Abstract
Background
Interleukin-17 (IL-17), a pleiotropic cytokine, plays a significant role in the inflammatory diseases. By a pilot study with small population, IL-17 polymorphisms (IL-17A rs2275913 and IL-17F rs763780) showed a more potential risk factor in knee osteoarthritis (OA) in our recruited subjects. In the current study, the association between IL-17A rs2275913 and IL-17F rs763780and the risk of OA in a Chinese population is studied.
Methods
The IL-17A rs2275913 and IL-17F rs763780 polymorphisms were determined in 594 knee OA cases and 576 healthy controls, using polymerase chain reaction-restriction fragment length polymorphism assay. The relationship between genotype distribution and disease risk, as well as OA severity was analyzed by Chi-square test and multivariate logistic regression.
Results
The experimental results indicated that the polymorphism in IL-17 gene rs2275913 site were related to knee OA risk after the adjustment of BMI, sex, age, smoking and drinking status (AA vs. GG: odds ratio (OR), 1.411; 95% confidence interval (CI), 1.021–1.950; P = 0.040; A allele vs. G allele: OR, 1.192; P = 0.037; 95% CI, 1.012–1.404;). Similarly, subjects who are bearing the rs763780 variant genotypes (TC and CC) and C allele also had a higher susceptibility to knee OA compared with those who are bearing the TT genotype (TC vs. TT, OR: 1.312; P = 0.039; 95% CI: 1.017–1.692; CC vs. TT, OR: 2.812, P = 0.006, 95% CI: 1.338–5.909; C allele Vs. T allele, OR:1.413, P = 0.002, 95% CI:1.141–1.751). In the meantime, one high-risk haplotypes, AC (OR was 7.22, P < 0.01) was found. Both two polymorphisms do not correlated with OA severity based on Kellgren-Lawrence (K&L) scales. Finally, serum IL-17 levels of knee OA patients were greatly higher than those of controls (P = 0.001).
Conclusions
With the limited size sample, our study shows that IL-17 gene polymorphisms possibly related to the high-risk knee OA occurrence.
Keywords: IL-17F, IL-17A, Inflammation, Polymorphism, Osteoarthritis
Background
Osteoarthritis (OA), is featured by diverse changes and cartilage breakdown in the subchondral bone, resulting in chronic pain, deformity, joint swelling, and whole joint abnormalities. However, the exact cause of OA remains unknown. According to previous studies, OA has a complicated pathogenesis due to the interaction with complex pathogenic factors, including mechanical, biochemical, genetic, endocrine, metabolic, and environmental factors [1–5]. Moreover, the bony outgrowths (osteophytes), inflammation of the synovial membrane, and its triggering catabolic and pro-inflammatory mediators such as prostaglandin E2, neuropeptides, cytokines, and nitric oxide are essential to OA formation [5–7]. It is estimated that 18% of females and 9.6% of males older than 60 years suffer from symptomatic OA worldwide [8–11].
Although OA is considered a non-inflammatory condition earlier, with in-depth research on its physiopathology, now it is widely accepted that synovial inflammation is a feature of OA, and characterized by immune cell infiltration and cytokine secretion [12]. Both Innate immune system activation [13] and T-cell mediated inflammatory pathway play central roles in disease development and progression [14]. IL-17 is essential to both the adaptive and innate immune systems. It has five confirmed receptors (IL-17RA-RD and SEF) and six members (IL-17A-F) [15, 16]. IL-17A and IL-17F are secreted by Th17 cells, which is a distinct lineage of CD4+ effector cells [17]. Moreover, IL-17, as a pro-inflammatory cytokine, can trigger the release of chemokines, cytokines, antimicrobial peptides, and matrix metalloproteinases from mesenchymal and myeloid cells [18]. Previous studies also demonstrate that IL-17 is expressed in synovial tissues, and could contribute to cartilage breakdown and synovial infiltration in OA by inducing the release of chemokines by chondrocytes [19] and augments nitric oxide production in OA cartilage via nuclear factor kappa B activation [20].
Studies have reported that genetic polymorphisms of IL-17 are related to the susceptibility of a scope of inflammation-related diseases including ulcerative colitis, gastric cancer, breast cancer, and rheumatoid arthritis [21–24]. Although there is no relative report about association of IL-17 polymorphism with OA in the publicly available Genome wide association study database, we still hypothesize that IL-17 is a potential risk factor for OA pathogenesis. The correlation between the polymorphisms of IL-17A and IL-17F and the risk of knee osteoarthritis among the Chinese population has not been reported. By a pilot study with small population, IL-17A rs2275913 and IL-17F rs763780 shows more potential risk factor in OA in our recruited subjects. The present study focusses on these two SNPs to investigate their relationship with OA morbidity and severity.
Methods
Subjects
With all criteria adhered to the Declaration of Helsinki, this study was permitted by the Ethics and Research Committee of the Central Hospital of Cangzhou City, China. All participants provided written informed consent with gender, age, weight and body mass index (BMI) included. All participants were older than 40 years. Out of the 594 patients diagnosed with primary knee OA, 576 age-matched healthy controls free from symptoms or signs of OA. Other types of arthritis or joint diseases were enrolled in this study.
The criteria of the American College of Rheumatology was utilized in diagnosing knee OA [25], including primary OA with any symptom and radiographic sign of OA as per the Kellgren-Lawrence (K&L) scale (≥2 scale). Two independent examiners blind to the clinical data carried out the clinical examination and radiological assessment. The sex and age matched control subjects were free from personal or family history of OA. Other etiologies that cause knee diseases, such as inflammatory arthritis, post septic arthritis, and posttraumatic, developmental or skeletal dysplasia were not included.
Sample collection
Peripheral blood was collected by venipuncture from participants into EDTA tube for DNA isolation. In addition, the biochemical parameters of the serum were detected.
Genomic DNA isolation
DNA for genomic studies was extracted with commercial DNA Blood Mini Kit (QIAGEN, Hilden, Germany) from 200 μL of whole blood following the manufacturer’s instructions. DNA concentration was measured by spectrophotometry (NanoDrop 2000, Thermo Scientific) diluted to nearly 40 ng/μL.
DNA sequencing analysis
IL-17 copies in the samples were measured by PCR through amplification. The primers of SNP rs2275913 were as follows: forward 5’-ATTTCTGCCCTTCCCATTTT-3′ and reverse 5′- CCAGGAGTCATCGTTGTTT-3′. For SNP rs763780, the primers used were: forward 5’-GCAGAGCACTGGGTAAGGAG-3′ and reverse 5’-CTGCATCAATGCTCAAGGAA-3′. Sequencing was carried out by a Bio-Informatic company (Life technology, Shang Hai, China).
Serum IL-17A and IL-17F levels
Serum samples were collected from all patients and healthy subjects. The blood was allowed to clot for 30 min at 4 °C before centrifugation at 3000 rpm for 10 min at 4 °C. Total serum was separated and stored at − 20 °C until they were ready to be used. Sandwich ELISA was used to measure the concentrations of serum IL-17A and F following the manufacturer’s instructions. The intra-assay coefficients of variation came was set as 10%.
Statistical analysis
The demographic and clinical data were shown as Mean ± SD and compared among groups by the Student’s t-test. The genotype and allelic frequencies were assessed by Hardy-Weinberg equilibrium (HWE) and compared by Chi-square test and Fisher’s exact test by the online calculator tool (http://www.oege.org/software/hardy-weinberg.html). Association between the SNP and the OA risk was evaluated by calculating the Odds ratio (OR) and 95% confidence interval (CI) for the additive model, the dominant model, the recessive model by using SPSS 19.0 software (IBM SPSS, USA). P values < 0.05 were regarded as statistically significant.
Results
Features of two groups are consistent
The characteristics of the two groups of 594 knee OA patients and 576 control subjects were summarized in Table 1. The mean ages of the control group and knee OA group were 58.3 ± 9.6 and 59.5 ± 8.9 years respectively. There was no significant difference in mean age, sex, drinking and smoking status between these two groups, indicating subjective matching. The mean BMI of the OA group (26.2 ± 3.8) was greatly higher compared with that of the control group (24.6 ± 3.1) (P = 0.0478), which is consistent with previous findings that reported high BMI in OA [26, 27].
Table 1.
Demographic characteristics of the study population
| Variables | Healthy control (n = 576) | Knee osteoarthritis patients (n = 594) | P value |
|---|---|---|---|
| Age (mean ± SD) | 58.3 ± 9.6 | 59.5 ± 8.9 | 0.214 |
| Gender | 0.517 | ||
| Male | 174 | 201 | |
| Female | 402 | 393 | |
| Body mass index (kg/m2) | 24.6 ± 3.1 | 26.2 ± 3.8 | 0.0478 |
| Smoking | |||
| Yes | 183 | 213 | 0.155 |
| No | 393 | 381 | |
| Drinking | 0.790 | ||
| Yes | 69 | 75 | |
| No | 507 | 519 | |
| IL-17A concentration (x ± S, pg/mL) | 2.98 ± 0.14 | 3.91 ± 0.27 | 0.0325 |
| IL-17F concentration (x ± S, pg/mL) | 104.5 ± 12.6 | 142.8 ± 14.9 | 0.0249 |
A allele of rs2275913 and C allele of rs763780 increase the risk of knee OA
The distribution of each allele and genotype is shown in Table 2. Both SNPs were within the Hardy-Weinberg equilibrium. In terms of the genotype and allele frequencies in rs2275913 polymorphism, there was a remarkably difference between OA patients and healthy controls, which was shown in Table 2. In addition, all subjects carrying AA genotype have significantly higher risks of OA compared with GG genotype (P < 0.05). The results revealed that subjects with the A allele were more likely to get knee OA compared with those bearing the G allele (OR, 1.1192; 95% CI, 1.012–1.404; p = 0.037). Further analysis demonstrated the higher risk of AA genotype and A allele mainly existed in female sub-population (P < 0.05), but not in male population (P > 0.05, Table 2).
Table 2.
Distributions of IL-17 SNPs genotypes in each group and analyses of associations between these polymorphisms and knee OA risk
| Genotype | Overall (N) | Female (N) | Male (N) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | OA | OR (95% CI) | P | control | OA | OR (95% CI) | P | control | OA | OR (95% CI) | P | |
| Additive model | ||||||||||||
| rs2275913 | 576 | 594 | 402 | 393 | 174 | 201 | ||||||
| GG | 207 | 189 | 1.00 | 1.000 | 145 | 122 | 1.00 | 1.00 | 62 | 67 | 1.00 | 1.000 |
| GA | 265 | 271 | 1.12 (0.864–1.452) | 0.427 | 184 | 179 | 1.156 (0.842–1.587) | 0.376 | 81 | 92 | 1.051 (0.666–1.659) | 0.907 |
| AA | 104 | 134 | 1.411 (1.021–1.950) | 0.04 | 73 | 92 | 1.498 (1.014–2.213) | 0.048 | 31 | 42 | 1.254 (0.703–2.236) | 0.466 |
| Allele | ||||||||||||
| G | 679 | 649 | 1.00 | 1.000 | 474 | 423 | 1.00 | 1.00 | 205 | 226 | 1.00 | 1.000 |
| A | 473 | 539 | 1.192 (1.012–1.404) | 0.037 | 330 | 363 | 1.233 (1.011–1.503) | 0.043 | 143 | 176 | 1.116 (0.835–1.493) | 0.460 |
| rs763780 | ||||||||||||
| TT | 411 | 380 | 1.00 | 1.000 | 287 | 246 | 1.00 | 1.000 | 124 | 134 | 1.00 | 1.000 |
| TC | 155 | 188 | 1.312 (1.017–1.692) | 0.039 | 108 | 131 | 1.415 (1.042–1.923) | 0.029 | 47 | 57 | 1.122 (0.711–1.772) | 0.643 |
| CC | 10 | 26 | 2.812 (1.338–5.909) | 0.006 | 7 | 16 | 2.667 (1.079–6.588) | 0.033 | 3 | 10 | 3.085 (0.830–11.468) | 0.093 |
| Allele | ||||||||||||
| T | 977 | 948 | 1.00 | 1.000 | 682 | 623 | 1.00 | 1.000 | 295 | 325 | 1.00 | 1.000 |
| C | 175 | 240 | 1.413 (1.141–1.751) | 0.002 | 122 | 163 | 1.463 (1.129–1.894) | 0.004 | 53 | 77 | 1.319 (0.899–1.935) | 0.176 |
| Dominant model | ||||||||||||
| rs2275913 | ||||||||||||
| GG + GA | 472 | 460 | 1.00 | 0.059 | 329 | 301 | 1.00 | 0.089 | 143 | 159 | 1.00 | 0.514 |
| AA | 104 | 134 | 1.322 (0.993–1.761) | 73 | 92 | 1.378 (0.976–1.944) | 31 | 42 | 1.219 (0.727–2.042) | |||
| rs763780 | ||||||||||||
| TT + TC | 566 | 568 | 1.00 | 0.01 | 395 | 377 | 1.00 | 0.058 | 171 | 191 | 1.00 | 0.098 |
| CC | 10 | 26 | 2.591 (1.238–5.422) | 7 | 16 | 2.395 (0.974–5.886) | 3 | 10 | 2.984 (0.808–11.023) | |||
| Recessive model | ||||||||||||
| rs2275913 | ||||||||||||
| GG | 207 | 189 | 1.00 | 0.139 | 145 | 122 | 1.00 | 0.154 | 62 | 67 | 1.00 | 0.664 |
| GA + AA | 369 | 405 | 1.202 (0.943–1.532) | 257 | 271 | 1.253 (0.933–1.684) | 112 | 134 | 1.107 (0.722–1.697) | |||
| rs763780 | ||||||||||||
| TT | 411 | 380 | 1.00 | 0.007 | 287 | 246 | 1.00 | 0.01 | 124 | 134 | 1.00 | 0.372 |
| TC + CC | 165 | 214 | 1.403 (1.096–1.795) | 115 | 147 | 1.491 (1.107–2.008) | 50 | 67 | 1.240 (0.798–1.926) | |||
OA osteoarthritis, OR odds ratio, CI confidence interval
With regards to the rs763780 polymorphism, similar results also observed. There was a significant difference in the genotype and allele frequencies between the two groups (knee OA patients and control subject groups), which was shown in Table 2. In addition, C allele rather than T allele can increase the risk of developing knee OA (Table 2). Further study also demonstrated that C allele more caused higher risk than in T allele in female subpopulation (P = 0.004). In the male population, the CC genotype and C allele did not induce significant higher risk for OA. Dominant and recessive model also demonstrated the similar results that A allele of rs2275913 and C allele of rs763780 increase the risk of knee OA.
IL-17 polymorphism is not related to OA severity
All included OA patients were over 2 Kellgren-Lawrence Grading Scales, and the typical CT-scan figures were showed per scale (Fig. 1). SNP distribution among healthy control (scale 0) and OA patients of different scale (2–4) were shown in Table 3. There is no significant difference among all the SNPs and alleles ranging from scale 2 to 4, suggesting no effect of IL-17 polymorphism on OA severity.
Fig. 1.
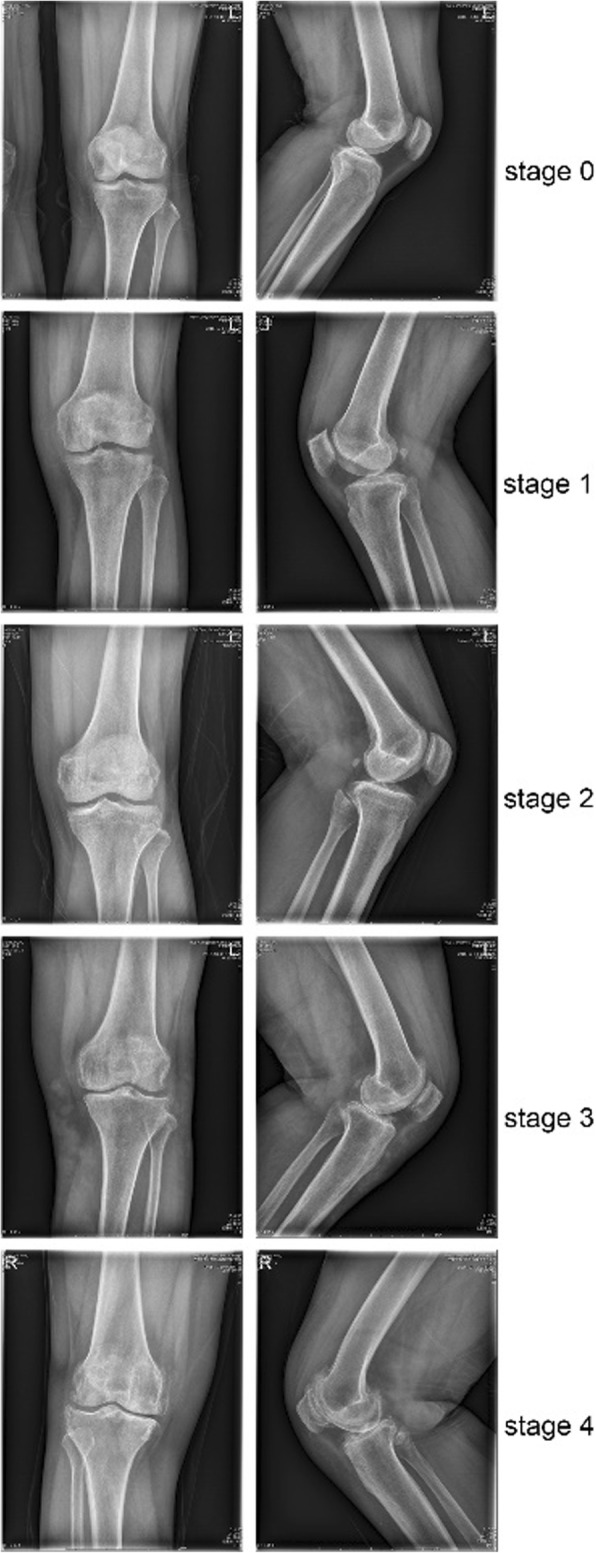
Representive photos of different severity of knee OA based on Kellgren-Lawrence (K&L) scale (from stage 0 to stage 4)
Table 3.
The association of different genotypes and alleles of various polymorphisms with the severity of knee OA
| Genotype | N | scale 2 | scale 3 | scale 4 | P value |
|---|---|---|---|---|---|
| rs2275913 | 594 | 0.116 | |||
| GG | 189 | 63 | 57 | 69 | |
| GA | 271 | 78 | 97 | 96 | |
| AA | 134 | 29 | 44 | 61 | |
| Allele | 0.057 | ||||
| G | 649 | 204 | 211 | 234 | |
| A | 539 | 136 | 185 | 218 | |
| rs763780 | 0.272 | ||||
| TT | 380 | 135 | 141 | 104 | |
| TC | 188 | 55 | 68 | 65 | |
| CC | 26 | 6 | 10 | 10 | |
| Allele | 0.077 | ||||
| T | 948 | 325 | 350 | 273 | |
| C | 240 | 67 | 88 | 85 |
Serum IL-17A/F levels are higher in OA patients
The median serum concentration of IL-17A was 2.98 ± 0.14 pg/ mL and 3.91 ± 0.27 pg/ mL in healthy controls and OA patients, respectively (Table 1). The serum levels of IL-17A in OA patients were higher than that of healthy controls (p = 0.0325). The serum levels of IL-17F in OA patients were also higher than that of healthy controls (P = 0.0249). However, when we further summarized the average serum concentration of IL-17A and IL-17F in each genotype, there was no significant difference among all three genotypes for these two SNPs (Table 4).
Table 4.
The serum levels of IL-17A and IL-17F in the different genotypes
| Genotype | IL-17A concentration (x ± S, pg/mL) | IL-17F concentration (x ± S, pg/mL) | P value |
|---|---|---|---|
| rs2275913 | > 0.05 | ||
| GG | 3.92 ± 0.48 | 142.9 ± 45.6 | |
| GA | 3.76 ± 0.35 | 134.5 ± 21.5 | |
| AA | 3.95 ± 0.50 | 143.4 ± 32.3 | |
| rs763780 | > 0.05 | ||
| TT | 3.81 ± 0.44 | 138.3 ± 43.4 | |
| TC | 3.75 ± 0.38 | 143.1 ± 52.1 | |
| CC | 3.94 ± 0.39 | 140.8 ± 43.8 |
Discussion
Previous studies implicated that IL-17/IL23 pathway and TH17 cells play an important role in inflammation-related diseases [28], and several meta-analysis examined the relationship between IL-17A (rs2275913) and IL-17F (rs763780) gene polymorphisms and the risk of inflammatory diseases, including periodontitis, rheumatoid arthritis (RA), and inflammatory bowel disease, and all these documents demonstrate that IL-17 polymorphisms may increased the RA risk, but dependent with race and ethnic groups [29]. Beside RA, IL17A (rs2275913) polymorphism has been reported to be associated with the development of rheumatic heart disease in south Indian population [30], and a variant of IL17F (rs763780) may contribute to the development of necrotizing enterocolitis [31]. All these publications suggest that IL17 polymorphism may widely to be associated with immune mediated diseases.
Although IL-17A and IL-17F have been reported to be associated with collagen-induced arthritis, and arthritis in experimental animal models [32], there is no study about relationship between rs2275913 (IL-17A SNP) and rs763780 (IL-17F SNP) with OA risk. Our study might be first time to investigate the involvement of IL-17 gene polymorphisms in knee OA and whether these are correlated with serum levels of IL-17. Up to date, we have yet to show whether these two SNPs affect the IL-17 secretion in the human plasma. But from data of Table 4, these two SNPs seem not influence the plasma concentration of IL-17A and IL-17F. Our results suggested that the IL-17A rs2275913 polymorphism has a significant impact on the risk of knee OA and that persons carrying the rs2275913 A allele are at a higher risk of developing knee OA as compared with those carrying the G allele. Additionally, rs763780 C allele, was also related to a greatly increased risk of developing knee OA. Further assessment of the effect of IL-17 polymorphisms on knee OA risk was stratified by sex, and showed a significant association in female patients’ subgroups.
In spite of the positive relationship between rs2275913 and rs763780 polymorphisms and the risk of knee OA observed in this study, IL-17 serum levels had no great differences in allele subgroups. These data did not show any association between IL-17 serum levels and IL-17 polymorphisms in the present study, however, a higher serum concentration was found in overall patients when it was compared with the healthy control. The limitation is that we did not test the concentration of IL-17A and IL-17F in knee joint (mainly from knee articular cavity and synovial tissues) of each patient, and local concentration of IL-17 may be more important than systemic IL-17 in plasma. Therefore, it is possible that SNPs of IL-17 might cause alteration of the IL-17 concentration in knee joint microenvironment, including knee articular cavity and synovial tissues, further influence the risk of OA development.
Patients carrying the A (rs2275913 G/A) and C (rs763780 T/C) alleles were related to increased risk for knee OA when compared to individuals who are carrying the wild-type alleles. Furthermore, we demonstrated that the correlation between the two polymorphisms under study and knee OA risk appeared significant among the female subjects; however, this evidence is just an association but not cause and effect. There is a possibility that this finding is as a result of the larger quantity of female subjects (n = 393) compared to the male subjects (n = 201), which may lead to a limited statistical power and robustness. This might also be attributed to other important factors related to women such as estrogen-relate effects and the fact that women are less exposed to tobacco smoking and heavy alcohol drinking [33, 34]. However, a larger sample size is needed to further investigate the underlying mechanisms of this association.
Aside the small sample size, we acknowledge other limitations such as the study population. With the study population confined to the population of He Bei province, the findings may not apply to other population. It would of great significance if the role of IL-17 polymorphisms in patient susceptibility to knee OA from other ethnic population is studied. Also, the present research investigated only two SNPs in the IL-17 gene. It would be of more interest to define more SNPs and investigate their role in knee OA. More importantly, population stratification—allele frequency differences between cases and controls due to systematic ancestry differences—can cause spurious associations in disease studies, and possible false positive results [35]. Therefore, more detailed statistical analysis is needed to do to make results more accurately.
Conclusions
In conclusion, we have successfully demonstrated that functional polymorphisms of IL-17 are significantly related to the risk of knee OA. Also, the variant alleles rs2275913 AA and rs763780 CC were related to the increased risk of knee OA but not the wild-type alleles. Serum IL-17 levels significantly correlated with increased risk of knee OA which taking together might facilitate defining high risk subjects to prevent the initial development of knee OA.
Acknowledgements
Not available.
Funding
No funding was obtained for this study.
Availability of data and materials
Data and material are available from the corresponding author if required.
Abbreviations
- BMI
Body mass index
- CI
Confidence interval
- HWE
Hardy-Weinberg equilibrium
- IL-17
Interleukin-17
- K&L
Kellgren-Lawrence
- OA
Osteoarthritis
- OR
Odds ratio
Authors’ contributions
BY draft the manuscript, LY is responsible for the experimental design and guidance; JS did statistical analysis; ZH and SK is in charge of sample collection, PCR amplification and other experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study is followed with all criteria adhered to the Declaration of Helsinki, and has been permitted by the Ethics and Research Committee of the Central Hospital of Cangzhou City, China. All participants signed an informed consent letter.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shijun Gao, Email: gao_shijundr@163.com.
Ying Liu, Phone: 86-317-2075651, Email: liuying3317@163.com.
References
- 1.Meulenbelt I. Osteoarthritis year 2011 in review: genetics. Osteoarthr Cartil. 2012;20(3):218–222. doi: 10.1016/j.joca.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Reynard LN. Analysis of genetics and DNA methylation in osteoarthritis: what have we learnt about the disease? Semin Cell Dev Biol. 2017;62:57–66. doi: 10.1016/j.semcdb.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Zou T, Yang L, Lee AM, Tan X, Wong E, Bai HX. Can genetics explain the higher risk of worsening knee pain in offspring of people with total knee replacement for severe primary knee osteoarthritis? Ann Rheum Dis. 2016;75(7):e45. doi: 10.1136/annrheumdis-2016-209380. [DOI] [PubMed] [Google Scholar]
- 4.Chapman K, Valdes AM. Genetic factors in OA pathogenesis. Bone. 2012;51(2):258–264. doi: 10.1016/j.bone.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Torres J, Hernandez-Diaz C, Espinosa-Morales R, Camacho-Galindo J, Galindo-Sevilla Ndel C, Lopez-Macay A, Zamudio-Cuevas Y, Martinez-Flores K, Santamaria-Olmedo MG, Pineda C, et al. Polymorphic variation of hypoxia inducible factor-1 a (HIF1A) gene might contribute to the development of knee osteoarthritis: a pilot study. BMC Musculoskelet Disord. 2015;16:218. doi: 10.1186/s12891-015-0678-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin MH, Lee SJ, Kee SJ, Song SK, Kweon SS, Park DJ, Park YW, Lee SS, Kim TJ. Genetic association analysis of GDF5 and ADAM12 for knee osteoarthritis. Joint Bone Spine. 2012;79(5):488–491. doi: 10.1016/j.jbspin.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Shi D, Zheng Q, Chen D, Zhu L, Qin A, Fan J, Liao J, Xu Z, Lin Z, Norman P, et al. Association of single-nucleotide polymorphisms in HLA class II/III region with knee osteoarthritis. Osteoarthr Cartil. 2010;18(11):1454–1457. doi: 10.1016/j.joca.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol. 2014;28(1):5–15. doi: 10.1016/j.berh.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 9.De Filippis L, Gulli S, Caliri A, Romano C, Munao F, Trimarchi G, La Torre D, Fichera C, Pappalardo A, Triolo G, et al. Epidemiology and risk factors in osteoarthritis: literature review data from "OASIS" study. Reumatismo. 2004;56(3):169–184. doi: 10.4081/reumatismo.2004.169. [DOI] [PubMed] [Google Scholar]
- 10.Pelaez-Ballestas I, Sanin LH, Moreno-Montoya J, Alvarez-Nemegyei J, Burgos-Vargas R, Garza-Elizondo M, Rodriguez-Amado J, Goycochea-Robles MV, Madariaga M, Zamudio J, et al. Epidemiology of the rheumatic diseases in Mexico. A study of 5 regions based on the COPCORD methodology. J Rheumatol Suppl. 2011;86:3–8. doi: 10.3899/jrheum.100951. [DOI] [PubMed] [Google Scholar]
- 11.Woolf AD, Pfleger B. Burden of major musculoskeletal conditions. Bull World Health Organ. 2003;81(9):646–656. [PMC free article] [PubMed] [Google Scholar]
- 12.de Lange-Brokaar BJ, Ioan-Facsinay A, van Osch GJ, Zuurmond AM, Schoones J, Toes RE, Huizinga TW, Kloppenburg M. Synovial inflammation, immune cells and their cytokines in osteoarthritis: a review. Osteoarthr Cartil. 2012;20(12):1484–1499. doi: 10.1016/j.joca.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Scanzello CR, Plaas A, Crow MK. Innate immune system activation in osteoarthritis: is osteoarthritis a chronic wound? Curr Opin Rheumatol. 2008;20(5):565–572. doi: 10.1097/BOR.0b013e32830aba34. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura H, Yoshino S, Kato T, Tsuruha J, Nishioka K. T-cell mediated inflammatory pathway in osteoarthritis. Osteoarthr Cartil. 1999;7(4):401–402. doi: 10.1053/joca.1998.0224. [DOI] [PubMed] [Google Scholar]
- 15.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21(4):467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK. IL-17 cytokine family. J Allergy Clin Immunol. 2004;114(6):1265–1273. doi: 10.1016/j.jaci.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 17.Rutitzky LI, Lopes da Rosa JR, Stadecker MJ. Severe CD4 T cell-mediated immunopathology in murine schistosomiasis is dependent on IL-12p40 and correlates with high levels of IL-17. J Immunol. 2005;175(6):3920–3926. doi: 10.4049/jimmunol.175.6.3920. [DOI] [PubMed] [Google Scholar]
- 18.Quan Y, Zhou B, Wang Y, Duan R, Wang K, Gao Q, Shi S, Song Y, Zhang L, Xi M. Association between IL17 polymorphisms and risk of cervical cancer in Chinese women. Clin Dev Immunol. 2012;2012:258293. doi: 10.1155/2012/258293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honorati MC, Bovara M, Cattini L, Piacentini A, Facchini A. Contribution of interleukin 17 to human cartilage degradation and synovial inflammation in osteoarthritis. Osteoarthr Cartil. 2002;10(10):799–807. doi: 10.1053/joca.2002.0829. [DOI] [PubMed] [Google Scholar]
- 20.Attur MG, Patel RN, Abramson SB, Amin AR. Interleukin-17 up-regulation of nitric oxide production in human osteoarthritis cartilage. Arthritis Rheum. 1997;40(6):1050–1053. doi: 10.1002/art.1780400609. [DOI] [PubMed] [Google Scholar]
- 21.Nordang GB, Viken MK, Hollis-Moffatt JE, Merriman TR, Forre OT, Helgetveit K, Kvien TK, Lie BA. Association analysis of the interleukin 17A gene in Caucasian rheumatoid arthritis patients from Norway and New Zealand. Rheumatology (Oxford) 2009;48(4):367–370. doi: 10.1093/rheumatology/ken512. [DOI] [PubMed] [Google Scholar]
- 22.Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Nakamura M, Yoshioka D, Arima Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28(1):44–49. doi: 10.1007/s10875-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 23.Hou C, Yang F. Interleukin-17A gene polymorphism is associated with susceptibility to gastric cancer. Int J Clin Exp Pathol. 2015;8(6):7378–7384. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Jiang Y, Zhang Y, Wang Y, Huang S, Wang Z, Tian B, Yang Y, Jiang W, Pang D. Association analysis of IL-17A and IL-17F polymorphisms in Chinese Han women with breast cancer. PLoS One. 2012;7(3):e34400. doi: 10.1371/journal.pone.0034400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki T, Inoue K, Ushiyama T, Fukuda S. Assessment of the American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the knee. Ryumachi. 1998;38(1):2–5. [PubMed] [Google Scholar]
- 26.Kawaguchi M, Takahashi D, Hizawa N, Suzuki S, Matsukura S, Kokubu F, Maeda Y, Fukui Y, Konno S, Huang SK, et al. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. J Allergy Clin Immunol. 2006;117(4):795–801. doi: 10.1016/j.jaci.2005.12.1346. [DOI] [PubMed] [Google Scholar]
- 27.Jin W, Dong C. IL-17 cytokines in immunity and inflammation. Emerg Microbes Infect. 2013;2(9):e60. doi: 10.1038/emi.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louahchi S, Allam I, Berkani L, Boucharef A, Abdesemed A, Khaldoun N, Nebbab A, Ladjouze A, Djidjik R. Association study of single nucleotide polymorphisms of IL23R and IL17 in rheumatoid arthritis in the Algerian population. Acta Reumatol Port. 2016;41(2):151–157. [PubMed] [Google Scholar]
- 29.Eskandari-Nasab E, Moghadampour M, Tahmasebi A. Meta-analysis of risk association between interleukin-17A and F gene polymorphisms and inflammatory diseases. J Interf Cytokine Res. 2017;37(4):165–174. doi: 10.1089/jir.2016.0088. [DOI] [PubMed] [Google Scholar]
- 30.Poomarimuthu M, Elango S, Solomon PR, Soundrapandian S, Mariakuttikan J. Association of IL17 and IL23R gene polymorphisms with rheumatic heart disease in south Indian population. Immunol Investig. 2018:1–11. [DOI] [PubMed]
- 31.Tian J, Liu Y, Jiang Y, Zhou H, Zhu T, Zhao X, Peng L, Yan C. Association of single nucleotide polymorphisms of IL23R and IL17 with necrotizing enterocolitis in premature infants. Mol Cell Biochem. 2017;430(1–2):201–209. doi: 10.1007/s11010-017-2972-6. [DOI] [PubMed] [Google Scholar]
- 32.Ishigame H, Kakuta S, Nagai T, Kadoki M, Nambu A, Komiyama Y, Fujikado N, Tanahashi Y, Akitsu A, Kotaki H, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30(1):108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Seng KC, Seng CK. The success of the genome-wide association approach: a brief story of a long struggle. Eur J Hum Genet. 2008;16(5):554–564. doi: 10.1038/ejhg.2008.12. [DOI] [PubMed] [Google Scholar]
- 34.Andersson A, Grahnemo L, Engdahl C, Stubelius A, Lagerquist MK, Carlsten H, Islander U. IL-17-producing gammadeltaT cells are regulated by estrogen during development of experimental arthritis. Clin Immunol. 2015;161(2):324–332. doi: 10.1016/j.clim.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and material are available from the corresponding author if required.


