Abstract
To better understand the real-world effects of pharmacogenomic (PGx) alerts, this study aimed to characterize alert design within the eMERGE Network, and to establish a method for sharing PGx alert response data for aggregate analysis. Seven eMERGE sites submitted design details and established an alert logging data dictionary. Six sites participated in a pilot study, sharing alert response data from their electronic health record systems. PGx alert design varied, with some consensus around the use of active, post-test alerts to convey Clinical Pharmacogenetics Implementation Consortium recommendations. Sites successfully shared response data, with wide variation in acceptance and follow rates. Results reflect the lack of standardization in PGx alert design. Standards and/or larger studies will be necessary to fully understand PGx impact. This study demonstrated a method for sharing PGx alert response data and established that variation in system design is a significant barrier for multi-site analyses.
Keywords: pharmacogenomics, pharmacogenetics, clinical decision support, precision medicine, process outcomes
BACKGROUND AND SIGNIFICANCE
Previous literature identified clinical decision support (CDS) tools integrated into the electronic health record (EHR) as a critical vector for disseminating pharmacogenomic (PGx) knowledge and improving drug prescribing accuracy.1–6 A number of organizations have published on their experiences implementing PGx CDS tools and have often taken very different design approaches.7–15 There has been little work examining how live PGx CDS systems affect physician experience and behavior, and those studies that do evaluate PGx CDS are generally limited to a single institution.16–18
Clinical genomics is difficult to study at a single institution, in part due to limited sample sizes. This is one reason a number of institutions have formed multi-center collaborations to integrate genetic and genomic knowledge into clinical care, including the Electronic Medical Records and Genomics (eMERGE) Network19–21 and the Clinical Sequencing Exploratory Research (CSER) Consortium.22 Prior work from the eMERGE Network demonstrated successful PGx CDS implementations, with ten sites successfully implementing at least one rule in a clinical environment.15 However, technical matters are only one aspect of successful IT implementation—the solution must also be effective in addressing the intended issue. A first step towards determining whether PGx CDS will be effective in improving prescribing decisions is to examine process outcome measures, such as physician uptake and response.
To-date, multi-site analysis of PGx CDS process outcomes has not been well documented in the literature. Multi-site data aggregation may enable studies that would not be possible at a single site, but it remains unclear whether such aggregated analyses are feasible, due to technical and design differences. Current EHR systems may make it possible to perform such studies through the use of logging and auditing data.
OBJECTIVE
This study aimed to demonstrate the feasibility of multi-site PGx CDS process outcome analysis across the eMERGE Network. The first objective was to understand and characterize PGx CDS design differences that could affect data aggregation. The second was to develop, and perform a pilot evaluation of, a common data dictionary that would enable multi-site data collection of physician response to PGx CDS alerts.
MATERIALS AND METHODS
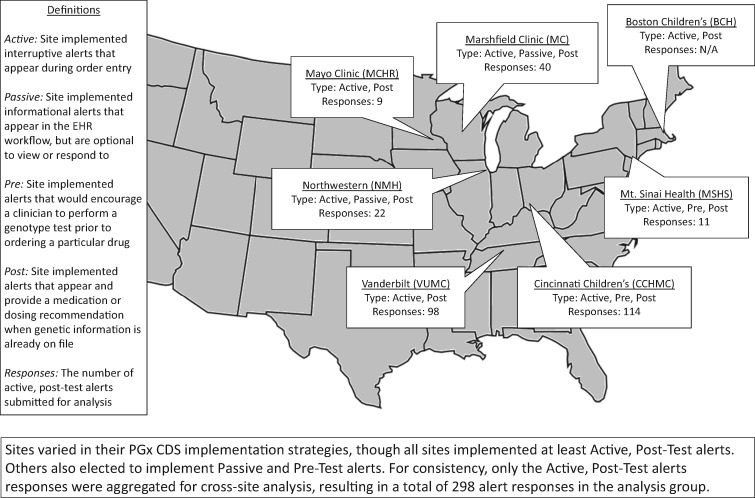
The study population included seven sites from the eMERGE-PGx workgroup. Major site characteristics are summarized in Figure 1 with additional details in Supplementary Tables SA and SB. Due to variation in EHR and CDS infrastructure,15 it was necessary to establish standards for data sharing. Over the course of regular workgroup calls, the group reached consensus on a standardized data dictionary. This included data fields that described alert scenarios implemented at each site, as well as data that described individual instances in which alerts fired in the EHR during clinical encounters. Each site queried its local database for the requested fields, formatted and de-identified data to the agreed specifications, and then submitted it to the first author for aggregation and analysis. Data were divided into three primary categories (the complete data dictionary is described in Supplementary Table SC):
Figure 1.
Site characteristics.
Alert Scenario: Describes the structure and design of a particular alert, including a name, description, and the recommendation the alert provides.
Alert Instance: Describes a specific instance of an Alert Scenario, including a scenario ID, the time the alert was seen by a provider, how the provider responded, and what medications were ordered after it was viewed.
Providers: Describes the provider(s) who interacted with an Alert Instance, including title and specialty.
Objective 1: analysis of PGx CDS implementation characteristics
TMH and JP conducted informational discussions with subject matter experts at each site to determine general themes of PGx CDS design within eMERGE. These discussions informed development of a data collection tool addressing implementation strategies. In addition to alert characteristics, we also collected operational characteristics. One representative from each site submitted responses via e-mail. Additional implementation characteristics were assessed through review of the submitted Alert Scenario data.
Alert characteristic analysis focused on the drug–gene interactions (DGI) for which alerts were defined, use of active or passive alerts, use of pre-test or post-test alerts, use of patient genotype or phenotype as CDS input, type of recommendation, and available alert actions. Active alerts were defined as interruptive alerts that appeared during order entry. Detailed definitions of these terms can be found in Figure 1 and Supplementary Table SA.
Objective 2: cross-site data collection and pilot outcomes analysis
Alert data were collected via the Alert Instance fields in the submitted datasets. These data were combined with the Alert Scenario and Providers data to analyze clinician response. We determined clinician alert response rate by site and by DGI. Clinician response to the alert consisted of the alert response and clinical response (see Table 2 for detailed definitions of these terms). While alert response values could be mapped directly to EHR logging data, local workflow variation required each site to report clinical responses based on local definitions. Because only two sites implemented passive alerts or pre-test alerts, quantitative analysis was limited to active, post-test alerts that contained a prescribing recommendation.
Table 2.
Pilot study – provider responses by site and by DGI
| Alert Response |
Clinical Response |
|||||||
|---|---|---|---|---|---|---|---|---|
| Site / DGI | Total Alerts | Accept | Override | Ignore | Unknown | Followed | Not Followed | No Action |
| Cincinnati Children’s | 114 | 102 (89%) | 0 (0%) | 10 (9%) | 2 (2%) | 69 (61%) | 18 (16%) | 27 (24%) |
| Marshfield Clinic | 40 | 39 (98%) | 0 (0%) | 1 (3%) | 0 (0%) | 17 (43%) | 23 (58%) | 0 (0%) |
| Mayo Clinic | 9 | 4 (44%) | 0 (0%) | 5 (56%) | 0 (0%) | 4 (44%) | 5 (56%) | 0 (0%) |
| Mt. Sinai | 11 | 8 (73%) | 3 (27%) | 0 (0%) | 0 (0%) | 8 (73%) | 1 (9%) | 2 (18%) |
| Northwestern | 22 | 9 (41%) | 0 (0%) | 13 (59%) | 0 (0%) | 5 (23%) | 14 (64%) | 3 (14%) |
| Vanderbilt | 98 | 21 (21%) | 72 (73%) | 5 (5%) | 0 (0%) | 21 (21%) | 65 (66%) | 12 (12%) |
| Total by Site | 294 | 183 (62%) | 75 (26%) | 34 (12%) | 2 (1%) | 124 (42%) | 126 (43%) | 44 (15%) |
| Codeine | 114 | 102 (89%) | 0 (0%) | 10 (9%) | 2 (2%) | 69 (61%) | 18 (16%) | 27 (24%) |
| Clopidogrel | 65 | 14 (22%) | 46 (71%) | 5 (8%) | 0 (0%) | 10 (15%) | 40 (62%) | 15 (23%) |
| Simvastatin | 24 | 22 (92%) | 1 (4%) | 1 (4%) | 0 (0%) | 11 (46%) | 13 (54%) | 0 (0%) |
| Warfarin | 91 | 45 (49%) | 28 (31%) | 18 (20%) | 0 (0%) | 34 (37%) | 55 (60%) | 2 (2%) |
| Total by DGI | 294 | 183 (62%) | 75 (26%) | 34 (12%) | 2 (1%) | 124 (42%) | 126 (43%) | 44 (15%) |
Alert response varied across sites. Variation in response rate is likely due to significant differences in alert design. As a result, it is difficult to draw any broad conclusions about how physicians are responding to PGx CDS alerts in general. Instead, it will be necessary to analyze alert response in relation to the different design choices made at each site to understand how those choices influence physician behavior.
Definitions.
Alert Response: How the clinician responded to the actual alert that appeared in the EHR interface.
Clinical Response: How the clinician behaved in caring for the patient after viewing the alert.
Accept: Provider explicitly used the CDS-provided “accept” functionality to dismiss an alert.
Override: Provider explicitly used the CDS “acknowledge” or “override” functionality to dismiss an alert.
Cancel: Provider closed an alert without interacting with its contents.
Followed: Provider performed the recommended action within a pre-defined time window after seeing an alert.
Not Followed: Provider performed a clinically relevant, but not recommended, action within a pre-defined time window after seeing an alert.
No Action: Provider performed no clinically relevant action within a pre-defined time window after seeing an alert.
RESULTS
Objective 1: analysis of PGx CDS implementation characteristics
All seven participating sites (Figure 1) submitted information describing the nature of their PGx CDS alert designs. There was consistency in several of the alert characteristic categories, particularly DGI, active vs. passive, and pre- vs. post-test alert strategies. Adult and pediatric sites differed in which DGIs they implemented. All five adult sites implemented alerts to assist in prescribing at least two out of three of the following drugs: clopidogrel (CYP2C19), simvastatin (SLCO1B1), and warfarin (CYP2C9, VKORC1). The two pediatric sites chose to focus on other DGIs more common in children. Cincinnati Children's Hospital Medical Center (CCHMC) implemented alerts for codeine, oxycodone, hydrocodone, and tramadol, all associated with CYP2D6. Boston Children’s Hospital implemented alerts for phenytoin, which is also associated with CYP2C9. Additional detail is in Supplementary Table SA.
All seven sites implemented some form of active alerts. Five of the seven sites implemented only post-test prescribing alerts. CCHMC implemented pre-test alerts to encourage clinicians, when attempting to order a relevant drug, to first order an appropriate genotype test. In addition, they also implemented an alert that would inform clinicians when a genotype test result already exists, if they are attempting to order a duplicate test. Mt. Sinai Health System implemented both pre- and post-test alerts for warfarin only.
There were sixteen total DGIs with alerts defined across the seven sites. Of those: seven algorithms utilized specific genotypes (eg, CYP2C19*1/*2) stored in the patient record; five utilized PGx phenotypes in the form of computed observations2,4 (eg, intermediate metabolizer); two could utilize either specific genotypes or phenotypes; and one utilized the simple existence of a genotype test result, regardless of its contents.
The mechanism by which recommendations were generated varied by site and specific DGI. For clopidogrel and simvastatin, all sites used static text to relay the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommendations in their alerts, though Marshfield Clinic also supplemented these recommendations with dynamically calculated values. Warfarin alerts demonstrated the greatest variability: two sites displayed only dynamically calculated doses, one site supplemented dynamically calculated doses with static CPIC recommendations, one site supplemented dynamically calculated doses with static FDA recommendations, and one site did not calculate a dose, but directed clinicians to the WarfarinDosing.org website. There was significant variation among the sites on which actions were available to providers on the pop-up alerts. Details are in Table 1 and Supplementary Table SA.
Table 1.
Available alert actions by site and DGI
| Site | Gene | Drug | Accept with No Update | Accept with Update | Explicit Cancel | Implicit Cancel | View Educational Materials | Override with Reason | Override with No Reason | Other |
|---|---|---|---|---|---|---|---|---|---|---|
| Cincinnati Children’s | CYP2D6 | codeine | X | X | ||||||
| oxycodone | ||||||||||
| hydrocodone | ||||||||||
| tramadol | ||||||||||
| Marshfield Clinic | CYP2C19 | clopidogrel | X | X | X | |||||
| SLCO1B1 | simvastatin | X | X | X | ||||||
| CYP2C9, VKORC1 | warfarin | X | X | X | ||||||
| Mayo Clinic | CYP2C19 | clopidogrel | X | X | X | |||||
| SLCO1B1 | simvastatin | X | X | X | ||||||
| CYP2C9, VKORC1 | warfarin | X | X | X | ||||||
| Mt. Sinai | CYP2C19 | clopidogrel | X | X | X | X | X | X | X | |
| SLCO1B1 | simvastatin | X | X | X | X | X | X | X | ||
| CYP2C9, VKORC1 | warfarin | X | X | X | X | X | X | X | X | |
| Northwestern | CYP2C19 | clopidogrel | X | X | X | X | ||||
| SLCO1B1 | simvastatin | X | X | X | X | |||||
| CYP2C9, VKORC1 | warfarin | X | X | X | X | |||||
| Vanderbilt | CYP2C19 | clopidogrel | X | X | X | X | ||||
| CYP2C9, VKORC1 | warfarin | X | X |
Sites varied in the actions that they provided to users on PGx CDS alerts, which affected the Accept, Override, and Ignore rates found upon aggregation. Table includes only the 6 sites that submitted data for aggregation in Table 2.
Objective 2: cross-site data collection and pilot outcomes analysis
Six sites participated in the alert-level analysis and submitted 294 active, post-test alert instances across all DGIs, using the agreed-upon data structure. Alert responses were aggregated by site and DGI (Table 2). CCHMC and Vanderbilt University Medical Center accounted for 212 of the 294 (72%) alerts, with submissions ranging from 9 to 114 instances. The most common alerts were for codeine and warfarin, accounting for 70% of submitted instances. Overall, 62% of alerts were accepted and 26% overridden. Clinically, 42% were followed and 43% not followed. By DGI, simvastatin (92%) and codeine (89%) alerts were most likely to be accepted, clopidogrel most likely to be overridden (71%), and warfarin most likely to be ignored (20%). Codeine alerts were most likely to be followed (60%), while clopidogrel alerts were most likely to be not followed (62%). Codeine (24%) and clopidogrel (23%) alerts were also most likely to have no action taken.
DISCUSSION
This study successfully characterized the PGx CDS alert design strategies within eMERGE-PGx and demonstrated the ability of the eMERGE-PGx sites to share and aggregate PGx CDS alert data from disparate systems.
The eMERGE Network provided participating sites the freedom to implement PGx CDS as they each saw fit. This study found wide variation in implementation strategies, but also revealed an emerging consensus on how to structure PGx CDS alerts. The most common alerts implemented were active, post-test, and were based on CPIC recommendations, confirming a preference for a central authority of PGx recommendations,2 and in line with other studies of PGx implementation.23 There is also a trend towards preferring dynamically calculated recommendations for warfarin dosing.
Despite these areas of commonality, there is significant variation in how PGx CDS alerts are designed throughout the eMERGE Network. This demonstrates the current lack of EHR standards for storing and using genomic data and lends support to previous calls for such standards.24 (Other work has shown that this sort of variation is not unique to PGx alerts.25) Informal follow-up discussions with participating sites revealed a variety of site-specific factors that led to these differences. Common themes in those discussions included differences in: EHR functionality; availability of informatics and programming expertise; laboratory data formats; level of resources (funding and personnel); level of clinician buy-in and participation; previous experience with CDS; and concerns about risk of alert fatigue. Previous publications shed some light on site-specific decision making.8,13,15
Sites were almost evenly split as to whether to use genotype or phenotype as algorithm input. Northwestern Memorial Hospital expressed a preference for use of phenotype in CDS to simplify programming logic and to provide results that were more easily interpreted by clinicians. MCHR reported that they were influenced by the reporting format and interface from their testing laboratory. Their algorithms prioritized phenotype where available and defaulted to genotype if a phenotype was not present. Most sites showed a preference for dynamic warfarin dosing, and commonly accepted algorithms require genotype as input. Concordantly, all sites that developed warfarin alerts used genotype, even when using phenotype elsewhere.
Sites also varied significantly in what responses they allowed to pop-up alerts. For instance, some sites used explicit “cancel” buttons, while others did not. Some sites allowed users to “accept” an alert recommending a medication change without automatically updating the order, while other sites would automatically update the order. In some cases, these sorts of variations occurred across DGIs within the same site.
While the pilot study was able to aggregate PGx CDS alert data from disparate systems, the variations in design and small sample size constrain generalized conclusions about physician response to PGx CDS. For instance, accepting an alert is not necessarily a meaningful act if it is the only explicit action available. Therefore, the results reported in Table 2 should not be viewed as a definitive representation of the current state of PGx CDS adoption. Though, with that caveat, it is worth noting that override rates for PGx CDS alerts were lower than in studies of other medication-related CDS.26–28 Results from other PGx CDS studies that examined effectiveness are limited and varied, making them difficult to compare, but it seems likely that local design differences influence effectiveness.9,11,16,29 These results should be interpreted in context as a pilot study, providing strong evidence that comprehensive studies of PGx CDS may be more complex than expected, due to design variation. Additionally, the relatively low firing rate of the majority of alerts in this study suggests that a larger sample size will be needed to draw definitive conclusions. The consolidation of the EHR market, and the inclusion of PGx CDS as a built-in EHR feature, may reduce inter-site variability and increase PGx CDS usage, thus facilitating multi-site aggregation in the future. Until then, multi-site studies must be carefully focused on areas where sites share common design approaches.
Aside from the issues of site variability and sample size, the pilot study is limited in that it did not examine the relevance of PGx CDS alerts, such as alerts potentially firing in inappropriate situations or providing incorrect recommendations. Nor did it assess the correctness of the clinical actions taken. Sample sizes also made it difficult to determine how often PGx CDS alerts fire on a per patient basis or how clinician demographics affect alert response. Such questions, and others relating to usability and user-acceptance, are unanswered by this study and are the subject of both previous17,18,30,31 and in-progress work within the eMERGE Network.
The eMERGE Network is comprised of large, research-oriented institutions. Their priorities, experience levels, and resources dedicated to genomics may not represent national norms for the use of PGx CDS.
CONCLUSION
This study serves as a proof of concept for multi-site PGx CDS analysis. We successfully characterized PGx CDS design across the eMERGE Network. We found heterogeneity in design choices, with notable consensus around the use of active, post-test alerts relaying CPIC recommendations. We found variation in whether sites utilized phenotype or genotype in CDS algorithms, and in the actions that users may perform on any particular alert. This variation was unsurprising and likely due to a lack of genomic medicine standards in the EHR. We successfully created a data dictionary and shared alert logging data across sites with different EHR infrastructures. Finally, a pilot analysis using shared data showed that system variations currently make it difficult to quantitatively analyze physician response to PGx CDS across sites. Such studies should be narrowly focused on areas of commonality until PGx CDS designs become further standardized.
FUNDING
The eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center).
Contributors
All authors contributed to the concept and design of this project. TMH collected, analyzed, and interpreted the data and drafted the manuscript. All other authors provided feedback and final approval of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Maureen Smith and Dr Laura Rasmussen-Torvik for their valuable feedback during the writing and editing process.
Conflict of interest statement. TMH and JBS founded Ancillary Genomic Systems, LLC to explore the commercial potential of omic ancillary systems. They have not received any payments for the work in this manuscript and are not currently marketing any products. All other authors have no competing interests to report.
REFERENCES
- 1. Ginsburg GS, Willard HF.. Genomic and personalized medicine: foundations and applications. Transl Res 2009; 1546: 277–87. [DOI] [PubMed] [Google Scholar]
- 2. Herr TM, Bielinski SJ, Bottinger E, et al. A conceptual model for translating omic data into clinical action. J Pathol Inform 2015; 61: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013; 154: 258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Starren J, Williams MS, Bottinger EP.. Crossing the omic chasm: a time for omic ancillary systems. JAMA 2013; 30912: 1237–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Welch BM, Kawamoto K.. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc 2013; 202: 388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caraballo PJ, Bielinski SJ, Sauver JLS, Weinshilboum RM.. Electronic medical record-integrated pharmacogenomics and related clinical decision support concepts. Clin Pharmacol Ther 2017; 1022: 254–64. [DOI] [PubMed] [Google Scholar]
- 7. Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 2015; 551: 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caraballo PJ, Hodge LS, Bielinski SJ, et al. Multidisciplinary model to implement pharmacogenomics at the point of care. Genet Med 2017; 194: 421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc 2014; 21 (e1): e93–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gottesman O, Scott SA, Ellis SB, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013; 942: 214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldspiel BR, Flegel WA, DiPatrizio G, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J Am Med Inform Assoc 2014; 213: 522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014; 166C1: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peterson JF, Bowton E, Field JR, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med 2013; 1510: 833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shuldiner AR, Palmer K, Pakyz RE, et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am J Med Genet C Semin Med Genet 2014; 166C1: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herr TM, Bielinski SJ, Bottinger E, et al. Practical considerations in genomic decision support: The eMERGE experience. J Pathol Inform 2015; 61: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Donnell PH, Wadhwa N, Danahey K, et al. Pharmacogenomics-based point-of-care clinical decision support significantly alters drug prescribing. Clin Pharmacol Ther 2017; 1025: 859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Overby CL, Devine EB, Abernethy N, McCune JS, Tarczy-Hornoch P.. Making pharmacogenomic-based prescribing alerts more effective: a scenario-based pilot study with physicians. J Biomed Inform 2015; 55: 249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peterson JF, Field JR, Shi Y, et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J 2016; 164: 393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 2013; 1510: 761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther 2014; 964: 482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Green RC, Goddard KA, Jarvik GP, et al. Clinical sequencing exploratory research consortium: accelerating evidence-based practice of genomic medicine. Am J Hum Genet 2016; 991: 246.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luzum JA, Pakyz RE, Elsey AR, et al. The pharmacogenomics research network translational pharmacogenetics program: outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clin Pharmacol Ther 2017; 1023: 502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 2017; 192: 215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kawamoto K, Del FG, Lobach DF, Jenders RA.. Standards for scalable clinical decision support: need, current and emerging standards, gaps, and proposal for progress. Open Med Inform J 2010; 41: 235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin CP, Payne TH, Nichol WP, Hoey PJ, Anderson CL, Gennari JH.. Evaluating clinical decision support systems: monitoring CPOE order check override rates in the Department of Veterans Affairs’ Computerized Patient Record System. J Am Med Inform Assoc 2008; 155: 620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nanji KC, Slight SP, Seger DL, et al. Overrides of medication-related clinical decision support alerts in outpatients. J Am Med Inform Assoc 2014; 213: 487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Sijs H, Aarts J, Vulto A, Berg M.. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc 2006; 132: 138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. St Sauver JL, Bielinski SJ, Olson JE, et al. Integrating pharmacogenomics into clinical practice: promise vs reality. Am J Med 2016; 12910: 1093–1099.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Devine EB, Lee CJ, Overby CL, et al. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: a mixed methods approach. Int J Med Inform 2014; 837: 473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peterson JF, Field JR, Unertl K, et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin Pharmacol Ther 2016; 1001: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.