Abstract
The central dogma processes of DNA replication, transcription, and translation are responsible for the maintenance and expression of every gene in an organism. An orthogonal central dogma may insulate genetic programs from host regulation and allow expansion in the roles of these processes within the cell.
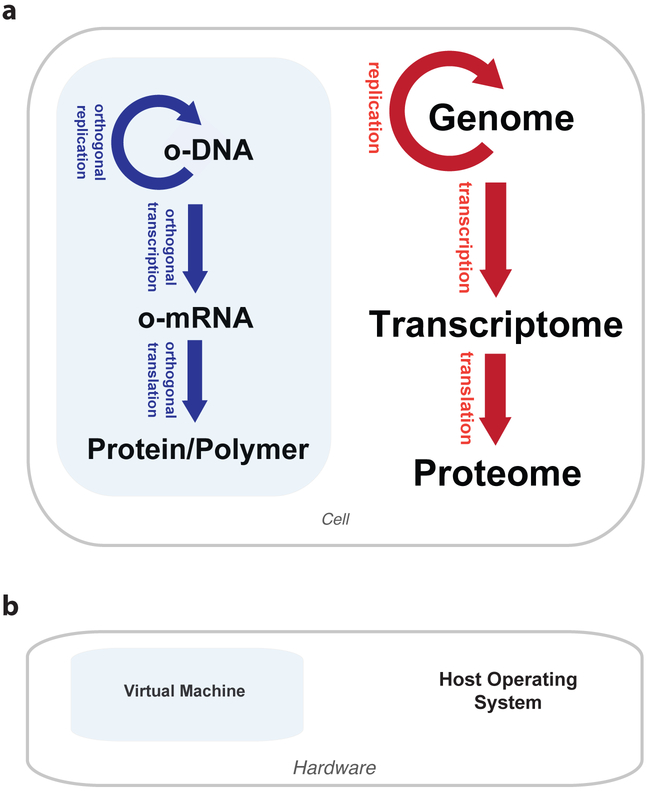
Much like computer programs rely on host operating systems to run, synthetic genes and genetic programs rely on the host cell’s central dogma processes – DNA replication, transcription, and translation – for propagation and expression. Synthetic biologists therefore write genetic programs with the host organism in mind and accept both the rigidities and regulatory complexities associated with host central dogma systems. This dependency creates two broad challenges for genetic engineering. First, a genetic program developed in the model organism that is most tractable for engineering is not easily transferred into the production host or cell type most suitable for application. Similarly, a computer program written for one operating system doesn’t run in a different operating system. Second, a wide range of desirable functions that could result from reengineering central dogma processes themselves, such as rapid mutagenesis or the repurposing of protein translation for generalized polymer synthesis and evolution, are inaccessible, because substantial changes to host central dogma processes would harm how host genes are read and expressed. Analogously, large modifications to an operating system in service of a specific program will prevent other programs from running properly. We therefore argue for the construction of orthogonal central dogma systems that act as specialized platforms for replicating, transcribing, and translating synthetic DNA in vivo (Fig. 1a). From this architecture, we should be able to minimize the impact of host-specific nuances on synthetic genes by encoding them for our independent central dogma system and gain unprecedented freedom to engineer the mechanisms of the central dogma to expand cellular function. Indeed, an orthogonal central dogma shares certain similarities with processes implemented on a virtual machine, where separation from large and unwieldy host operating systems achieves portability and the potential for considerable specialization (Fig. 1b). These are two highly desirable properties for synthetic biology that have been difficult to achieve in any general manner, and they motivate our basic argument for an orthogonal central dogma.
Figure 1: The orthogonal central dogma concept.
(a) Unlike host systems, orthogonal central dogma processes can be engineered for specialized purposes and may be a platform for portable genetic programs. (b) An orthogonal central dogma shares similarities to virtual machines in computer science. Blue boxes indicate isolation from host systems.
We define an orthogonal system as a network of (engineered) components (e.g. proteins, RNAs, DNAs, and small molecules) that interact with each other to achieve a specific function without impeding or being impeded by the native functions of the host cell. The components making up an orthogonal system are characteristically strongly connected to each other but weakly connected to the rest of the cell, except in strategic ways chosen by the biological engineer. The power of orthogonal systems derives from this “isolated hub” network design, which gives us the ability to selectively abstract the workings of heterologous processes from host processes. Applied to the central dogma, we envision an engineered set of macromolecular machines (e.g. DNA polymerases, RNA polymerases, ribosomes, tRNAs, and aminoacyl-tRNA synthetases) exclusively dedicated to the replication and expression of genes encoded on special DNA or mRNA templates unrecognized by the host. The orthogonal machines and templates therefore form an isolated genetic hub within which the rules of replication and expression can be predictable and engineerable in service of reliable and expanded function by the genes encoded on the orthogonal templates.
An orthogonal central dogma will need numerous components, and each one carries potential undesired interactions with the rest of the cell. Although direct molecular interactions with host components can be engineered away, such interactions may be difficult to identify in the first place. Furthermore, the demands that an orthogonal central dogma may place on the rest of the cell’s resources is a challenge that will need to be addressed. Therefore, the extent to which such large orthogonal networks can be constructed is uncertain. This is especially true of protein translation components, as we will later discuss. Yet recent progress has already led to examples of an orthogonal DNA replication system, several orthogonal transcription systems, and orthogonal translation components. Natural systems such as mitochondria and chloroplasts already have dedicated replication, transcription, and translation machinery, suggesting that such systems are possible and acting as potential platforms for further engineering. Here, we highlight progress on orthogonal versions of each central dogma component and consider the possibilities of, and the potential paths to, an integrated orthogonal central dogma. We do so with the understanding that we are outlining a platonic ideal, towards which any meaningful progress will be useful, as well as a grand goal that our field should attempt to realize.
Orthogonal Replication
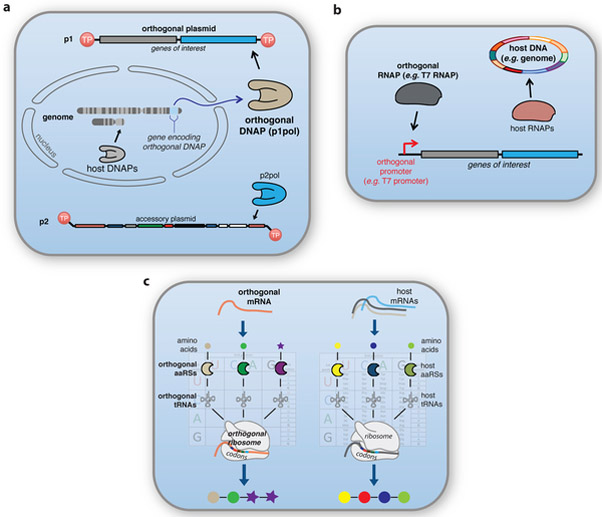
DNA replication is arguably the most basic process of life and as such, host replication systems are only minimally manipulable in vivo without harming the cell. An orthogonal DNA replication system, specifically a DNA polymerase (DNAP) exclusively dedicated to the replication of a DNA plasmid that cannot be replicated by native host systems, was recently established in yeast.1 This orthogonal DNA replication system exploits a selfish cytoplasmic plasmid system from Kluveromyces lactis that stably propagates two DNA plasmids, called pGKL1 and pGKL2 or p1 and p2 (ref. 2), each using a dedicated DNA polymerase and associated shared machinery. It was shown that the DNAP encoded on p1 is responsible for replicating p1 and that engineered changes in the DNAP’s error rate result in an elevation of p1’s error rate but not the genome’s (Fig. 2a).1 We have since substantially expanded the error rates accessible to the orthogonal system, achieving per-base substitution rates ~100,000-times higher than those of the host genome, and have demonstrated recombination, copy number control, and generality in various host yeast strains. We expect orthogonal replication to enable a variety of applications, including rapid continuous evolution of target genes, continuous barcoding, and the implementation of new genetic alphabets, all in vivo. Indeed, many biotechnological successes have been driven by the manipulation of DNA replication in vitro. For example, various forms of PCR and PCR-based diagnostics, DNA sequencing, gene diversification and library synthesis, the evolution of chemically non-natural nucleic acid polymers, and even concepts in molecular recording rely on DNAPs with heavily engineered properties.3-6 Although it is possible to expand the properties of DNA replication in vivo using native host DNAPs (e.g. bacteria whose native DNAPs can stably propagate DNA with non-natural nucleotides7 or even a non-natural base pair8), orthogonal DNA replication should enable a new level of DNA and DNAP-based engineering in vivo.
Figure 2: Component orthogonal central dogma systems.
(a) Orthogonal DNA replication. The current instantiation of orthogonal replication involves an autonomous plasmid system replicated by a dedicated DNA polymerase (p1pol) that does not replicate the host genome. p2 is an accessory plasmid to p1 replication and encodes several proteins involved in DNA replication and transcription, including its own DNA polymerase (p2pol). (b) Orthogonal transcription. Orthogonal transcription requires an orthogonal RNAP/promoter pair, which can be derived from many sources (e.g. viral systems). (c) Orthogonal translation. Ultimately, orthogonal translation requires two separate translation systems, including aminoacyl-tRNA synthetases, tRNAs, ribosomes, and ribosome-specific mRNAs. Together, these can in principle lead to distinct genetic codes running in the same cell and the mRNA-templated polymerization of unnatural amino acids or other building blocks into new biological polymers.
Orthogonal Transcription
Historically, the commonness of orthogonal transcription owes to bacteriophage RNAPs, which have evolved to recognize only their cognate promoters whose sequences are distinct from host promoters. These RNAP–promoter pairs form an isolated network that carries out transcription of target genes. So far, this orthogonality has proven useful mainly in three ways. First, bacteriophage-based RNAP–promoter pairs are easy to use for synthetic biology – they are encoded as a single gene, can drive very high transcription levels, and are compatible with a multitude of organisms – but they can lead to growth defects, because they are decoupled from host cell translation.9 Because phage RNAPs are orthogonal, however, one can introduce mutations that reduce abortive cycling, decrease their speed and processivity, use tightly regulated systems to control RNAP expression level, or use control theory to incorporate feedback loops that maintain transcription levels at homeostasis.10-13 Since the resulting tweaked transcription systems are orthogonal, they have minimal effect on transcription of host genes. Second, from a synthetic biology perspective, orthogonal transcription systems are essential for building synthetic gene regulation and custom genetic circuits: the ability to control one gene’s transcription without interfacing excessively with host transcription systems is how synthetic transcription-based regulation systems might be predictably combined, tuned, and robust to species transfer (Fig. 2b). Indeed, it has been possible to systematically expand the number of orthogonal RNAP-promoter pairs for the purpose of regulatory systems that require independent control of multiple genes and to engineer a split RNAP that can act as a logic gate.14 In conjunction with the recent explosion in sequence-programmable DNA-binding proteins, fueled largely by advances in ZFPs, TALEs, and CRISPRs, orthogonal transcription systems should result in ever-more reliable genetic circuits for cellular engineering.15 Finally, cells use native RNAPs and a set of proteins (e.g., σ factors in bacteria) that direct them to different promoters in response to environmental and stress conditions. With orthogonal transcription systems and non-native promoters controlling synthetic genes, synthetic genetic programs are better insulated against variations in growth, nutrients, and other signals that directly regulate host transcription.
Orthogonal Translation
Substantial progress has been made towards orthogonal translation through the engineering of translational components (Fig. 2c). Orthogonal aminoacyl-tRNA synthetases (aaRSs) that specifically charge cognate engineered tRNAs have been created and evolved to incorporate hundreds of unnatural amino acids and other unnatural building blocks site-specifically into proteins; an orthogonal ribosome has been created that selectively reads an orthogonal mRNA via reprogrammed interactions between an engineered small subunit ribosomal RNA and a corresponding mRNA leader sequence; and the orthogonal ribosome has been evolved to no longer recognize release factor, enabling the recoding of the amber stop codon as a sense codon and the efficient reading of quadruplet codons that may be assigned to unnatural monomers.16 Orthogonal translation pathways have been created in which unnatural amino acids are loaded onto orthogonal tRNAs and selectively decoded on the orthogonal ribosome in response to codons on the orthogonal message, enabling the efficient incorporation of multiple unnatural amino acids into a single polypeptide.17 Complementary efforts to make more codons available for encoding new monomers include work aimed at compressing the number of sense codons used for natural protein synthesis from the genome18,19 and work aimed at adding new codons by expanding the genetic alphabet.8 Elegant experiments have demonstrated that the large subunit ribosomal RNA can be evolved to accommodate β-amino acids and D-amino acids, and evolved ribosomes have recently been used for β-amino acid incorporation in vivo.20-22 Creating orthogonal ribosomes in which both the large and small ribosomal subunits are selectively directed to an orthogonal message, and do not cross-assemble with endogenous ribosomal subunits, might expand the sequence space that can be explored for large subunit evolution, and potentially the scope of monomers that can be accommodated by ribosomal translation.23 Progress towards these goals may be accelerated by the observation that the ribosomal RNA of the large subunit can be circularly permuted,24 which has enabled the creation of functional ribosomes in which the subunits are covalently linked through RNA.25,26
Towards Integration
As we envision it, an integrated orthogonal central dogma would encode the components for orthogonal transcription and translation on an orthogonal DNA replication system. The result would be a platform for the predictable design and transfer of genetic programs as well as a versatile genetic subsystem for general reengineering and building up of new chemical life. The promise of an orthogonal central dogma system is large. One of the most defining goals of synthetic biology is to achieve predictable and portable operation of genes and genetic programs across host species, but variation in how different hosts read DNA has limited progress towards this goal. For example, in the common problem of gene cluster engineering, synthetic biologists often need to complete multiple promoter matching and gene recoding experiments in order to minimize the effects of host regulation and environmental changes on a gene cluster’s desired activity or to transfer the gene cluster to new hosts. A particularly challenging version of gene cluster engineering is engineering across different domains of life. For instance, eukaryotes express genes differently than prokaryotes – eukaryotes don’t translate polycistronic mRNAs, don’t rely on transcriptional–translational coupling, use distinct promoter architectures, have a suite of posttranscriptional modifications, have an elaborate cell cycle, etc. – so it is difficult to reengineer prokaryotic gene assemblies for functional expression in eukaryotic cells. If we had orthogonal central dogma components that followed prokaryotic rules but that operated in a eukaryote or vice-versa, we could avoid this case-by-case need for refactoring. More generally, if we simply had a compact orthogonal central dogma process operating with well-defined rules that are buffered from environmental variation, we would only need to “write” genetic programs once, as we could expect these programs to be similarly “read” in any host into which we installed the molecular machinery to implement the orthogonal central dogma. Indeed, much like the virtual machine idea (Fig. 1b), orthogonal central dogmas may enhance portability and predictability.
Another defining goal of synthetic biology is building up new chemical life, by which we mean synthetic genetic and functional polymer systems that can replicate and evolve in vivo. Towards this goal, we view an orthogonal central dogma as one that can be gradually engineered with unnatural building blocks and associated components, using the host cell as a scaffold. The basic science implications of the semi-synthetic living systems are considerable, as these organisms would provide points of comparison to natural life forms, which all descend from a common ancestor using the same set of four nucleotides and twenty canonical amino acids. The practical applications could be equally extensive, and include expanded genetic information encoding capacity, unnatural polymer engineering, and biocontainment.
Progress on individual orthogonal systems has already encouraged efforts at their integration. The p1/p2-based orthogonal DNA replication system is already a combined orthogonal replication and transcription system, since p2 encodes a special RNAP that only initiates transcription from special promoters driving genes encoded on p1 and p2; these promoters are not recognized by host transcription systems even if encoded on nuclear plasmids.2 Orthogonal transcription and orthogonal translation have also been combined to create genes that are unreadable by the host, but selectively transcribed and translated by an orthogonal RNAP and ribosome. By making the transcription of orthogonal ribosomal RNA dependent on the orthogonal RNAP, it has been possible to create new types of transcription–translation logic.27 The success of these early efforts combining two of three central dogma processes suggest that the full integration of orthogonal replication, transcription, and translation may indeed be possible.
Going forward, there are four critical challenges that we need to overcome to arrive at an integrated orthogonal central dogma system. The first is to understand whether orthogonal replication systems have the capacity to encode large collections of genes representing all the pieces of orthogonal transcription and translation in addition to synthetic genes that will be read by the orthogonal central dogma. With the p1/p2-based orthogonal DNA replication system, this may be possible through additional engineering. Although the natural size of p1 is only 8.9 kilobases, we have found that we can expand p1 to at least 16 kilobases. Moreover, there are multiple copies of p1 and p2, each copy of which could contain different genes. In addition, it is likely that p1 replication is mutually orthogonal to p2 replication, and not just orthogonal to host genome replication. This mutual orthogonality is due to specific recognition of the different replication origins distinguishing p1 from p2, suggesting the possibility of creating additional versions of p1 and p2 that constitute mutually orthogonal sets.
Second, we need a better understanding of the minimal set of components necessary for protein translation. Despite rapid progress, orthogonal translation systems are far from fully orthogonal. They still share many translation factors with the host, including initiation factors, elongation factors, release factors, recycling factors, ribosome biogenesis factors, and ribosome modifying enzymes. In addition, orthogonal ribosomes still use more than 50 proteins of the host cell’s ribosome, and are therefore unlikely to be immediately functional in diverse hosts. Nonetheless, it is possible that with a coordinated engineering effort, ribosomal protein operons can be tweaked to function in multiple hosts, as ribosomal proteins across all kingdoms are conserved, with most eukaryotic ribosomal proteins being enlarged by extensions of a bacterial core.28,29 It may also be possible to engineer or evolve synthetic rRNAs that require fewer ribosomal proteins.
Third, it remains to be seen whether aaRS–tRNA engineering can scale. Current orthogonal translation systems use natural synthetases and tRNAs alongside orthogonal ones, whereas a fully orthogonal system would need an entire set of aaRS‒tRNAs. In addition, the tRNAs would need to interact only with the orthogonal ribosome. While strategies for creating mutually orthogonal aaRS–tRNA pairs have been reported,30,31 the scalability of these strategies remain untested, and the tRNAs still utilize host ribosomes.
Fourth, the extent to which biological interactions are orthogonal is finite, so even though we have described component central dogma systems that are orthogonal, it is unclear a priori whether orthogonality in replication, transcription, and translation will scale to the creation of a system that enforces an entire orthogonal central dogma. Host strain adaptation may be required to fully accommodate an integrated orthogonal central dogma once the key components are functionally installed.
One possible approach to circumvent some of these engineering difficulties might be to repurpose intracellular organelles or parasites that have their own replication, transcription, and translation systems. The mitochondrion is a good example, as it uses a dedicated DNA polymerase, RNA polymerase, ribosome and associated translational components to propagate and express a small set of genes that are non-essential in certain hosts and conditions.32 Alluring shortcuts aside, we predict that the approach of systematically encoding components for orthogonal transcription and translation onto existing orthogonal replication systems, testing function along the way will prevail, as both intermediate successes and the ultimate goal will yield new genetic systems for predictable design of genetic programs and the synthesis of new biological function.
Acknowledgements
We thank members of our group and reviewers for valuable comments. C.C.L. acknowledges the Defense Advanced Research Projects Agency (DARPA-14-49-AS-BRICS-FP-017), the National Institutes of Health (1DP2GM119163-01), and the Arnold and Mabel Beckman Foundation. M.C.J. acknowledges the Army Research Office (W911NF-16-1-0372), the National Science Foundation (MCB-1716766), the Air Force Research Laboratory Center of Excellence (FA8650-15-2-5518), the David and Lucile Packard Foundation, and the Camille-Dreyfus Teacher-Scholar Program. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of Air Force Research Laboratory or the U.S. Government. J.W.C. acknowledges the Medical Research Council, UK (MC_U105181009 and MC_UP_A024_1008) and the European Research Council (ERC Advanced Grant (SGCR)). C.A.V. acknowledges the Office of Naval Research Multidisciplinary University Research Initiative (N00014-16-1-2388).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Ravikumar A, Arrieta A, & Liu CC, An orthogonal DNA replication system in yeast. Nat Chem Biol 10, 175–177 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Klassen R, Linear protein-primed replicating plasmids in eukaryotic microbes. Microbial Linear Plasmids (2007). [Google Scholar]
- 3.Kranaster R, & Marx A, Engineered DNA polymerases in biotechnology. Chembiochem 11, 2077–2084 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Zhang S, & Chaput JC, Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat Chem 4, 183–187 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Pinheiro VB, et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser JI, et al. Statistical Analysis of Molecular Signal Recording. PLoS Comput Biol 9, e1003145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruveiller S, Bouzon M, & Mutzel R, Chemical Evolution of a Bacterium’s Genome. Angewandte Chemie Int Ed Engl 50, 7109–7114 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Malyshev DA, et al. A semi-synthetic organism with an expanded genetic alphabet. Nature 509, 385–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowrishankar J, & Harinarayanan R, Why is transcription coupled to translation in bacteria? Mol Microbiol 54, 598–603 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Guillerez J, Lopez PJ, Proux F, Launay H, & Dreyfus M, A mutation in T7 RNA polymerase that facilitates promoter clearance. Proc Natl Acad Sci USA 102, 5958–5963 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonner G, Lafer EM, & Sousa R Characterization of a set of T7 RNA polymerase active site mutants. J Biol Chem 269, 25120–25128 (1994). [PubMed] [Google Scholar]
- 12.Becskei A, & Serrano L, Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Bleris L, et al. Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol Syst Biol 7, 519–519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shis DL, & Bennett MR, Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc Natl Acad Sci USA 110, 5028–5033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaj T, Gersbach CA, & Barbas CF, ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin JW, Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem 83, 379–408 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Wang K, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem 6, 393–403 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maini R, et al. Protein Synthesis with Ribosomes Selected for the Incorporation of β-Amino Acids. Biochemistry 54, 3694–3706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dedkova LM, et al. β-Puromycin selection of modified ribosomes for in vitro incorporation of β-amino acids. Biochemistry 51, 401–415 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Melo Czekster C, Robertson WE, Walker AS, Söll D & Schepartz A In Vivo Biosynthesis of a β-Amino Acid-Containing Protein. J Am Chem Soc 138, 5194–5197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Kim DS, & Jewett MC, Repurposing ribosomes for synthetic biology. Curr Opin Chem Biol 40, 87–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitahara K, & Suzuki T The Ordered Transcription of RNA Domains Is Not Essential for Ribosome Biogenesis in Escherichia coli. Mol Cell 34, 760–766 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Orelle C, et al. Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Fried SD, Schmied WH, Uttamapinant C, & Chin JW, Ribosome Subunit Stapling for Orthogonal Translation in E. coli. Angew Chem Int Ed Engl 54, 12791–12794 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An W, & Chin JW, Synthesis of orthogonal transcription-translation networks. Proc Natl Acad Sci USA 106, 8477–8482 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Neumann H, Slusarczyk AL, & Chin JW, De novo generation of mutually orthogonal aminoacyl-tRNA synthetase/tRNA pairs. J Am Chem Soc 132, 2142–2144 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Chatterjee A, Xiao H, & Schultz PG Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc Natl Acad Sci USA 109, 14841–14846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernardi G Lessons from a small, dispensable genome: the mitochondrial genome of yeast. Gene 354, 189–200 (2005). [DOI] [PubMed] [Google Scholar]