Abstract
Purpose:
To test whether a miRNA panel may serve as an alternative biomarker of FGFR TKI sensitivity in lung cancer.
Methods:
Histologically diverse lung cancer cell lines were submitted to assays for ponatinib and AZD4547 sensitivity. miRNAs, FGFR1 mRNA, gene copy number and protein expression were detected using the method of RT-PCR, FISH and immunoblotting in 34 lung cancer cell lines respectively.
Results:
Among 34 cell lines, 14 exhibited ponatinib sensitivity and 20 exhibited AZD4547 sensitivity (IC50 values < 100 nmol/L). 39 out of 377 miRNAs set were initially identified from the 4 paired ponatinib sensitive or insensitive cell lines to have at least an 8-fold differential expression, and then were detected in the whole of 34 cell lines. A predictive panel of three miRNAs (let-7c, miRNA155 and miRNA218) was developed that had an area under the curve (AUC) of 0.886 with sensitivity of 71.4% and specificity of 77.3% to predict response to ponatinib. The miRNA panel performed similar to FGFR1 protein expression (AUC = 0.864) and mRNA expression (AUC = 0.939), and superior to FGFR1 amplification (AUC = 0.696). Furthermore, we validated this panel using data for sensitivity to AZD4547 in the cell line cohort with an AUC of 0.931 with sensitivity of 73.3 % and specificity of 76.2% respectively.
Conclusions:
The developed miRNA panel (let-7c, miRNA155 and miRNA218) may be useful in predicting response to FGFR TKIs, either ponatinib or AZD4547 in lung cancer cell lines, and warrants further validation in the clinical setting.
Keywords: miRNA, FGFR inhibitor, lung cancer, cell lines
Micro-abstract:
We investigate whether miRNAs may serve as alternative biomarker of FGFR TKI sensitivity in lung cancer cell lines, and found a miRNA panel (let-7c, miRNA155 and miRNA218) could be useful in predicting response to FGFR TKIs, either ponatinib or AZD4547.
INTRODUCTION
The fibroblast growth factor receptor (FGFR) pathway is an important oncogenic driver in malignant cancer (1). It controls cellular processes such as cell proliferation, differentiation, migration, cycle progression, metabolism, and survival. In NSCLC, the most frequent alteration of the FGFR pathway is represented by FGFR1 amplification, which is reported as occurring in up to 20% of squamous NSCLC; other, less frequent alterations include point mutations or translocations of the genes encoding for FGFR1–4 (2).
Currently, several FGFR inhibitors are being investigated in phase 1–3 clinical trial in solid tumors with FGFR amplification or protein over-expression (2), such as nintedanib (Boehringer Ingelheim; Ingelheim am Rhein, Germany) (3), Ponatinib (Ariad Pharmaceuticals; Cambridge, MA) (4), AZD4547 (AstraZeneca; London, UK) (5), and BJG398(Novartis, Switzerland) (6). In these studies, preliminary results showed that only a subset of patients with FGFR amplification or protein over-expression responded to the FGFR TKIs AZD4547 and BJG003(5–6). The response rates did not reach those observed for other lung cancer driver mutation genes such as mutant EGFR or ALK/ROS1 fusion, suggesting that the biomarkers used for enrolling into the FGFR TKI trials were inaccurate (7). In a set of 58 lung cancer cell lines, sensitivity to ponatinib was correlated with FGFR1 amplification, mRNA and protein expression, as well as mRNA expression of FGF2 and FGF9 (8). This study reported better correlation of FGFR1 TKI sensitivity with FGFR1 mRNA or protein expression as compared to FGFR1 amplification. These data clearly identify the need for further investigation to find additional biomarkers which may be better able to predict response to FGFR inhibitors in the clinical setting.
MicroRNAs (miRNAs) are small, non-coding, stable sequences of RNA with regulatory functions exerted through inhibition of crucial mRNA (9). Recent studies demonstrated that pathologic conditions, such as solid tumors, are associated with specific intracellular miRNA patterns and are also able to affect circulating miRNA. Based on this assumption, several studies have identified specific miRNAs or groups of miRNAs (miRNA signatures) with a potential diagnostic or prognostic role in solid tumors (10–11). Some miRNA, such as miR-34bc, are currently considered promising predictors of poor outcome for early-stage lung cancer, apparently due to a correlation between their target genes inactivation and an aggressive phenotype (12). Another study suggested the existence of a circulating miRNA signature able to detect lung cancer (13). Furthermore, our previous study also found miRNA signatures were also reported as able to predict the sensitivity of lung cancer to EGFR-TKIs (14–15). In this present study, we performed a comprehensive analysis of miRNAs in a panel of human lung cancer cell lines that were previously characterized for sensitivity to two FGFR1 TKIs (8). We developed a 3-miRNA panel that accurately predicts the sensitivity to ponatinib in 34 cell lines and the chemically-distinct TKI, AZD4547.
METHODS
Cell culture
All cell lines were cultured in RPMI-1640 growth medium supplemented with 10% FBS at 37°C in a humidified 5% CO2 incubator. The following cell lines were available in our laboratories and submitted to DNA fingerprint analysis for authentication: H1703, HCC95, NE-18, DMS-114, SKMES-1, H460, SW1573, H520, H661, H125, HCC44, H1299, H157, Colo699, H1581, HCC15, H2126, H1869, H1435, and H441. The remaining 14 cell lines were obtained directly from the University of Colorado Cancer Center Tissue Culture Core (Aurora, CO). The core laboratory routinely performs DNA fingerprint analyses on all banked cell lines to ensure their authenticity.
IC50 analysis to ponatinib and AZD4547 in cell lines
Sensitivity to ponatinib was defined as an IC50≤ 100 nM/L in a cohort of cell lines that were previously characterized for FGF ligand and FGFR receptor-mediated autocrine signaling status (8,16). The IC50 for AZD4547 was also determined in the lung cancer cell lines, including 13 ponatinib-sensitive cell lines and 44 ponatinib-resistant cell lines by using the IC50 cutoff value of 100 nM/L. Cell line information is presented in Table S1.
RT-qPCR for miRNA analysis
We used a comprehensive, commonly employed commercial panel (Life Technologies, TaqMan® Array Human MicroRNA A Card v2.0) to perform the miRNA expression level detection; this panel utilizes a 384-well microfluidic card, enabling to test 377 miRNA plus three provided endogenous controls and one non-human negative control.
Initially, 4 lung cancer cell lines determined to be sensitive to ponatinib (COLO699, H1581, SW1573, and H520) were selected as well as 4 insensitive cell lines (H125, NE-18, H2126, and SK-MES-1) and were analyzed for miRNA expression. RNA was extracted from treatment-naïve cell cultures, and real-time quantitative polymerase chain reaction (RT-qPCR) was achieved using a 7900HT Fast Real-Time cycler (Applied Biosystems). Cycle thresholds (ΔCt) were normalized using U6 small nuclear RNA, as per the manufacturer’s protocol. Each cell line was analyzed in triplicate and averaged data generated. In order to determine the ΔCt, the threshold was set at 0.2, and baseline data collected at cycles 3–15. Based on the results of the RT-qPCR, a statistical analysis of microarrays (SAM) plot analysis was performed (Stanford University; http://statweb.stanford.edu/~tibs/SAM/) which identified the 39 most differentially expressed miRNAs from the original set of 377. Here miRNA which exhibited an 8-fold expression difference between sensitive and insensitive cell lines were included. Following selection, the panel of 39 miRNAs were then assayed across the 34 lung cancer cell lines (8).
FGFR1 protein, mRNA expression and gene copy number detection
Selected lung cancer cell lines were submitted to the University of Colorado Cancer Center Molecular Pathology shared resource for evaluation of FGFR1 gene copy number by FISH analysis. FGFR1 protein and mRNA levels were measured by immunoblotting with an antibody against the carboxyl-terminus of FGFR1 and quantitative PCR with primers annealing to sequences within the invariant second immunoglobulin domain. The methods for FISH, protein immunoblotting and QPCR are described previously (8).
Transfection of let-7c inhibitor
Based on the observed data, the interaction between the most relevant miRNA, let-7c and the status of FGFR1 mRNA expression were further explored through an additional experiment. We included 11 ponatinib-sensitive cell lines, treated with let-7c inhibitor and its negative control oligonucleotide (Invitrogen; Carlsbad, CA). Cells were added into 6-well plates (2×105 cells/well) and transfected with miRNA inhibitor using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer’s protocol. After 72 hours, cells were harvested and tested for alternations in the levels of specific miRNA and FGFR1 mRNA by qRT-PCR.
Statistical analysis
We assessed the expression of miRNAs for classification of lung cancer cells as sensitive or insensitive to ponatinib using logistic regression by computing the receiver operating characteristic (ROC) curve and used the area under the ROC curve (AUC) as an accuracy index. Among potential biomarkers, miRNAs characterized by a statistical p< 0.1 in the univariate analysis were selected to enter into multivariable logistic regression, which were selected in a combination of biomarkers potentially able to predict sensitivity to ponatinib. We further validated the biomarker panel for a distinct FGFR inhibitor, AZD4547, in the same cell line cohort. All P values were two-sided, with values < 0.05 considered statistically significant. SAS 9.3 (SAS Institute Inc., Cary, NC, USA) and SPSS (Version 21.0, IBM, Chicago, IL) were used for the statistical analyses.
RESULTS
Cell lines and their sensitivity to ponatinib and AZD4547
The IC50 for ponatinib and AZD4547 are listed in Table S1. We used our previous criteria to distinguish ponatinib sensitive cell lines (IC50≤ 100 nM) from resistant cell lines and identified 14 ponatinib-sensitive cell lines and 20 ponatinib-resistant cell lines [11]. As for AZD4547, the IC50 was not available for the H1048 cell line. Thus, when we use the IC50 cutoff of 100 nM, we identified 13 AZD4547-sensitive cell lines and 20 AZD4547 insensitive cell lines. Among the 33 cell lines, there were 4 cell lines, H211, H1703, H1993, H125 that showed an opposite sensitivity to ponatinib and AZD4547.
Developing a miRNA panel to predict sensitivity to ponatinib
The RT-qPCR analysis enabled us to identify the median ΔCt for each miRNA and to compare them between 4 paired ponatinib-sensitive and ponatinib-resistant cell lines. Notably, a lower ΔCt indicates a higher concentration of each specific miRNA, as the amplification takes place earlier in presence of increased nucleic acid. Full data regarding the initial 377 miRNAs set, including ΔCt for sensitive and resistant cell lines are reported in table S2 and table S3 respectively. SAM analysis identified 39 miRNAs that have more than an 8-fold difference in expression which were used in subsequent investigations in the 34 cell lines (see Table 1).
Table 1.
Logistic regression analysis of sensitivity to ponatinib according to the 39 miRNAs selected through SAM analysis.
| N# | VARIABLE | ESTIMATE | P |
|---|---|---|---|
| 1 | let7c | 7.073 | 0.008 |
| 2 | miR22 | 0.836 | 0.360 |
| 3 | miR23b | 0.804 | 0.370 |
| 4 | miR34c | 2.033 | 0.154 |
| 5 | miR99a | 0.172 | 0.678 |
| 6 | miR100 | 0.451 | 0.502 |
| 7 | miR105 | 0.543 | 0.461 |
| 8 | miR127 | 0.158 | 0.691 |
| 9 | miR134 | 1.188 | 0.276 |
| 10 | miR141 | 2.547 | 0.110 |
| 11 | miR146a | 1.560 | 0.212 |
| 12 | miR155 | 3.602 | 0.058 |
| 13 | miR200a | 1.245 | 0.264 |
| 14 | miR200b | 2.769 | 0.096 |
| 15 | miR200c | 3.576 | 0.059 |
| 16 | miR203 | 1.778 | 0.182 |
| 17 | miR204 | 0.396 | 0.529 |
| 18 | miR205 | 1.188 | 0.276 |
| 19 | miR218 | 4.060 | 0.044 |
| 20 | miR221 | 0.984 | 0.321 |
| 21 | miR224 | 1.513 | 0.219 |
| 22 | miR335 | 0.869 | 0.351 |
| 23 | miR3375p | 0.844 | 0.358 |
| 24 | miR3383p | 4.514 | 0.034 |
| 25 | miR376a | 0.438 | 0.508 |
| 26 | miR376c | 1.153 | 0.283 |
| 27 | miR379 | 3.169 | 0.075 |
| 28 | miR382 | 1.708 | 0.191 |
| 29 | miR411 | 2.094 | 0.148 |
| 30 | miR429 | 2.840 | 0.092 |
| 31 | miR452 | 2.434 | 0.119 |
| 32 | miR487b | 1.146 | 0.284 |
| 33 | miR492 | 0.054 | 0.817 |
| 34 | miR495 | 1.624 | 0.203 |
| 35 | miR539 | 1.325 | 0.250 |
| 36 | miR5425p | 0.004 | 0.949 |
| 37 | miR655 | 0.431 | 0.511 |
| 38 | miR8863p | 0.674 | 0.412 |
| 39 | miR8865p | 1.148 | 0.284 |
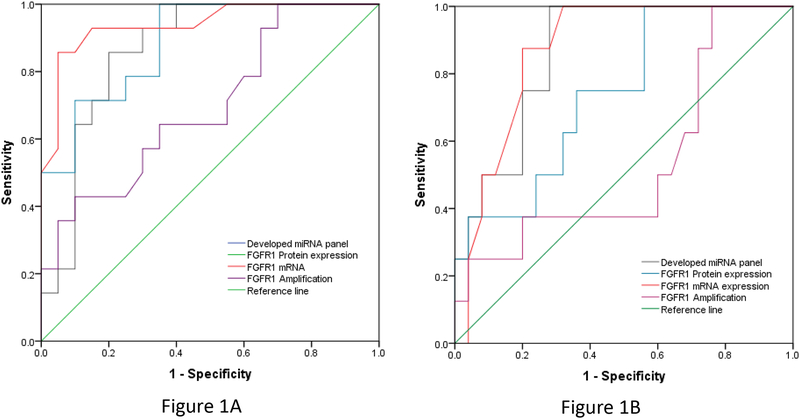
Univariate analysis identified 8 distinct miRNAs whose concentration was significantly or marginally significantly associated with sensitivity to ponatinib, including Let-7c, miR155, miR218, miR3383p, miR200b, miR379, miR200c, miR429(see Table 1). Due to the limited cell lines included in this study, we performed the ROC curve analysis with the above 8 individual miRNAs and found that miR200c or miR429 alone was not significantly predictive of the sensitivity to ponatinib in cell lines. Finally, Let-7c, miR155, miR218, miR3383p, miR200c, miR379 and miR200b were evaluated by multivariate analysis and allowed the identification of a miRNA panel associated with sensitivity to ponatinib, including increasing let-7c, increasing miR-218 and decreasing miR-155. This 3-miRNA panel was associated with a model AUC= 0.886 with a sensitivity of 71.4% and specificity of 77.3% to predict the response to ponatinib. The ROCs are depicted in Figure 1.
Figure 1.
Receiver operating curves (ROCs) for the developed miRNA (let-7c, miR-155, and miR-218) together with FGFR1 mRNA, protein expression and amplification to predict the sensitivity of ponatinib (Figure 1A, on the left) and AZD4547 (Figure 1B, on the right) by using the IC50 cutoff values 100 nmol/l.
Comparing the developed miRNA panel with other biomarkers including FGFR mRNA, protein expression and amplification
In a previous study, we found that FGFR mRNA or FGFR protein expression, but not FGFR amplification were associated with sensitivity to ponatinib in lung cancer cell lines (8). These biomarkers were studied in all 34 cell lines deployed in the present study. Similar to our previous reports, we found that FGFR1 mRNA or FGFR1 protein expression could predict sensitivity to ponatinib with an AUC of 0.939 and 0.864 respectively. We determined that FGFR amplification predicts sensitivity to ponatinib with an AUC of 0.696. The 3-miRNA panel developed in this study has an AUC of 0.886, which is similar to the sensitive biomarkers of FGFR1 mRNA or FGFR1 protein expression in the study (see Figure 1).
Validating the 3-miRNA panel and evaluating other biomarkers for sensitivity to AZD4547 in the cell line cohort
Data for sensitivity to AZD4547 were established for 33 out of 34 study cell lines. Here we chose to evaluate the same concentration for AZD4547 that was used for ponatinib (100 nM), resulting in the identification of 13 sensitive and 20 resistant cell lines for the validation cohort. The 3-miRNA panel developed for ponatinib predicted cell line sensitivity to AZD4547 with an AUC of 0.931 and a sensitivity of 73.3 % and specificity of 76.2%. We further investigated the biomarkers of FGFR1 mRNA, protein expression and amplification in the setting of the 34 cell lines cohort. Analysis revealed that FGFR1 mRNA or FGFR1 protein expression could predict the sensitivity of AZD4547 with an AUC of 0.871 and 0.735 respectively, while FGFR amplification was less able to predict the sensitivity of ponatinib with a AUC of 0.585 (see Figure 1).
Comparing the distinguished yield of the miRNA panel using different IC50 cutoff
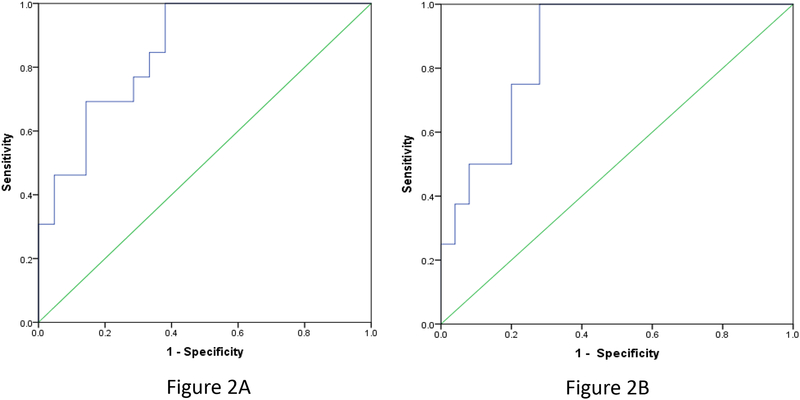
We further investigated whether the miRNA panel exhibits a consistent distinguished yield by using a different IC50 cutoff. When 50 nM was used as the IC50 cutoff value for both ponatinib and AZD4547, 13 ponatinib sensitive and 21 resistant cell lines were identified, while 8 sensitive and 25 resistant cell lines were identified for AZD4547. This 3-miRNA panel was associated with a model AUC= 0.853 to predict the response to ponatinib. The 3-miRNA panel developed for ponatinib predicted cell line sensitivity to AZD4547 with an AUC of 0.865. These results using the cutoff value of 100 nM were consistent to those for both ponatinib and AZD4547 (see Figure 2).
Figure 2.
Receiver operating curves (ROCs) for the developed miRNA (let-7c, miR-155, and miR-218 to predict the sensitivity of ponatinib (Figure 2A, on the left, AUC=0.853) and AZD4547 (Figure 2B, on the right, AUC=0.865) by using the IC50 cutoff values 50 nmol/l.
The relationship of let-7c with FGFR1 mRNA expression
Since let-7c was the most robust miRNA in the univariate analysis (in terms of p value), it was considered the best candidate for studying interactions with FGFR1 mRNA. Experiments employed transient transfection with a let-7c silencing RNA (Life Technologies) to explore effects of let-7c levels on FGFR1 mRNA expression. In these experiments, the high mobility group AT-hook 2 (HMGA2) gene served as a surrogate measurable target of repression exerted by let-7c (17–18). After 72 hours, 6 out of 11 cell lines showed an increase in HMGA2 levels (reflecting a decrease in let-7c levels); of these 6 cell lines, 5 also showed a reduction in FGFR1 mRNA levels from baseline (Table 2).
Table 2.
modifications in FGFR1 mRNA after silencing let-7c in ponatinib-sensitive cell lines.
| CELL LINE | Base-line LET-7C | Base-line FGFR1 mRNA (qPCR) | HGMA2 | FGFR1 mRNA | ||||
|---|---|---|---|---|---|---|---|---|
| Change trend | Response | % change | Change trend | Response | % change | |||
| H1581 | 6.7 | 0.6 | None | N/A | N/A | None | N/A | N/A |
| H226 | 12.8 | 0.26 | Increase | sat | 395.00% | None | N/A | N/A |
| H2066 | 11.6 | 0.49 | None | N/A | N/A | None | N/A | N/A |
| Colo699 | 12.1 | 1 | Increase | dose | 575.00% | Decrease | dose | 36.10% |
| H1563 | 11 | 0.28 | Increase | sat | 542.00% | Decrease | dose | 40.80% |
| H661 | 8.3 | 0.13 | None | N/A | N/A | None | N/A | N/A |
| H522 | 7.9 | 1.1 | Increase | sat | 436.00% | Decrease | dose | 50.10% |
| H1734 | 8.5 | 0.26 | None | N/A | N/A | None | N/A | N/A |
| SW1573 | 13.7 | 0.24 | Increase | sat | 182.00% | Decrease | dose | 33.40% |
| DMS-114 | 10.8 | 0.4 | None | N/A | N/A | None | N/A | N/A |
| H520 | 10 | 0.49 | Increase | dose | 170.00% | Decrease | dose | 29.40% |
DISCUSSION
Precision therapy, guided by biomarkers of response, has dramatically improved the prognosis of patients with advanced lung cancer (19). Examples include mutant EGFR that can be effectively inhibited by EGFR TKIs and deficient mismatch repair (MMR), high microsatellite instability (MSI), are susceptible to anti-PD-1 immunotherapy respectively (20–22). In this study, we investigated the role of miRNAs as a biomarker to predict sensitivity to FGFR inhibitors. Analysis revealed differences in the miRNA expression between cell lines determined to be sensitive and resistant to ponatinib with identification of a predictive miRNA panel including let-7c, miRNA155 and miRNA218. Analysis of this panel identified an AUC of 0.886 to predict sensitivity to ponatinib, which is as good as other predictive biomarkers including FGFR1 mRNA and protein expression as previously reported. Furthermore, the predictive role of this miRNA panel was validated when comparing sensitivity of AZD4547 in the cell line cohort. Moreover, we found that the mechanism of let-7c to predict the sensitivity of ponatinib may involve regulating the mRNA expression of FGFR1.
After the successful development of molecular targeted drugs in lung adenocarcinoma, substantial efforts have been made to provide similar targeted drugs in lung squamous carcinoma (23). Since FGFR gene alteration is the most frequent occurrence in squamous carcinoma, targeted therapies for FGFR including ponatinib, AZD4547 and BJG003 are being evaluated (4–6). Preliminary clinical data found that only a small subset of enrolled patients respond to these FGFR1 TKIs. One potential explanation may be due to the improper use of FGFR amplification as a patient selection biomarker (7). As identified in our previous study, FGFR1 mRNA or protein expression, but not gene copy number better predicts FGFR TKI sensitivity across all lung cancer cell lines studies (8). This previous data suggested the need for better biomarkers to predict tumor sensitivity to targeted drugs.
Currently, efforts to identify patients who are likely to experience anti-cancer treatment failure are ongoing, and evaluations of miRNA dysregulation to support this endeavor have been reported (24–25). Lim EL et al further used RNA sequencing to comprehensively analysis the miRNA difference expression between primary and refractory pediatric AML samples and found three candidate miRNAs, indicating that they may be associated with treatment resistance (26). In this study, we firstly developed a miRNA panel to predict sensitivity to FGFR inhibitors through comprehensive analysis of miRNA expression in 34 thoracic cancer cell lines, which showed consistent results both in ponatinib and AZD4547 cell line cohorts. While this work was performed exclusively in cell lines, further studies on tumor tissue specimens from patients must be performed to validate the potential role for this miRNA panel. However, liquid biopsy showed obvious advantage comparing with tumor biopsy in the areas of efficacy surveillance and relapse monitoring because liquid biopsy is nearly non-invasive (27). Thus, our miRNA panel might show superiority than FGFR1 mRNA or protein expression if this miRNA panel in circulating miRNA was identify as a reliable predictive factor for screening patients for FGFR1 inhibitor, which is also our next step work in the near future. Besides that, 34 cell lines included into this study contained different histological subtypes such as adenocarcinoma, squamous, large cell and small cell cell lines without known oncogenic mutations, which might not represent the whole lung cancer population. However, since FGFR1 inhibitor are rarely effective in patients with EGFR/ALK/ROS1 mutation, investigation the biomarker for FGFR1 inhibitor in oncogenic driver pan-negative cell lines will be helpful to identify the potential one for clinical implication.
Additionally, since let-7c was the most robust miRNA in the univariate analysis to predict the sensitivity to ponatinib, we further investigated the correlation between let-7c and FGFR1 mRNA. We found that let-7c silencing was significantly associated with decreased expression of FGFR1, suggesting that let-7c predicts, but also participates in regulation of FGFR1 mRNA levels and thereby sensitivity to ponatinib. It has been reported that Let-7 has been demonstrated to be a direct regulator of RAS (28) and high mobility group A2 (HMGA2) expression (29) in human cells through binding sequences in their 3’UTRs. However, the detailed mechanism of let-7c and correlated changes in FGFR1 mRNA is still unknown and warrants further investigation.
Conclusion
This study comprehensively evaluated the predictive role of miRNA to FGFR inhibitors in 34 cell lines and developed a miRNA panel (let-7c, miRNA155 and miRNA218), which was validated with sensitivity to AZD4547 in the cell line cohort. Similar to FGFR1 mRNA or protein expression, the miRNA panel predicted the sensitivity of ponatinib and AZD4547. Since miRNA panel could also be detected by a non-invasive liquid biopsy, we will further explore its role for predicting sensitivity to FGFR-TKIs in the clinical setting.
Supplementary Material
Supplementary table S1. Lung cancer cell lines and their IC50 values to ponatinib or AZD4547. ADC: adenocarcinoma; Meso: malignant pleural mesothelioma; NOS: not otherwise specified; NSCLC: non-small cell lung cancer; SCC: squamous cell carcinoma; SCLC: small cell lung cancer. (*): cell lines included in the starting analysis of 377 miRNAs.
Supplementary table S2. Preliminary RT-qPCR panel of the initial 377 miRNAs for ponatinib-sensitive cell lines. The ΔCt for each cell line was conducted in three replicates and the average value for each cell line was accounted for the calculation of median ΔCt for each miRNA. The miRNAs reported in green are the ones selected through SAM analysis as having significantly lower expression in sensitive cell lines, while the miRNAs reported in red are the ones selected through SAM analysis as having significantly higher expression in sensitive cell lines.
Supplementary table S3. Preliminary RT-qPCR panel of the initial 377 miRNAs for ponatinib-resistant cell lines. The ΔCt for each cell line was conducted in three replicates and the average value for each cell line was accounted for the calculation of median ΔCt for each miRNA. The miRNAs reported in green are the ones selected through SAM analysis as having significantly higher expression in resistant cell lines, while the miRNAs reported in red are the ones selected through SAM analysis as having significantly lower expression in resistant cell lines.
Clinical Practice Points.
The improper use of FGFR amplification as a patient selection biomarker might be the potential explanation that preliminary clinical data found that only a small subset of enrolled patients respond to these FGFR1 TKIs.
Our previous study, FGFR1 mRNA or protein expression, but not gene copy number better predicts FGFR TKI sensitivity across all lung cancer cell lines studies.
This study found that a miRNA panel (let-7c, miRNA155 and miRNA218) could predict the sensitivity of FGFR-TKIs, not only ponatinib but also AZD4547.
Acknowledgments
The authors acknowledge technical contributions of Kim Ellison (Hirsch lab) for FGFR protein staining and preparation of slides. The studies were supported by the NIH (CA127105, Lung SPORE P50 CA58187, UC Cancer Center Support Grant P30 CA046934), IASLC Fellowship awards (S. Ren and C. Genova) and a research contract from ARIAD Pharmaceuticals to L.E. Heasley.
Footnotes
Disclosure
None
References
- 1.Jiang T, Gao G, Fan G, et al. FGFR1 amplification in lung squamous cell carcinoma: a systematic review with meta-analysis. Lung Cancer. 2015. January;87(1):1–7. [DOI] [PubMed] [Google Scholar]
- 2.Desai A, Adjei AA .FGFR Signaling as a Target for Lung Cancer Therapy. J Thorac Oncol. 2016. January;11(1):9–20. [DOI] [PubMed] [Google Scholar]
- 3.Hibi M, Kaneda H, Tanizaki J, et al. FGFR gene alterations in lung squamous cell carcinoma are potential targets for the multikinase inhibitor nintedanib. Cancer Sci. 2016. November;107(11):1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012. March;11(3):690–9. [DOI] [PubMed] [Google Scholar]
- 5.Paik PK, Shen R, Berger MF, et al. A Phase Ib Open-Label Multicenter Study of AZD4547 in Patients with Advanced Squamous Cell Lung Cancers. Clin Cancer Res. 2017;23(18):5366–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nogova L, Sequist LV, Perez Garcia JM, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1–3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol. 2017. January 10;35(2):157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paik PK, Rudin CM. Missing the mark in FGFR1-amplified squamous cell cancer of the lung.Cancer. 2016;122(19):2938–40. [DOI] [PubMed] [Google Scholar]
- 8.Wynes MW, Hinz TK1, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20(12):3299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legras A, Pécuchet N, Imbeaud S, et al. Epithelial-to-Mesenchymal Transition and MicroRNAs in Lung Cancer. Cancers (Basel). 2017;9(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truini A, Coco S, Alama A, et al. Role of microRNAs in malignant mesothelioma. Cell Mol Life Sci. 2014;71(15):2865–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeBlanc VC, Morin P. Exploring miRNA-Associated Signatures with Diagnostic Relevance in Glioblastoma Multiforme and Breast Cancer Patients. J Clin Med. 2015;4(8):1612–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadal E, Chen G, Gallegos M, et al. Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res. 2013;19(24):6842–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadal E, Truini A, Nakata A, et al. A Novel Serum 4-microRNA Signature for Lung Cancer Detection. Sci Rep. 2015;5:12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryant JL, Britson J, Balko JM, et al. A microRNA gene expression signature predicts response to erlotinib in epithelial cancer cell lines and targets EMT. Br J Cancer. 2012;106(1):148–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Li X, Ren S, et al. miR-200c overexpression is associated with better efficacy of EGFR-TKIs in non-small cell lung cancer patients with EGFR wild-type. Oncotarget. 2014;5(17):7902–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marek L, Ware KE, Fritzsche A, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009. January;75(1):196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motoyama K, Inoue H, Nakamura Y, et al. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res. 2008;14(8):2334–40 [DOI] [PubMed] [Google Scholar]
- 18.Peng Y, Laser J, Shi G, et al. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol Cancer Res. 2008;6(4):663–73. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L.Lung cancer: current therapies and new targeted treatments. Lancet. 2017. January 21;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011. August;12(8):735–42. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 21.Gong J, Wang C, Lee PP, et al. Response to PD-1 Blockade in Microsatellite Stable Metastatic Colorectal Cancer Harboring a POLE Mutation. J Natl Compr Canc Netw. 2017. February;15(2):142–147. [DOI] [PubMed] [Google Scholar]
- 22.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–1833. [DOI] [PubMed] [Google Scholar]
- 23.Heist RS, Sequist LV, Engelman JA. Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol. 2012;7(5):924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment.J Clin Med. 2016. March 9;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gasparini P, Cascione L, Landi L, et al. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS-driven lung cancers.Proc Natl Acad Sci U S A. 2015. December 1;112(48):14924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim EL, Trinh DL, Ries RE, et al. MicroRNA Expression-Based Model Indicates Event-Free Survival in Pediatric Acute Myeloid Leukemia. J Clin Oncol. 2017. October 25:JCO2017747451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2016. October 1;34(28):3375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005. March 11;120(5):635–47. [DOI] [PubMed] [Google Scholar]
- 29.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007. March 16;315(5818):1576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table S1. Lung cancer cell lines and their IC50 values to ponatinib or AZD4547. ADC: adenocarcinoma; Meso: malignant pleural mesothelioma; NOS: not otherwise specified; NSCLC: non-small cell lung cancer; SCC: squamous cell carcinoma; SCLC: small cell lung cancer. (*): cell lines included in the starting analysis of 377 miRNAs.
Supplementary table S2. Preliminary RT-qPCR panel of the initial 377 miRNAs for ponatinib-sensitive cell lines. The ΔCt for each cell line was conducted in three replicates and the average value for each cell line was accounted for the calculation of median ΔCt for each miRNA. The miRNAs reported in green are the ones selected through SAM analysis as having significantly lower expression in sensitive cell lines, while the miRNAs reported in red are the ones selected through SAM analysis as having significantly higher expression in sensitive cell lines.
Supplementary table S3. Preliminary RT-qPCR panel of the initial 377 miRNAs for ponatinib-resistant cell lines. The ΔCt for each cell line was conducted in three replicates and the average value for each cell line was accounted for the calculation of median ΔCt for each miRNA. The miRNAs reported in green are the ones selected through SAM analysis as having significantly higher expression in resistant cell lines, while the miRNAs reported in red are the ones selected through SAM analysis as having significantly lower expression in resistant cell lines.