Significance
Our discoveries provide evidence that posttranslational modification of Dicer1 drives aging, infertility, and metabolic disorder in mammals. These data suggest that phosphorylation of Dicer1 is a critical regulatory mechanism that dictates Dicer1 function and localization and provides a selective bias in miRNA processing. In addition, this work highlights the potential relevance of Dicer1 phosphorylation clinically, as Dicer1 mutations and expression levels do not account for all cases of Dicer1-associated disease.
Keywords: ERK signaling, microRNA, infertility, mouse model, aging
Abstract
DICER1 gene alterations and decreased expression are associated with developmental disorders and diseases in humans. Oscillation of Dicer1 phosphorylation and dephosphorylation regulates its function during the oocyte-to-embryo transition in Caenorhabditis elegans. Dicer1 is also phosphorylated upon FGF stimulation at conserved serines in mouse embryonic fibroblasts and HEK293 cells. However, whether phosphorylation of Dicer1 has a role in mammalian development remains unknown. To investigate the consequence of constitutive phosphorylation, we generated phosphomimetic knock-in mouse models by replacing conserved serines 1712 and 1836 with aspartic acids individually or together. Dicer1S1836D/S1836D mice display highly penetrant postnatal lethality, and the few survivors display accelerated aging and infertility. Homozygous dual-phosphomimetic Dicer1 augments these defects, alters metabolism-associated miRNAs, and causes a hypermetabolic phenotype. Thus, constitutive phosphorylation of Dicer1 results in multiple pathologic processes in mice, indicating that phosphorylation tightly regulates Dicer1 function and activity in mammals.
Dicer1 is an essential riboendonuclease that processes pre-miRNAs into functional miRNAs. DICER1 mutations are associated with developmental defects and diseases in humans (1–3). Patients with DICER1 syndrome carry germline heterozygous mutations in DICER1 (missense and truncating). These patients present with multinodular goiter, macrocephaly, global developmental delay, lung cysts, overgrowth, and increased risk for a variety of cancers (3, 4). DICER1 somatic heterozygous mutations are also frequently observed in human cancers (5–8). These data highlight the relevance of DICER1 in human disease.
Multiple mechanisms regulate Dicer1 function and activity. For example, DICER1 is positively regulated by the universal transcription factor SOX4 in melanoma cells (9). Following melanocyte differentiation, DICER1 is regulated in a lineage-specific manner by the melanocyte master transcriptional regulator MITF (10). Additionally, miRNA-103/107, miRNA-192, and the let-7 miRNA family control Dicer1 transcript levels in a negative feedback loop in cultured cells (11–15). Alterations in DICER1 mRNA and protein levels are associated with macular degenerative blindness, infertility, aging, and cancer (16, 17). These findings emphasize that Dicer1 is globally expressed in cells and tissues, and its function is under tight regulatory control.
Recently, we discovered that ERK-dependent phosphorylation of Caenorhabditis elegans DCR1 at Ser (S) 1705 and Ser1833 is essential for oogenesis, whereas dephosphorylation is essential for progression of embryogenesis (18). Thus, oscillation of Dicer1 phosphorylation and dephosphorylation is essential for oogenesis and embryogenesis. This discovery highlighted that Dicer1 function is under tight posttranslational control during development. In mammals, we found that Dicer1 is phosphorylated at the conserved serines in mouse embryonic fibroblasts (MEFs) and human embryonic kidney cell lines upon FGF stimulation (18). Moreover, Dicer1 is phosphorylated at Ser1712 and Ser1836 in the developing mouse uterine glands in vivo (18). In all of these cases, phosphorylation of Dicer1 was coupled with its translocation from the cytoplasm to the nucleus (18). Burger et al. (19) identified an additional serine that is phosphorylated by ATM/ATR during DNA damage response, resulting in translocation of Dicer1 from cytoplasm to the nucleus in cultured human cells. These results suggest that phosphorylation of Dicer1 is a conserved event; however, the role of Dicer1 phosphorylation and dephosphorylation in vivo in mammals remains unknown.
To test whether phosphorylation of Dicer1 regulates its function and activity in mammals, we generated and characterized three phosphomimetic Dicer1 knock-in mouse models. These models test the physiological impact of constitutive and unregulated Dicer1 phosphorylation. We demonstrate that phosphomimetic Dicer1 at Ser1712 (DicerS1712D/S1712D) is nonphenotypic. A phosphomimic at Ser1836 (DicerS1836D/S1836D) results in highly penetrant early postnatal lethality. The few survivors display infertility and accelerated aging. Dual phosphomimic at both serines (Dicer2SD/2SD) phenocopies the DicerS1836D/S1836D mutant with increased severity. We find that DicerS1712D is primarily cytoplasmic, whereas DicerS1836D is partially nuclear and Dicer2SD is primarily nuclear, suggesting that nuclear translocation is associated with the phenotypes observed. Additionally, Dicer2SD homozygosity drives a hypermetabolic phenotype in MEFs and mice, suggesting that an increased rate of metabolism may be the earliest defect in these animals. This defect is coupled with an alteration of a small subset of metabolic miRNAs. Together, these results demonstrate that regulation of Dicer1 phosphorylation in vivo in mammals is likely under tight control for balanced Dicer1 function and normal development.
Results
Phosphorylation at Ser1836 Impairs Dicer1 Function.
To examine the role of phosphorylation-mediated regulation of Dicer1 function in mammals, we generated three phosphomimetic Dicer1 knock-in mouse models (Fig. 1 A and B and SI Appendix, Fig. S1). Mice with Ser1712 replaced with aspartic acid (DicerS1712D) were generated by using CRISPR/Cas9 technology (SI Appendix, Fig. S1D). Mice with Ser1836 replaced with aspartic acid (DicerS1836D) and Ser1712 and Ser1836 replaced with aspartic acids (Dicer2SD) were obtained by using the traditional embryonic stem (ES) cell technology. Additionally, CRISPR/Cas9-mediated targeting also resulted in generation of Dicer1 alleles with frame-shift truncation starting at position 1712 (Dicer−) and 3-bp in-frame deletion of codon 1712 (Dicer∆1712).
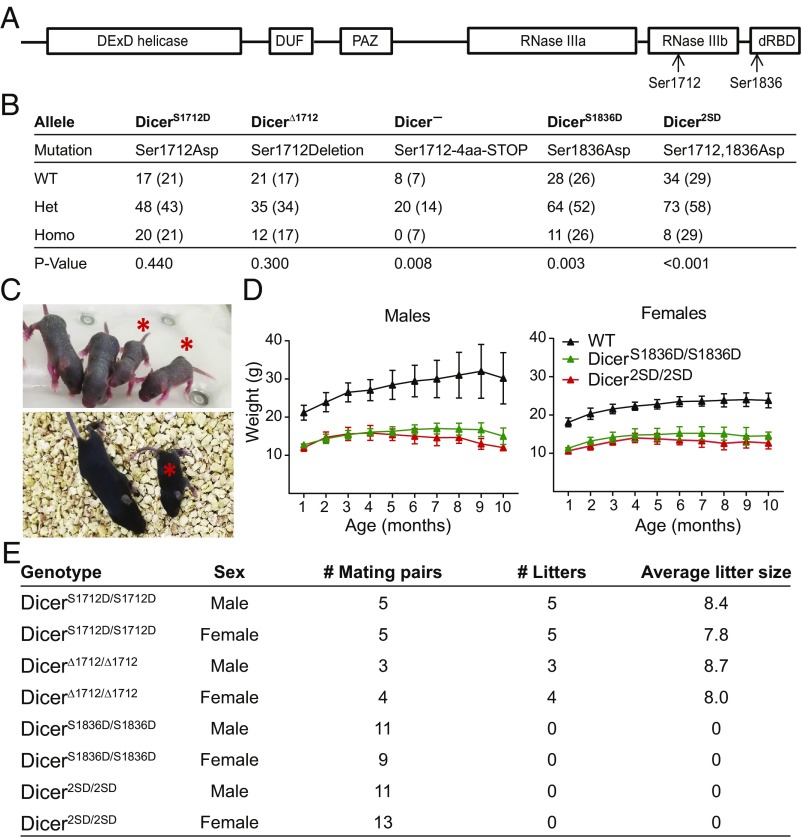
Fig. 1.
Dicer1 alleles and phenotype of homozygous mutants. (A) Sites of Dicer1 phosphorylation are shown. dRBD, dsRNA binding domain; DUF, domain of unknown function; PAZ, Piwi Argonaute and Zwille binding domain. (B) Homozygous viability of all Dicer1 mutants was tested by inbreeding heterozygous mutants (nomenclature and associated mutations are listed). Observed and expected numbers (in parentheses) of all genotypes are listed. Het, heterozygous mutants; Homo, homozygous mutants. χ2 tests were performed, and P values are listed. (C) Representative images of 1-wk-old (Top) and 2-wk-old (Bottom) Dicer2SD/2SD mice (asterisk) and normal littermates. (D) One- to 10-mo-old WT, DicerS1836D/S1836D, and Dicer2SD/2SD male (Left; n = 12, n = 12, and n = 10, respectively) and female (Right; n = 12, n = 9, and n = 11, respectively) mice were weighed regularly. Average weights with SDs at multiple time points (in months) are presented. At least three mice were present for all genotypes at the endpoint (some animals died before reaching the endpoint). (E) Fertility of all homozygous mutants was tested by mating with WT mice. Number of litters and average litter size for all mutants (males and females) are listed.
Dicer1-null mice are embryonic-lethal at embryonic day (E) 7.5 (2). To determine if phosphomimetic Dicer1 alleles mimicked loss of Dicer1 activity, we assayed for lethality in homozygous mutant mice for each allele generated. DicerS1712D/S1712D and DicerΔ1712/Δ1712 mice were born at the expected Mendelian ratios (Fig. 1B). Dicer−/− mice, as expected, were embryo-lethal. DicerS1836D/S1836D mutants were born, but only 11% of progeny (11 of 103) were homozygous mutants at weaning instead of the expected 25% frequency indicating partial lethality (P = 0.003; Fig. 1B). Dicer2SD/2SD mice, like the DicerS1836D/S1836D mice, were also partially lethal, as only 7% of progeny (8 of 115) were homozygous mutants at weaning (P < 0.001). Examination of E18.5 embryos from Dicer+/2SD heterozygous crosses revealed that homozygous mutant embryos were viable. Of the 42 embryos examined, 10 were WT, 22 were heterozygous, and 10 were homozygous mutants. Histopathologic examination of E18.5 embryos did not reveal any defects in homozygous mutants; however, we observed dead pups within 4 d after birth that were genotyped as homozygous mutants, suggesting that lethality is postnatal. Combined, these data reveal that constitutive phosphorylation at Ser1836 impairs Dicer1 function and causes postnatal lethality.
Next, we examined if mutant Dicer1 alleles are functionally hypomorphic by introducing a null allele over each mutant background. DicerS1836D/− and Dicer2SD/− mice were nonviable, whereas the DicerS1712D/− and DicerΔ1712/− mutants were viable (SI Appendix, Fig. S2). These data demonstrate that constitutive phosphomimic or deletion of Ser1712 has seemingly no impact on Dicer1 functions, whereas phosphomimic at Ser1836 is sufficient to alter Dicer1 functions with lethal consequences.
Studies have shown that Dicer1 loss activates the p53 pathway (20, 21). We therefore genetically tested whether postnatal lethality in Dicer2SD/2SD mice is p53-dependent by concomitantly deleting p53 in the Dicer2SD/2SD background. Of the 72 progeny born from Dicer+/2SD p53+/− intercrosses, none were Dicer2SD/2SD p53−/− (P = 0.01). This result indicates that loss of p53 does not rescue the postnatal lethality in the Dicer2SD/2SD mice, suggesting that a distinct pathway is responsible for lethality.
Constitutive Dicer1 Phosphorylation at Ser1836 Leads to Growth Retardation and Infertility.
To determine whether the DicerS1836D/S1836D and Dicer2SD/2SD mice that survived postnatal lethality displayed developmental defects, we extensively characterized survivors and found multiple striking defects. The most obvious of these defects was growth retardation. Male and female DicerS1836D/S1836D and Dicer2SD/2SD survivors displayed a 35–50% reduction in size compared with their WT sex-matched littermates, and the growth retardation phenotype persisted throughout their lifespan (Fig. 1 C and D). DicerS1836D/S1836D and Dicer2SD/2SD mice also displayed fertility defects as assayed via housing homozygous mutant males with WT females and vice versa. DicerS1712D/S1712D and DicerΔ1712/Δ1712 mice were fertile and produced eight to nine progeny per litter (Fig. 1E); however, DicerS1836D/S1836D and Dicer2SD/2SD mice (males and females) did not give rise to any progeny (Fig. 1E). These results suggest that constitutive Dicer1 phosphomimic at Ser1836 is sufficient to drive early postnatal lethality, retard growth, and cause infertility. Dicer2SD/2SD mice display similar phenotypes with greater magnitude (i.e., increased postnatal lethality and growth retardation) compared with DicerS1836D/S1836D mice. Thus, we focused on Dicer2SD/2SD mice from this point forward.
Constitutive Dicer1 Phosphorylation at Serines 1712 and 1836 Drives Accelerated Aging Phenotypes.
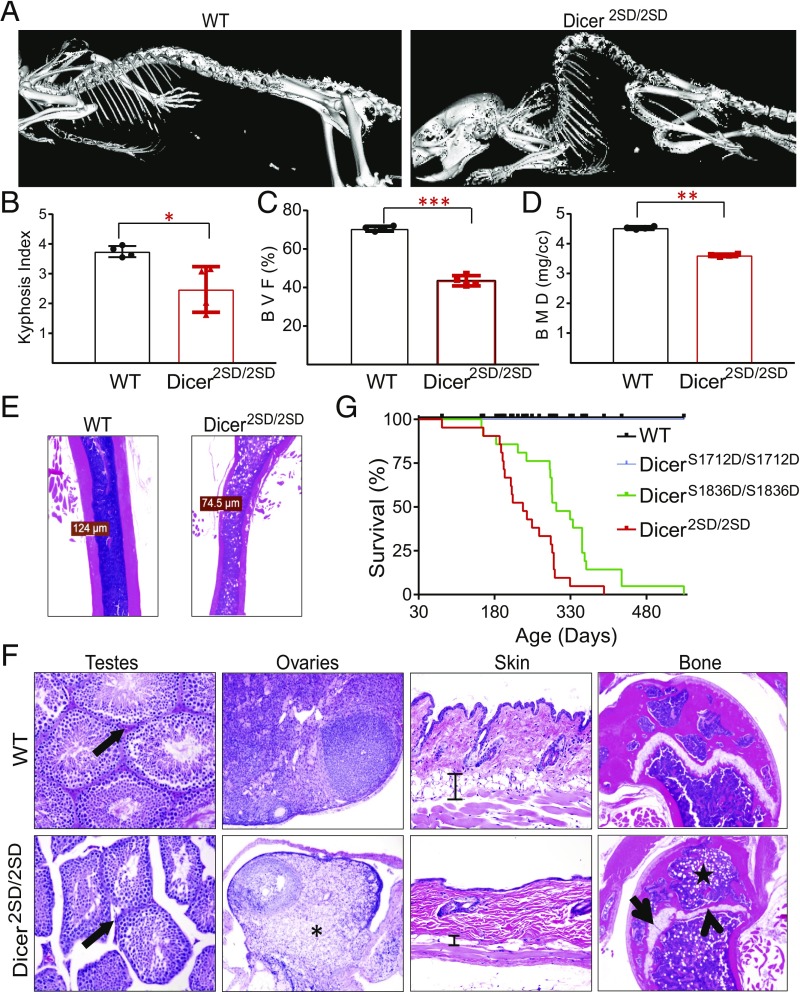
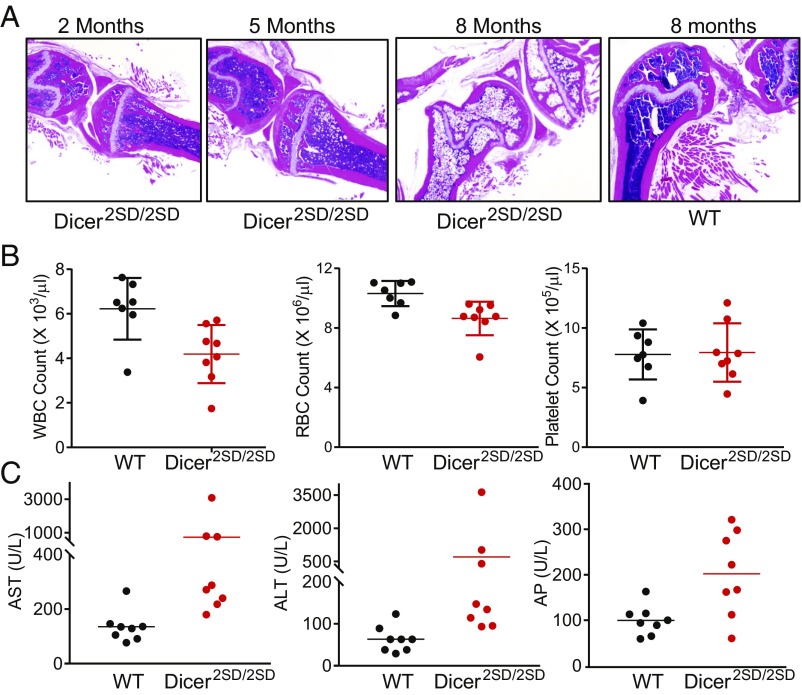
Aging phenotypes in mice are assessed by several criteria, including growth retardation, infertility, reduced survival, kyphosis (i.e., curvature in the spine), osteoporosis, skin atrophy, cardiomyopathy, muscular dystrophy, reduced fat mass, anemia, and alopecia (22–26). As Dicer2SD/2SD mice display retarded growth, infertility, and early lethality, which suggests accelerated aging, we examined other phenotypes of accelerated aging in these mice. Examination of mutant and WT mice as they aged revealed pronounced hunched posture in Dicer2SD/2SD mice starting at 6 mo of age compared with normal posture in WT littermates (Fig. 2A). Quantitative micro-CT scans of 7–9-mo-old Dicer2SD/2SD and WT mice confirmed significant kyphosis in mutant mice relative to WT mice (kyphosis indices of 2.5 and 3.7 respectively; P = 0.02; Fig. 2 A and B). Micro-CT scans also revealed severe osteoporosis in Dicer2SD/2SD mice with significant reduction in bone volume fraction (BVF; 38% vs. 65%; P = 0.0003; Fig. 2C) and bone mineral density (BMD; 347 mg/cm3 vs. 420 mg/cm3; P = 0.008; Fig. 2D) in the spine of mutant mice compared with WT mice. Histopathologic examination of the femur and tibia showed that the bone marrow was infiltrated by adipocytes in mutant mice as early as 2 mo of age, and the adipocytes replaced most of the hematopoietic cells by 9 mo (Figs. 2 E and F and 3A). We also observed bone loss throughout the skeletons of mutant mice (Fig. 2 A and E). As hematopoietic cells are predominantly found in the bone marrow in mice, we examined the mutant animals for compromised hematopoiesis. Complete blood count analysis revealed no change in platelet count (P = 0.89), a 33% reduction in total white blood cell (WBC) count (4.18 × 103/μL vs. 6.22 × 103/µL; P = 0.01), and a 15% reduction in red blood cell (RBC) count (8.6 × 106/μL vs. 10.3 × 106/µL; P = 0.007) in Dicer2SD/2SD mice compared with WT (Fig. 3B). These results suggest that phosphomimetic Dicer1 dramatically impacts the skeletal integrity and bone marrow adipogenesis with an associated reduction of hematopoiesis.
Fig. 2.
Accelerated aging phenotypes in Dicer2SD/2SD mice. (A) Representative micro-CT scans (isosurface threshold, 1,000) of WT and Dicer2SD/2SD mouse skeletons. (B) Kyphosis index for WT (n = 4) and Dicer2SD/2SD mice (n = 4) was quantified and averaged and is presented with SD (*P = 0.02, Student’s t test). (C) BVF in the spine of WT (n = 4) and Dicer2SD/2SD mice (n = 4) was quantified and averaged and is presented with SD (***P = 0.0003, Student’s t test). (D) BMD of spine for WT (n = 4) and Dicer2SD/2SD mice (n = 4) was quantified and averaged and is presented with SD (**P = 0.008, Student’s t test). (E) Representative H&E longitudinal sections of the femoral diaphysis (1.25×) from 6-mo-old WT and Dicer2SD/2SD mice. Measurements are shown for the thinnest section of the cortical bone. (F) Representative H&E sections of testes (magnification 20×), ovaries (10×), skin (20×), and femoral epiphyses and metaphyses (5×) from 8-mo-old WT and Dicer2SD/2SD mice. Leydig cells (arrow) in testes section, ovarian parenchyma (asterisk), s.c. adipose tissue (scale bar) in skin, irregular thickness of growth plate cartilage (arrow), and replacement of physeal cartilage by bone (arrowhead) and adipocyte infiltration (star) in the femoral epiphyseal bone marrow are marked. (G) Survival curve of WT (n = 57), DicerS1712D/S1712D (n = 15), DicerS1836D/S1836D (n = 21), and Dicer2SD/2SD (n = 21) mice followed for 560 d.
Fig. 3.
Hematological phenotypes of Dicer2SD/2SD mice. (A) Representative H&E sections of the femur and tibia from 2–8-mo-old Dicer2SD/2SD mice and an 8-mo-old WT mouse. (B) Complete blood counts of 2–8-mo-old WT (n = 7) and Dicer2SD/2SD littermate mice (n = 8). Average measurement with SD is shown (P = 0.01 for WBC, P = 0.007 for RBC, and P = 0.89 for platelets). Each dot represents one mouse. (C) Serum enzyme levels in the blood of 2–8-mo-old WT (n = 7) and Dicer2SD/2SD littermate mice (n = 8). Average measurement with SD is shown [P = 0.11 for AST, P = 0.16 for ALT, and P = 0.01 for alkaline phosphatase (AP)]. Each dot represents one mouse.
We examined multiple organs from Dicer2SD/2SD and WT mice microscopically to assay tissue degeneration. Consistent with our previous observation of infertility in the mutant mice, sections of testes from 2–7 mo-old mutant mice lacked mature spermatozoa and had reduced numbers of Leydig cells compared with WT mice (Fig. 2F). Similarly, the ovaries from homozygous mutant mice were reduced in size and contained fewer follicles, and the majority of ovarian parenchyma was composed of vacuolated cells (Fig. 2F). The epidermis in mutant mice was thin with abnormal tissue architecture and reduced amounts of s.c. adipose tissue (Fig. 2F). Mutants older than 6 mo of age consistently lacked abdominal fat (SI Appendix, Fig. S3A). Young Dicer2SD/2SD mice (1–2 mo old) also developed neutrophilic conjunctivitis, and, as the mice aged, they occasionally developed corneal lesions (SI Appendix, Fig. S3 B and C). Additionally, reproductive tissues including the prostate, seminal vesicles, and uterus were reduced in size, and the accessory sex glands contained less secretory material in mutants. We also observed chronic cardiomyopathy, membranoproliferative glomerulonephritis, hepatocellular karyomegaly, and pulmonary bronchus-associated lymphoid tissue (BALT) hyperplasia in some mutant mice. The BALT hyperplasia and membranoproliferative glomerulonephritis indicate nonspecific chronic immune stimulation. Because these lesions indicated chronic immune stimulation in multiple organs of Dicer2SD/2SD mice, we suspected that the mutant mice might have altered serum enzyme levels in circulation. Blood chemistry analysis revealed elevated levels of aspartate aminotransferase (AST; 729 vs. 136 U/L), alanine aminotransferase (ALT; 703 vs. 63 U/L), and alkaline phosphatase (202 vs. 100 U/L) in mutant mice as compared to WT mice (Fig. 3C). All phenotypes present in Dicer2SD/2SD mice were also evident in DicerS1836D/S1836D mice, albeit with less severity. These results together suggest that phosphomimetic Dicer1 disrupts normal tissue function, resulting in infertility, tissue atrophy, and changes often associated with aging, including osteoporosis, reduced hematopoiesis, and increased bone marrow adipogenesis.
The end result of accelerated aging is shortened lifespan. Whereas all Dicer+/+ and DicerS1712D/S1712D mice were alive by the 540-d time point of this study, Dicer2SD/2SD and DicerS1836D/S1836D mice had median survival times of 236 d and 302 d, respectively (P = 0.0001; Fig. 2G). In line with previous observations that Dicer2SD/2SD mice display phenotypes with greater magnitude than DicerS1836D/S1836D mice, the difference in overall survival between DicerS1836D/S1836D and Dicer2SD/2SD mice was also statistically significant (P = 0.02). Together, these results reveal that constitutive Dicer1 phosphomimic at Ser1836 or dual phosphomimic drives accelerated aging phenotypes, and further support our hypothesis that phosphorylation alters Dicer1 functions.
Phosphorylation Alters Dicer1 Localization.
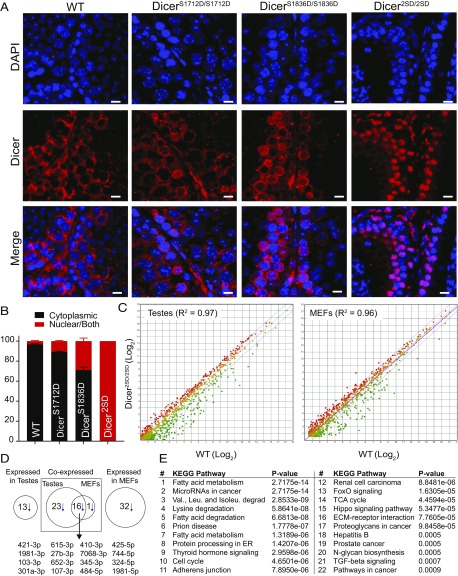
As phosphorylation leads to nuclear localization of Dicer1 in C. elegans, we examined Dicer1 localization in mutant and WT testes. Immunofluorescence staining using a Dicer1 antibody on testes sections from WT and mutant mice revealed that Dicer1 protein accumulated in the nucleus in 3% of WT, 10% of DicerS1712D/S1712D, 30% of DicerS1836D/S1836D, and 100% of Dicer2SD/2SD spermatocytes that were positive for Dicer1 staining (Fig. 4 A and B). By using stringent criteria, spermatocytes with low levels of Dicer1 protein in the nucleus (<50% of nuclear surface) were considered negative for nuclear accumulation, and cells that were not stained with the Dicer1 antibody were excluded during quantification (SI Appendix, Fig. S5). Nuclear accumulation of Dicer2SD was also confirmed in kidney and liver sections (SI Appendix, Fig. S4). This result provides two insights. First, phosphorylation at both sites is essential for efficient nuclear translocation of Dicer1 protein. Second, phosphomimetic mutation at Ser1836, with some nuclear Dicer1 protein in most cells, was sufficient to drive the observed phenotypes in mice. This suggests that phosphomimic at Ser1836 is sufficient to impair Dicer1 functions, irrespective of the degree of nuclear localization.
Fig. 4.
Phosphorylation alters Dicer1 localization and miRNA profile. (A) Immunofluorescence with an anti-Dicer1 antibody on representative testes sections (20×) from WT, DicerS1712D/S1712D, DicerS1836D/S1836D, and Dicer2SD/2SD mice. Dicer1 is stained red and nuclei are stained blue with DAPI. (Scale bars: 10 µm.) (B) Percentage of spermatocytes (excluding spermatids and spermatozoa) with nuclear Dicer1 was calculated in the seminiferous tubules of three WT, DicerS1712D/S1712D, DicerS1836D/S1836D, and Dicer2SD/2SD testes. Cells were binned as cytoplasmic or nuclear/both. Average was calculated with SD (SI Appendix, Fig. S5). (C) Differential expression of miRNAs in testes (n = 3 for Dicer2SD/2SD and n = 2 for WT) and MEFs (n = 4 for Dicer2SD/2SD and n = 3 for WT) from WT and Dicer2SD/2SD mice. Green, red, and yellow dots represent down-regulated, up-regulated, and unaffected miRNAs, respectively, in Dicer2SD/2SD samples compared with WT samples. Blue lines represent twofold cutoff. (D) Number of down-regulated miRNAs (>10 reads per million and more than twofold difference) and overlap in mutant testes and MEFs. Overlapping miRNAs are listed. (E) Pathway analysis of down-regulated miRNAs in mutant testes was performed by using DIANATOOLS. The top 22 pathways on the list are presented.
Constitutive Dicer1 Phosphorylation at Serines 1712 and 1836 Alters a Subset of Metabolic miRNAs.
To determine whether phosphorylation of Dicer1 at serines 1712 and 1836 altered its canonical miRNA processing function, we identified the mature miRNAs by using next-generation deep sequencing in MEFs and testes of WT and Dicer2SD/2SD mice. MEFs were profiled to determine the earliest molecular changes before manifestation of gross pathologic processes. Testes were profiled because we saw clear morphological changes in this tissue as the animal aged. We found that a subset of miRNAs were down-regulated in the MEFs and testes samples from Dicer2SD/2SD mice compared with WT samples (Fig. 4C). Of the 359 and 368 miRNAs expressed in MEFs and testes, respectively, 49 miRNAs in the mutant MEFs (14%) and 52 miRNAs in the mutant testes (14%) were down-regulated more than twofold (more than eightfold on average; Datasets S1 and S2). Of the 49 down-regulated miRNAs in mutant MEFs, only 17 miRNAs were expressed in testes, and 16 of these 17 miRNAs (94%) were down-regulated in both tissues (Fig. 4D). Similarly, of the 52 down-regulated miRNAs in mutant testes, 39 were expressed in MEFs, and 16 of these 39 miRNAs (41%) were down-regulated in both tissues (Fig. 4D). This overlap strongly suggests that phosphomimetic mutations in Dicer1 modulate the generation of a subset of miRNAs. Pathway analysis on the differentially down-regulated miRNAs in mutant testes highlighted metabolic pathways as most affected (Fig. 4E). Pathway analysis on differentially down-regulated miRNAs from MEFs also led to the identification of the metabolic pathways, further underscoring the specificity of dysregulated processing of the affected miRNAs (SI Appendix, Fig. S6). As aging is linked with altered metabolism, these results suggest that down-regulation of metabolism-associated miRNAs may be the driving force behind the accelerated aging phenotypes in mutant mice (27, 28). This led us to investigate whether dual phosphomimetic Dicer1 mice have altered metabolism.
Constitutive Dicer1 Phosphorylation at Serines 1712 and 1836 Drives a Hypermetabolic Phenotype.
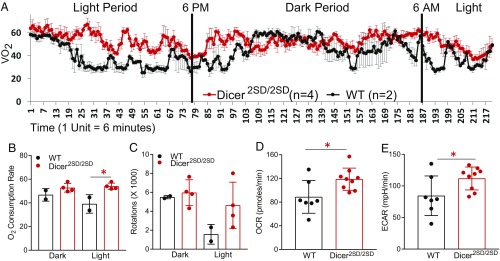
Our identification of down-regulated miRNAs in Dicer2SD/2SD mutants as players in metabolic pathways suggested that metabolism may be altered in these mice. To determine whether metabolic changes may be underlying the accelerated aging phenotypes of Dicer2SD/2SD mutants, we used the Comprehensive Lab Animal Monitoring System (CLAMS) test to measure activity and rates of respiration in 3–4-mo-old mutant mice and WT littermates. We observed that the mutant mice had a significant increase (24%) in average oxygen consumption rate (OCR) relative to WT mice (52.6 vs. 42.3 mL/kg/min; P = 0.04; Fig. 5 A and B). This difference was more pronounced during the light period when the mice typically rest (38% increase relative to WT, 52.7 vs. 38.1 mL/kg/min; P = 0.02; Fig. 5B). Similar to the increase in respiration rates, the mutant mice displayed a 50% increase in wheel activity vs. WT mice (10,555 vs. 7,057 rotations), and the difference was 2.5 fold during the light period (4,605 vs. 1,853 rotations; Fig. 5C). Together, these results indicate that Dicer2SD/2SD mice exhibit an increased metabolic rate and hyperactivity during the resting period relative to WT mice.
Fig. 5.
Higher metabolism in Dicer2SD/2SD mice and MEFs. (A) CLAMS test was used to measure OCR of WT (n = 2) and Dicer2SD/2SD mice (n = 4) from 10 AM to 9 AM. Rate of oxygen consumption (VO2) in milliliters per kilogram per minute with SD is shown for every measurement. Vertical lines separate the light period (6 AM to 6 PM) and dark period (6 PM to 6 AM) during the experimental measurements. On the x-axis, 1 unit is equal to 6 min from the beginning of measurement. (B) OCR rates were averaged and grouped into light and dark periods (*P = 0.02 for the light period, Student’s t test). (C) CLAMS test was used to measure wheel activity of WT (n = 2) and Dicer2SD/2SD (n = 4) mice. Measurements were combined and grouped into light and dark periods. Each dot represents one mouse. (D) Seahorse mitochondrial stress test assay was used to measure OCR of WT (n = 7) and Dicer2SD/2SD (n = 9) MEFs. Average measurements with SD are plotted (*P = 0.02, Student’s t test). (E) Seahorse glycolytic stress test assay was used to measure extracellular acidification rates (ECAR) of WT (n = 7) and Dicer2SD/2SD (n = 9) MEFs. Average measurements with SD are plotted (*P = 0.04).
To investigate whether metabolic changes precede the phenotypic manifestation, we assayed MEFs from mutant and WT mice and measured their glycolytic and mitochondrial respiration rates by using the Seahorse Bioanalyzer. Consistent with the results from the CLAMS test, Dicer2SD/2SD MEFs displayed a statistically significant increase in OCR (118.7 vs. 88.6 pmol/min; P = 0.02) and extracellular acidification rate (111.8 vs. 84.4 mpH/min; P = 0.04) compared with WT MEFs (Fig. 5 D and E). These results indicate that the increased metabolic rates are likely the earliest defects presented in Dicer2SD/2SD mice during embryonic development and influence the defects that manifest later. Taken together, these results demonstrate that phosphomimetic Dicer1 regulates metabolic pathways and suggest that increased metabolic rates may be the underlying mechanism behind accelerated aging phenotypes of Dicer2SD/2SD mice.
Discussion
Dicer1 is critical for normal vertebrate development because mutations in any of the core functional domains in DICER1 are associated with severe human diseases (3, 4, 29–43). For example, patients with DICER1 syndrome with heterozygous germline mutations in DICER1 (missense and truncating) present with multinodular goiter, macrocephaly, global developmental delay, lung cysts, overgrowth, and increased risk for a variety of cancers (3, 4). Similarly, conditional mouse models with loss of Dicer1 in a tissue-specific manner point to Dicer1’s role in regulating insulin resistance, one aspect of aging (17, 41, 44), spermatogenesis (35, 36), oocyte meiotic maturation (38), melanogenesis (10), and a variety of cancers (5–8). Our studies show that constitutive phosphorylation of Dicer1 also causes disease. We find that a phosphomimetic mutation at Ser1836 as well as the dual phosphomimetic mutations at serines 1712 and 1836 significantly impact Dicer1 function in mice. Phosphorylation at Ser1836 partially drives its nuclear localization, and severely compromises its function, whereas dual phosphorylation at Ser1712 and Ser1836 results in primarily nuclear localization of Dicer1 and a stronger phenotype than phosphorylation at Ser1836 alone. In addition, dual phosphorylation of Dicer1 leads to down-regulation of a subset of miRNAs and a phospho-Dicer1–specific miRNA signature. These data lead us to speculate that not all DICER1-associated human diseases may be caused by mutations in DICER1, as they may also arise from mutations in the phosphatases that dephosphorylate DICER1. Thus, phosphorylated DICER1 may serve as a biomarker for human disease.
Our observations that dual phosphorylation leads to nuclear accumulation of Dicer1 and associated aging phenotypes led us to propose that Dicer1 nuclear accumulation impairs the cytoplasmic functions of Dicer1 and potentiates its nuclear functions, resulting in cellular stress. In addition to accumulation of unwanted protein in the nucleus, phosphomimetic Dicer1 may interfere with other cellular processes by carrying out potential nuclear functions even when it is not required, resulting in activation of stress response that may contribute to the phenotypes. Unlike naturally phosphorylated Dicer1, phosphomimetic Dicer1 cannot be dephosphorylated in the nucleus and exported to cytoplasm. However, even nonphenotypic DicerS1712D/S1712D mutants show nuclear accumulation of phospho-Dicer1 in 10% of cells (spermatocytes), and DicerS1836D/S1836D mutants display phenotypes despite lacking nuclear accumulation of Dicer1 in most cells. Additionally, a previous study in MEFs shows that approximately 5% of Dicer1 is nuclear, suggesting that the phenotypes are primarily driven by constitutive phosphorylation, and mislocalization only partially contributes to exacerbate the phenotypes.
In yeast, Dicer1 shuttles between cytoplasm and nucleus under normal homeostasis, and nuclear accumulation is obligatory for the assembly of heterochromatin at centromeric repeats (45). ERK-mediated phosphorylation and dephosphorylation at Ser1712 and Ser1836 drives nuclear import and export, respectively, during oocyte-to-embryo transition in C. elegans. In this context, nuclear accumulation is likely driven by increased phosphorylation or inhibition of dephosphorylation at Ser1712 and Ser1836. Studies in human transformed cells reveal that DICER1 phosphorylation at S1728 and S1852 is required in DNA damage response, and nuclear accumulation was driven by an additional phosphorylation at Ser1016 mediated by ATM/ATR (19). During DNA damage response, phosphorylation at Ser1016 likely delays/prevents nuclear export of Dicer1, independent of dephosphorylation at Ser1712 and Ser1836, resulting in nuclear accumulation. Thus, nuclear accumulation and activity of Dicer1 is mediated by different posttranslational events under different stress conditions (18, 19).
During DNA damage response, phosphorylated DICER1 processes nuclear dsRNA for DNA damage repair. In C. elegans, reduction of Dicer1 function upon phosphorylation alters 26G endo-siRNAs for oocyte-to-embryo transition, whereas miRNAs are unaffected likely as a result of maternal rescue (18). In mice, we show that a subset of miRNAs is down-regulated in a physiological context upon constitutive Dicer1 phosphorylation. Whether Dicer1 regulates dsRNA or miRNAs in different contexts will need further mechanistic studies. Together, these studies unveil the role of Dicer1 phosphorylation in modulating the small RNA landscape in multiple contexts across evolution and in vivo.
Our studies show that phosphomimetic Dicer1 affects a broader spectrum of tissues and phenotypes than have been previously described for Dicer1 hypomorphic alleles. Previous reports show that reduction of Dicer1 transcripts to below 20% causes severe embryonic lethality (similar to the null allele), whereas mutant mice with >20% Dicer1 transcript levels survive with minimal phenotypes (43, 46, 47). Relative to the Dicer1-null and hypomorphic mice (with >20% Dicer1 transcript levels), which display a fully penetrant embryonic lethality (2, 43, 46), many DicerS1836D/S1836D and Dicer2SD/2SD mice survive a partial postnatal lethality. In addition, one copy of each mutant allele over a null allele is not viable, whereas Dicer1 heterozygous mice are viable and normal. These results demonstrate that phosphorylation is a critical regulator of Dicer1 function in mammals, and phosphorylation-mediated alteration in Dicer1 function and the associated phenotypes are different from Dicer1-null or hypomorphic alleles.
Human patients with DICER1 alterations are prone to developmental defects, childhood mortality, and cancer (3, 4). In addition, DICER1 alterations have been associated with diseases such as aging, infertility, and blindness. As homozygosity of phosphomimetic Dicer1 drives early postnatal lethality, accelerated aging phenotypes, and infertility, we suspect that dysregulation of this controlled posttranslational modification process occurs in patients. We propose that dysregulation of Dicer1 phosphorylation may be associated with multiple human pathologic conditions, and pharmacological intervention to control this process may have far-reaching benefits.
Methods
Construct Design and Generation of Mouse Models.
The targeting construct was designed to generate a double-phosphomimetic mutant (S1712D and S1836D in the same allele) with possibilities to obtain single mutants. The construct contains a floxed neomycin-resistance gene (neoR) inserted into intron 24 of Dicer1 with a 5.9-Kbp 5′ homologous arm and a 2.7-Kbp 3′ homologous arm. The 5′ arm contains S1712D mutation (TCT → GAT) and the 3′ arm contains S1836D mutation (TCT → GAT). The S1712D mutation on the 5′ arm divides the homologous region into a 4.2-Kb outer region and 1.7-Kb inner region (considering neoR as the center). Similarly, the S1836D mutation divides the 3′ arm into a 2.0-Kb outer region and 0.7-Kb inner region. When homologous recombination (HR) occurs at the outer region of both homologous arms, both mutations will be introduced and a double mutant will be generated. On the contrary, if HR occurs at the outer region of one homologous arm (i.e., mutation is introduced) and inner region of the other homologous arm (i.e., mutation is not introduced), single mutants will be generated.
The construct was fully sequenced and successfully electroporated into TC1 mouse ES cells. Neomycin-resistant clones were screened by Southern blot analysis by using 5′ and 3′ external probes. SpeI-digested genomic DNA was used for Southern blot screening. Positive clones were confirmed by PCR and sequencing of targeted region. Two lines of ES clones that were accurately generated were injected into blastocysts, which were then implanted into pseudopregnant females to generate chimeras. Male chimeras were crossed to C57BL/6 females for germline transmission of the mutant allele. One male progeny carrying the mutant allele was crossed with C57BL/6 female mice carrying Zp3-Cre transgene to remove neoR. Male progeny carrying the mutant allele (without neoR) were crossed with WT C57BL/6 females for two more generations.
CRISPR/Cas9-Based Gene Targeting.
As previously described (48), a target site was selected, sgRNA was synthesized, and fertilized zygotes were injected with sgRNA, Cas9 mRNA, and donor oligo containing the mutation. Briefly, a 43-base sequence (20 bases on either side of the targeted codon) was used to score all possible target sites by using the Zhang Lab Guide Design Resources (zlab.bio/guide-design-resources) tool. A site with high score and containing the mutagenesis site in the seed region of sgRNA was selected for targeting. Six possible off-target sites with a PAM sequence were also selected for screening of unintended mutations.
The template for sgRNA synthesis was generated by annealing two complementary oligonucleotides. The T7 promoter sequence is underlined, and sgRNA target sites are in bold:
5′GAAATTAATACGACTCACTATAGACCCACGGCAGCATTCTCCGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT3′
Five micrograms of each oligo were mixed in nuclease-free water, boiled for 5–10 min, and cooled at room temperature for 2 h to overnight. To hundred to four hundred nanograms of the template was used to synthesize sgRNAs by using the MEGAshortscript Kit (AM1354; Invitrogen), and sgRNAs were purified by acid phenol-chloroform extraction and ethanol precipitation (Invitrogen), resuspended in 70 µL of RNase-free water, and further purified by using Biospin P30 chromatography columns (no. 732–6223; Biorad) per manufacturer’s protocol. A final injection solution containing 10 ng/µL Cas9 mRNA (PrecisionX hspCas9 SmartNuclease mRNA; System Biosciences), 7.5 ng/µL of sgRNA, and 20 ng/µL of donor oligo was prepared in Tris-EDTA buffer (5 mM Tris, 0.1 mM EDTA). The final solution was injected into the pronucleus of 200–250 zygotes. The zygotes were then implanted into pseudopregnant mice (n = 20–25 per animal). All injections and implantations were done in the MD Anderson Cancer Center genetically engineered mouse facility.
Selection and Screening of Off-Target Sites.
List of all possible off-target sites were obtained from the Zhang Lab Guide Design Resources site. Previously described criteria (48) were used to select the sites for screening. Primers are designed approximately 500 bases upstream and 500 bases downstream of the selected sites, and the region is PCR amplified and Sanger-sequenced:
OTS1 forward, GCTCAGTGGCTGGCATATATG; OTS1 reverse, CCCAACTGCAGCTCCTTTG;
OTS2 forward, GCCGTGATTTGGGACAAAAAG; OTS2 reverse, CCCCATAGGTGGGTTTGTATC;
OTS3 forward, CCCCCATAGCTGGTTCAAAC; OTS3 reverse, GGCAACAAGGGCAGATACATG;
OTS4 forward, CCTCTAGGCCTGTGATCAGAATAG; OTS4 reverse, GGCGGGTTAATGACTCATACAG;
OTS5 forward, GGCTGGACATAGTTGTCTGTTG; OTS5 reverse, GCTCCACCTGGCTTCATTATC; and
OTS6 forward, GAGGACCGATGGTTGTGAAAAATC; OTS6 reverse, CCTGGCACCTAGGAGAATTTAG.
Mouse Breeding, Maintenance, Screening and Genotyping.
Mice were maintained in >90% C57BL/6 background. All mouse studies were conducted in compliance with an institutional animal care and use committee protocol. Live mice were weaned at the age of 3 wk, and ear biopsy specimens were collected. The tissues were digested by lysis buffer (1× PCR buffer, 1% Triton X, 250 μg/μL Proteinase K) at 55 °C overnight and heated at 95 °C for 15 min to denature Proteinase K. Two microliters of the lysed tissue extract was used for PCR to amplify a 1-Kb region of the targeted site. The PCR product is gel purified and Sanger-sequenced to identify any indels at the target site.
S1712D screening primer forward, GTGCCAGGGATGTAGAAGAC;
S1712D screening primer reverse, GGGCTGCAGGAATTCGATATC;
S1836D screening primer forward, CACGTGGTACCTTAAGATGCATG; and
S1836D screening primer reverse, GCGGGTGACTTGAACCTAAG.
Homozygous Viability Test.
For each mutant allele, heterozygous mice were inbred and the progeny were genotyped at weaning. From five Dicer+/S1712D mating pairs, 10 litters with a total of 85 progeny were genotyped at weaning. From four Dicer+/∆1712 mating pairs, eight litters with a total of 68 progeny were genotyped at weaning. From three Dicer+/− mating pairs, five litters with a total of 28 progeny were genotyped at weaning. From seven Dicer+/S1836D mating pairs, 14 litters with a total of 103 progeny were genotyped at weaning. From 10Dicer+/2SD mating pairs, 18 litters with a total of 115 progeny were genotyped at weaning.
E18.5 Embryo Dissection.
Dicer+/2SD and Dicer+/S1836D heterozygous animals were inbred, and plugs were examined every morning. Females positive for plug (E0.5) were euthanized 18 d from that date (E18.5). Uteri were dissected, and each decidua was separated and placed in a 10-cm plate with PBS buffer. Each decidua is dissected to recover the embryos. Embryos were examined for viability, weighed, and euthanized, and tail biopsy specimens were collected and genotyped.
Weight Measurements.
Weaned WT (12 males and 12 females), DicerS1836D/S1836D (12 males and 9 females), and Dicer2SD/2SD (10 males and 11 females) mice were weighed (to 0.1 g) every week from 4 wk after birth by using an electronic scale. When a mutant mouse was euthanized, its littermate WT mouse was also killed. At least three mice were present at all time points (males and females) from all genotypes. An end point of 10 mo was selected because only two Dicer2SD/2SD mice (one male and one female) survived past this age.
Fertility Test.
Eight- to 12-wk-old homozygous mice (males and females) of all mutant alleles were mated with WT mice (8–16 wk old). The mating pairs were monitored daily and maintained until one or two litters were born. Litter size was counted and recorded at the time of weaning. In the absence of any progeny, WT animals were replaced with younger (8–16 wk old) WT mice every 2 mo for 6 mo.
Micro-CT Scan.
Micro-CT scans were performed at the small animal imaging facility of MD Anderson Cancer Center as described at https://www.mdanderson.org/research/research-resources/core-facilities/small-animal-imaging-facility/equipment/micro-ct_.html. Briefly, four Dicer2SD/2SD mice (two males and two females, 6–8 mo old) and their four WT littermates (two males and two females) were anesthetized using isoflurane, immobilized, and scanned using the Explore Locus RS preclinical in vivo scanner (GE Medical Systems), which is a cone-beam volume CT system that permits live imaging of rodents. The X-ray source and CCD-based detector gantry is rotated around the subject in approximately 1.0° increments. Geometric models were produced by the system. MicroView software was used to create isosurface with thresholds of 700 and 1,000. Quantifications of BMDs and BVFs of spine were done at a threshold of 700. Kyphosis index was calculated as previously described (49).
Histopathology and Immunofluorescence Staining.
As previously described, tissues harvested from mice were fixed in 10% neutral buffered formalin and paraffin-embedded. Four-micrometer sections were stained with H&E and examined by light microscopy. Tissue processing, paraffin embedding, sectioning, and H&E staining were performed by the MD Anderson Department of Veterinary Medicine & Surgery Histology Laboratory. Selected unstained sections were analyzed by immunofluorescence with respective antibodies. Total Dicer1 antibody (1:30, MABN461; EMD Millipore) and phospho-Dicer–specific antibody [1:100, generated by us (18)] were used. Quantification of cells with nuclear Dicer1 in testes sections was performed manually (SI Appendix, Fig. S5). Tissue sections stained with total Dicer1 antibody and DAPI were used. Three 20× images of each testis section (different regions; three testes sections from WT, DicerS1712D/S1712D, DicerS1836D/S1836D, and Dicer2SD/2SD mice) were taken. Images were opened by using ImageJ software, and total number of spermatocytes (round nuclear staining by DAPI, spermatids and spermatozoa excluded) were counted per seminiferous tubule. Cells with Dicer1 staining in >50% of nucleus were considered positive for nuclear staining. To avoid bias, a blinded study was performed.
Survival Curve.
A cohort of DicerS1712D/S1712D (n = 15), DicerS1836D/S1836D (n = 21), Dicer2SD/2SD (n = 21), and WT (n = 57) littermate mice was generated and monitored every day after weaning (3 wk after birth). Moribund animals were euthanized, and their tissues were harvested. When the mutant mice were euthanized, the littermate control mice were also euthanized. Mice that had to be euthanized for nonmoribund reasons were excluded from the study.
MEF Preparation and Cell Culture.
Dicer+/2SD heterozygous animals were inbred, and plugs were examined every morning. Females positive for plug (E0.5) were euthanized 13 d from that date (E13.5). Uteri were dissected, and each decidua was separated and placed in a 10-cm plate with PBS buffer. Each decidua is dissected to recover the embryos. Fetal liver, head, and limbs (collected for genotyping) were removed from each embryo, and the remaining tissues were chopped and treated with trypsin for 5–10 min at 37 °C. Five milliliters DMEM media supplemented with 10% FBS and penicillin/streptomycin was added to each sample, and the cells were dissociated by pipetting up and down. Cells were centrifuged, washed with PBS solution, resuspended in 10 mL media, transferred to 10-cm plates, and incubated at 37 °C. Media was changed every 24 h, and the cells were harvested and replated (into three to four plates) when they reached confluence. Early-passage (2–3) MEFs were used for analysis.
Small RNA Sequencing and Pathway Analysis.
RNA from testes and MEFs were extracted and purified per manufacturer’s protocol (Direct-zol RNA Miniprep plus; ZYMOL) and submitted to the MD Anderson Cancer Center sequencing core facility. Barcoded libraries were prepared by using the Illumina TruSeq small RNA sequencing kit. The libraries were sequenced on a NextSeq 500 system. Quality assessment of the Illumina raw reads was performed with FASTQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/).
A total of 1,915 mature miRNA sequences from the mouse genome assembly GRCm38 were obtained from the miRBase and used as reference for subsequent short-read mapping (50). Contaminating adapter sequences were removed by using the adapter scan function implemented in SeqMan NGen version 14 (DNASTAR). Sequences with median quality lower than a phred of 30 were discarded from the analysis. Short-read mapping to reference mature miRNAs was also performed with SeqMan NGen with the following specifications: mer size (the minimum length of a mer, overlapping region of a fragment read, in bases, required to be considered a match when assembling reads into contigs) 15, minimum match percentage (minimum percentage of matches in an overlap that are required to join two sequences in a contig) 95%, and minimum aligned length (minimum length of at least one aligned segment of a read after trimming) 18. Duplicate sequences were marked and combined.
Statistical analysis of mapped data from technical and biological replicates was performed with ArrayStar (DNASTAR). Briefly, normalization of mapped data was done by using the reads per million (RPM) method, and fold change in miRNA abundance between the WT and mutant samples was calculated from log2 of total RPM (51). Significance of observed differences was determined with the Student’s t test, and the Benjamini–Hochberg correction was employed for multiple hypothesis testing; false discovery rate was set at 0.05. Hierarchical clustering was performed with the Euclidean distance.
Finally, all miRNAs with less than 10 reads per million in both groups (WT and mutant) were filtered out. Differentially expressed miRNAs with less than twofold difference and CI of <95% were filtered out. From the final list, pathway analysis on differentially expressed miRNAs was performed by using DIANATOOLS mirPath version 3 software. List of miRNAs were uploaded into the program, and analysis was performed by using three different target prediction tools (TarBase, TargetScan, and microT-CDS).
CLAMS Test.
CLAMS test assays were performed at the Baylor College of Medicine as described elsewhere (https://www.bcm.edu/research/advanced-technology-core-labs/lab-listing/mouse-metabolic-research-unit/instruments/clams). Briefly, four Dicer2SD/2SD mice (two males and two females) and two WT mice (one male and one female) were housed in individual metabolic cages and acclimatized for 2 h, and measurements were made every 1 min for 24 h. The calorimetry system monitored OCR and carbon dioxide production rate. The free running wheels in each cage recorded the number of revolutions and the time of day when the activity occurred. The CLAMS system also monitored the amount of food consumed by the mouse and the frequency and duration of each meal. All animals were weighed at the beginning and end of the experiments.
Seahorse Metabolic Assays.
Per manufacturer’s protocol (XFe96 training manual; Seahorse Bioscience), passage-two Dicer2SD/2SD (n = 3) and WT (n = 2) MEFs were trypsinized, counted, and resuspended to a concentration of 200 cells per microliter in DMEM media supplemented with 10% FBS and penicillin/streptomycin. Eighty microliters of each MEF sample (six technical replicates) were plated into a 96-well plate and incubated at 37 °C overnight. Sensor cartridge was hydrated with XF calibrant overnight at 37 °C. The next day, the cells were washed and incubated with assay-specific media (described in the aforementioned manual) for 1 h at 37 °C in a non-CO2 incubator. Sensor cartridge was loaded into the machine for precalibration, and the plates with cells are loaded when ready. Measurements (OCR and extracellular acidification rate) were made three to five times. The experiment was repeated with new Dicer2SD/2SD (n = 6) and WT (n = 5) MEFs. For data analysis, averages of technical replicates were used and the two experiments were combined.
Statistical Analysis.
Student’s t tests and Kaplan–Meier survival analyses were performed by using Prism 7 software (GraphPad Software). P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This study was supported in part by National Institutes of Health Grants GMR0198200 (to S.A.) and Cancer Center Support Grant CA016672. S.A. is an Andrew Sabin Family Foundation Fellow at The University of Texas MD Anderson Cancer Center. N.K.A. is a Gigli Family Endowed Scholar at The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814377116/-/DCSupplemental.
References
- 1.Nicholson RH, Nicholson AW. Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm Genome. 2002;13:67–73. doi: 10.1007/s00335-001-2119-6. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 3.Khan NE, et al. Macrocephaly associated with the DICER1 syndrome. Genet Med. 2017;19:244–248. doi: 10.1038/gim.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein S, et al. Expanding the phenotype of mutations in DICER1: Mosaic missense mutations in the RNase IIIb domain of DICER1 cause GLOW syndrome. J Med Genet. 2014;51:294–302. doi: 10.1136/jmedgenet-2013-101943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshikawa T, et al. Unique haploinsufficient role of the microRNA-processing molecule Dicer1 in a murine colitis-associated tumorigenesis model. PLoS One. 2013;8:e71969. doi: 10.1371/journal.pone.0071969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambertz I, et al. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrate MP, et al. MicroRNA biogenesis is required for Myc-induced B-cell lymphoma development and survival. Cancer Res. 2010;70:6083–6092. doi: 10.1158/0008-5472.CAN-09-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jafarnejad SM, Ardekani GS, Ghaffari M, Martinka M, Li G. Sox4-mediated Dicer expression is critical for suppression of melanoma cell invasion. Oncogene. 2013;32:2131–2139. doi: 10.1038/onc.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy C, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurzynska-Kokorniak A, et al. The many faces of Dicer: The complexity of the mechanisms regulating Dicer gene expression and enzyme activities. Nucleic Acids Res. 2015;43:4365–4380. doi: 10.1093/nar/gkv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg-Gorenshtein G, et al. MiR-192 directly binds and regulates Dicer1 expression in neuroblastoma. PLoS One. 2013;8:e78713. doi: 10.1371/journal.pone.0078713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Tarallo V, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori MA, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake M, et al. A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev Cell. 2014;31:614–628. doi: 10.1016/j.devcel.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burger K, et al. Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. J Cell Biol. 2017;216:2373–2389. doi: 10.1083/jcb.201612131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudhasani R, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swahari V, et al. Essential function of Dicer in resolving DNA damage in the rapidly dividing cells of the developing and malignant cerebellum. Cell Rep. 2016;14:216–224. doi: 10.1016/j.celrep.2015.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurkar AU, Niedernhofer LJ. Comparison of mice with accelerated aging caused by distinct mechanisms. Exp Gerontol. 2015;68:43–50. doi: 10.1016/j.exger.2015.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harkema L, Youssef SA, de Bruin A. Pathology of mouse models of accelerated aging. Vet Pathol. 2016;53:366–389. doi: 10.1177/0300985815625169. [DOI] [PubMed] [Google Scholar]
- 24.Kõks S, et al. Mouse models of ageing and their relevance to disease. Mech Ageing Dev. 2016;160:41–53. doi: 10.1016/j.mad.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Reiling E, et al. The progeroid phenotype of Ku80 deficiency is dominant over DNA-PKCS deficiency. PLoS One. 2014;9:e93568. doi: 10.1371/journal.pone.0093568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treiber N, et al. Accelerated aging phenotype in mice with conditional deficiency for mitochondrial superoxide dismutase in the connective tissue. Aging Cell. 2011;10:239–254, and erratum (2011) 10:912. doi: 10.1111/j.1474-9726.2010.00658.x. [DOI] [PubMed] [Google Scholar]
- 27.Barzilai N, Huffman DM, Muzumdar RH, Bartke A. The critical role of metabolic pathways in aging. Diabetes. 2012;61:1315–1322. doi: 10.2337/db11-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 29.Pugh TJ, et al. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene. 2014;33:5295–5302. doi: 10.1038/onc.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakheja D, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buza-Vidas N, et al. Dicer is selectively important for the earliest stages of erythroid development. Blood. 2012;120:2412–2416. doi: 10.1182/blood-2011-10-383653. [DOI] [PubMed] [Google Scholar]
- 32.Chen JF, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frezzetti D, et al. The microRNA-processing enzyme Dicer is essential for thyroid function. PLoS One. 2011;6:e27648. doi: 10.1371/journal.pone.0027648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103:2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim GJ, et al. Dicer is required for Sertoli cell function and survival. Int J Dev Biol. 2010;54:867–875. doi: 10.1387/ijdb.092874gk. [DOI] [PubMed] [Google Scholar]
- 36.Korhonen HM, et al. Dicer is required for haploid male germ cell differentiation in mice. PLoS One. 2011;6:e24821. doi: 10.1371/journal.pone.0024821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, He X, Feng J. Dicer is essential for neuronal polarity. Int J Dev Neurosci. 2012;30:607–611. doi: 10.1016/j.ijdevneu.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Liu HC, Tang Y, He Z, Rosenwaks Z. Dicer is a key player in oocyte maturation. J Assist Reprod Genet. 2010;27:571–580. doi: 10.1007/s10815-010-9456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morita S, et al. Dicer is required for maintaining adult pancreas. PLoS One. 2009;4:e4212. doi: 10.1371/journal.pone.0004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reis FC, et al. Fat-specific Dicer deficiency accelerates aging and mitigates several effects of dietary restriction in mice. Aging (Albany NY) 2016;8:1201–1222. doi: 10.18632/aging.100970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 43.Yang WJ, et al. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 44.Anderson RM. A role for dicer in aging and stress survival. Cell Metab. 2012;16:285–286. doi: 10.1016/j.cmet.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emmerth S, et al. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell. 2010;18:102–113. doi: 10.1016/j.devcel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 46.Otsuka M, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Otsuka M, et al. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118:1944–1954. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aryal NK, Wasylishen AR, Pant V, Riley-Croce M, Lozano G. Loss of digestive organ expansion factor (Diexf) reveals an essential role during murine embryonic development that is independent of p53. Oncotarget. 2017;8:103996–104006. doi: 10.18632/oncotarget.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laws N, Hoey A. Progression of kyphosis in mdx mice. J Appl Physiol. 2004;97:1970–1977. doi: 10.1152/japplphysiol.01357.2003. [DOI] [PubMed] [Google Scholar]
- 50.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.