Abstract
SCHLAP1 is a long-noncoding RNA that is reported to function by depleting the SWI/SNF complex from the genome. We investigated the hypothesis that SCHLAP1 affects only specific compositions of SWI/SNF. Using several assays we found that SWI/SNF is not depleted from the genome by SCHLAP1, and that SWI/SNF is associated with many coding and non-coding RNAs, suggesting SCHLAP1 may function in a SWI/SNF independent manner.
Keywords: SWI/SNF, SCHLAP1, lncRNA, chromatin remodeling, prostate cancer, genomics
The long non coding RNA (lncRNA) Second Chromosome Locus Associated with Prostate cancer 1 (SCHLAP1) is a promising biomarker for metastatic prostate cancer 1,2. SCHLAP1 is proposed to function by antagonizing the SWI/SNF complex through direct interaction leading to complete disruption of SWI/SNF genomic occupancy 1. Evidence for this mechanism comes from the reported loss of SMARCB1 occupancy measured by ChIP-seq 1. SWI/SNF is a large multi-subunit chromatin remodeling complex that can be combinatorially assembled to yield hundreds to thousands of biochemically unique complexes3–5. We investigated the alternate hypothesis that distinct forms of SWI/SNF are affected by SCHLAP1 expression. However, using a variety of biochemical and genomics assays we demonstrate that SWI/SNF occupancy is unaffected by SCHLAP1 expression, in contrast to previously reported results. We show that SWI/SNF binds coding and non-coding RNA, raising the possibility that SCHLAP1 function is SWI/SNF independent. Consistent with the report by Prensner et. al. we observed an interaction between SMARCB1 and SCHLAP1 (Fig. 1A). We next generated SCHLAP1 over-expressing benign prostate epithelial cells (RWPE1;SCHLAP1) or control cells (RWPE1;LACZ) 1 (SCHLAP1 gift of A. Chinnayian). This model is the same used to originally suggest global depletion of SMARCB1 by SCHLAP1, and we confirmed their phenotype with respect to SWI/SNF expression and growth (Fig 1B Supplementary. Fig 1A-C, Supplementary Fig. 2)1. Additionally, we confirmed the key result that SCHLAP1 increased invasion (Fig 1C).
Figure 1. SCHLAP1 does not evict SWI/SNF from chromatin.
A. Native RIP for SMARCB1, asterisk denotes p-value 0.02 (two-sided t-test) between SCHLAP1 and U1 primer sets for SMARCB1 IP, n = 3 independent experiments). ErrorbBars represents average and whiskers extend to the standard deviation B. SCHLAP1 expression in RWPE1;LACZ and RWPE1;SCHLAP1 cells quantitative relative to GAPDH, n = 3 indpendent cell lines, error bars represent mean and standard deviation. C. Invasion assay of RWPE1;LACZ and RWPE1;SCHLAP1 cells quantitative by fluorescent intensity (Licor). Representative from n = 3 independent invasion assays run on one cell line, p-value = 0.007 (two-sided t-test). Error bars indicate mean and standard deviation. D. Chromatin fractionation showing SWI/SNF subunit presence in cytoplasm (Cytoplasm), nuclear (N), or chromatin (Ch) fractions from RWPE1;LACZ and RWPE1;SCHLAP1 cells.Representative images from 2 independent experiments E. Quantitation of G401 growth inhibition (n = 4 two independent cell lines plated in duplicate, error bars equal mean +/− standard deviation). F. Example browser images for SMARCA4, SMARCA2, and SMARCB1 from RWPE1;SCHLAP1 cells ChIP-seq experiments. Aggregate data from two independent ChIP-seq experiments is shown. G. Occupancy for SMARCA4, SMARCA2, and SMARCB1, centered on all transcription start sites (GENCODE), aligned by expression in RWPE1 cells.
To investigate which SWI/SNF subunits were depleted from chromatin upon SCHLAP1 expression, we fractionated RWPE1;LACZ and RWPE1;SCHLAP1 based on subcellular localization or salt extraction. Surprisingly, all SWI/SNF subunits assayed remained strongly enriched in the chromatin or high salt fractions (Fig. 1D, Supplementary Fig. 3A, Supplementary Fig. 4, Supplementary Fig. 5). Consistent with these biochemical experiments, we found that SMARCA4 and SMARCB1 localization was not affected in RWPE1;SCHLAP1 cells by immunofluorescence (Supp. Fig. 3B). Immunoprecipitation of SMARCA4 or SMARCB1 demonstrated that the SWI/SNF complex remains intact in the presence of SCHLAP1 (Supplementary. Fig. 3C, Supplementary Fig. 6). Finally, we used a malignant rhabdoid tumor cell line with inducible SMARCB1, that when expressed causes growth arrest (Kind gift of B. Weissman) 6. We reasoned that if SCHLAP1 disrupted SMARCB1 chromatin occupancy, overexpression of SCHLAP1 should allow G401 to proliferate following induction of SMARCB1. However, SMARCB1 induction led to growth arrest in a dose dependent manner (Fig. 1E, Supplementary. Fig. 7A,B, Supplementary Fig. 8). Together these results demonstrate that SCHLAP1 does not induce changes to SWI/SNF composition or association with chromatin.
We then performed ChIP-seq for three SWI/SNF subunits in RWPE1; SCHLAP1 cells (SMARCB1, SMARCA2, and SMARCA4). In contrast to the previous report, we identify robust binding for all three subunits in RWPE1 cells expressing SCHLAP1 (Fig. 1F). In RWPE1;SCHLAP1 cells we identified 6490, 22185, and 51505 peaks for SMARCB1, SMARCA2, and SMARCA4 respectively (Supplementary Table 1). This large number of peaks is in contrast to the original report which identified ~6500 SMARCB1 peaks in RWPE1; LACZ cells and nearly zero peaks in the SCHLAP1 expressing cells. The numbers of peaks are consistent with previous work from our lab showing 30,000–45,000 SMARCA4 peaks 4 Additionally, we and others have reported a large number of SWI/SNF peaks for a variety of subunits3,4,7,8. The majority of SMARCA2 peaks overlapped a SMARCA4 peak (Supp. Fig. 9A), and SWI/SNF peaks were predominantly located at promoters (45%−75% Supp Fig. 9B).4. SWI/SNF binding was most prominent at highly expressed genes with little to no occupancy at non-expressed genes (Fig. 1G, expression data - GSE988989). These results demonstrate that SCHLAP1 does not function by disrupting SWI/SNF occupancy genome-wide and raises the question of how SCHLAP1 functions to promote invasion and progression to metastatic disease.
We investigated whether SCHLAP1 expression induced chromatin changes, we performed ATAC-seq on RWPE1;LACZ and RWPE1;SCHLAP1 cells 10. We identified 273 and 3167 sites that open and close respectively (Supplementary Table 2, Supplementary Fig. Fig 10A, B). The sites that opened were more likely to be located distally or in introns of genes (Supplementary. Fig. 10A, B). Sites that open upon SCHLAP1 expression were enriched for distinct motifs from those that close (Supplementary. Fig 11C, Supplementary Table 3, 4). Open sites were enriched in motifs for TEAD and AP1 transcription factors, which are known to play a role in defining oncogenic enhancers (Supplementary. Fig 11A, Supplementary Table 3, 4)11. To test if these sites became activated enhancers, we performed ChIP-seq for H3K4me1, H3K4me3, and H3K27ac in RWPE1;LACZ and RWPE1;SCHLAP1 cells (Supplementary Fig. 10B), however, we did not find differences in any histone modifications. Additionally, SMARCA2, SMARCA4, and SMARCB1 remain highly enriched on sites that decrease chromatin openness. (Supplementary. Fig. 10D). GO analysis of the open sites revealed pathways involved in responses to NFKB signaling, epithelial to mesenchymal transitions, and nucleotide metabolism (Supplementary. Fig 11B), while closed sites were enriched for pathways involved in cell adhesion and signal transduction (Supplementary. Fig 11B). Although we noted changes in gene expression associated with ATAC-seq changes, we did not detect a significant association with the direction of altered chromatin openness (Supplementary. Fig. 12A. 12B)1. These data suggest SCHLAP1 could modestly influence chromatin, but is not sufficient to induce histone modification changes.
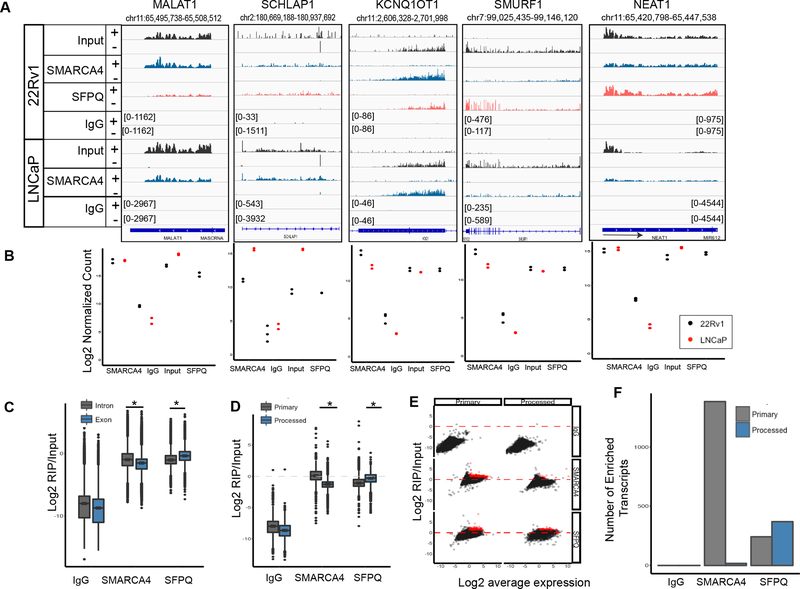
Recent evidence suggests that chromatin regulators display widespread interactions with RNA 12–17. Therefore, we investigated whether SWI/SNF interacts with other lncRNA. using crosslinked RIP-seq for SMARCA4 and a general splicing factor as a control (SFPQ) (Fig. 2A, B) 18. Consistent with previous observations of chromatin regulators, we saw widespread enrichment of SMARCA4 on most expressed transcripts relative to an IgG control (Supplementary Table 5, 6) 18. Hendrickson et. al. demonstrated using the same fRIP-seq that chromatin regulators could display either exonic or intronic enrichment. The pattern of SWI/SNF enrichment appeared uniform throughout the transcript, suggesting SMARCA4 associates frequently with primary transcripts (Fig 2C-F). In contrast, SFPQ was mostly enriched at exons and high levels of enrichment at the 3’ UTR of many transcripts. We observed SMARCA4 and SFPQ signal at SCHLAP1, although the level of enrichment was not markedly different between the proteins. Among the enriched transcripts was NEAT1, which is a known interacting partner of both SWI/SNF 13 and SFPQ 19–21. Additionally, we identified high levels of SMARCA4 associated with MALAT1, while SFPQ was not enriched (Fig 2A,B). A recent report showed a functional interaction between SMARCA4, HDAC9, and MALAT1, further supporting the specificity of our result 17. Given the widespread binding of SMARCA4 on many RNAs, and the modest enrichment of SCHLAP1 relative to bona fide SWI/SNF interactions such as MALAT 1, we interpret these data as evidence that the SCHLAP1-SWI/SNF interaction is the result of a widespread nonspecific interaction between SWI/SNF and transcribing RNA. This may be functional as RNA has been shown to inhibit activity of SWI/SNF and PRC2.16,22
Fig 2. SMARCA4 binds many RNAs.
A. Example loci associated with SMARCA4 or SFPQ in 22Rv1 and LNCaP cells. Strand specific RNA is shown (+/−) at each loci. Within cell lines and on the same strand scales are equal. RIP-seq was performed two times for each antibody in each cell line. B. Quantitation of signal at each of the genes in panels A for each antibody and each cell line. C. Enrichment of reads mapping to exons compared to introns for each IP, asterisk denotes p-value < 2.2e-16, wilcoxon test. Boxplots show first, second, and third quartile, and whiskers extend to 1.5X the interquartile range. D. Enrichment of reads mapping to primary compared to processed transcripts, asterisk denotes p-value = 0 for both tests by < 2.2e-16, two-sided wilcoxon test. Boxplots show first, second, and third quartile, and whiskers extend to 1.5X the interquartile range. E. Log2 fold change relative to input is plotted against the average expression of a transcript for both the primary and processed transcripts for the three antibodies tested. Red points indicate those genes greater than log2 fold change of 1 and an average log2 fold change greater than 0. F. Number of transcripts assigned to the red points in panel E.
Together these results demonstrate that the interaction between SWI/SNF and SCHLAP1 does not lead to a global depletion of SWI/SNF from the genome as previously reported 1. We confirm that SCHLAP1 induces RWPE1 cells to become more invasive, and our results suggest that this phenotype may be driven by a SWI/SNF independent mechanism.
Methods
Cell Culture
RWPE1 cells were purchased from ATCC ( CRL-11609, Lot 61840713 ) and grown in Keratinocyte Serum Free Medium (Life Technologies ) supplemented with penicillin and streptomycin. Cells were STR profiled by ATCC before purchase, and all experiments were performed on low passage cells (< 10 passages). Lenti6-SCHLAP1 was a kind gift of A. Chinnayian 1. Lenti6-Lacz was generated by removing SCHLAP1 from the above plasmid and cloning LACZ from pLentipuro-LACZ into pLenti6 from which SCHLAP1 was removed by EcoRI-BamHI digest. Lentiviral particles were produced using psPAX2 and pMD2.G to package plasmids in 293T cells. RWPE1;Lacz and RWPE1;SCHLAP1 cells were then generated by transduction with lentivirus and selecting with 2.5ug/mL blasticidin for 1 week. 22RV1 and LNCaP Cells were purchased from ATCC (CRL-2505, Lot 60437301; CRL-1740, Lot 62129998) and grown in RPMI 1640 with 10% Fetal Bovine Serum supplemented with penicillin and streptomycin. Cells were STR profiled by ATCC prior to purchase and were used at early passages ( < 10 passages). All cells were tested and free of mycoplasma contamination.
Antibodies
SMARCA2 (Cell Signaling 11966) Lot 2; SMARCA4 (Abcam ab110641) - GR150844–42; SMARCB1 (Abcam ab192864) - GR315927–2; BAF53A (Abcam ab3882) - GR95633–1; BAF60A ( BD Biosciences 611728) - 5199785; BAF155 ( Cell Signaling D7F8S) - Lot 2; BAF180 (Bethyl A301–591A) Lot 1; SFPQ ( Genetex GTX114209 ) - 40828; NCL (Bethyl A300–711A) - Lot 1; ARID1A (Abcam ab182560 ) - GR3186289–2; ARID1B (Abcam ab57461) - GR308911–3; ARID2 (ThermoFisher PA5–35857) SF24034426; H3K4me3 (Abcam ab8580); H3K4me1 (Active Motif 39635) 16611004; H3K27ac (Active Motif 39133) 20017049; H3 (epicypher 13–0001) NA gift of Brian Strahl, available from Epicyphyer
Invasion/Migration Assay
Invasion was assessed using a Corning BioCoat Matrigel Invasion Assay with 8μm pore size. Cells were seeded into Boyden Chambers at 100,000 cells/well in 100μl of normal growth media. The lower chamber was filled with either normal growth media supplemented with 10% FBS or PBS as a negative control. Cells were allowed to invade for 40.25 hours after which the top chamber was swabbed to remove non-invading cells. Invading cells were fixed and stained with crystal violet. Membranes were imaged using a Zeiss Axio Imager for representative images or scanned using a Licor fluorescent imager for quantitation. Results are reported as the average of three biological replicates. For each replicate, crystal violet fluorescence was measured and background fluorescence was subtracted. Background fluorescence of the membranes was obtained from a negative control.
RNA Immunoprecipitation
RIP experiments were performed using a modified version of a protocol from Hendrickson et al. 18
Fixation -
22Rv1 and LNCaP cells were fixed in 0.3% methanol free formaldehyde for 30 minutes at 4°C Formaldehyde was quenched with 125mM glycine for 5 minutes at room temperature. Plates were washed 3 times in room temperature PBS and cells were scraped in 1mM PMSF in PBS. Cells were snap frozen in liquid nitrogen and stored at −80°C.
Fragmentation and IP -
Cells were resuspended in 0.5mL RIPA buffer (50mM Tris-HCl pH 8, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 5mM EDTA, 150mM KCl) + 0.5mM DTT with 1X protease inhibitors (Sigma ) and 2.5uL RNAsin and incubated on ice for 10 minutes prior to lysing using a Bioruptor (Diagenode) for two cycles of 30 seconds on and 1 minute off, followed by centrifugation at 4°C for 10 minutes at max speed. Protein A magnetic beads (Biorad, ) were pre-conjugated with antibody for 2 hours at 4 degrees. Cells were then incubated overnight at 4 degrees with antibody conjugated beads. Beads were washed consecutively with fRIP buffer (25mM Trix-HCl pH 7.5, 5mM EDTA, 0.5% Ipegal CA-630, 150mM KCl), 3 times in ChIP buffer (50mM Tris-HCl pH 7.5, 140mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), 1x in high salt buffer (ChIP buffer, but with 500mM NaCl) and 1x in fRIPbuffer. All washes were performed for 5 minutes at 4 degrees. After final wash, beads were resuspended in 56uL water and 33uL of 3X reverse crosslinking buffer (3x PBS, 6% N-lauroyl sarcosine, 30MM EDTA ) supplemented with 5mM DTT, add 20uL proteinase K, and 1uL RNAsin and incubated 1 hour at 42 degrees, 1 hour at 55 degrees, and 30 minutes at 65 degrees. Following elution and crosslink reversal, trizol was used to extract RNA. Finally, RNA in the aqueous phase was supplemented with 1 volume of ethanol and purified using a Zymo-spin IC column, including the on-column DNAse digestion, per the manufacturer’s instruction. RNA was eluted in 15uL ddH20 and used to prepare RNA-seq libraries or to synthesize cDNA for qpCR. RNA-seq libraries were prepared by using equal volumes of IPs and included 1uL of 1:250uL dilution of ERCC spike-in mix 1 (Life Technologies). Input libraries were prepared from the same amount of RNA as the IP with the most RNA, and spike-ins were added as above. Libraries were then prepared using the Kapa Ribo-zero kit per manufacturers instructions, pooled, and sequenced on using single-end 75bp reads on a Nextseq 500.
RlP-seq Analysis
Reads were aligned using STAR to hg38 with gencode v27 annotations that included ERCC spike-ins and using --quantMode GeneCounts. In parallel an alignment-free quantitation method (Salmon version 0.8) was used to quantitate against gencode v27 containing ERCC spike ins and an extra transcript for each gene that contained the whole genomic DNA locus to represent an unspliced transcript. Browser tracks were generated by converting BAM files to bigWig files using Deeptools (v 2.5.2) and scaling tracks according to the 75% percentile of the ERCC-spike ins. Enrichment relative to IgG was calculated using DESeq2 with sizeFactors generated from only the ERCC spike-in data. For visualization relative to Input in Figure 4C,D,E normalized count values were used and log2 (RIP/Input) was calculated for each gene. RIP-seq data are available under GEO accession GSE114393
Chromatin Fractionation
Chromatin fractionation was performed as previously described 23. Approximately 1e7 cells were washed and resuspended in 200uL Buffer A (10mM Hepes pH 7.9, 10mM KCl, 1.5mM MgCl2, 0.34M Sucrose, 10% Glycerol, 1mM DTT, supplemented with 1X protease inhibitors and 1mM PMSF). Triton X-100 was then added to 0.1% final concentration and cells were incubated for 8 minutes on ice. Cells were then centrifuged at 1300g × 5 minutes at 4 degrees. The supernatant was saved as the cytosolic fraction and the pellet (nuclei) was washed once with Buffer A. Cells were lysed for 30 minutes on ice in 100uL Buffer B (3mM EDTA, 0.2MM EGTA, 1mM DTT) supplemented with 1X protease inhibitors and 1mM PMSF and centrifuge at 1700g × 5 minutes at 4 degrees. The supernatant was saved as the nucleoplasmic fraction and the insoluble material was resuspended in 1X laemmli buffer, and sonicated twice for 30 seconds on high power (Bioruptor). All lysates were boiled for 10 minutes in 1X laemmli buffer with 0.1M DTT and used in Western blot analysis.
Salt Extraction of Nuclei
Salt extraction of chromatin was performed as previously described 24. Cells were harvested from plates prior to protocol and snap frozen on liquid nitrogen and stored at −80 prior to extraction. Frozen cells were thawed on ice and gently resuspended in 1mL hypotonic buffer (10mM Hepes pH 7.9, 1.5mM MgCl2, 10mM KCl, 1mM PMSF, 0.5mM DTT) and incubated 30 minutes on ice, dounced 40 times with a tight pestle and an aliquot was set aside as whole cell extract. Cells were spun at 1500g × 5 minutes and the supernatant removed. Next 400uL of Buffer III.A (10mM Tris pH 7.4, 2mM MgCl2, 1mM PMSF, 5mM CaCl2) was added gently with a wide orifice p1000 tip and 1uL MNase (NEB, 2000 U) was added. Nuclei were incubated for 30 minutes at 37 degrees in a water bath, mixing every 10 minutes. To stop MNase digestion 25uL of ice-col 0.1M EGTA was added to nuclei and an aliquot was set aside as the nuclei fraction. Nuclei were then centrifuged at 400g × 10 minutes at 4 degrees and the supernatant was kept as the MNase fraction. Nuclei were then washed with 400uL Buffer III.B (Same as Buffer III.A, but without CaCh). Chromatin fractions were isolated by adding 400uL of IV.80, incubating for 30 minutes and spinning as above. The supernatant following each spin was saved, and the next salt buffer was added (IV.80, IV.150, IV.300, IV.600, 10mM Tris pH 7.4, 2mM MgCh, 2mM EGTA, 0.1% Triton X100, with NaCl added per buffer name). All samples were prepared for western blot by boiling in 1X laemmli buffer containing 0.1M DTT.
Immunofluorescence
RWPE1;LACZ and RWPE1;SCHLAP1 cells were grown on 0.1% gelatin-coated glass coverslips, fixed in 2% paraformaldehyde, blocked in antibody dilution buffer (goat serum PBS) and immunostained for SMARCA4 (abcam, 1:500 dilution) and SMARCB1 (abcam, 1:400 dilution) overnight at 4 degrees in antibody dilution buffer. Coverslips were washed 3 × 5 minutes in PBS and then stained in secondary antibody (goat anti rabbit, Alexa Fluor 568, 1:500 in antibody dilution buffer) for 45 minutes at RT. Coverslips were again washed 3 × 5 minutes in PBS and mounted with Prolong Gold containing DAPI and imaged on a Zeiss Axio Imager 2.
Growth Assays
2D growth assays were performed by plating 1000 cells/well in 96 well plate and were assayed at 1, 3, and 5 days post plating by Cell Titer Glo (Promega). 3D Growth assays were performed by plating 10 or 100 cells per well in 96-well plates and counting cell growth by Cell Titer Glo 3D (Promega) at 4 days following plating.
Immunoprecipitation
Nuclear lysates and co-immunoprecipitation were performed as previously described 4. Cells were washed with PBS and then centrifuged at 1300 rpm for 10 min at 4 C. Cells were washed with 20 packed cell volumes with hypotonic cell lysis buffer (10 mM HEPES-KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT plus protease inhibitors) and placed on ice for 10 min to swell. Cells were then centrifuged at 1300 rpm for 10 min at 4 C. Cells were dounced with B pestle in 2 packed cell volumes of hypotonic cell lysis buffer. Nuclei were pelleted at 1300 rpm for 10 min at 4 C, washed with 10 packed cell volumes with hypotonic cell lysis buffer and centrifuged at 5000 rpm for 10 min. Extractions were performed twice with 0.6 volume nuclear lysis buffer (20 mM HEPES-KOH pH 7.9, 25% glycerol, 420 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM and protease inhibitors). Lysates were clarified at 14,000 rpm for 10 min at 4 C between extractions. Lysates were diluted with storage buffer (20 mM HEPES-KOH pH 7.9, 20% glycerol, 0.2 mM EDTA, 0.2 mM DTT) to bring final KCl concentration to 150 mM and stored at − 80.
Prior to beginning the IP, we washed protein A magnetic beads three times with PBS + 0.5% BSA at 4 C. We resuspended beads in 400 uL of 1X + 0.5% BSA, then added 4–10ug of antibody and incubated overnight at 4 C. The following day, we thawed the nuclear lysates on ice. Lysates were added to antibody-conjugated beads and incubated overnight. Beads were washed four times with BC-150 (20 mM HEPES-KOH pH 7.9, 0.15 M KCL, 10% glycerol, 0.2 mM EDTA ph 8.0, 0.1% Tween-20, 0.5 mM DTT and protease inhibitors), 2 × BC-100 (20 mM HEPES-KOH pH 7.9, 0.1 M KCL, 10% glycerol, 0.2 mM EDTA ph 8.0, 0.1% Tween-20, 0.5 mM DTT and protease inhibitors), 1 × BC-60 (20 mM HEPES-KOH, pH 7.9, 60 mM KCl, 10% glycerol, 0.5 mM DTT and protease inhibitors). Proteins were eluted from beads using 2X Laemmli buffer with 100 mM DTT for 10 min at 95 C.
ChlP-seq
ChIP experiments were performed as previously described 4. Cells were fixed for 30 min at 4 C in 0.3% methanol-free formaldehyde, quenched for 5 min with 125 mM glycine, washed three times and snap-frozen in liquid nitrogen and stored at − 80 C. Frozen pellets were thawed for 30 min on ice, resuspended each pellet in 1 mL swelling buffer (25 mM HEPES + 1.5 mM MgCl2 + 10 mM KCl + 0.1% IGEPAL CA-630 containing 1 mM PMSF and 1X protease inhibitor cocktail, Roche) and incubated 10 min at 4 C. Cells were dounced 20 strokes with a ‘B’ pestle, and then, nuclei were pelleted at 2000 rpm for 7 min at 4 C. The nuclei were washed with 10 mL MNase digestion buffer (15 mM HEPES pH 7.9, 60 mM KCl, 15 mM NaCl, 0.32 M sucrose) and pelleted at 2000 rpm for 7 min at 4 C. The pellet was then resuspended in 1 mL MNase digestion buffer per 4e7 cells + 3.3 uL 1 M CaCh per mL + PMSF (1 mM) and protease inhibitor cocktail (1X, Roche) and then incubated for 5 min at 37 C to warm. We added MNase (NEB M0247S 2000 U/uL) at 0.5 uL/1e7 cells and incubated for 15 min at 37 C with agitation. Following digestion, the MNase was chelated using 1/50 volume 0.5 M EGTA on ice for 5 min. We added 1 volume of 2X IP buffer (20 mM TrisCl pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA), then passed the sample for a 21G needle five times and added Triton X-100 to 1% final concentration. The sample was cleared at 13,000 RPM for 15 min at 4°, and chromatin was used incubated with antibody overnight at 4° [SMARCA4 (Abcam ab110641)/SMARCA2 (Cell signaling 11966) / SMARCB1 (Abcam )]. Antibody/chromatin complexes were captured with protein A magnetic beads (Bio-Rad) for 2 h at 4° and washed 5 times with Agilent RIPA (50 mM HEPES pH 7.9/500 mM LiCl/1 mM EDTA/1% IGEPAL-ca-630/0.7% Na-deoxycholate) and once with 10 mM Tris/1 mM EDTA/50 mM NaCl. DNA was eluted at 65 C with agitation using 100 uL 1% SDS + 100 mM NaHCO3 made freshly. Cross-links were reversed overnight by adding 5 uL of 5 M NaCl and incubating at 65 C. DNA was treated with 3 uL RNaseA for 30 min at 37 C and then proteinase K for 1 h at 56 C and purified with Zymo Clean and Concentrator ChIP Kit and quantified using qubit before library preparation (Kapa Hyperprep).
ChlP-seq Analysis
Reads were aligned to hg38 using bowtie2 25 using the sensitive parameters, and duplicates were removed using SAMtools 26. For visualization, bigwig tracks were generated using DeepTools 27(version 2.5.2), bamCoverage tool with a binsize of 30 bp and extending fragments to the approximate nucleosome size of 150 bp. Tracks can be visualized using IGV 28, and bigwig files are available in GEO Accession number GSE114392.
Peak calling
Peaks were called using Macs2 (version 2.1.0 29) using the narrowpeak mode using the following parameters. Qval = 0.05 -keep-dup-all --fix-bimodal -nomodel -extsize 150. Additionally, we filtered the peaks against the ENCODE blacklist regions. Peaks were then annotated for the nearest transcription start site using ChIPPeakAnno 30 relative to GENCODE v26.
ATAC-seq
ATAC-seq was performed as previously described 10 with modifications. Briefly, 50,000 cells were harvested and lysed in a buffer containing 0.05% Ipegal CA-630 before transposition with a Nextera library prep kit containing TN5 transposomes. Libraries were amplified using 6–8 PCR cycles to enrich for TN5 products and add indexes and sequenced as paired-end 50bp libraries on a Hiseq 2500. ATAC-seq data are available under GEO Accession number GSE114391.
ATAC-seq analysis
Nextera adapter sequences were trimmed using trim_galore and reads were aligned to hg38 using bowtie2 with the -X 2000 setting 25. We removed any reads mapping to the mitochondrial genome and filtered any reads with a mapping quality less than 20 using SAMtools 26. Peaks were called using Macs2 (version 2.1.029) using the narrowpeak mode using default settings and --keep-dup-all. Differential openness was identified use csaw 31 with window size 150 and background window size 5000 and an adjusted FDR of 0.05 for the combined windows. Motif analysis was performed using HOMER and comparing the open or closed ATAC sites to the background set of static sites 32.
Supplementary Material
Acknowledgements
We thank members of the Magnuson Lab for helpful comments and discussion. We thank Bernard Weissman for the G401;SNF5 inducible cell line and Arul Chinnaiyan for the SCHLAP1 expression construct. This work was supported by grants to JRR from The University of North Carolina at Chapel Hill University Cancer Research Fund (Tier 1 Development Award) and North Carolina Translational and Clinical Sciences Institute (2KR851610). Additionally, this work was supported by Grants from The National Institute of Child Health and Development (TM 5R01HD036655) and The National Institute of General Medical Sciences (JRR 5F32GM108367 and JMC R01GM121806).
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Accession Codes
GEO accession number GSE114394
Data Availability
All data are available from GEO under accession number GSE114394. ATAC-seq and ChIP-seq read depth information can be found in Supplementary Table 7.
Code AvailabilityComputer Code
AllAny commands, settings, or programs used to generate figures are described in the analysis section above. Any additional code used to generate figures is available on request.
References
- 1.Prensner JR et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet 45, 1392–1398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prensner JR et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 15, 1469–1480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raab JR, Resnick S & Magnuson T Genome-Wide Transcriptional Regulation Mediated by Biochemically Distinct SWI/SNF Complexes. PLoS Genet. 11, e1005748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raab JR, Runge JS, Spear CC & Magnuson T Co-regulation of transcription by BRG1 and BRM, two mutually exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin 10, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadoch C & Crabtree GR Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuwahara Y, Wei D, Durand J & Weissman BE SNF5 reexpression in malignant rhabdoid tumors regulates transcription of target genes by recruitment of SWI/SNF complexes and RNAPII to the transcription start site of their promoters. Mol. Cancer Res 11, 251–260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris SA et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol 21, 73–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakayama RT et al. SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet (2017). doi: 10.1038/ng.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Z, Rhie SK, Lay FD & Farnham PJ A Prostate Cancer Risk Element Functions as a Repressive Loop that Regulates HOXA13. Cell Rep. 21, 1411–1417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanconato F et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol 17, 1218–1227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng S, Wang L, Deng C-H, Du S-C & Han Z-G ARID1A represses hepatocellular carcinoma cell proliferation and migration through lncRNA MVIH. Biochem. Biophys. Res. Commun (2017). doi: 10.1016/j.bbrc.2017.07.072 [DOI] [PubMed] [Google Scholar]
- 13.Kawaguchi T et al. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. U. S. A 112, 4304–4309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16, 413–425 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Zhu Y, Rowley MJ, Böhmdorfer G & Wierzbicki AT A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell 49, 298–309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cajigas I et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development 142, 2641–2652 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lino Cardenas CL et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat. Commun 9, 1009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.G Hendrickson D, Kelley DR, Tenen D, Bernstein B & Rinn JL Widespread RNA binding by chromatin-associated proteins. Genome Biol. 17, 28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemson CM et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose T et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online References
- 21.Naganuma T et al. Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31, 4020–4034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko S, Son J, Shen SS, Reinberg D & Bonasio R PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol 20, 1258–1264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Méndez J & Stillman B Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol 20, 8602–8612 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann C, Avgousti DC & Weitzman MD Differential Salt Fractionation of Nuclei to Analyze Chromatin-associated Proteins from Cultured Mammalian Cells. Bio Protoc 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol 29, 24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu LJ et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lun ATL & Smyth GK csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44, e45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.