Abstract
Soluble oligomers of the β-amyloid peptide, Aβ, are associated with the progression of Alzheimer’s disease. Although many small molecules bind to these assemblies, the details of how these molecules interact with Aβ oligomers remain unknown. This paper reports that crystal violet, and other C3 symmetric triphenylmethane dyes, bind to C3 symmetric trimers derived from Aβ17–36. Binding changes the color of the dyes from purple to blue, and causes them to fluoresce red when irradiated with green light. Job plot and analytical ultracentrifugation experiments reveal that two trimers complex with one dye molecule. Studies with several triphenylmethane dyes reveal that three N,N-dialkylamino substituents are required for complexation. Several mutant trimers, in which Phe19, Phe20, and Ile31 were mutated to cyclohexylalanine, valine, and cyclohexylglycine, were prepared to probe the triphenylmethane dye binding site. Size exclusion chromatography, SDS-PAGE, and X-ray crystallographic studies demonstrate that these mutations do not impact the structure or assembly of the triangular trimer. Fluorescence spectroscopy and analytical ultracentrifugation experiments reveal that the dye packs against an aromatic surface formed by the Phe20 side chains and is clasped by the Ile31 side chains. Docking and molecular modeling provide a working model of the complex in which the triphenylmethane dye is sandwiched between two triangular trimers. Collectively, these findings demonstrate that the X-ray crystallographic structures of triangular trimers derived from Aβ can be used to guide the discovery of ligands that bind to soluble oligomers derived from Aβ.
Graphical Abstract

Introduction
Fluorescent dyes have longstanding history as probes to study the molecular basis of amyloid disorders, such as Alzheimer’s disease, Parkinson’s disease, and type 2 diabetes. The dye Congo red has been used to stain amyloid plaques in tissue for nearly a century, while thioflavin T and thioflavin S have been used for several decades as both probes of amyloid plaques and tools to monitor the aggregation of amyloid-forming proteins into fibrils.1–5 New life has been breathed into these chemical probes through the creative application of their selectivity for amyloid plaques. For example, homologues of Congo red have been used to block amyloid fibril formation, radio-labeled homologues of thioflavin T are currently used in PET imaging diagnostic tests of Alzheimer’s disease, and combinatorial fluorescent molecular sensors based around thioflavin T have enabled differentiation among aggregates of Aβ.6–9 High-resolution structures of fibrils and fibril-dye complexes, coupled with molecular dynamics simulations, have revealed structural bases of this molecular recognition, wherein planar aromatic dyes lie parallel to the fibril axis and pack against hydrophobic residues displayed on the fibril surface.10–19 These structures have guided the design of other small molecules that bind to amyloid fibrils.20
Although a handful of chemical probes are used to study the formation of the biologically inert amyloid fibrils, there are few that specifically detect neurotoxic oligomers.21–27 The specific oligomer structures that these chemical probes recognize remain unknown. Chemical probes that detect amyloid oligomers are urgently needed to help study the structures and function of amyloid oligomers.28,29 The current lack of high-resolution structures of amyloid oligomers hinders the design of chemical probes that detect these assemblies.30
While studying the self-assembly of peptides derived from the β-amyloid peptide, Aβ, we have discovered that triphenylmethane dyes bind to assemblies of covalently stabilized oligomers formed by peptides derived from Aβ17–36.31 This discovery was guided by the complementary shape and symmetry of triphenylmethane dyes and the oligomeric assemblies our laboratory has characterized by X-ray crystallography.31 This binding produces marked changes to the color of the dyes, from purple in the unbound state to blue in the bound state. Binding also induces pronounced red fluorescence upon excitation with green light. Here we report the discovery of triphenylmethane dyes that bind to triangular trimers derived from Aβ.
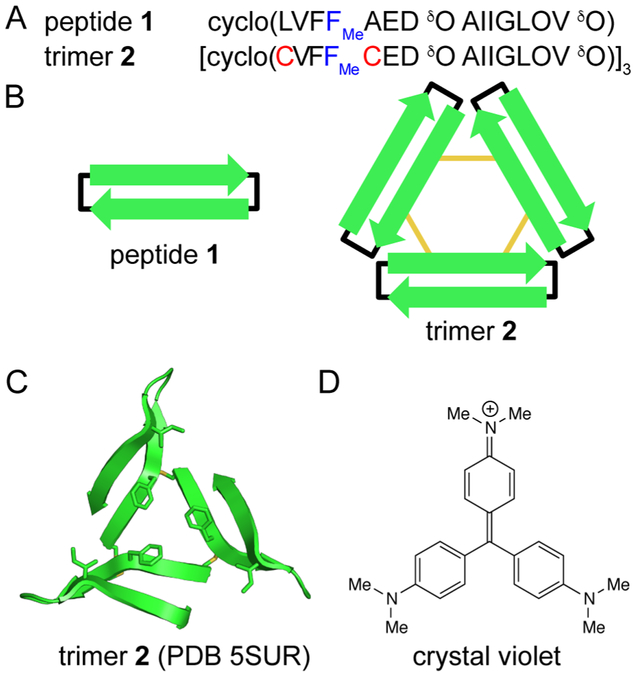
Our laboratory previously synthesized macrocyclic β-hairpin peptide 1 to mimic a βhairpin formed by Aβ17–36 (Figure 1).32 Peptide 1 contains the heptapeptide β-strands Aβ17–23 and Aβ30–36 connected by two δ-linked ornithine turn units. Peptide 1 bears a single Nmethyl group, on Phe20, which limits uncontrolled aggregation of the peptide. The X-ray crystallographic structure of peptide 1 revealed a triangular trimer motif wherein three copies of peptide 1 assemble to form an equilateral triangle (PDB 4NW9). To ascertain the biological and biophysical properties of this triangular trimer, we covalently stabilized this oligomer, resulting in trimer 2.31 Trimer 2 is stabilized by disulfide crosslinks, which we engineered into the three vertices of the triangular trimer. In our initial 2017 report of trimer 2,33 we found that it induces apoptosis in the human neuroblastoma SH-SY5Y cell line and reacts with the amyloid oligomer-specific antibody A11,34 thus mimicking the biological properties of oligomers formed by full-length Aβ.35
Figure 1:
Chemical models of oligomers formed by Aβ. (A) Amino acid sequences of peptide 1 and trimer 2. (B) Cartoon of peptide 1 and trimer 2. Black lines represent δ-linked ornithine turn units; yellow lines represent disulfide bonds. (C) X-ray crystallographic structure of trimer 2 (PDB 5SUR). The side chains of Cys17, Phe20, Cys21, and Ile31 are shown as sticks. (D) Chemical structure of crystal violet.
The discovery and development of compounds that interact with fibrils have been guided by high resolution structures of fibrils and fibril-like assemblies.17,20,36,37 The X-ray crystallographic structure of trimer 2 (PDB 5SUR) provided us with a starting point for discovering chemical probes that bind the triangular trimers that we have observed crystallographically and that we postulate to be formed by Aβ. We hypothesized that C3 symmetric dyes would interact with the C3 symmetric trimer. Specifically, we envisioned that the three side chains of Phe20, arrayed around the C3 symmetry axis of trimer 2, would interact with C3 symmetric small molecules (Figure 1C). We began our exploration with crystal violet, a C3 symmetric triphenylmethane dye (Figure 1D).
Results and Discussion
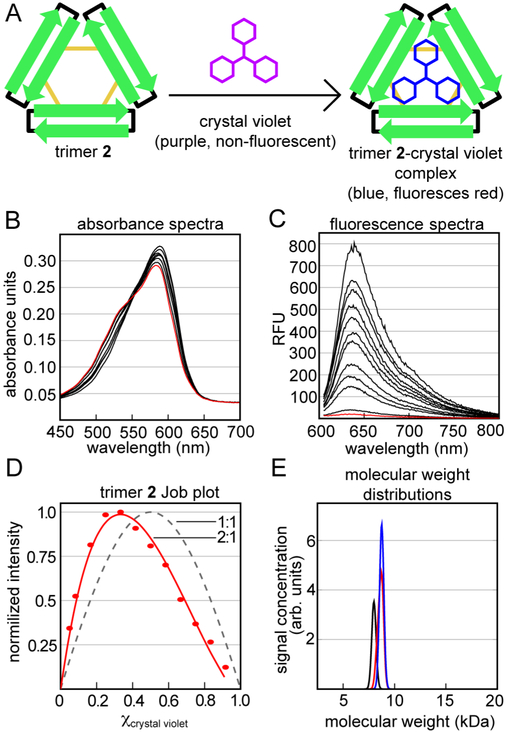
Trimer 2 forms a complex with crystal violet in aqueous solution (Figure 2). The visible absorption spectrum of crystal violet is sensitive to changes in the local environment surrounding the dye.38 The resulting color change has been used to measure Complexation of crystal violet by proteins, including bovine serum albumin.39–41 We also observe color changes, from violet to blue, upon addition of trimer 2 to crystal violet. The visible spectrum of crystal violet displays an absorption maximum at 590 nm and a shoulder at ca. 530 nm in 100 mM aqueous acetate buffer (pH 4.6). When trimer 2 is added to crystal violet at low micromolar concentrations, the maximum shifts to longer wavelengths and becomes more intense, while the shoulder disappears (Figure 2B). The spectra that result from titration of crystal violet with trimer 2 do not exhibit a well defined isosbestic point, suggesting the formation of multiple trimer 2-crystal violet complexes in solution. The complexation also enhances the fluorescence spectra of crystal violet, causing it to fluoresce red when irradiated with green light (Figure 2C). Crystal violet is largely non-fluorescent in aqueous solution due to non-irradiative modes of relaxation that result from bond rotations in the excited state.38 Complexation by trimer 2 likely restricts these rotations, resulting in the enhanced fluorescence of crystal violet.42,43
Figure 2:
Crystal violet binds to trimer 2 in aqueous buffer. (A) Cartoon of the color and fluorescence changes that occur upon binding. (B and C) Absorbance and fluorescence spectra of crystal violet (red) titrated with increasing amounts of trimer 2 (black) in buffer comprising 50 mM sodium acetate and 50 mM acetic acid (pH 4.6). Fluorescence spectra were acquired with a 590 nm excitation wavelength. (D) Job plot analysis of the stoichiometry of the trimer 2-crystal violet complex. The dashed line is a 1:1 (trimer 2-crystal violet) fit of the data, while the solid line is a 2:1 fit of the data. (E) Molecular weight distributions calculated from sedimentation velocity analytical ultracentrifugation experiments. The distributions are calculated from mixtures of 7 μM crystal violet with 14 μM (black), 42 μM (red), and 56 μM (blue) trimer 2.
To determine the stoichiometry of the complex between crystal violet and trimer 2, we used Job’s method of continuous variation (Figure 2D).44–47 In this experiment, we mixed crystal violet and trimer 2 at varying ratios while maintaining the total concentration of the two components, and we measured the fluorescence intensities of the resulting mixtures. The resulting fluorescence spectra display a maximum intensity at a 0.33 mole fraction (χ) of crystal violet, which suggests that two molecules of trimer 2 bind to one molecule of crystal violet. The data do not fit a simple 1:1 trimer-dye binding model, wherein the maximum fluorescence intensity would occur at a 0.5 mole fraction of crystal violet. From the 2:1 fit of the data, the dissociation constant is ca. 0.94·10−9 M2, which corresponds to a complex that dissociates around 30 μM. The 2:1 stoichometry is consistent with our laboratory’s previously reported observation that trimer 2 assembles to form a dimer in solution.31
To corroborate the apparent 2:1 stoichiometry, we performed sedimentation velocity analytical ultracentrifugation (AUC) experiments. In these experiments, we mixed 7 μM crystal violet with one, two, four, six, or eight molar equivalents of trimer 2 in sodium acetate buffer, and monitored the sedimentation of crystal violet by measuring the visible absorbance at 593 nm. Crystal violet does not sediment upon ultracentrifugation in the absence of trimer 2, because it is too low in molecular weight (Figure S1). When crystal violet is mixed with two or more equivalents of trimer 2, it sediments as a ca. 9 kDa species (Figure 2E). This molecular weight is largely consistent with crystal violet binding to two molecules of trimer 2, each weighing 5.3 kDa. These experiments support the Job plot analysis that trimer 2 forms a 2:1 complex with crystal violet.
To further corroborate the formation of the 2:1 trimer 2-crystal violet complex, we performed isothermal titration calorimetry (ITC) experiments. In these experiments, we added aliquots of a 500-μM crystal violet solution to a solution of 100 μM trimer 2 and recorded the heat of association. Although the reaction is clearly exothermic, the association constant is too low to permit precise analysis of the titration data (Figure S2). The limited solubility of trimer 2 in sodium acetate buffer and the propensity of crystal violet and trimer 2 to selfassociate at higher concentrations complicate a rigorous analysis of binding constants and thermodynamic parameters by ITC. At the concentrations used in the titration, the data fit well to a 2:1 binding model, with a dissociation constant of ca. 1.4·10−9 M2 (Figure S2B and C). This dissociation constant is similar to that estimated by Job’s method of continuous variation. The data deviate from a simple 1:1 binding model, where the residuals display sinusoidal deviation similar to the deviation in the attempted 1:1 fit of the Job plot data in Figure 2D.48 The fit to a 2:1 binding model shows substantially smaller deviations.
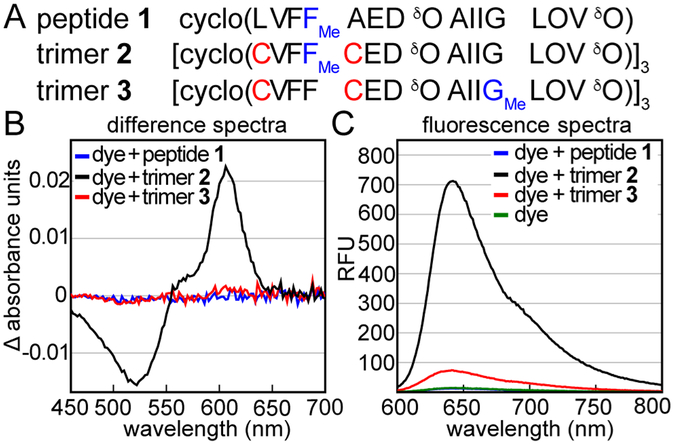
Crystal violet is known to interact with numerous proteins, such as BSA and acetylcholine receptors.39–41,49–53 For these reasons, we were concerned that the interaction of crystal violet and trimer 2 might not reflect the specific recognition of the three-fold symmetric dye by the triangular trimer motif. To determine if the association of crystal violet with trimer 2 requires the triangular trimer motif, we studied the interaction of crystal violet with peptide 1 (Figure 3). Peptide 1 does not assemble into a triangular trimer at the low micromolar concentrations (5–150 μM) at which trimer 2 complexes crystal violet.31 No significant changes occur in the absorbance or fluorescence spectra of crystal violet upon addition of peptide 1 (Figure 3B and C). ITC experiments further corroborate that peptide 1 does not bind crystal violet at concentrations where trimer 2 does (Figure S3). The association of crystal violet with macrocyclic β-hairpins derived from Aβ appears to require a binding site that is unique to the triangular trimer.
Figure 3:
Interaction of other Aβ-derived peptides with crystal violet. (A) Amino acid sequences of peptide 1, trimer 2, and trimer 3. Residues bearing N-methyl groups are highlighted in blue. (B) Difference absorbance spectra of 25 μM crystal violet with 75 μM peptide 1, 25 μM trimer 2, or 25 μM trimer 3. Difference spectra are calculated by subtracting the absorbance spectrum of 25 μM crystal violet from the absorbance spectra of the mixed samples. (C) Fluorescence spectra of 25 μM crystal violet or 25 μM crystal violet with 75 μM peptide 1, 25 μM trimer 2, or 25 μM trimer 3. Fluorescence spectra were acquired with a 550 nm excitation wavelength. Crystal violet exhibits near-baseline fluorescence alone or with peptide 1. Experiments were performed in buffer comprising 50 mM sodium acetate and 50 mM acetic acid (pH 4.6).
To further test if the interaction between crystal violet and trimer 2 results from specific molecular recognition of the trimer 2 surface, we studied the interaction between crystal violet and trimer 3 (Figure 3). Trimer 3 is almost identical to trimer 2, differing only in the location of the three N-methyl groups. The N-methyl groups are on Phe20 in trimer 2, while they are on Gly33 in trimer 3. The surfaces of trimer 3 differ from those of trimer 2 because the two trimers have different hydrogen-bonding patterns between the cross-linked monomers that comprise the trimers (Figure S4). Absorbance spectroscopy, fluorescence spectroscopy, and ITC experiments indicate that trimer 3 does not appreciably associate with crystal violet at low micromolar concentrations. No significant changes occur to the absorbance spectrum of crystal violet when crystal violet and trimer 3 are mixed at 25 μM concentrations (Figure 3B). In contrast, trimer 2 visibly alters the color of crystal violet under the same conditions. A small enhancement does occur to the fluorescence spectrum of crystal violet when mixed with trimer 3, but this enhancement is far smaller than that which occurs when crystal violet is mixed with trimer 2 (Figure 3C). ITC experiments further corroborate that trimer 3 does not bind crystal violet under conditions wherein trimer 2 does (Figure S5). These titration data do suggest that trimer 3 weakly associates with crystal violet, with an association constant that corresponds to a complex that dissociates in the hundred-micromolar regime. However, the limited solubility of trimer 3 precludes the accurate measurement of this weak association constant by ITC. The interaction of crystal violet with trimer 2 but not trimer 3, under these conditions, suggests that the surfaces displayed by trimer 2 are primed to interact with crystal violet.
To understand why trimer 2 interacts with crystal violet while other Aβ-derived peptides we have studied do not, we performed docking simulations. In these simulations, we used AutoDock to propose a model for the recognition of crystal violet by trimer 2.54 We performed these docking studies because X-ray crystallography and NMR spectroscopy have been unsuccessful in illuminating the details of the trimer 2-crystal violet complex: We have been unable to co-crystalize crystal violet with trimer 2, and the NMR spectra of trimer 2 in the presence or absence of crystal violet are broad and uninterpretable. Docking simulations, on the other hand, have enabled us to generate testable models for the molecular recognition between trimer 2 and crystal violet (Figure 4). In these simulations, we docked crystal violet to a single copy of trimer 2, extracted from the X-ray crystallographic structure of trimer 2 (PDB 5SUR). These simulations suggest that the three side chains of Phe20 complement the three aryl rings of crystal violet, and that the three side chains of Ile31 are positioned to pack against the three N,N-dimethylamino substituents on crystal violet. We proceeded to probe this model by studying the association of crystal violet homologues with trimer 2, as well as the association of crystal violet with trimer 2 homologues.
Figure 4:
Representative molecular model of the trimer 2-crystal violet complex generated in AutoDock. The side chains of Phe20 and Ile31 are depicted as spheres.
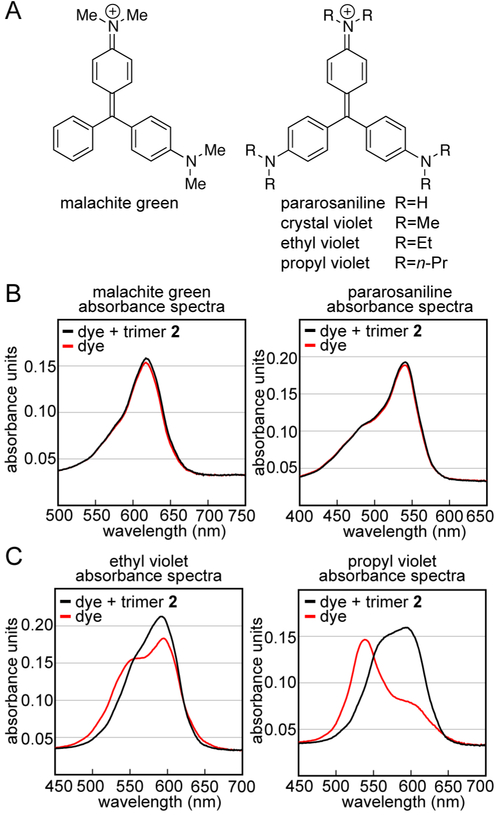
The model proposed in Figure 4 suggests that the N,N-dimethylamino substituents of crystal violet pack against the side chains of Ile31. To test the role of the N,N-dimethylamino substituents on complex formation, we studied the association of crystal violet homologues with trimer 2 (Figures 5 and S6). We studied two dyes with smaller substituents than crystal violet and two dye with larger substituents than crystal violet (Figure 5A): malachite green contains only two N,N-dimethylamino groups; pararosaniline contains three amino groups; ethyl violet contains three N,N-diethylamino groups; propyl violet contains three N,N-dipropylamino groups. No significant changes occur in the visible spectrum of either malachite green or pararosaniline upon mixing with trimer 2 (Figure 5B). No enhancement occurs to the fluorescence spectrum of pararosaniline upon mixing with trimer 2 (Figure S6A). A small enhancement does occur to the fluorescence spectrum of malachite green, but it is much smaller than that which occurs to the fluorescence spectrum of crystal violet. ITC experiments corroborate that malachite green does not associate with trimer 2 at concentrations that crystal violet does (Figure S6B).
Figure 5:
Interaction of other triphenylmethane dyes with trimer 2. (A) Chemical structures of crystal violet and related triphenylmethane dyes. (B) Absorbance spectra of 25 μM malachite green and 25 μM pararosaniline in the presence (black) or absence (red) of equimolar trimer 2. (C) Absorbance spectra of 25 μM ethyl violet and 20 μM propyl violet in the presence (black) or absence (red) of equimolar trimer 2. Experiments were performed in buffer comprising 50 mM sodium acetate and 50 mM acetic acid (pH 4.6).
ITC experiments suggest that pararosaniline associates weakly with trimer 2, with an association constant that corresponds to a complex that dissociates in the hundred-micromolar regime (Figure S6C). The limited solubility of trimer 2 precludes the accurate measurement of this association constant by ITC. In contrast to malachite green and pararosaniline, dramatic changes occur to both the visible and fluorescence spectra of ethyl violet and propyl violet upon mixing with trimer 2 (Figures 5C and S6D). It is difficult to characterize the complexes of ethyl violet and propyl violet with trimer 2 using Job’s method of continuous variation, AUC, and ITC, because the dyes aggregate strongly in aqueous solution. Collectively these studies with crystal violet homologues suggest that three N,N-dialkylamino substituents are required for complexation.
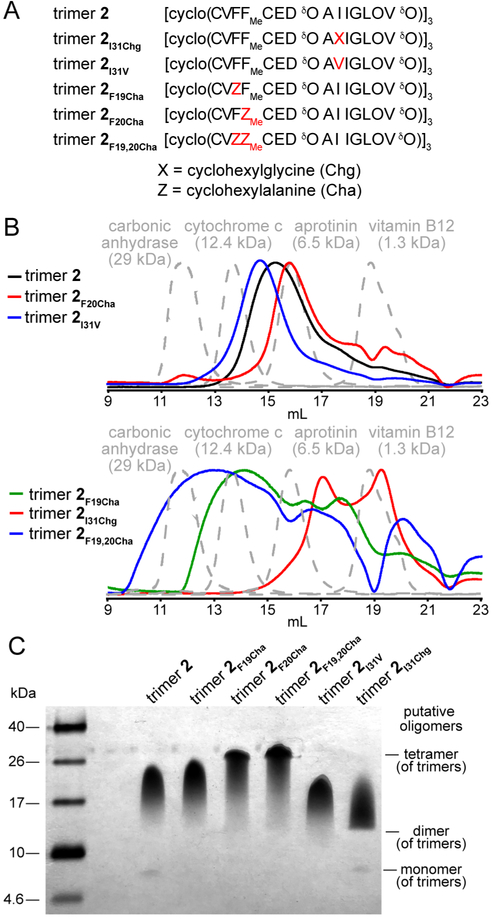
To further test the molecular model proposed in Figure 4, we prepared five homologues of trimer 2 containing cyclohexylalanine (Cha), valine, and cyclohexylglycine (Chg) at positions 19, 20, and 31. Cha is the fully reduced (non-aromatic) analogue of phenylalanine; Chg is a bulkier analogue of both valine and isoleucine. We prepared trimers 2I31V and 2I31Chg to test if crystal violet packs against the side chain of Ile31, and we prepared trimers 2F19Cha, 2F20Cha, and 2F19,F20Cha to test if crystal violet packs against the side chain of Phe20 (Figure 6A). We prepared these trimer 2 homologues using the same procedures our laboratory has previously used to prepare other crosslinked trimers.31,55
Figure 6:
Trimer 2 homologues. (A) Amino acid sequence of trimer 2 homologues. Mutations relative to trimer 2 are colored red. (B) SEC chromatograms of trimer 2 and its homologues. SEC was performed on 1 mg/mL (ca. 150 μM) solutions of trimer dissolved in buffer comprising 50 mM sodium acetate and 50 mM acetic acid on a Superdex 75 10/300 column. (C) Silver-stained SDS-PAGE gel of trimer 2 and its homologues. Aliquots (5 μL) of 0.1 mg/mL solutions of each trimer were loaded into the corresponding lanes in the gel.
To determine the impact of these five mutations on the oligomerization state of trimer 2, we compared the oligomerization state of these trimer 2 homologues to that of trimer 2 using size exclusion chromatography (SEC) and SDS-PAGE. In SEC, we found that some of the trimer 2 homologues elute as well defined oligomers, while others elute as broad peaks that contain multiple features, some of which suggest adsorption to the column (Figure 6B). In 100 mM acetate buffer (pH 4.6), trimer 2 elutes between the cytochrome C (12.4 kDa) and aprotinin (6.5 kDa) size standards, which elute at 13.6 and 15.8 mL, respectively. The retention volume (15.6 mL) is consistent with the assembly of trimer 2 into a dimer (10.6 kDa).31 Trimer 2 shows pronounced tailing in its elution, which suggests that trimer 2 may adsorb to the column. Trimer 2F20Cha and trimer 2I31V both elute with retention volumes similar to that of trimer 2, and exhibit tailing similar to that present in the elution of trimer 2 (14.6 and 15.7 mL). These retention volumes suggest that both of these trimer 2 homologues assemble into dimers, like that formed by trimer 2.
In contrast, trimers 2F19Cha, 2F19,20Cha, and 2I31Chg, elute as broad peaks with multiple distinct features. Trimer 2I31Chg elutes as two peaks with retention volumes of 17.0 and 19.1 mL. It is possible that these two peaks correspond to a dimer and monomer that both adsorb to the column. In contrast, trimers 2F19Cha and 2F19,20Cha elute as broad heavily-featured bands with retention volumes as early as ca. 12 and 14 mL and as late as 22 mL. The early retention volumes are consistent with these hydrophobic mutations promoting the solution-phase assembly of oligomers larger than the dimers formed by trimers 2, 2F20Cha, and 2I31V, while the late retention volumes are consistent with the adsobption of these hydrophobic trimers to the column. Mutating Phe19 to cyclohexylalanine in full-length Aβ40 also enhances its propensity to aggregate.56
In contrast to the SEC studies, SDS-PAGE studies indicate that the trimer 2 homologues assemble in a fashion similar to that of trimer 2 (Figure 6C). These 5.0–5.4 kDa trimers migrate as bands between the 17 and 26 kDa size standards, consistent with the trimers forming tetramers under these conditions. These putative tetramer bands show pronounced downward streaking, indicating an equilibrium with lower weight oligomers. The degree of this downward streaking varies among the mutants, likely reflecting the mutations stabilizing or destabilizing the tetrameric assemblies in SDS. Mutating Phe20 to Cha (trimers 2F20Cha and 2F19,20Cha) appears to stabilize the putative tetrameric assemblies. In contrast, mutating Phe19 to Cha (trimer 2F19Cha) or mutating Ile31 to Val (trimer 2I31V) appears to have little impact on the stabilities of the tetramers relative to trimer 2. Mutating Ile31 to Chg (trimer 2I31Chg) appears to disrupt the tetramer and instead stabilize a putative dimer in the presence of SDS. This dimer band shows pronounced upward streaking, rather than downward streaking, suggesting an equilibrium with higher weight oligomers such as trimeric and tetrameric assemblies.
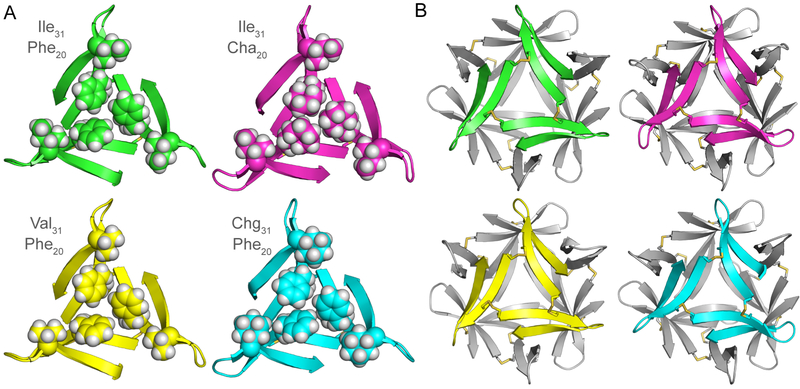
To further characterize the impact of these mutations on the structure and assembly of trimer 2, we determined their structures using X-ray crystallography. Of the five trimer 2 homologues, only trimer 2F19,20Cha failed to grow crystals. The four other trimer 2 homologues grew crystals with hexagonal morphologies, similar to crystals of trimer 2. To remove any possible model bias from using molecular replacement to generate the electron density maps, we determined the X-ray crystallographic phases of each mutant using sulfur single-wavelength anomalous diffraction (S-SAD) phasing.57–62 Table S1 summarizes the crystallographic properties, crystallization conditions, data collection, and model refinement statistics for trimers 2I31V, 2I31Chg, and 2F20Cha.63
The X-ray crystallographic structures of trimer 2I31V (PDB 6DR4), trimer 2I31Chg (PDB 6DR5), and trimer 2F20Cha (PDB 6DR6) are nearly identical to that of trimer 2 (PDB 5SUR, Figure 7). The mutations do not appear to alter the proposed crystal violet binding site, depicted in Figure 4. The mutant trimers differ mainly in the conformations of Asp23, Ala30, and the δ-linked ornithine turn that connects them.64 These mutations do not significantly alter the supramolecular assembly of the cross-linked trimers; the three trimer 2 homologues assemble into ball-shaped tetramers which resemble the ball-shaped tetramer formed by trimer 2 (Figure 7B). The tetramers pack together in the lattice to form dimer interfaces. Although the specific details of these dimer interfaces vary amongst the trimer 2 homologues, the modes of assembly are similar to that of trimer 2, which forms tetramers that pack in the lattice through dimer interfaces.31
Figure 7:
X-ray crystallographic structures of trimer 2 (PDB 5SUR, green), trimer 2F20Cha (PDB 6DR6, magenta), trimer 2I31V (6DR4, yellow), and trimer 2I31Chg (PDB 6DR5, cyan). (A) Comparison of the triangular trimers. The side chains of Phe20, Cha20, Ile31, Val31, and Chg31 are shown as spheres; the side chains of Cys17 and Cys21 are shown as sticks. (B) Comparison of the ball-shaped tetramers formed by trimer 2 and its homologues in the crystal lattices.
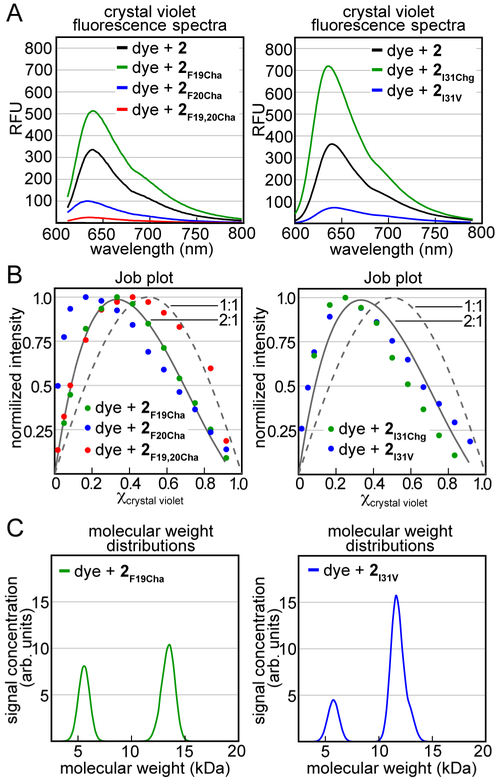
To ascertain the impact of these mutations on the molecular recognition of crystal violet, we studied the interaction of crystal violet with the five trimer 2 homologues using fluorescence spectroscopy (Figure 8). We compared the fluorescence spectra of crystal violet in the presence of the five trimer 2 homologues to the fluorescence spectrum of crystal violet in the presence of trimer 2. Fluorescence spectroscopy reveals that the side chains of Phe20 and Ile31 are important for the recognition of crystal violet (Figure 8A). The fluorescence enhancements that occur upon mixing crystal violet with trimer 2F20Cha or 2F19,F20Cha are far smaller than those which occur upon mixing crystal violet with trimers 2 or 2F19Cha (Figure 8A, left). These results indicate that the aromatic surface formed by the Phe20 side chains is involved in the association of trimer 2 with crystal violet. The fluorescence enhancement that occurs upon mixing crystal violet with trimer 2I31Chg is greater than that which occurs upon mixing crystal violet and trimer 2, while the fluorescence enhancement that occurs upon mixing trimer 2I31V is far smaller that that which occurs upon mixing crystal violet with trimer 2 (Figure 8A, right). These results indicate that the steric bulk provided by the Ile31 side chains is involved in the association of trimer 2 with crystal violet and suggest that the bulkier Chg stabilizes the 2:1 trimer-dye complex while the smaller Val destabilizes the 2:1 trimer-dye complex.
Figure 8:
Complexation of crystal violet by trimer 2 homologues. (A) Fluorescence spectra from mixtures of crystal violet and trimers 2, 2F19Cha, 2F20Cha, and 2F19,20Cha (left) and trimers 2, 2I31Chg, and 2I31V (right). Fluorescence spectra where acquired from samples comprising 20 μM crystal violet and 20 μM of the indicated trimer using an excitation wavelength of 550 nm. (B) Job plot analyses for trimers 2F19Cha, 2F20Cha, and 2F19,20Cha (left) and trimers 2I31Chg and 2I31V (right). The dashed line is a 1:1 binding model, while the solid line is a 2:1 binding model. Job plot experiments where conducted with a total concentration of 30 μM crystal violet and the indicated trimer. (C) Molecular weight distributions calculated from sedimentation velocity analytical ultracentrifugation experiments for trimer 2F19Cha (left) and trimer 2I31V (right). the distributions are calculated from samples comprising 10 μM crystal violet and 60 μM of the indicated trimer. All experiments were performed in buffer comprising 50 mM sodium acetate and 50 mM acetic acid (pH 4.6).
We used Job’s method of continuous variation to determine the stoichiometry of the five trimer 2 homologue-crystal violet complexes.44–47 All five homologues deviate from a simple 1:1 trimer-dye binding model (Figure 8B). The Job plot for trimer 2F19Cha fits well to a 2:1 binding model, with a dissociation constant of ca. 0.92·10−9 M2. This dissociation constant is similar to that of the trimer 2-crystal violet dissociation constant determined by Job’s method of continuous variation (above). The Job plots for the other four trimer 2 homologues do not fit well to either a 2:1 or a 1:1 binding model. The deviations from these models may reflect more complex associations of crystal violet with these trimer 2 homologues. The maxima in these Job plots occur at χcrystal violet ranging from 0.2–0.4, and suggest that multiple molecules of the trimer 2 homologues associate with one molecule of crystal violet. Alternatively, the deviations may reflect unavoidable errors that result from the lower fluorescence intensity of these complexes coupled with the absorption of the emitted light by the dye in samples that contain higher concentrations of crystal violet (e.g., χcrystal violet >0.3).
To corroborate the apparent stoichiometries of the trimer 2 homologue-crystal violet complexes, we performed sedimentation velocity analytical ultracentrifugation experiments (Figure S7). In these experiments, we monitored the sedimentation of 10 μM crystal violet in the presence of 60 μM of each of the trimer 2 homologues. When mixed with trimers 2F19Cha and 2I31V, crystal violet sediments as a mixture of ca. 5 and 13 kDa species (Figure 8C). These molecular weights are largely consistent with crystal violet associating with one and two molecules of trimer 2F19Cha or trimer 2I31V. When mixed with trimer 2I31Chg, crystal violet sediments as high molecular weight aggregates (Figure S7A). In contrast, crystal violet does not sediment when mixed with trimers 2F20Cha or 2F19,20Cha (Figure S7E and F). This lack of sedimentation reflects the weak association of crystal violet with either of these mutants, as also seen in the reduction in fluorescence intensity relative to trimer 2 (Figure 8A).
Collectively, these studies support the model depicted in Figure 4, wherein the three aryl rings of crystal violet pack against the side chains of Phe20 and the three N,N-dimethylamino substituents pack against the side chains of Ile31. The complex likely resembles a sandwich, in which two copies of trimer 2 sandwich around crystal violet through the interactions described above. Such a model is consistent with the observation that two molecules of trimer 2 bind to one molecule of dye. This sandwich is likely in equilibrium with an “open-faced” sandwich, wherein only one molecule of trimer 2 binds crystal violet. Such an equilibrium is consistent with the lack of an isosbestic point in the transition from free to bound dye (Figure 2B), and the observation of crystal violet sedimenting as ca. 5 and 13 kDa species in the presence of trimers 2F19Cha and 2I31V (Figure 8C).
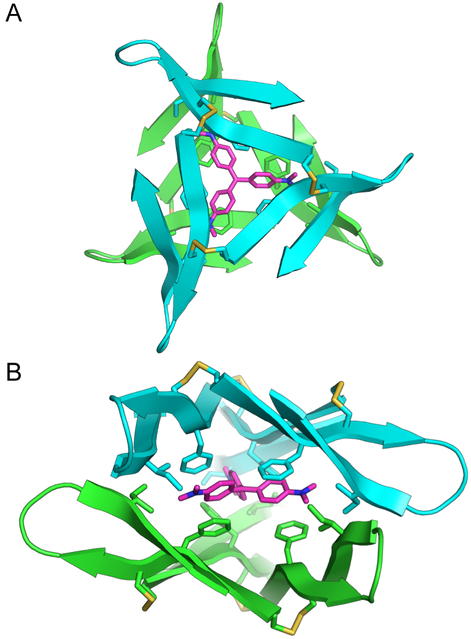
We used a combination of docking and molecular mechanics to construct a model for the sandwich complex (Figure 9). In this model, two molecules of trimer 2 envelop one molecule of crystal violet. The side chains from the six Phe20 residues sandwich the three aryl rings of crystal violet, and the side chains from two Ile31 residues clasp each N,N-dimethylamino group. These interactions are similar to the interactions between crystal violet and other proteins observed in high resolution X-ray crystallographic structures.50,52,53 These structures show that the aryl rings of crystal violet pack against aromatic residues and that the N,Ndimethylamino groups pack against hydrophobic residues. These co-crystal structures do not indicate that the three-fold symmetry of crystal violet is important for recognition, unlike the trimer 2-crystal violet model. The association of ethyl violet with an insulin hexamer does suggest that symmetry of the triphenylmethane dye can be important in complexation of an oligomer.65
Figure 9:
Molecular model of the 2:1 trimer 2-crystal violet complex. (A) Top view. (B) Side view. Crystal violet and the side chains of Cys17, Phe20, Cys21, and Ile31 are shown as sticks.
This working model of the trimer 2-crystal violet complex may explain the interaction of other triphenylmethane dyes with full-length Aβ. The triphenylmethane dye coomassie brilliant blue G associates with neurotoxic trimers and tetramers formed by Aβ.66 This triphenylmethane dye differs from those studied here in the substitution of the three nitrogen atoms and in methylation of the triphenylmethane core. Formation of these complexes detoxifies these neurotoxic oligomers.67 This triphenylmethane dye also alters the in vitro aggregation of Aβ and is neuroprotective in animal models of Alzheimer’s disease.68–70 It is possible that coomassie brilliant blue G is recognized by a surface similar to that which recognizes crystal violet in trimer 2. The selectivity of other oligomer-specific dyes has been rationalized through docking studies with the X-ray crystallographic structures of triangular trimers reported by our laboratory.22–24 BODIPY-based probes,22 developed by the Chang group, spiropyran-based probes,23 developed by the Yi group, and cyanine-based probes,24 developed by the Wong group, all specifically stain Aβ oligomers. To explain the oligomer-specific reactivity of these probes, all three groups performed docking simulations, in which the probes were docked on a triangular trimer formed by an Aβ-derived peptide (PDB 4NTR). These docking studies suggest that the side chains of three Phe19 residues pack against the probes and that the side chains of three Val36 residues clasp the probes. These proposed interactions are similar to those that occur between crystal violet and trimer 2, wherein crystal violet packs against an aromatic surface and is clasped by hydrophobic side chains. While these three fluorogenic probes are not C3-symmetric, they may recognize a surface similar to that which binds crystal violet in trimer 2.
Conclusion
Crystal violet and other triphenylmethane dyes have rich history in their use as metachromatic probes for amyloid deposition in tissue.71 The results described here suggest that these dyes may be repurposed to serve as metachromatic and fluorogenic probes for amyloid oligomers. The X-ray crystallographic structure of trimer 2 suggested that C3 symmetric small molecules might bind to the triangular trimer. Several different C3 symmetric triphenylmethane dyes associate with trimer 2 to form supramolecular complexes. Association with trimer 2 alters the color of the triphenylmethane dyes, and enhances their fluorescence. These changes and enhancements are specific to trimer 2 and several homologues, but do not occur in the presence of the unassembled monomer or in the presence of the related triangular trimer 3. The triphenylmethane dyes appear to pack against the aromatic surface formed by the three Phe20 side chains while being clasped by the three Ile31 side chains. These triphenylmethane dyes recognize a motif distinct from the amyloid fibrils recognized by planar aromatic dyes.
These studies suggest a blueprint for other small molecules that may bind to Aβ-derived triangular trimers, wherein C3-symmetric molecules position three aryl groups around the symmetry axis. We envision that such molecules may be identified in fluorescence- and absorbance-based competition assays in which small molecules compete with crystal violet for the binding site formed by Phe20 and Ile31.72,73 We also envision that the fluorescence enhancements of crystal violet may facilitate imaging trimer 2 and related triangular trimers in cellular environments.74–77 Triphenylmethane dyes, and other C3-symmetric small molecules, thus might prove a rich source of chemical probes to explore the structure and function triangular trimers.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health GM097562 and the National Science Foundation CHE-1507840 and CHE-1808096. The authors thank the Laser Spectroscopy Facility at University of California, Irvine for assistance with fluorescence measurements. The development of UltraScan was supported by NIH grant GM120600 and NSF grant NSF-ACI-1339649. Supercomputer calculations were performed on Comet at the San Diego Supercomputing Center, supported through NSF/XSEDE grant TG-MCB070039N, and on Lonestar-5 at the Texas Advanced Computing Center, supported through UT grant TG457201. We also acknowledge the support of the San Antonio Cancer Institute grant P30 CA054174 for support of the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center at San Antonio. P.J.S. thanks the UCI Training Program in Chemical and Structural Biology T32 GM108561 for training grant support as well as ARCS Foundation Orange County for additional support.
Footnotes
Supporting Information Available
Procedures for the synthesis of trimers 2; details of the crystallization, X-ray diffraction data collection, processing, and refinement of trimers 2; procedures for absorbance spectroscopy, fluorescence spectroscopy, analytical ultracentrifugation, isothermal titration calorimetry, SDS-PAGE, size exclusion chromatography, and Job’s method of continuous variation; crystallographic data in CIF format. Crystallographic coordinates of trimer 2I31V, trimer 2I31Chg, and trimer 2F20Cha were deposited into the Protein Data Bank with PDB codes 6DR4, 6DR5, and 6DR6.
This material is available free of charge via the Internet at http://pubs.acs.org/.
Notes
The authors declare no competing financial interest.
Notes and References
- (1).Vassar PS; Culling CF Fluorescent stains, with special reference to amyloid and connective tissue. Arch. Pathol 1959, 68, 487–498. [PubMed] [Google Scholar]
- (2).Naiki H; Higuchi K; Hosokawa M; Takeda T Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavine T. Anal. Biochem 1989, 177, 244–249. [DOI] [PubMed] [Google Scholar]
- (3).Westermark GT; Johnson KH; Westermark P Staining methods for identification of amyloid in tissue. Methods Enzymol. 1999, 309, 3–25. [DOI] [PubMed] [Google Scholar]
- (4).Frid P; Anisimov SV; Popovic N Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev 2007, 53, 135–160. [DOI] [PubMed] [Google Scholar]
- (5).Hawe A; Sutter M; Jiskoot W Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res 2008,, 25, 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gestwicki JE; Crabtree GR; Graef IA Harnessing chaperones to generate smallmolecule inhibitors of amyloid-β aggregation. Science 2004, 306, 865–869. [DOI] [PubMed] [Google Scholar]
- (7).Klunk WE; Engler H; Nordberg A; Wang Y; Blomqvist G; Holt DP; Bergström M; Savitcheva I; Huang GF; Estrada S; Ausén B; Debnath ML; Barletta J; Price JC; Sandell J; Lopresti BJ; Wall A; Koivisto R; Antoni G; Mathis CA; Långström B Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann. Neurol 2004, 55, 306–319. [DOI] [PubMed] [Google Scholar]
- (8).Yang L; Rieves D; Ganley C Brain amyloid imaging–FDA approval of florbetapir F18 injection. N. Engl. J. Med 2012, 367, 885–887. [DOI] [PubMed] [Google Scholar]
- (9).Hatai J; Motiei L; Margulies D Analyzing amyloid β aggregates with a combinatorial fluorescent molecular sensor. J. Am. Chem. Soc 2017, 139, 2136–2139. [DOI] [PubMed] [Google Scholar]
- (10).Scherzer-Attali R; Pellarin R; Convertino M; Frydman-Marom A; Egoz-Matia N; Peled S; Levy-Sakin M; Shalev DE; Caflisch A; Gazit E; Segal D Complete phenotypic recovery of an Alzheimer’s disease model by a quinone-tryptophan hybrid aggregation inhibitor. PLOS one. 2010, 5, e11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Landau M; Sawaya MR; Faull KF; Laganowsky A; Jiang L; Sievers SA; Liu J; Barrio JR; Eisenberg D Toward a pharmacophore for amyloid. PLOS Biol. 2011, 9, e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wu C; Scott J; Shea J-E Binding of Congo red to amyloid protofibrils of the Alzheimer Aβ9–40 peptide probed by molecular dynamics simulations. Biophys. J 2012, 103, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Skeby KK; Sørensen J; Schiøtt B Identification of a common binding mode for imaging agents to amyloid fibrils from molecular dynamics simulations. J. Am. Chem. Soc 2013, 135, 15114–15128. [DOI] [PubMed] [Google Scholar]
- (14).Scherzer-Attali R; Convertino M; Pellarin R; Gazit E; Segal D; Caflisch A Methylations of tryptophan-modified naphthoquinone affect its inhibitory potnetial toward Aβ aggregation. J. Phys. Chem. B 2013, 117, 1780–1789. [DOI] [PubMed] [Google Scholar]
- (15).Lemkul JA; Bevan DR The role of molecular simulations in the development of inhibitors of amyloid-β peptide aggregation for the treatment of Alzheimer’s disease. ACS Chem. Neurosci 2102, 3, 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Parikh ND; Klimov DK Molecular mechanisms of Alzheimer’s biomaker FDDNP binding to Aβ amyloid fibril. J. Phys. Chem. B 2015, 119, 11568–11580. [DOI] [PubMed] [Google Scholar]
- (17).Young LM; Ashcroft AE; Radford SE Small molecule probes of protein aggregation. Curr. Opin. Chem. Biol 2017, 39, 90–99. [DOI] [PubMed] [Google Scholar]
- (18).He H; Xu J; Cheng D-Y; Fu L; Ge Y-S; Jiang F-L; Liu Y Identification of binding modes for amino naphthalene 2-cyanoacrylate (ANCA) probes to amyloid fibrils from molecular dynamics simulations. J. Phys. Chem. B 2017, 121, 1211–1221. [DOI] [PubMed] [Google Scholar]
- (19).Sangwan S; Sawaya MR; Murray KA; Hughes MP; Eisenberg DS Atomic structures of corkscrew-forming segments of SOD1 reveal varied oligomer conformations. Protein Sci. 2018, 27, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jiang L; Liu C; Leibly D; Landau M; Zhao M; Hughes MP; Eisenberg DS Structure-based discovery of fiber-binding compounds that reduce the cytotoxicity of amyloid-β. ELife 2013, 2, e00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Maezawa I; Hong H-S; Liu R; Wu C-Y; Cheng RH; Kung M-P; Kung HF; Lam KS; Oddo S; LeFerla FM; Jin L-W Congo red and thioflavin-T analogs detect Aβ oligomers. J. Neurochem 2008, 104, 457–468. [DOI] [PubMed] [Google Scholar]
- (22).Teoh CL; Su D; Sahu S; Yun S-W; Drummond E; Prelli F; Lim S; Cho S; Ham S; Wisniewski T; Chang Y-T Chemical fluorescent probe for detection of Aβ oligomers. J. Am. Chem. Soc 2015, 137, 13503–13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lv G; Sun A; Wei P; Zhang N; Haichuang L; Yi T A spiropyran-based fluorescent probe for the specific detection of β-amyloid peptide oligomers in Alzheimer’s disease. Chem. Commun 2016, 52, 8865–8868. [DOI] [PubMed] [Google Scholar]
- (24).Li Y; Xu D; Sun A; Ho S-L; Poon C-Y; Chan H-N; Ng OTW; Yung KKL; Yan H; Li H-W; Wong MS Fluoro-substituted cyanine for reliable in vivo labelling of amyloid-β oligomers and neuroprotection against amyloid-β induced toxicity. Chem. Sci 2017, 8, 8279–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Yates EV; Meisl G; Knowles TPJ; Dobson CM An environmentally sensitive fluorescent dye as a multidimensional probe of amyloid formation. J. Phys. Chem. B 2016, 120, 2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kumar M; Hong Y; Thorn DC; Ecroyd H; Carver JA Monitoring early-stage protein aggregation by an aggregation-induced emission fluorogen. Anal. Chem 2017, 89, 9322–9329. [DOI] [PubMed] [Google Scholar]
- (27).Brahmachari S; Paul A; Segal D; Gazit E Inhibition of amyloid oligomerization into different supramolecular architectures by small molecules: mechanistic insights and design rules. Future Med. Chem 2017, 9, 797–810. [DOI] [PubMed] [Google Scholar]
- (28).Reinke AA; Gestwicki JE Insight into amyloid structure using chemical probes.Chem. Biol. Drug Des 2011, 77, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).LeVine H; Walker LC What amyloid ligands can tell us about molecular polymorphism and disease. Neurobiol. Aging 2016, 42, 205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Polyclonal and monoclonal antibodies have been developed as biochemical probes for amyloid oligomers. The structures of the oligomers that these antibodies recognize are also unknown: De Genst E; Messer A; Dobson CM Biochim. Biophys. Acta 2014, 1844, 1907–1919. [DOI] [PubMed] [Google Scholar]
- (31).Kreutzer AG; Yoo S; Spencer RK; Nowick JS Stabilization, assembly, and toxicity of trimers derived from Aβ. J. Am. Chem. Soc 2017, 139, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Spencer RK; Li H; Nowick JS X-ray crystallographic structures of trimers and higher-order oligomeric assemblies of a peptide derived from Aβ17–36. J. Am. Chem. Soc 2014, 136, 5595–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).See reference 31, wherein the molecule named “trimer 6” is the same molecule we call trimer 2 in this report.
- (34).Kayed R; Head E; Thompson JL; McIntire TM; Milton SC; Cotman CW; Glabe CG Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003, 300, 486–489. [DOI] [PubMed] [Google Scholar]
- (35).Benilova I; Karran E; De Stooper B The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat. Neurosci 2012, 15, 349–357. [DOI] [PubMed] [Google Scholar]
- (36).Sievers SA; Karanicolas J; Chang HW; Zhao A; Jiang L; Zirafi O; Stevens JT; Mu¨nch J; Baker D; Eisenberg D Structure-based design of nonnatural amino-acid inhibitors of amyloid fibril formation. Nature 2011, 475, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Seidler PM; Boyer DR; Rodriguez JA; Sawaya MR; Cascio D; Murray K; Gonen T; Eisenberg D Structure-based inhibitors of tau aggregation. Nat. Chem 2018, 10, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Duxbury DF The photochemistry and photophysics of triphenylmethane dyes in solid and liquid media. Chem. Rev 1993, 93, 381–433. [Google Scholar]
- (39).Baptista MS; Indig GL Effect of BSA on photophysical and photochemical properties of triarylmethane dyes. J. Phys. Chem. B 1998, 102, 4678–4688. [Google Scholar]
- (40).Bartlett JA; Indig GL Spectroscopic and photochemical properties of malachite green noncovalently bound to bovine serum albumin. Dyes Pigm. 1999, 43, 219–226. [Google Scholar]
- (41).Qin M; Yin T; Shen W The interaction between crystal violet and bovine serum albumin: spectroscopic and molecular docking investigations. J. Dispersion Sci. Technol 2015, 37, 1623–1629. [Google Scholar]
- (42).Hong Y; Lam JWY; Tang BZ Aggregation-induced emission. Chem. Soc. Rev 2011, 40, 5361–5388. [DOI] [PubMed] [Google Scholar]
- (43).Mei J; Leung NLC; Kwok RTK; Lam JWY; Tang BZ Aggregation-induced emission: Together we shine, united we soar! Chem. Rev 2015, 115, 11718–11940. [DOI] [PubMed] [Google Scholar]
- (44).Job P Formation and stability of inorganic complexes in solution. Ann. Chim 1928, 9, 113–203. [Google Scholar]
- (45).Bruneau E; Lavabre D; Levy G; Micheau JC Quantitative analysis of continuousvariation plots with a comparison of several methods. J. Chem. Educ 1992, 69, 833–837. [Google Scholar]
- (46).Olson EJ; Bühlmann P Getting more out of a Job Plot: determination of reactant to product stoichiometry in cases of displacement reactions and n:n complex formation. J. Org. Chem 2011, 76, 8406–8412. [DOI] [PubMed] [Google Scholar]
- (47).Truex NL; Nowick JS Coassembly of peptides derived from β-sheet regions of β-amyloid. J. Am. Chem. Soc 2016, 138, 13891–13900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ulatowski F; Dąbrowa K; Bałakier T; Jurczak J Recognizing the limited applicability of job plots in studying host-guest interactions in supramolecular chemistry. J. Org. Chem 2016, 81, 1746–1756. [DOI] [PubMed] [Google Scholar]
- (49).Lurtz MM; Pedersen SE Aminotriarylmethane dyes are high-affinity noncompetitive antagonists of the nicotinic acetylcholine receptor. Mol. Pharmacol 1999, 55, 159–167. [DOI] [PubMed] [Google Scholar]
- (50).Schumacher MA; Miller MC; Grkovic S; Brown MH; Skurray RA; Brennan RG Structural mechanisms of QacR induction and multidrug recognition. Science 2001, 294, 2158–2163. [DOI] [PubMed] [Google Scholar]
- (51).Arias HR; Bhumireddy P; Spitzmaul G; Trudell JR; Bouzat C Molecular mechanisms and binding site location for the noncompetitive antagonist crystal violet on nicotinic acetylcholine receptors. Biochemistry 2006, 45, 2014–2026. [DOI] [PubMed] [Google Scholar]
- (52).Yamasaki S; Nikaido E; Nakashima R; Sakurai K; Fujiwara D; Fujii I; Nishino K The crystal structure of multidrug-resistance regulator RamR with multiple drugs. Nat. Commun 2013, 4, 2078. [DOI] [PubMed] [Google Scholar]
- (53).Allgardson A; Andersson CD; Akfur C; Worek F; Linusson A; Ekström F An unusual dimeric inhibitor of acetylcholinesterase: Cooperative binding of crystal violet. Molecules 2017, 22, E1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Morris GM; Huey R; Lindstrom W; Sanner MF; Belew RK; Goodsell DS; Olson AJ Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem 2009, 30, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Salveson PJ; Haerianardakani S; Thuy-Boun A; Kreutzer AG; Nowick JS Controlling the oligomerization state of Aβ-derived peptides with light. J. Am. Chem. Soc 2018, 140, 5842–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Korn A; Surendran D; Krueger M; Maiti S; Huster D Ring structure modifications of phenylalanine 19 increase fibrillation kinetics and reduce toxicity of amyloid β (1–40). Chem. Commun 2018, 54, 5430–5433. [DOI] [PubMed] [Google Scholar]
- (57).Hendrickson WA; Teeter MM Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulphur. Nature 1981, 290, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Sarma GN; Karplus PA In-house sulfur SAD phasing: a case study of the effects of data quality and resolution cutoffs. Acta Crystallogr., Sect. D: Biol. Crystallogr 2006, 62, 707–716. [DOI] [PubMed] [Google Scholar]
- (59).Lie Q; Dahmane T; Zhang Z; Assur Z; Brasch J; Shapiro L; Mancia F; Hendrickson WA Structures from anomalous diffraction of native biological macromolecules. Science 2012, 336, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Liu Q; Liu Q; Hendrickson WA Robust structural analysis of native biological macromolecules from multi-crystal anomalous diffraction data. Acta Crystallogr., Sect. D: Biol. Crystallogr 2013, 69, 1314–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Liu Q; Hendrickson WA Crystallographic phasing from weak anomalous signals. Curr. Opin. Struct. Biol 2015, 34, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Liu Q; Hendrickson WA Contemporary use of anomalous diffraction in biomolecular structure analysis. Methods Mol. Biol 2017, 1607, 377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).We were unable to determine the X-ray crystallographic structure of trimer 2F19Cha using either S-SAD or molecular replacement.
- (64).The backbone RMSD overlay of the trimer 2 homologues with trimer 2 ranges from 0.9–1.1Å.
- (65).Mohanty J; Shinde MN; Barooah N; Bhasikuttan AC Reversible insulin hexamer assembly promoted by ethyl violet: pH-controlled uptake and release. J. Phys. Chem. Lett 2016, 7, 3978–3983. [DOI] [PubMed] [Google Scholar]
- (66).Jana MK; Cappai R; Ciccotosto GD Oligomeric amyloid-β toxicity can be inhibited by blocking its cellular binding in cortical neuronal cultures with addition of the triphenylmethane dye brilliant blue G. ACS Chem. Neurosci 2016, 7, 1141–1147. [DOI] [PubMed] [Google Scholar]
- (67).We have been unable to determine if crystal violet or its homologues can detoxify trimer 2, as these triphenylmethane dyes are toxic to the neuroblastoma SH-SY5Y cell line at concentrations at which the dyes associate with trimer 2.
- (68).Wong HE; Qi W; Choi H-M; Fernandez EJ; Kwon I A safe, blood-brain barrier permeable triphenylmethane dye inhibits amyloid-β neurotoxicity by generating nontoxic aggregates. ACS Chem. Neurosci 2011, 2, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Chen X; Hu J; Jiang L; Xu S; Zheng B; Wang C; Zhang J; Wei X; Chang L; Wang Q Brilliant blue G improves cognition in an animal model of Alzheimer’s disease and inhibits amyloid-β-induced loss of filopodia and dendrite spines in hippocampal neurons. Neuroscience 2014, 279, 94–101. [DOI] [PubMed] [Google Scholar]
- (70).Irwin JA; Erisir A; Kwon I Oral triphenymethane food dye analog, brilliant blue G, prevents neuronal loss in APPSwDI/NOS2−/− mouse model. Curr. Alzheimer Res 2016, 13, 663–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Bancroft JD Methyl green as a differentiator and counterstain in the methyl violet technique for the demonstration of amyloid. Stain Technol. 1983, 38, 336–337. [PubMed] [Google Scholar]
- (72).Dsouza RN; Pischel U; Nau WM Fluorescent dyes and their supramolecular host/guest complexes with macrocycles in aqueous solution. Chem. Rev 2011, 111, 7941–7980. [DOI] [PubMed] [Google Scholar]
- (73).Wu J; Kwon B; Liu W; Anslyn EV; Wang P; Kim JS Chromogenic/fluorogenic ensemble chemosensing systems. Chem. Rev 2015, 115, 7893–7943. [DOI] [PubMed] [Google Scholar]
- (74).Babendure JR; Adams SR; Tsien RY Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc 2003, 125, 14716–14717. [DOI] [PubMed] [Google Scholar]
- (75).Bhasikuttan AC; Mohanty J; Nau WM; Pal H Efficient fluorescence enhancement and cooperative binding of an organic dye in a supra-biomolecular host-protein assembly. Angew. Chem. Int. Ed 2007, 46, 4120–4122. [DOI] [PubMed] [Google Scholar]
- (76).Szent-Gyorgi C; Schmidt BF; Creeger Y; Fisher GW; Zakel KL; Adler S; Fitzpatrick JAJ; Woolford CA; Yan Q; Vasilev KV; Berget PB; Bruchez MP; Jarvik JW; Waggoner A Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat. Biotechnol 2008, 26, 235–240. [DOI] [PubMed] [Google Scholar]
- (77).Gotrik M; Sekhon G; Saurabh S; Nakamoto M; Eisenstein M; Soh HT Direct selection of fluorescence-enhancing RNA aptamers. J. Am. Chem. Soc 2018, 140, 3583–3591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.