example, many significant SNPs from the first half of coevolution could have lost significance during the second half due to increased population divergence (i.e., lack of parallel evolution). However, if we follow frequency changes of these particular SNPs in their replicate populations, then almost none of them changed the sign of selection, as required for aFDS. Consequently, the underlying genomic dynamics are likely more complex. It is conceivable that the phenotypic pattern is produced by nonparallel allele oscillations at different loci across the independently evolving replicate populations or, as a more parsimonious explanation consistent with the patterns identified in our genome data, by parallel-occurring incomplete sweeps, which occur at different loci during different phases of coevolutionary adaptation (Fig. 3C). Our findings are thus different from those of previous work, which, for example, suggested canonical allele oscillations at a single locus underlying aFDS in the D. magna model (29) or showed a series of complete allele fixations underlying RSS in (cyano)bacteria–phage experiments (12, 19).
Bacterial Coevolutionary Adaptation Is Driven by Copy Number Variation of a Toxin-Encoding Plasmid.
In parallel to the host, we assessed genome sequence variation in six coevolved and six control B. thuringiensis populations. These populations were from the same time points and replicates as those of the host, analyzed above. For the pathogen, all replicate populations were founded by a single clone, and thus, adaptation should rely on only de novo mutations. As the phenotypic response was consistent across replicates from a specific evolution treatment, we again expected to find patterns of parallel genomic evolution. Our analysis identified a total of 24 point mutations and small insertions and deletions (indels) during coevolution and 49 mutations in the control treatments (SI Appendix, Tables S18–S20). Eighteen mutations are nonsilent and are predicted to have functional consequences. Three mutations in the coevolving populations affected genes involved in sporulation initiation (dnaA, kinB, spo0B; SI Appendix, Figs. S20 and S21). For the control treatments, we also found mutations affecting sporulation, although in a different set of genes (e.g., spo0B, abrB, kinE, ynzD). These results suggest that changes in the process of sporulation are an adaptation to the general experimental evolution protocol. It cannot be excluded that the specific genes affected under coevolution contribute, at least to some extent, to the observed pattern of genotype-specific temporal coadaptation (SI Appendix, Figs. S2D and S9). Apart from the sporulation initiation genes, no other mutations showed parallelism in the coevolving populations.
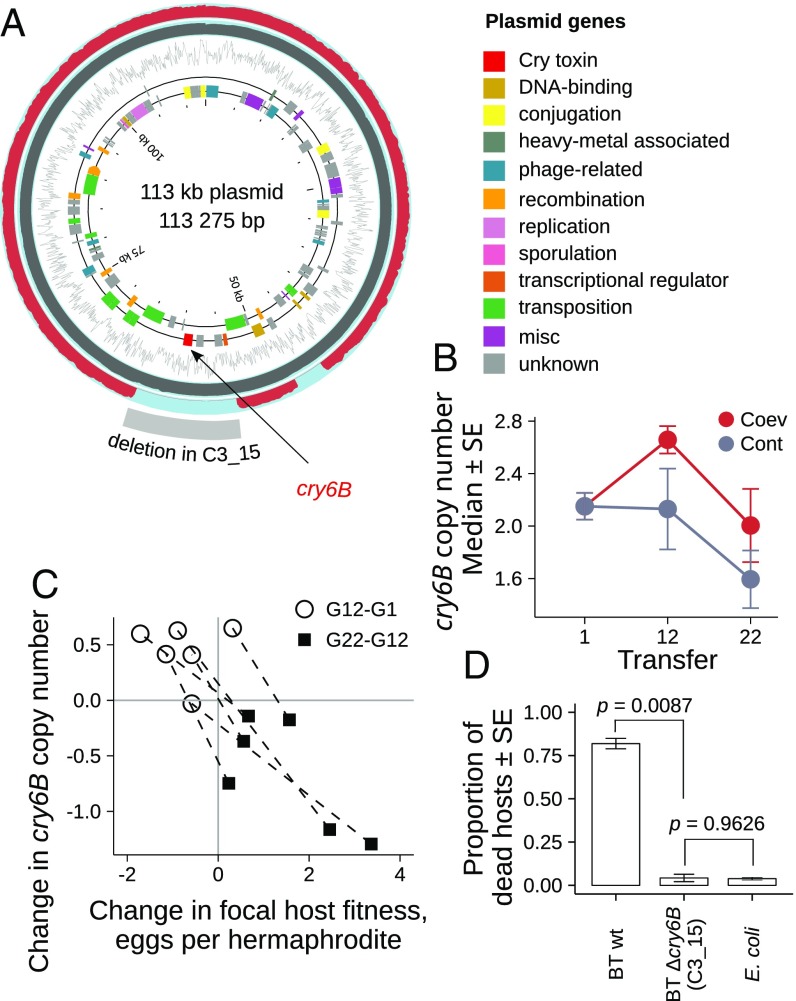
As structural genome changes may also underlie evolutionary adaptation, we next sought the presence of copy number variations (Fig. 4). We found that a specific plasmid differed in copy number dynamics between control and coevolved populations (Fig. 4B and SI Appendix, Table S21). More precisely, a 113-kb plasmid carrying the cry6B toxin gene decreased in copy number across time in the control populations. In contrast, in the coevolving populations, the same plasmid first increased in copy number at transfer 12 and then returned to initial frequencies by transfer 22 (Fig. 4B and SI Appendix, Table S21). The encoded Cry6B belongs to the cry6 family of pore-forming crystal toxins that cause C. elegans gut damage and mortality and reduced reproduction (30, 31). The general decrease of this plasmid in the control treatment is thus consistent with the observed reduced virulence (SI Appendix, Fig. S5), whereas oscillating copy number changes during coevolution indicates aFDS. Importantly, the change in cry6B copy number correlates significantly with the pathogen’s effect on host fitness in the time-shift experiment (Fig. 4C, Spearman correlation ρS = −0.58, P = 0.0053). Thus, high copy numbers are associated with lowered host fertility, while low copy numbers lead to higher fertility in a host genotype-specific form during the time-shift experiments. This correlation thereby supports the involvement of copy number variation in the pathogen’s temporal adaptation to the coevolving host. Complete sequencing of the avirulent pathogen line C3_15 (Fig. 4D) revealed that the 113-kb plasmid was present at comparatively high frequency, yet it contained a 9.5-kb deletion encompassing the cry6B gene and five other ORFs (Fig. 4A), suggesting that the deletion caused the loss of virulence. Based on these results, we conclude that cry6B gene copy number variation largely determines the pathogen’s response during coevolutionary adaptation.
Fig. 4.
Coevolving Bacillus thuringiensis show copy number changes in a toxin-encoding plasmid. (A) The 113-kb plasmid encoding Cry-toxin gene cry6B. Circles starting from the inside: coding DNA sequences on positive and negative strands, with colors indicating gene functions, GC content, sequence coverage for the virulent ancestor (gray on blue background), coverage for evolved avirulent pathogen line C3_15 (red on blue background), and deleted region in C3_15. Pathogen line C3_15 is a replicate population from the coevolution treatment that completely lost virulence after 23 transfers. (B) Change in cry6B-encoding plasmid copy number (relative to the chromosome) under coevolution and control conditions (six lines per treatment). Avirulent line C3_15 is not included. (C) The cry6B copy number differences covary with the effect of the pathogen on the fitness of hosts from transfer 12 in the time-shift experiment (see the results of the full time-shift experiment in SI Appendix, Fig. S8). Dashed lines connect two observations for the same lines. (D) Virulence of pathogen line C3_15 lacking cry6B relative to the ancestor (Wilcoxon rank sum test; P values adjusted with the Holm–Bonferroni method for performing three nonindependent comparisons). BT wt, B. thuringiensis wild-type strain MYBT18247.
Our findings point to the general importance of plasmid copy number variation as a mechanism underlying rapid adaptation of pathogenic bacteria. Here, we used an initially clonal pathogen and genetically diverse host. Such asymmetric levels of genetic diversity are rarely considered in coevolution experiments (SI Appendix, Table S1). However, under natural conditions, pathogens are often clonal and subjected to bottlenecks during the infection process and/or more generally during transmission, while the host population is usually genetically diverse. For the pathogen, this leads to reduced standing genetic variation and a limited supply of de novo mutation (24). Under such commonly occurring conditions of low genetic diversity, it is unclear how pathogens acquire the relevant changes to quickly adapt to their host. Our findings now suggest that plasmid copy number variation could provide the means to respond fast, even when pathogens are initially clonal. Our results further demonstrate that copy number variations can cause aFDS-like dynamics during coevolution. Interestingly, our previous work already indicated the importance of copy number variation of a toxin-encoding plasmid in B. thuringiensis adapting to C. elegans, although for a genomically very distinct strain (MYBT18679) with a distinct set of Cry toxin genes and without any evidence for temporal coadaptation or aFDS dynamics (21). It is conceivable that such copy number changes also determine rapid adaptation in the large variety of other pathogens that similarly possess plasmid-encoded virulence factors (32).
Conclusions
Based on a controlled experimental approach, we demonstrated that rapid coevolutionary adaptations between C. elegans and its pathogen are consistent with aFDS at the phenotypic level. This finding highlights the potential importance of antagonistic frequency dependence for shaping coevolutionary dynamics in eukaryotic host systems. Most surprisingly, our genome sequencing results did not conform to the classic aFDS model with several alleles cycling at a single locus. Instead, we found that the phenotypic pattern of aFDS is determined by more complex changes at the genomic level and that these changes are clearly distinct in the two antagonists. In the case of B. thuringiensis, rapid adaptation of the initial clone was mediated by copy number variation of the cry6B toxin gene rather than oscillations of different alleles. Such copy number variations may generally enhance fast evolutionary responses (33) and may explain why many virulence factors are associated with mobile genetic elements (32). In the case of C. elegans, the phenotypic pattern of aFDS is likely produced by changes in different genome regions. These may occur independently across the replicate populations, possibly each still mirroring aFDS. Alternatively, they may occur in parallel, but for each genome region at different time periods of coevolution, thus yielding the identified pattern of multiple incomplete sweeps. Interestingly, the primary trait under selection in the experiment, host fertility, is known to have a quantitative genetic basis in this nematode, controlled by multiple loci of small effect (26). The quantitative nature of this trait may explain the involvement of several genome regions during coevolutionary adaptation. It is a clearly different response than that previously observed for aFDS dynamics in Daphnia water fleas, in which the interaction with the coevolving pathogen was found to be controlled by one or a few host loci (29, 34). In fact, to date, it is unclear what type of selection dynamics emerge if coevolution is mediated by changes in a quantitative trait (35) and/or by copy number variation. Our results highlight that both can simultaneously shape coevolution and cause rapid coadaptation of the two antagonists.
Methods
All methods are described in more detail in the SI Appendix. The evolution experiment was started with a previously established, genetically diverse C. elegans population (36) and the nematocidal B. thuringiensis strain MYBT18247 (21, 22, 37). Experimental evolution was performed in a viscous medium which allowed us to maintain C. elegans in microtiter plates in a homogeneous environment, where they can mate and reproduce at rates comparable to those on agar plates. All protocols for C. elegans maintenance and manipulation (e.g., synchronization, feeding, worm transfer, cryopreservation, infection protocol) were specifically optimized for this medium (SI Appendix, Material and Methods). The nematode populations were preadapted to the experimental evolution protocol over 22 wk. The evolution experiment was run over 23 wk with weekly transfers and included three treatments (SI Appendix, Fig. S3; 16 biological replicates per treatment): a coevolution treatment, in which evolving hosts and pathogens were forced to coadapt to each other, and two control treatments, in which either C. elegans or B. thuringiensis was subjected to the passaging in the absence of the antagonist. Host populations were maintained at a constant population size of 3,000 individuals, while B. thuringiensis was always added at a defined concentration of 2 × 108 spores/mL at each transfer step. Host and pathogen could be effectively separated by either a bleaching protocol (only survived by host eggs) or a pasteurization step (only survived by pathogen spores), allowing full experimental control over the (co)evolutionary process. Host and pathogen populations were cryopreserved at almost every second transfer. After the evolution experiment, the cryopreserved material was subjected to phenotypic analysis in the presence of either the ancestral antagonist or the respective coadapted antagonists from different time points. We characterized changes in the following traits: (i) host fertility (or the pathogen’s effect on host fertility), (ii) pathogen competitive ability inside the host (using the red fluorescent evolved bacteria in combination with a green fluorescent ancestral bacterium), and (iii) host resistance (or, analogously, pathogen virulence).
Complete genome sequences were generated with the Illumina HiSeq2500 technology for entire host or pathogen populations at three time points across the evolution experiment (transfers 1, 12, and 22; always six biological replicates per treatment and time point). The paired-end sequence reads were processed using a custom pipeline, including mapping to reference genomes for B. thuringiensis (37) and C. elegans (version WS235) and calling SNPs and indels (SI Appendix, Material and Methods). Plasmid copy number estimation was based on read coverage of the plasmid relative to the average of the corresponding chromosome. The selection coefficient s was estimated for two time intervals (transfers 1–12 and 12–22) based on the change in allele frequencies of biallelic SNPs using a tailored statistical modeling approach. This approach allowed us to simultaneously estimate s for a given time interval and the associated P value based on information from all replicate populations. Moreover, we evaluated genetic differentiation of the evolved populations from the ancestor with FST statistics using a sliding window approach as implemented in PoPoolation2 (38).
The data are supplied as Datasets S1 and S2. Sequence data are available from the National Center for Biotechnology Information (NCBI) BioProject PRJNA475030 (39).
Supplementary Material
Acknowledgments
We thank the members of the Priority Program Schwerpunktprogramm (SPP) 1399 on Host–Pathogen Coevolution, funded by the German Science Foundation, especially J. Kurtz, R. Schulte-Iserlohe, H. Ließegang, J. Hollensteiner, B. Milutinovic, S. Cremer, G. Jansen, and C. Rafaluk, and also the members of the H.S. group for advice and support. We also thank S. Gandon for comments on the manuscript. Our work was funded within the SPP 1399, Grants SCHU 1415/8 and SCHU 1415/9 (to H.S.) and RO 2994/3 (to P.C.R.). T.G. and H.T. were supported by a grant from the Agence Nationale de la Recherche (Grant ANR-14-ACHN-0032-01). A.P. and W.Y. were additionally supported by the International Max Planck Research School for Evolutionary Biology; P.C.R. and H.S. were supported by infrastructural funding from the Deutsche Forschungsgemeinschaft (DFG) Excellence Cluster EXC 306 Inflammation at Interfaces, and H.S. was supported by a Max Planck Fellowship.
Footnotes
This article is a PNAS Direct Submission.
Data deposition: The sequence data reported in this paper are available from the National Center for Biotechnology Information (NCBI) under BioProject accession no. PRJNA475030 (https://www.ncbi.nlm.nih.gov/bioproject).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810402116/-/DCSupplemental.
References
- 1.Van Valen L. A new evolutionary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 2.Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 3.Lively CM, Apanius V. Genetic diversity in host-parasite interactions. 1995. Ecology of Infectious Diseases in Natural Populations, Publications of the Newton Institute, eds Grenfell BT, Dobson AP (Cambridge Univ Press, Cambridge, UK), Vol 7, pp 421–449.
- 4.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 6.Jokela J, Dybdahl MF, Lively CM. The maintenance of sex, clonal dynamics, and host-parasite coevolution in a mixed population of sexual and asexual snails. Am Nat. 2009;174(Suppl 1):S43–S53. doi: 10.1086/599080. [DOI] [PubMed] [Google Scholar]
- 7.Morran LT, Schmidt OG, Gelarden IA, Parrish RC, 2nd, Lively CM. Running with the Red Queen: Host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host-parasite interactions. Trends Ecol Evol. 2008;23:678–685. doi: 10.1016/j.tree.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaba S, Ebert D. Time-shift experiments as a tool to study antagonistic coevolution. Trends Ecol Evol. 2009;24:226–232. doi: 10.1016/j.tree.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Gandon S, Buckling A, Decaestecker E, Day T. Host-parasite coevolution and patterns of adaptation across time and space. J Evol Biol. 2008;21:1861–1866. doi: 10.1111/j.1420-9101.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 11.Betts A, Kaltz O, Hochberg ME. Contrasted coevolutionary dynamics between a bacterial pathogen and its bacteriophages. Proc Natl Acad Sci USA. 2014;111:11109–11114. doi: 10.1073/pnas.1406763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockhurst MA, Morgan AD, Fenton A, Buckling A. Experimental coevolution with bacteria and phage. The Pseudomonas fluorescens–Phi2 model system. Infect Genet Evol. 2007;7:547–552. doi: 10.1016/j.meegid.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Marston MF, et al. Rapid diversification of coevolving marine Synechococcus and a virus. Proc Natl Acad Sci USA. 2012;109:4544–4549. doi: 10.1073/pnas.1120310109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14:635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 15.Lopez Pascua L, et al. Higher resources decrease fluctuating selection during host-parasite coevolution. Ecol Lett. 2014;17:1380–1388. doi: 10.1111/ele.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frickel J, Sieber M, Becks L. Eco-evolutionary dynamics in a coevolving host-virus system. Ecol Lett. 2016;19:450–459. doi: 10.1111/ele.12580. [DOI] [PubMed] [Google Scholar]
- 17.Decaestecker E, et al. Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 18.Thrall PH, et al. Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol Lett. 2012;15:425–435. doi: 10.1111/j.1461-0248.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts A, Gray C, Zelek M, MacLean RC, King KC. High parasite diversity accelerates host adaptation and diversification. Science. 2018;360:907–911. doi: 10.1126/science.aam9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paez-Espino D, et al. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. MBio. 2015;6:e00262-15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masri L, et al. Host-pathogen coevolution: The selective advantage of Bacillus thuringiensis virulence and its cry toxin genes. PLoS Biol. 2015;13:e1002169. doi: 10.1371/journal.pbio.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc Natl Acad Sci USA. 2010;107:7359–7364. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci. 2002;269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papkou A, Gokhale CS, Traulsen A, Schulenburg H. Host-parasite coevolution: Why changing population size matters. Zoology (Jena) 2016;119:330–338. doi: 10.1016/j.zool.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Duneau D, Luijckx P, Ben-Ami F, Laforsch C, Ebert D. Resolving the infection process reveals striking differences in the contribution of environment, genetics and phylogeny to host-parasite interactions. BMC Biol. 2011;9:11. doi: 10.1186/1741-7007-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noble LM, et al. Polygenicity and epistasis underlie fitness-proximal traits in the Caenorhabditis elegans multiparental experimental evolution (CeMEE) panel. Genetics. 2017;207:1663–1685. doi: 10.1534/genetics.117.300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes S, et al. The C. elegans TPR containing protein, TRD-1, regulates cell fate choice in the developing germ line and epidermis. PLoS One. 2014;9:e114998. doi: 10.1371/journal.pone.0114998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pees B, Kloock A, Nakad R, Barbosa C, Dierking K. Enhanced behavioral immune defenses in a C. elegans C-type lectin-like domain gene mutant. Dev Comp Immunol. 2017;74:237–242. doi: 10.1016/j.dci.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Bento G, et al. The genetic basis of resistance and matching-allele interactions of a host-parasite system: The Daphnia magna-Pasteuria ramosa model. PLoS Genet. 2017;13:e1006596. doi: 10.1371/journal.pgen.1006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dementiev A, et al. The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins. BMC Biol. 2016;14:71. doi: 10.1186/s12915-016-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei J-Z, et al. Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci USA. 2003;100:2760–2765. doi: 10.1073/pnas.0538072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams V, et al. Virulence plasmids of spore-forming bacteria. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.PLAS-0024-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millan AS, Escudero JA, Gifford DR, Mazel D, MacLean RC. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol. 2017;1:0010. doi: 10.1038/s41559-016-0010. [DOI] [PubMed] [Google Scholar]
- 34.Routtu J, Ebert D. Genetic architecture of resistance in Daphnia hosts against two species of host-specific parasites. Heredity (Edinb) 2015;114:241–248. doi: 10.1038/hdy.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nuismer SL, Ridenhour BJ, Oswald BP. Antagonistic coevolution mediated by phenotypic differences between quantitative traits. Evolution. 2007;61:1823–1834. doi: 10.1111/j.1558-5646.2007.00158.x. [DOI] [PubMed] [Google Scholar]
- 36.Teotonio H, Carvalho S, Manoel D, Roque M, Chelo IM. Evolution of outcrossing in experimental populations of Caenorhabditis elegans. PLoS One. 2012;7:e35811. doi: 10.1371/journal.pone.0035811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hollensteiner J, et al. Complete genome sequence of the nematicidal Bacillus thuringiensis MYBT18247. J Biotechnol. 2017;260:48–52. doi: 10.1016/j.jbiotec.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Kofler R, Pandey RV, Schlötterer C. PoPoolation2: Identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papkou A, et al. 2018 Caenorhabditis elegans and Bacillus thuringiensis coevolution. National Center for Biotechnology Information. Available at http://www.ncbi.nlm.nih.gov/bioproject?term=PRJNA475030. Deposited June 7, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.