Abstract
A large proportion of biomedical research and the development of therapeutics is focused on a small fraction of the human genome. In a strategic effort to map the knowledge gaps around proteins encoded by the human genome and to promote the exploration of currently understudied, but potentially druggable, proteins, the US National Institutes of Health launched the Illuminating the Druggable Genome (IDG) initiative in 2014. In this article, we discuss how the systematic collection and processing of a wide array of genomic, proteomic, chemical and disease-related resource data by the IDG Knowledge Management Center have enabled the development of evidence-based criteria for tracking the target development level (TDL) of human proteins, which indicates a substantial knowledge deficit for approximately one out of three proteins in the human proteome. We then present spotlights on the TDL categories as well as key drug target classes, including G protein-coupled receptors, protein kinases and ion channels, which illustrate the nature of the unexplored opportunities for biomedical research and therapeutic development.
Target selection and prioritization are common goals for academic and commercial drug research organizations. While motivations differ, in all cases, the target selection task is fundamentally one of resource allocation in the face of incomplete information. Consequently, target selection strategies (and metric-based approaches to assess their success) remain complex1 and are hindered by multiple bottlenecks. Some bottlenecks pertain to the data themselves, such as disjointed, disparate data and metadata standards, data recording errors and accessibility issues; overcoming these issues will require human and computational efforts and coordination across multiple communities. Another set of bottlenecks pertains to the scientists involved. These include a tendency to focus on a small subset of well-known genes2 and the tendency to avoid riskier research paths, driven by poor research funding climates3.
For the purposes of this article, we define knowledge as the consensus of information aggregated from dif-ferent sources and information as structured data, with a contextual layer that supports a broad range of data analytics. Data have quantity, quality and dimensionality (for example, genomic knowledge is defined in relation to associations with distinct entities such as molecular probes and disease concepts). Data, like facts, may also have an expiration date (Supplementary Box S1), and thus knowledge is subject to change. Yet, within a given time frame, knowledge provides context for interpretation and integration of emergent data, information and models.
Data-driven drug discovery strategies rely on the integration of proprietary and internal data with third-party resources — both public databases, such as PubMed, PubChem4, ChEMBL5 and The Cancer Genome Atlas (TCGA6), and commercial databases, such as Integrity. This integration requires fusion and reconciliation of heterogeneous and sometimes conflicting data sources and types. Although many of these resources are already partially interlinked, data heterogeneity, complexity and incompleteness, as well as contextual information and metadata capture, pose substantial barriers to reliable systematic analyses of all data required to address biomedical research questions, such as target prioritization in drug discovery1.
With the increasing scale and variety of data generation, collection and curation in the biomedical sciences, there is an unmet need for in-depth, accurate and truthful integration of multiple scientific domains across disciplines. Once successful, these data and knowledge integration efforts enable us to ask both global and fundamental questions about genes, proteins and the processes they are involved in. Integrated resources also allow us to address aspects of reproducibility7 via concordance of similar data types from unrelated sources and deficits in our knowledge of biological systems and their function. More generally, data integration facilitates our ability to quantify knowledge using an evidence-based approach.
Illuminating the Druggable Genome.
“The reluctance to work on the unknown” (REF. 2) is inherent to the scientific endeavour, partly due to our subconscious tendency to choose research subjects more likely to confirm what we already know or believe8. In a deliberate, strategic attempt to map the knowledge gaps around potential drug targets and to prompt exploration of currently understudied but potentially druggable proteins, the US National Institutes of Health (NIH) launched the Illuminating the Druggable Genome (IDG) initiative in 2014. As part of this broad, multimillion-dollar initiative, the IDG Knowledge Management Center (KMC) aims to systematize general and specific biomedical knowledge by processing a wide array of genomic, proteomic, chemical and disease-related resources (BOX 1), with the explicit goal of supporting target hypothesis generation and subsequent knowledge creation, especially for genes and proteins that are not well studied.
Box 1 | Overview of the Illuminating the Druggable Genome Knowledge Management Center
Knowledge management implies the ability to structure data into information88 while combining low-volume, high-quality data, such as thorough analyses of experimental data (for example, high-resolution Xray crystallographic structures) or evidence-based systematic reviews (for example, the Cochrane Collaboration), with high-volume (and perhaps lower quality) data such as genome-wide association studies (GWAS) or high-throughput screening data sets. As the overall scientific process requires the archiving, evaluation and re-interpretation of sometimes conflicting data, the Illuminating the Druggable Genome Knowledge Management Center (IDG KMC) faces similar challenges. Consensus emerges based on repeated independent experiments, robustness of the results (for example, modified reagents or conditions, or model organisms), increased domain expertise and qualitative judgement. To this end, the IDG KMC automates algorithmic processing of structured data by extracting and processing expression and functional data related to proteins and genes, molecular probes such as small molecules and antibodies, small-molecule bioactivities, GWAS, disease associations and launched drug information (among other data types) into the Target Central Resource Database (TCRD)89. TCRD content is presented via Pharos, a multimodal web interface89 (see below).
TCRD–Pharos is not unique in providing integrated content: ChEMBL, DrugBank90 and UniProt91 are excellent examples of drug discovery integration systems, for example, for chemical structure and drug bioactivity data and protein and disease information, largely focused on a specific knowledge domain. CiViC92 combines multiple resources with a specific goal, for example, to enable clinical interpretation of gene variants. The only resource that parallels the scope of IDG KMC is OpenTargets93, a consortium focused on disease-specific target validation efforts. The KMC collates evidence about all human proteins from multiple domains, supporting research on understudied proteins and new biology, and includes the following resources.
Target Central Resource Database
TCRD is the central open-access data repository for the IDG KMC and is the primary data source for the IDG KMC project-wide web portal Pharos89. TCRD integrates 55 heterogeneous data sets, with over 85 million gene and/or protein attributes. Special emphasis is placed on four families that were of interest to the pilot phase of the IDG programme: G protein-coupled receptors, ion channels, kinases and nuclear receptors (TABLE 1). The focus on this fraction of the proteome is justified by historical evidence, which indicates that these four protein families are among the most consistently successful druggable target classes (see also TABLE 2). TCRD is available under the CC-BY-SA 4.0 licence. Programmatic access to TCRD is also available via a REST application programme interface (API).
Pharos
Access to TCRD content is via the web portal Pharos89, which is a Java platform that supports efficient and intuitive search queries and browsing of all TCRD data. Features include search filters to reduce lists of targets, query-saving capability for sharing, and dossier functionality to collate data during searching or browsing. Pharos provides an extensive REST API to support programmatic access and inclusion in pipelining tools.
Harmonizome
Given the wide variety of experimental data that is generated on individual proteins, it is useful to characterize the total availability of data types around individual targets. This Harmonizome is a resource developed for KMC17 that contains a collection of processed data sets from 70 major online resources, abstracted and organized into ~72 million functional associations between genes and proteins and their attributes. Such attributes could be physical relationships with other biomolecules, expression in cell lines and tissues, genetic associations with knockout mouse or human phenotypes or changes in expression after drug treatment.
These associations are stored in a relational database along with rich metadata for genes and proteins, their attributes and the original sources. To report overall levels of knowledge for each target, the Harmonizome computes a cumulative probability of a protein occurring within a given data set. With appropriate normalization, this results in an association score for a protein–data source pair, with values ranging from 0 to 1. When a source has no data associated with a target, its score is set to 0. Currently, 110 individual data sources (including supplementary files from publications and public repositories of omics data) are made available through the Harmonizome, resulting in a 110-element vector representation for each target. From this vector, we compute the data availability score as the sum of the 110 association scores.
The Harmonizome is available through a web portal, a web service and a mobile app for querying, browsing and downloading all data. The Harmonizome visualizes gene–gene and attribute–attribute similarity networks for all processed data sets.
DrugCentral
This online compendium provides chemical, pharmacological and regulatory information for active pharmaceutical ingredients and pharmaceutical products by linking chemical entities, multiple drug identification codes, drug mode of action and pharmacological action at the target level, and pharmaceutical formulation and product-specific information, as well as indications, contraindications and off-label indications10. DrugCentral links 4,509 active ingredients to 93,084 pharmaceutical products and is available under the CC-BY-SA 4.0 licence.
Drug Target Ontology
This is an interactive framework to integrate, navigate and analyse drug discovery data, based on formalized and standardized classifications and annotations of human proteins94, available under the CC-BY-SA 4.0 licence.
In this article, we first define objective, evidence-based criteria for tracking target development levels (TDLs) for human proteins, using multiple sets of current knowledge. We discuss the data collected by the KMC on TDLs, which show the existence of a substantial knowledge deficit concerning a large portion of the human proteome (one out of three proteins). Reflecting the goal of illuminating the druggable genome, we then present spotlights on the TDL categories, as well as on key target classes, including G protein-coupled receptors (GPCRs), protein kinases and ion channels.
Knowledge-based protein classification
Target development levels.
Most current protein classification schemes are based on structural and functional criteria. For any given protein, it is also possible to identify associated drugs and chemical or biologic modulators, and many types of experimental data can be associated with the protein, including publications, patents, gene expression data and experimental or modelled 3D structures.
For target prioritization and therapeutic development, it is useful to understand the quantity and diversity of data that are available for a given protein and to assign a qualitative knowledge metric that characterizes the degree to which a target is comparatively well studied or unstudied. To address this, we developed the TDL classification scheme, which categorizes proteins into four groups — Tclin, Tchem, Tbio and Tdark — with respect to the depth of investigation from a clinical, chemical and bio-logical standpoint (FIG. 1; TABLE 1). Except for Tclin, TDL assignments were performed without human curation. Formal definitions for the TDL categories are as follows.
Tclin (clinic) proteins are drug targets linked to at least one approved drug (that is, an active pharmaceutical ingredient) by mechanism of action (MoA) (this criterion supersedes any of the other parameters). Classification into this TDL category was achieved through exhaustive manual querying of primary literature and drug labels for MoA assignments with respect to molecular (protein) targets9; drug targets annotated as MoA-related proteins are categorized as Tclin (see further discussion below)
Tchem (chemistry) proteins lack MoA-based links to approved drugs but are known to bind to small mole-cules with high potency. The interactions between proteins and small molecules (and sometimes approved drugs) are usually studied in the context of a disease and often arise from medicinal chemistry efforts. For inclusion in the Tchem category, we required the bio-activity of at least one small molecule to be above a specific cut-off chosen to include about 90% of the bioactivity values of drugs with a confirmed MoA for a target from that protein family (Supplementary Figure S2). Currently chosen thresholds are ≤30 nM for kinases, ≤100 nM for GPCRs and nuclear receptors, ≤10 μM for ion channels and ≤1 μM for other target families. Bioactivity values were extracted from ChEMBL5 and DrugCentral10.
Tbio (biology) refers to those proteins that have a confirmed Mendelian disease phenotype in the Online Mendelian Inheritance in Man (OMIM) database11 (that is, at least two publications), have Gene Ontology (GO)12 leaf term annotations based on experimental evidence or meet two of the following three conditions: a fractional PubMed publications count13 above five; three or more National Center for Biotechnology Information (NCBI) Gene Reference Into Function (RIF) annotations; or 50 or more commercial antibodies, counted from data made available by the Antibodypedia database14. Tbio assignments imply that these proteins are not MoA-related drug targets (these are Tclin proteins). However, it does not follow that these proteins lack associations with bioactive molecules, including approved small-molecule drugs and biologics. It does, however, imply that given current levels of evidence, associated bioactivity values and clinical observations did not meet Tchem or Tclin criteria, respectively.
Tdark (dark genome) refers to the remaining proteins that have been manually curated at the primary sequence level in UniProt15 yet do not meet any of the criteria for Tclin, Tchem or Tbio. Even for this category, evidence may be available concerning genome-wide association studies (GWAS), tissue location, dysregulation, inferred function via homology, etc. Many proteins in the Tdark category are not contextless sequences. However, these are proteins for which there is the least current knowledge and a low number of specific molecular probes available, and some represent unexplored opportunities within the druggable human genome. While evidence that approved drugs interact with some Tdark proteins may be available, the above criteria were observed for all Tdark assignments (Supplementary Table S3).
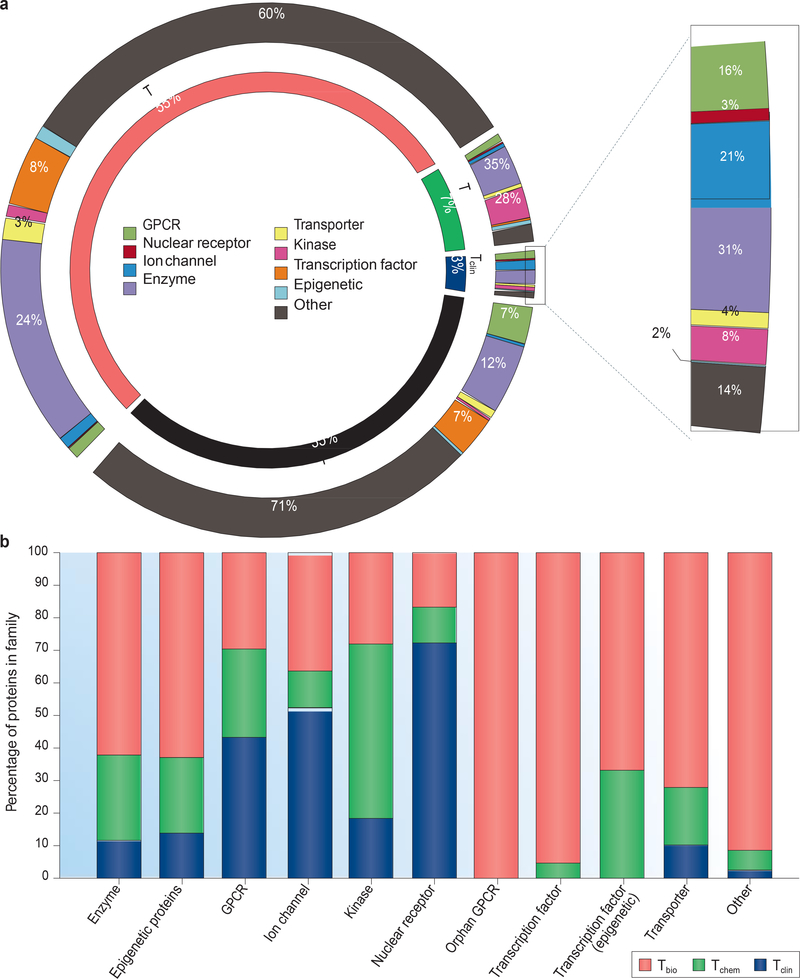
Figure 1 |. Target development level categories applied to the human proteome.
a | Percentages of the whole proteome are shown in the inner ring. Percentages of each target development level (TDL) category for selected major protein families are shown in the outer ring, with the Tclin category expanded. Inner ring colours are as follows: Tdark, black; Tbio, red; Tchem, green; and Tclin, blue. b | TDL distribution across protein families, coloured by TDL category. Data show 3,644 proteins that have a confirmed disease association according to the Online Mendelian Inheritance in Man (OMIM) database. The enzyme category excludes kinases, which are considered separately. GPCR, G protein-coupled receptor.
Table 1 |.
Current distribution of TDL categories by protein family for the druggable genome
| Tdark | |||||
|---|---|---|---|---|---|
| GPCRs (non-olfactory) | 406 | 96 | 113 | 145 | 52 |
| Olfactory GPCRs | 421 | 0 | 0 | 8 | 413 |
| Kinases | 634 | 50 | 390 | 163 | 31 |
| Ion channels | 355 | 126 | 44 | 150 | 35 |
| Nuclear receptors | 48 | 18 | 19 | 11 | 0 |
| Transporters | 473 | 26 | 46 | 287 | 114 |
| Transcription factors | 1,400 | 0 | 27 | 866 | 507 |
| Epigenetic proteinsa | 280 | 12 | 53 | 178 | 37 |
| Enzymesb | 4,146 | 186 | 493 | 2,607 | 860 |
| Others | 11,957 | 87 | 217 | 6,671 | 4,982 |
| Total | 20,120 | 601 | 1,402 | 11,086 | 7,031 |
GPCR, G protein-coupled receptor; TDL, target development level.
Includes 40 transcription factors not already counted with those in the transcription factor category.
Excludes kinases.
The knowledge deficit.
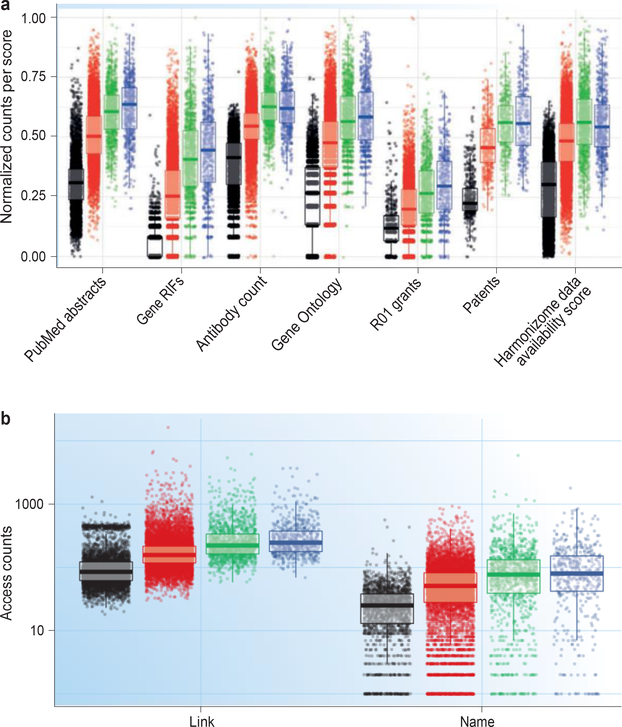
FIGURE 2a summarizes the varying degree of available data (represented using a normalized count of occurrence) for seven different data types associated with individual targets and grouped by TDL. The first three groups illustrate category differences for three TDL defining criteria discussed above — namely, the fractional count of protein and/or gene mentions in PubMed abstracts, NCBI Gene RIF counts and antibody counts per protein. ‘GO terms’ examines the distribution of GO12 annotation counts per protein using data from UniProt15. ‘R01 grants’ examines the distribution of textmined R01 grant counts detected for each protein using NIH RePORTER data (see below for further discussion). ‘Patents’ examines the distribution of text-mined granted patents for each protein using SureChEMBL16 data. Finally, the data availability score summarizes experimental information density per protein obtained from Harmonizome17 data — a resource developed independently for the KMC that provides an abstract representation of the many types of data associated with all human genes and proteins (BOX 1).
Figure 2 |. Patterns of target development level distribution across different data: visualizing the knowledge deficit.
a | The three criteria used in establishing the target development level are to the left, and their independent validation by four other data types are to the right. For PubMed abstracts, Gene Reference Into Function (RIF) annotations, antibodies, Gene Ontology, R01 grants and patents, the score for each target is the count of those entities associated with the target, normalized between 0 and 1. The values for the Harmonizome data availability score were computed differently, as described in the main text. See FIG. 1 for colour codes and Supplementary Table S4 for further details. b | Patterns of scientific curiosity: STRING database access counts by target development level (January–December 2016).
Whereas the first three data types were used to assign the TDL category for proteins in the Tbio and Tdark categories, the other four data types — derived from separate text corpora and repositories — provide independent validation of our criteria for categorization overall. Distribution trends within TDL categories are consistently reproduced across all data types in FIG. 2a and have statistically significant differences (Supplementary Table S4). Tdark proteins have the least amount of data associated with them regardless of source.
Increasing amounts of data are observed for proteins when progressing through the categories from Tdark to Tbio, Tchem and Tclin. For example, Tdark proteins tend not to be the object of study for many funded NIH R01 grants and are significantly less discussed in patents compared with proteins in other TDLs. Statistical significance breaks down when comparing Tclin and Tchem, but because successful clinical trials are required for the Tchem-to-Tclin progression, this evidence may not be well captured by the four data types highlighted in Supplementary Table S4. However, this is less surprising from a knowledge management perspective, since on average, the biochemistry and pharmacology of a protein are likely to be well studied upon reaching the Tchem development stage. It is important to note that the Tchem stage can be completely bypassed for targets of therapeutic antibodies and other biologics.
In summary, all the data, information and knowledge aggregated and processed within the IDG KMC archive (partially illustrated in FIG. 2a) confirm the existence of a knowledge deficit about many proteins, some of which could have therapeutic relevance. The bias towards well-described proteins2 is confirmed not only with respect to publications but also with respect to patents, NIH funding patterns, GWAS and mouse phenotype data (data not shown), availability of molecular probes such as antibodies and small molecules, and even queries in the STRING18database (see below and FIG. 2b). Because of this bias, one out of three human proteins (Tdark) have been largely unstudied. Although the NIH acknowledged that illumination should directly target understudied proteins, scientists engaged in target selection are likely to remain risk-averse and perhaps systematically less inclined to study Tdark proteins.
Our classification provides overall insight into the current illumination levels and sizes the opportunity for drug targets from well-established and precedented druggable protein families. The natural progression is for proteins of potential therapeutic interest to migrate from Tdark to Tclin over time, and TDL monitors knowledge accumulation using multiple types of clinical, chemical and biological evidence, while providing an easily interpretable ranking scheme. We argue that proteins in Tdark and Tbio are understudied and more in need of illumination, and we discuss approaches for achieving this later in the article, after first overviewing knowledge on proteins in the Tclin and Tchem categories.
Spotlight on Tclin and Tchem
Evaluating protein target druggability — the ability of a protein to be therapeutically modulated by medicines —can involve complex assessments of a range of protein characteristics. Structural biology and computational and medicinal chemistry assessments of druggability largely focus on forecasting whether a target protein can bind to drug-like small molecules with high affinity and specificity19. However, druggability literature rarely mentions biologics, antibodies and other protein therapeutics, radiotherapy (Supplementary Box S5), gene therapy or stem cells. In this section, we discuss Tclin and Tchem proteins in the context of small-molecule drug discovery.
Tclin proteins.
Ideally, unequivocal Tclin assignment (that is, identification of molecular drug targets) would require several layers of evidence: a full matrix of in vitro bio-activity for all prodrugs, drugs and active metabolites (active ingredients) assayed against all relevant human and non-human (for example, bacterial and viral) targets (such as the half-maximal inhibitory concentration (IC50), effector concentration for half-maximum response (EC50), inhibitory constant (Ki) and the dissociation constant (Kd)); on-off rate constants and other kinetic measure-ments performed at appropriately relevant concentra-tions in the tissue or tissues relevant for that particular disease context, preferably with matching in vivo data in humanized animal models (although human data are preferable); and phenotypic confirmation supported by pharmacodynamic data. In animal disease models lacking the gene or genes responsible for the MoA of the drug, the drug should lack therapeutic effect. Meeting these criteria would be needed in order to attribute the desired clinical outcome to a specific drug target interaction mechanism.
Because the above criteria are difficult to implement by automation, a previous analysis carefully curated MoA data from approved drug labels as well as primary literature, based on a rigorous definition of a drug target9. This ongoing process, performed in parallel by three teams, is anticipated to improve our ability to link drug responses to genetic variation and to help us understand the molecular basis of clinical efficacy, safety and adverse events. The interplay between target tissue expression under disease-specific conditions and the local concentration of the drug or its active metabolites at the relevant disease site is often difficult to ascertain, which is why we attributed a higher weight of evidence to data derived from multiple drugs belonging to the same therapeutic class. Indeed, we anticipate that efficacy target annotations will become more precise as our capability to colocalize target, disease and drug increases.
From this analysis, Tclin currently consists of ~600 protein targets9, which is at the lower end of the original estimate of between 600 and 1,500 targets for the intersection between proteins in the druggable genome and disease-modifying genes by Hopkins and Groom20 (note, however, that Tclin includes targets of biologic drugs as well as small molecules, while the estimate was for small-molecule drug targets only20). So far, proteins in the Tclin category thus represent only a small fraction (3%) of the human proteome (FIG. 1a). From a commercial perspective, it is also noteworthy that most of the global revenues of the pharmaceutical industry are derived from drugs that target a relatively small number of the proteins in the Tclin category (BOX 2; TABLE 2). The majority (259 or 79.7%) of these targets are single proteins, whereas 39 (12%) are complex multiprotein targets. Only 25 targets (8.3%) are comprised of multiple proteins for non-selective drugs; these include the muscarinic, α-adrenergic and oestrogen receptors, as well as cyclooxygenases and histone deacetylases.
Box 2 | Financial spotlight on the human proteome.
Analyses of drug sales focus on pharmaceutical products95 and the companies authorized to market them96. Here, we ask a target-centric question — what are the most financially valuable therapeutic targets — by exploring IMS Health (now known as IQVIA) data from their MIDAS™ platform. We used IMS Health MIDAS drug sales data from 75 countries covering Europe, North America, Australia and Japan, aggregated for the 2011–2015 period. MIDAS tracks products from most therapeutic classes, estimating product volumes, trends and market share through retail and non-retail channels. We chose quinquennial aggregation over annual sales data as it diminishes the importance of factors less relevant to this analysis, such as fluctuations in currency exchange rates. Because active ingredients lose patent coverage and become generic, annual sales figures can abruptly drop from one year to the next.
After excluding traditional medicines, including botanicals and animal products, the MIDAS set comprised 51,095 unique pharmaceutical products, including small molecules and biologics. As most anti-infective and antiparasitic drugs target non-human proteins (with the notable exception of maraviroc, which targets the host (human) CC-chemokine receptor type 5 (CCR5)), we removed these drugs because their targets are outside of the scope of this analysis. The remainder were mapped to 1,182 active pharmaceutical ingredients (APIs) from DrugCentral10, which were first normalized by the number of APIs per pharmaceutical formulation, then by the number of manually curated mechanism of action (MoA) targets9 per API. Thus, we used 581 Tclin proteins and 1,096 APIs, a subset of the 893 human and pathogenic biomolecules through which 1,578 previously analysed approved drugs act9.
By linking global drug sales data to drug targets, we sought to assess a snapshot of their commercial value and to evaluate the market value of human MoA targets. The top 20 MoA targets ranked by aggregated sales data, together with National Institutes of Health (NIH) R01 funding for the same period, are shown in TABLE 2. The entire set covers 325 drug targets, comprising 581 Tclin proteins, totalling over US$3,417 billion in global drug sales (Supplementary Table S10). These data indicate that the cytokine tumour necrosis factor (TNF) is the most valuable target, and cytokines are the only target class comprised entirely of biologic drugs in this analysis for the 2011–2015 period. analysis for the 2011–2015 period (GPCRs) are the most valuable class of druggable targets, with total aggregated sales nearing $917 billion over the 5‑year period. This spotlight covers 72 of the 108 druggable GPCRs reviewed elsewhere97. Kinases ($263 billion, with 45 drugs acting on 43 targets) and cytokines ($242 billion, with 17 drugs acting on 12 targets) are the only two target classes with an extremely active ratio of ongoing versus completed projects, particularly for emerging mechanism– indication pairs98. Finally, combining financial data with targets organized by family and Anatomical, Therapeutic and Chemical (ATC) classification system level 2 codes shows that the top revenue categories are antineoplastics and immunomodulators, followed by nervous system targets (see box figure; a larger version is available as Supplementary Figure S11; see also Supplementary Table S10).
The commercial outcomes of target selection and product-led validation can also be analysed with respect to research funding. NIH RePORTER data (see the NIH ExPORTER website) were processed using the same text-mining methods described earlier for FIG. 2. During the same period, 2011–2015, the NIH funded 42,924 R01 grant applications, at a total cost of $32 billion. These projects discuss up to 7,851 human proteins (see Supplementary Table S10). For example, R01 grants associated with oestrogen receptors were awarded $101 million, compared with $50 billion in sales earned by 18 drugs acting through oestrogen receptors during that same period (TABLE 2). Some targets, having over 30 drugs each, are also top-earning and well funded, for example, the μ-opioid (OPRM1) and the glucocorticoid (NR3C1) receptors. Other top-earning targets with over 30 drugs each, such as the β2 adrenergic receptor and cyclooxygenase 1, are not as well funded. We found no relationship between MIDAS global drug target sales and NIH R01 funding during the 2011–2015 period, even when factoring in the number of APIs per target. Overall, $4.2 billion was awarded to study 496 Tclin proteins, representing 13% of the R01 budget and 6% of all R01-funded proteins. Another 615 proteins (485 Tbio and 67 Tdark) had just one funded R01 project dedicated to their study during 2011–2015, and 8,857 proteins were not associated with any NIH funding for this time frame. AT1 receptor, angiotensin II type 1 receptor; COX2, cyclooxygenase 2; DPP4, dipeptidyl peptidase 4; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA.
Table 2 |.
Financial (sales and NIH funding) activity for the top 20 mechanism-of-action targets
| Target | Target class | Top two drugs | Number of APIs | MIDAS global sales, 2011–2015 per target (US$) | NIH R01 funding, 2011–2015 per target (US$) | Target type | Protein count |
|---|---|---|---|---|---|---|---|
| TNF | Cytokine | Adalimumab and etanercept | 5 | 163 billion | 165 million | Single | 1 |
| INSR | Kinase | Insulin glargine and insulin aspart | 7 | 144 billion | 2 million | Single | 1 |
| NR3C1 | NHR | Fluticasone propionate and budesonide | 36 | 143 billion | 52 million | Single | 1 |
| HMGCR | Enzyme | Rosuvastatin andatorvastatin | 8 | 123 billion | 1 million | Single | 1 |
| H+/K+-ATPase | Transporter | Esomeprazole and omeprazole | 10 | 118 billion | 5 million | Complex | 2 |
| AGTR1 | GPCR | Valsartan and olmesartan medoxomil | 9 | 100 billion | 17 million | Single | 1 |
| ADRB2 | GPCR | Salmeterol and salbutamol | 36 | 90 billion | 8 million | Single | 1 |
| OPRM1 | GPCR | Oxycodone and fentanyl | 34 | 88 billion | 51 million | Single | 1 |
| COX2 | Enzyme | Paracetamol and diclofenac | 40 | 84 billion | 8 million | Single | 1 |
| DRD2 | GPCR | Aripiprazole and quetiapine | 48 | 75 billion | 17 million | Single | 1 |
| Muscarinic acetylcholine receptors | GPCR | Tiotropium bromide and solifenacin | 40 | 64 billion | 65 million | Multiple | 5 |
| SLC6A4 | Transporter | Duloxetine and escitalopram | 26 | 59 billion | 46 million | Single | 1 |
| HTR2A | GPCR | Aripiprazole and quetiapine | 27 | 58 billion | 13 million | Single | 1 |
| L-Type calcium channels | Ion channel | Amlodipine and nifedipine | 23 | 57 billion | 21 million | Complex | 3 |
| SLC6A2 | Transporter | Duloxetine and methylphenidate | 36 | 56 billion | 5 million | Single | 1 |
| VEGFA | Cytokine | Bevacizumab and ranibizumab | 4 | 55 billion | 162 million | Single | 1 |
| HRH1 | GPCR | Olopatadine and cetirizine | 56 | 54 billion | 1 million | Single | 1 |
| Interferon α–β receptor | Membrane receptor | Interferon β-1a and interferon β-1b | 5 | 51 billion | 7 million | Multiple | 2 |
| Voltage-gated sodium channels | Ion channel | Lidocaine and lamotrigine | 39 | 51 billion | 40 million | Multiple, complex | 10 |
| Oestrogen receptors | NHR | Ethinyloestradiol and oestradiol | 18 | 50 billion | 101 million | Multiple | 2 |
Targets are ranked by aggregated API sales data. The number of drugs is the number of MoA-target-associated APIs used in this data set. Target type: single, one protein; multiple, more than one target; complex, more than one protein per target. Protein count, number of proteins associated with that target. ADRB2, β2-adrenoceptor; AGTR1, AT1 receptor; API, active pharmaceutical ingredient; HMGCR, hydroxymethylglutaryl-CoA reductase; DRD2, dopamine D2 receptor; GPCR, G protein-coupled receptor; HRH1, histamine H1 receptor; HTR2A, serotonin 5-HT2A receptor; INSR, insulin receptor; MoA, mechanism of action; NHR, nuclear hormone receptor; NIH, National Institutes of Health; NR3C1, glucocorticoid receptor; OPRM1, μ-opioid receptor; COX2, cyclooxygenase 2; SLC6A2, sodium-dependent noradrenaline transporter; SLC6A4, sodium-dependent serotonin transporter; TNF, tumour necrosis factor; VEGFA, vascular endothelial growth factor A.
Among the factors contributing to the small fraction of each major protein family in Tclin so far, one factor is that not all members of a protein family have drug-compatible or ligandable21 binding sites; for example, some nuclear receptors lack an (endogenous) ligand-binding domain or do not appear to be amenable to small-molecule perturbation. Another factor is that not all proteins can (or will) alter the course of disease via therapeutic intervention, perhaps in some cases owing to our lack of understanding of the underlying pathology.
Kubinyi pointed out that single proteins combine in vivo in ways that could lead to many more drug target combinations across multiple pathways — that is, a ‘druggable proteome’ (REF. 22) — and there is now experimental evidence that alternative splicing, posttranslational modification and heterogeneous oligomers produce functional isoforms with different interaction profiles, which may further result in increased diversity of the proteome23. It is also important to note that for many drugs, the precise MoA and contributing molecular targets remain cryptic, especially when polypharmacology (the simultaneous modulation of multiple targets by drugs) occurs. Shedding light on this would require data completeness24, namely, experiments across all proteins, in relevant physiological conditions, for all approved drugs. This remains a resource-intensive and costly task, which was partially accomplished25 NIH Molecular Libraries Initiative26.
Tchem proteins.
Assignment to Tchem is based on compound activity thresholds originating from binding experiments for small molecules (Supplementar y Figure S2). Selectivity, though important both in vivo and in vitro, could not be factored in for all Tchem targets (Supplementary Box S6 and Supplementary Table S7). Because, by definition, Tclin attribution requires supporting evidence for the MoA, many proteins known to interact with approved drugs, even with high affinity, remain in the Tchem category. Additional bioactivity data from, for example, patent literature and papers currently not indexed in ChEMBL may progress more targets to Tchem.
Many compounds that have reported activity against Tchem targets are also candidate drugs undergoing clinical trials. Based on an in-depth analysis of clinical trial data combined with data from ChEMBL, PubMed, the IUPHAR Guide to Pharmacology27 and ChemIDplus, we mapped 144 Tchem proteins to 356 clinical (phase I-III trial) candidates, for a total of 701 unique target-clinical candidate pairs. For 175 (25%) of these pairs, therapeutic indication data extracted from ChEMBL highlight the different distribution among protein families (TABLE 3). Most targeted proteins are kinases (93 unique enzymes), followed by GPCRs (31 unique receptors) and ion channels (13), with seven targets from other families, which is similar to the prior observation that most clinical candidates target the most druggable target families28. Analysis of the target-clinical candidate subset in which anticipated therapeutic indications are available shows that most of the kinase-targeting clinical candidates are aimed at oncology applications, whereas GPCRs and ion channel-targeting clinical candidates are aimed at central nervous system disorders.
Table 3 |.
Summary of clinical candidates (phase I–III) with activity against Tchem proteins
| Disease category | GPCRs | Ion channels | Kinases | Other |
|---|---|---|---|---|
| Cancer | - | - | 41 (35) | 1 (1) |
| Central nervous system disorders | 7 (14) | 8 (5) | - | - |
| Inflammation and immune disorders | 5 (5) | 1 (1) | 5 (5) | - |
| Respiratory disorders | 3 (7) | 1 (1) | 1 (1) | - |
| Metabolic disorders | 3 (2) | - | - | - |
| Other | 3 (3) | - | 7 (6) | 1 (1) |
| Unmapped | 28 (58) | 11 (23) | 87 (175) | 5 (14) |
Numbers in brackets indicate the number of unique clinical candidates. GPCRs, G protein-coupled receptors.
As noted above, target druggability is frequently estimated based on the ability to bind to small molecules20, and expectations of druggability typically diminish as the size of the binding pocket increases, with affinity and selectivity being major concerns. The challenge is even greater when the binding pocket is shallow and highly exposed to a solvent or when the therapeutic strategy involves disrupting the interaction between the targeted protein and other proteins. One approach to evaluating druggability is to focus on protein domains mined from the InterPro database29 and then to prioritize proteins that contain domains known to interact with approved drugs20 or bioactive small molecules30. Others have explored target druggability by evaluating side-effect similarity for known drugs31 or by performing combined chemical and target similarity queries32, followed by experimental confirmation of novel drug targets derived from clinical observation or computation. It is possible that induced binding sites in proteins in which a druggable pocket is not initially found may enable them to be targeted with drug-like small molecules, but identifying these binding sites with structural approaches is likely to be challenging, and phenotypic screens may be more useful.
An emerging approach harnessing so-called PROTACs (proteolysis-targeting chimaeras) may help substantially in addressing the issue of undruggability33, at least for proteins that are capable of selectively binding a smallmolecule ligand, although not necessarily at a typical binding site34. Essentially, this strategy harnesses the endogenous ubiquitin-proteasome system to promote targeted degradation of desired proteins following binding of the PROTAC35; the mechanism for ternary ligase-PROTAC-target complex activation has been recently elucidated36. This technology may also be subcellular location-specific, which could be an additional advantage in some (but not other) cases. However, the oral bioavailability of PROTAC molecules may be constrained by their size.
Spotlight on Tbio and Tdark
A critical effort in addressing the knowledge deficit about Tbio and Tdark proteins is being undertaken by the Monarch Initiative37, which relies on informatics methods to iden-tify phenotypically relevant disease models in research and diagnostic contexts based on integrated model organism and clinical research data. One of the main sources for the Monarch Initiative is phenotype data from the International Mouse Phenotype Consortium (IMPC), which was set up to generate and phenotypically characterize mouse knockout lines. Their recent analysis of 1,751 unique gene knockouts found that human disease genes are enriched for essential genes38.
The IDG KMC incorporates gene-centric mouse phenotype data and maps these data to the respective human orthologues. IDG coordinated with IMPC production centres to prioritize production of knockout mouse strains for druggable genes. As of November 2017, 568 new knockout strains had been produced: 166 GPCRs, 141 ion channels, 238 kinases and 23 nuclear receptors (see Supplementary Table S8). When ignoring olfactory GPCRs, these represent a little more than one-third of the druggable genes in these protein families. Phenotype data are available for 80% of these strains, with abnormalities detected in numerous biological systems, including those related to development, immune function, metabolism and behaviour. These IMPC strains provide evidence for biological systems that may be affected when a drug targets a gene with little-known function.
Of the 119 Tdark genes (51 GPCRs, 36 ion channels and 32 kinases) submitted by IDG to IMPC, 45 mouse lines were produced, with 41 phenotypes observed. For example, knockouts of the Tdark kinase gene Alpk3 have increased embryonic and perinatal lethality, with the surviving adults displaying severe heart defects (see Further information). Of 482 Tbio genes submitted by IDG (135 GPCRs, 133 ion channels, 200 kinases and 14 nuclear receptors), 184 mouse lines were produced, with 145 phenotypes observed. For example, knockouts of the Tbio GPCR gene Adgrd1 display reproductive defects, such as female infertility, and skeleton phenotype defects, such as decreased bone mineral density (see Further information).
Among 2,788 genes phenotyped in mice at the IMPC, 953 have at least one significant behavioural, neurological or other nervous system-related phenotype observation. Target Central Resource Database (TCRD) data from the GWAS Catalog39, OMIM11 and text-mined DISEASES13 databases confirmed human disease phenotypes for 191 (20%) of these 953 genes, ranging from neurological (for example, seizure disorders) to cognitive (for example, tauopathy) and psychotic affective disorders. Because only 9 of the 953 genes lack confirmed expression in any of the 34 neuro-related tissues tracked by IDG KMC (for example, GTEx40,41, HPA42 and HPM43), these data suggest that the remaining 80% of this set have the potential to be associated with human neurobehavioural phenotypes, paving the way for new research avenues in this direction (see Supplementary Table S8). Production of IMPC strains is set to continue for several more years, and so further knockout strains for druggable genes and their phenotype data are anticipated.
To further explore the characteristics of Tdark and Tbio proteins, we analysed their distribution in the L1000 gene set, as annotated in TCRD (Supplementary Table S3).L1000 is a set of 978 landmark genes that have been selected for their ability to predict a large portion of the total variability seen in large sets of microarray experiments. The proportion of Tdark proteins in the L1000 set (79 of 978; 8%) is substantially smaller than would be anticipated based simply on the a priori distribution of Tdark proteins (which make up 35% of the proteome), whereas Tbio targets (671 of 978; 69%) are more common than expected, as these make up 53% of the proteome. The proportions of Tclin proteins (41 of 978; 4%) and Tchem proteins in the L1000 set (187 of 978; 19%) are also higher than expected, as these make up 3% and 7% of the proteome, respectively. The L1000 TDL distribution data support the existence of a knowledge deficit.
To some extent, the data on Tbio and Tdark suggest a causality dilemma: are Tdark proteins underfunded because there is no scientific interest in this category, or is the lack of knowledge perpetuated by lack of funding? Although our data do not allow us to establish a causal relationship, we suggest that the absence of high-quality, well-characterized molecular probes is a root cause for this situation. Lack of tools leads to lack of interest, and lack of interest diminishes the probability of such tools being developed. A bibliometric evaluation by Edwards and colleagues2 examined how many newly sequenced proteins from several protein families were the subject of new studies 10 years after the completion of the human genome sequencing project. This analysis concluded that the process of druggable target selection is conservative and incremental and that limited progress has been observed with respect to understanding newly discovered proteins.
“If you don’t know very much to begin with, don’t expect to learn a lot quickly” (REF. 44). Anecdotal evidence (summarized in TABLE 4) suggests that it is possible for proteins to migrate from Tdark to Tclin within 12-20 years. Data on the six protein targets highlighted indicate that proteins for which little information was available two decades ago (effectively Tdark) became attractive from a drug discovery perspective following key papers, namely, deorphanization and protein- disease association studies. Five of these six targets are modulated by at least one approved drug, which places them in the Tclin category.
Table 4 |.
Examples of successful attempts of targeting the dark genome
| Gene name | Relevant study (year) | Study type and reference | Citation counta | API name | Therapeutic indication | Market approval (year) |
|---|---|---|---|---|---|---|
| LEPR | 1995 | Receptor deorphanization99 | 4,100 | Metreleptin | Lipodystrophy | 2014 |
| SMO | 1998 | Protein-diseaseassociation study100 | 1,195 | Vismodegib | Basal cell carcinoma | 2012 |
| S1PR1 | 1998 | Receptordeorphanization101 | 968 | Fingolimod | Multiple sclerosis | 2010 |
| HCRTR1 and HCRTR2 | 1998 | Receptor deorphanization102 | 4,608 | Suvorexant | Insomnia | 2014 |
| PCSK9 | 2003 | Protein-diseaseassociation study103 | 1,840 | Evolocumab | Hypercholesterolaemia | 2015 |
| GHSR | 1999 | Receptor deorphanization104 | 8,248 | Anamorelinb | Cachexia | Successful phase III clinical trial105 |
API, active pharmaceutical ingredient; GHSR, ghrelin receptor; HCTR1, orexin receptor type 1; LEPR, leptin receptor; PCSK9, proprotein convertase subtilisin/kexin type 9; S1PR1, sphingosine 1-phosphate receptor 1; SMO, smoothened homologue.
Citation count for the 'relevant study' reference, according to Google Scholar, as of 28 December 2017.
The European Medicines Agency refused marketing authorization for anamorelin in September 2017.
Successful ‘promotions’ across classes, such as those in TABLE 4, are currently rare. We expect the rate of knowledge accumulation for Tdark proteins to be low, at least initially. Well-studied proteins require multiple layers of management for diverse, rich sets of data, with information and knowledge stemming from corpora such as biomedical literature, patents and clinical trials. A paucity of data and lack of information for understudied proteins (Tbio and Tdark) affect both knowledge management and the decision-making process with respect to experimental planning, what research questions need to be asked (and in what order) and which methods may be better suited for each task. For example, we examined access counts for human proteins in STRING18 during 2016 (FIG. 2b). ‘Counts by name’ represents users that access the STRING website and type in a gene symbol. ‘Counts by link’ represents users accessing the network for a gene in STRING by linking to it from another resource (for example, GeneCards45 or UniProt15). Whereas ‘Counts by link’ shows a more comprehensive method to access the entire proteome, it also suggests that Tdark proteins have a lower probability of being recognized (input) by gene name. These data show a pattern similar to that observed in FIG. 2a: Tdark proteins are less likely to be the subject of scientific curiosity, which is a reflection of funding patterns and an overall lack of information and molecular probes. Indeed, the paucity of antibodies and small molecules (criteria that help define Tbio and proteins diminishes our ability to subject Tdark proteins to scientific inquiry.
Genomic and proteomic responses following radiation therapy are also understudied. One in vitro study46 suggested that as many as one-third of the 10,174 genes examined in immortalized B cells following ionizing radiation are radioresponsive (GSE26835 column46 in Supplementary Table S3). Of the 447 genes with significant fold changes in the GSE26835 set, only 26 are Tclin and 61 are Tchem, whereas the majority (268 Tbio, 92 Tdark) are understudied (see also Supplementary Box S5).
As many as 3,644 proteins have significant disease (confirmed OMIM11 phenotype) associations. Given their TDL assignments (335 Tclin, 543 Tchem and 2,766 Tbio), we examined the distribution of the TDL in relation to druggable protein family categories (FIG. 1b). It appears that Tbio-disease associations are quite rare for druggable families such as nuclear receptors, ion channels and GPCRs, as these families are more likely to be in the Tclin or Tchem category. Instead, Tbio assignments are quite frequent for transcription factors, epigenetic targets, transporters and unassigned protein families. The exception among druggable families are olfactory GPCRs, which appear to attract less interest from drug discovery programmes, despite some of these GPCRs being linked to metabolism and ageing47.
Concerted efforts focused on an entire target class have sometimes led to new drugs. For example, GlaxoSmithKline (GSK) had a comprehensive programme aimed at finding new ligands and characterizing the biology of nuclear receptors48. New insights into bile acid metabolism49 and xenobiotic transcription of cytochrome P450s50, mediated by nuclear receptors, were described. A bile acid receptor (FXR) agonist has reached the market since this programme started: obeticholic acid (Ocaliva), which was discovered by GSK in collaboration with the University of Perugia51 and subsequently developed by Intercept Pharmaceuticals. Currently, several FXR agonists are in clinical development52. Choosing the appropriate proteins as drug targets remains a complex process, where scientific factors need to be balanced against commercial factors (such as company investors and medical insurance companies) and societal factors (such as physicians and patients), as well as legal factors (such as the requirements of regulatory agencies)1.
Spotlight on G protein-coupled receptors
GPCRs are membrane-bound, cell-surface receptors that transduce signals via interactions with heterotrimeric G proteins, arrestins and other cellular transducers53,54.Alterations in GPCR signalling are implicated in the pathogenesis and treatment of neuropsychiatric55, immunological56, gastrointestinal57,58, cardiac59, renal, hormonal, infectious60 and many other disorders53,54. GPCRs represent the largest family of druggable targets in the human genome9, with between 20% and 30% of approved drugs acting on them53,54.
The number of publications per GPCR and the number of chemicals associated with that GPCR in ChEMBL were examined61 to determine which of the druggable, non-olfactory GPCRs are understudied: less than 100 citations and less than ten ChEMBL compounds define understudied, uninterrogated GPCRs. The number of publications, similar to the fractional PubMed publications count13, does not take into account large-scale (many proteins per paper) analyses. Counting ChEMBL compounds, a quantitative criterion similar to Tchem assignments, does not consider bioactivity values. However, this independent analysis validates the more general TDL criteria with respect to GPCR biological functions and corresponding chemical matter.
Tclin and Tchem G protein-coupled receptors.
Currently 827 GPCRs — including 421 olfactory GPCRs — are tracked by IDG KMC; of these, 96 are Tclin and 113 are Tchem (none of which are olfactory). Slightly more than half of the non-olfactory GPCRs have annotated drugs and small molecules targeting them53,54,61; see also TABLE 1. A recent analysis indicates, however, that a handful of GPCRs — mainly biogenic amine, muscarinic and opioid receptors — represent the most abundantly targeted receptors for FDA-approved medications53,54. GPCRs also represent important off-targets for kinase inhibitors62,63, ion channel modulators64, anti-infectives65 and other classes of drug-like molecules53,54. As with other druggable target classes, off-target actions within the GPCR class can be associated with severe and lifethreatening side effects. For example, valvular heart disease is associated with anorectic agents, such as fenfluramine, and antimigraine medications, such as ergot-amine, via serotonin 5-HT2B receptor agonism66. Recent successes in structure-guided and cheminformaticsdriven drug discovery show promise for creating safer and more effective medications targeting GPCRs.
Tbio and Tdark G protein-coupled receptors.
Although 52 non-olfactory GPCRs are categorized as Tdark, the availability of new screening platforms to discover chemical matter for these GPCRs has begun the process of illumination64,67,68. Of 62 GPCRs for which significant phenotype calls have been reported by IMPC (Supplementary Table S8), 24 are Tbio and 7 are Tdark; of these, 15 are associated with neurological and behavioural phenotypes. Including olfactory GPCRs, 618 proteins are classified as Tdark or Tbio; of these, 126 non-olfactory GPCRs and 51 olfactory GPCRs have significant associations with human diseases via OMIM, GWAS and text mining. Whereas the majority of these associations (nearly 59%) stem from text mining69, 48 GPCRs have confirmed associations from at least two information channels. For example, Adgrb2-mutant mice (Tbio) showed significant antidepressant-like behaviour compared with wild-type mice70, whereas the association71 between schizophrenia and Fzd3 mutants (T) remains controversial72,73.
Spotlight on protein kinases
Tclin and Tchem kinases.
The ~600-member human kinome (Supplementary Table S3) is made up of protein kinases, in addition to metabolic and lipid kinases, and is highly druggable using both competitive and allosteric small-molecule inhibitors. However, the functions of about one-third of the kinases in this family are poorly defined or unknown. The 634 human kinases were categorized as follows: Tclin, N = 50; Tchem, N = 390; Tbio, N = 163; and Tdark, N = 31 (TABLE 1). Tclin kinases are not exclusively protein kinases, and the number of FDA-approved small-molecule kinase inhibitors varies depending on inclusion criteria. Wu and colleagues74 found 38 small-molecule protein kinase inhibitors. Based on DrugCentral, we found 50 approved kinase inhibitors, including 40 small-molecule protein kinase inhibitors, of which 32 are FDA-approved, one FDA-approved protein kinase activator (ingenol mebutate) and the phosphoinositide 3-kinase subunit-δ (PIK3CD) small-molecule inhibitor idelalisib, which is also FDA-approved. An additional seven FDA-approved antibodies target the receptor tyrosine kinases human epidermal growth factor receptor 2 (HER2; also known as ERBB2), epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor 2 (VEGFR2) and platelet-derived growth factor receptor-α (PDGFRα), and there is also an FDA-approved HER2-targeting antibody-drug conjugate, trastuzumab emtansine (see Supplementary Table S9).
Tbio and Tdark kinases.
A number of Tbio and Tdark kinases are known to interact with FDA-approved multikinase inhibitors. According to data in DrugCentral, sorafenib inhibits 114 kinases, of which only 9 are associated MoArelated targets, whereas sunitinib inhibits 263 kinases, of which 9 are MoA-related targets. Given the current state of kinase inhibitor chemistry, it is very likely that Tbio and Tdark kinases can be effectively therapeutically targeted with highly selective small-molecule inhibitors. Some of the characteristics shared by understudied Tbio and Tdark kinases include poorly defined integration of the kinase in signalling networks, poorly defined function and regulation, lack of activation-loop phospho-antibodies or immunohistochemistry-grade antibodies, and lack of selective chemical tools for functional characterization. Primary tools for knockout and/or altered expression are RNA interference (RNAi) and CRISPR-Cas9, and cDNAs for overexpression; kinase knockout or altered expression rarely provides readily assayable phenotypes (for example, growth, migration, apoptosis or in vivo function in mouse organ physiology). Currently, the IMPC has targeted 238 kinases with 114 knockouts having a significant phenotype; of the latter, 22 are of current interest for phase 2 of the IDG programme.
Many Tchem, Tbio or Tdrak kinases are altered in expres-sion or mutated in TCGA. TABLE 5 shows ten Tbio or Tdark kinases whose amplification is observed in the TCGA database, together with their RNA expression in triple-negative breast cancer (TNBC) cells. These understudied kinases are frequently altered in breast cancer. The potential increased expression of many kinases in primary human tumours suggests these understudied kinases have important functions for the tumour cell phenotype that have not been characterized to date. These represent unexplored kinases with possible therapeutic utility.
Table 5 |.
Understudied kinases that are frequently altered in breast cancer
| Gene name | Protein name | TDL | SUM 159 averagea | Alteration frequencyb (%) | Kinase family |
|---|---|---|---|---|---|
| TRIB1 | Tribbles homologue 1 | Tbio | 342 | 24 | CAMK |
| RPS6KC1 | Ribosomal protein S6 kinase δ1 | Tdark | 463 | 23 | Other |
| UHMK1 | Serine/threonine-protein kinase Kist | Tbio | 1,308 | 23 | Other |
| NRBP2 | Nuclear receptor-binding protein 2 | Tdark | 350 | 21 | Other |
| PIP5K1A | Phosphatidylinositol 4-phosphate 5-kinase type1-α | Tbio | 2,246 | 19 | Metabolic |
| CDK12 | Cyclin-dependent kinase 12 | Tbio | 3,148 | 14 | CMGC |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 | Tbio | 455 | 11 | STE |
| STRADA | STE20-related kinase adapter protein-α | Tbio | 464 | 9 | STE |
| BCKDK | (3-methyl-2-oxobutanoate dehydrogenase(lipoamide)) kinase, mitochondrial | Tbio | 1,087 | 6 | Atypical |
| EEF2K | Eukaryotic elongation factor 2 kinase | Tbio | 1,176 | 6 | Atypical |
Expression levels in SUM159PT claudin-low triple-negative breast cancer (TNBC) cells.
Alteration frequency in breast cancer from The Cancer Genome Atlas (TCGA). TDL, target development level.
The potential therapeutic importance of Tbio and Tdark kinases in the kinome is highlighted by a recent clinical study that assessed the response to trametinib, a MEK1 and MEK2 inhibitor, in TNBC patients75. Pretreatment needle biopsies and surgical tumour resections following 7-day trametinib treatment were used for RNA sequencing (RNA-seq) to analyse tumour transcriptomic changes in response to the drug. Pretreatment biopsies matched to post-treatment surgical specimens showed overall concordance of the transcriptional kinase response to trametinib, with FRK (Tchem) exhibiting the highest mean increase and cytoplasmic BMX (also Tchem) exhibiting the highest mean decrease among patients in response to a 7-day drug treatment. Among the kinases transcriptionally altered in the TNBC tumours were several understudied kinases, including MRCK-γ (also known as CDC42BPG), PRKACB, STK32B and leukocyte tyrosine kinase receptor (LTK). These findings demonstrate that in TNBC tumours in patients, members of the understudied Tbio and Tdark kinome are co-regulated transcriptionally with kinases from the Tclin and Tchem category, in a dynamic adaptive response to targeted inhibition.
Spotlight on ion channels
Ion channels mediate signalling within cells, between cells, and between cells and their environment. Defects in ion channels underlie many major disorders in humans, also known as channelopathies, including neuronal disorders76, diabetes77 and heart failure78. This makes ion channels an attractive target class for drug development. Ion channels are mostly heteromeric complexes that require optimal interactions with ligands at specific locations. Currently, 355 ion channel pore-forming and auxiliary subunits are tracked by IDG KMC (TABLE 1; Supplementary Table S3). About 100 ion channel modulators, including auxiliary subunits, are reported, but to our knowledge, a systematic list of cell type-specific auxiliary subunits for all ion channels is not available.
Tclin and Tchem ion channels.
Many drugs are known to bind to ion channels. There are 217 drugs annotated in DrugCentral10 as acting through 125 (T) ion channels for the MoA9. The number of drugs increases to 497 when querying how many drugs are known to interact with ion channels outside of the MoA-related constraint. Some of these interactions are likely to be responsible for side effects such as cardiac toxicity. An accurate understanding of MoA and side-effect assignment at the target (molecular) level is required if we are to improve upon available drugs. For example, the anaesthetic ketamine, which has been postulated to act as a noncompetitive N-methyl-d-aspartate (NMDA) antagonist79, has been used off-label as an antidepressant80. However, in-depth analysis of the antidepressant effects of ketamine found that its active metabolite (2R,6R)-hydroxynorketamine (HNK) does not block the NMDA receptor. Instead, HNK displays sustained activation of α-amino-3hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors and lacks ketamine-related side effects81. This may pave the way for the development of novel, rapid-acting antidepressants. It is therefore conceivable that some ion channels currently categorized as Tclin or Tchem are in need of further illumination with respect to MoA and drug specificity. Indeed, the low bioactivity cut-off criterion for ion channels (≤10 μM) in Tchem (see also Supplementary Figure S2) may need revision, given that older drugs continue to reveal unexpected modes of action.
Tdark ion channels.
A relatively small number of ion channels (31) are categorized as Tdark (TABLE 1). Part of the difficulty in illuminating dark ion channels is the replication of physiological context and expression of proteins in the appropriate heteromeric, pore-forming functional complexes. Currently, there are no scalable systems available to study the localization of functional complexes. Moreover, most ion channels have paralogues that function redundantly. Gene redundancy increases the difficulty of revealing phenotype and precise localization, both important elements for understanding physiological functions other than ion channel activity. This considerably delays our progress in understanding ion channel function in vivo and their role in human health. Unlike GPCRs or kinases, neither pore-forming subunits nor auxiliary subunits share characteristic motifs. Lack of specific protein sequence motifs makes it difficult to flag candidate genes for further study, even with computational assertions. There could be other ion channels, which perhaps should be categorized as Tdark.dark, to reflect our complete lack of knowledge, even by computational means, regarding these proteins.
The list of ion channel pore-forming subunits, as well as auxiliary subunits, continues to grow. For example, leucine-rich repeat-containing protein 8 (LRRC8) heteromers form82 volume-regulated anion channels (VRACs), and ORAI proteins assemble to form83 calcium-releaseactivated calcium channels (CRACs), whereas anoctamins are olfactory calcium-activated chloride channels (CACCs)84. Currently, LRRC8B, LRRC8C and LRRC8D subunits are classified as Tdark, with the exception being the subunit LRRC8A (Tbio); ORAI1 is annotated as Tchem, wheras the ORAI2 and ORAI3 proteins are annotated as Tbio. With the exception of anoctamin 1 (Tclin), all other anoctamins are labelled Tbio. These, and all other Tdark proteins that lack computational assertions, are in need of systematic genomic-scale studies.
Conclusions
Modern medicine often employs artificial distinctions in terms of what and how biological systems are studied: segregated by organ (for example, ophthalmology and cardiology) or by disease (for example, oncology and infectious diseases), medical specialty separations carry over into the research arena, both in academia and industry. This distinction breaks down in nature, as we are likely to observe the interplay between the same genes and pathways regardless of organ, albeit in a contextspecific manner. These artificial divisions can prevent scientists from achieving a translational, integrative view of gene and protein function. We suspect this to be another reason why funding to study Tdark proteins is scarce: for functionally enigmatic proteins, or the ‘ignorome’ (REF. 85), anticipating which organ, disease or phenotype is relevant may be far from trivial. To address this limitation, the NIH launched a series of high-risk programmes via the Common Fund resource, aimed to catalyse transdisciplinary research. The IDG is one such Common Fund programme. The IDG programme’s ostensible goal is to encourage and track the illumination of relatively understudied and unstudied parts of the genome.This implicitly requires the construction of a knowledge base objectively and in an unbiased manner, asserting what is currently believed to be true (a process that is explored metaphorically in a classic book by Italo Calvino, ‘The Castle of Crossed Destinies’; see Supplementary Box S1). The IDG KMC enables us to quantitatively demonstrate the existence of a knowledge deficit with respect to dark and understudied proteins, which underscores the need for basic science and its major role in illuminating gene functions and roles in human disease. The TDL classification scheme provides a convenient way to partition human targets that highlights the focus (or lack thereof) of science and drug discovery efforts on different targets. Through the use of the TDL groupings, we can highlight knowledge accumulation, as well as deficits, for a variety of target families, with a common theme being that while much is known, there remains a large fraction of the proteome that is understudied. The IDG KMC, by collating and linking a plethora of disparate and diverse data sources and data types, aims to shed light on these dark regions with the hope that researchers will be empowered to use the data and knowledge presented by the KMC to jumpstart research programmes on these targets.
Confirmed associations with a specific disease, or receptor deorphanization (TABLE 4), remain major incentives to allocate resources and further study of Tdark proteins. As mentioned above, the only other deliberate targeted effort to study Tdark proteins in addition to IDG is the IMPC. As of March 2017, mouse lines corresponding to 4,165 human genes have been produced, with phenotypes available, 2,788 of which have resulted in statistically significant phenotype calls. Of these 2,788 proteins, 827 (436 Tdark) are not associated with any NIHfunded grants between 2000 and 2015 (Supplementary Table S8). By contrast, only 120 of the 1,961 proteins with significant IMPC phenotype calls and that are associated with NIH funding are Tdark. It was Edgar Allan Poe who once said, “the enormous multiplication of books in every branch of knowledge is one of the greatest evils of this age, since it presents one of the most serious obstacles to the acquisition of correct information”. Poe’s 19th century line of thought is remarkably apt in the context of current KMC activities, since the “acquisition of correct information” remains the largest challenge.
Another challenge relates to an area of knowledge largely neglected in the scientific literature: the large-scale capture of negative results. Due to confirmation bias8, scientists have a tendency to primarily publish successful accounts of research. Although there are attempts to overcome this problem86, we are not aware of the existence of an unbiased, easy mechanism to capture negative results. The aphorism “absence of evidence is not evidence of absence” illustrates practical limitations of knowledge management systems: does lack of evidence imply that the study was conducted, but nothing was found, or does it imply (more often) that the measurement was not carried out? Proper archiving of negative results (for example, “protein P is not expressed in cell type CT” or “gene mutation Gm does not play a role in disease D”) would benefit the community at large and would improve our specific knowledge about proteins. However, such non-positive facts fit poorly to current publishing and citation paradigms. One possibility for archiving such statements could be nanopublications87, which would be amenable to largescale integration into systems such as TCRD-Pharos.
Finally, a key challenge faced by IDG KMC, and perhaps by other data analysts working in drug discovery, is that of reliable predictions: when examining Tdark proteins in particular, experimentalists would like to know what experiment to do next, what phenotypic changes should be examined first and which pathway is relevant in a specific disease. These, and other similar questions, have yet to find a computer-driven, reliable answer. To paraphrase William Gibson, “The truth is already here — it’s just not very evenly distributed”.
Supplementary Material
Drug.
Externally administered, possibly endogenous but mostly xenobiotic, substances that are administered to patients in order to influence the outcome of a disease, syndrome or condition.
Drug targets.
Molecular entities present in living systems that, upon interaction with therapeutic agents or their by-products, result in modified biological responses that lead to therapeutic outcomes. The interaction between a drug and its target leads, directly or indirectly, to observable clinical outcomes.
Druggable genome Originally defined by Hopkins and Groom as the set of genes that encode proteins that could be modulated by an orally administered small molecule, as estimated by Lipinski’s ‘rule of five’ guidelines.
Mode of action Referred to as ‘mechanism of action’ when the molecular interactions are well understood; describes the way in which drugs exert their intended therapeutic action, resulting in the intended therapeutic outcome.
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grants U54 CA189205 and U24 224370 (Illuminating the Druggable Genome Knowledge Management Center (IDG KMC)) at the University of New Mexico, Novo Nordisk Foundation Center for Protein Research, European Bioinformatics Institute (EBI) and University of Miami, U54 CA189201 and U24 CA224260 (A.M., Mount Sinai), P30 CA118100 (T.I.O., G.N.G. and L.A.S., UNM) and UL1 TR001449 (T.I.O. and L.A.S.), UM1 HG006370 (International Mouse Phenotyping Consortium, T.F.M. and I.T.), U01 MH104974 (B.L.R.), U01 MH104984 (S.T.), U01 MH105028 (M.T.M.), U01 MH105026 (J.Q. and A.M., Baylor) and U01 MH104999, R01 CA177993 and U24 DK116204 (S.G. and G.L.J.) and by the European Molecular Biology Laboratory (EMBL) and Wellcome Trust Strategic Awards WT086151/ Z/08/Z and WT104104/Z/14/Z (A.G., A.H., A.R.L., A.K., J.P.O.,and G.P.); and by Novo Nordisk Foundation Denmark grantNNF14CC0001 (S.B., L.J.L. and D.W). R.G., A.J., D.T.N., A.S., N.S., and G.Z.K. were supported by the Intramural Research Program, National Center for Advancing Translational Sciences (NCATS) and by U54 CA189205. Dedicated to Francisc Schneider (1933-2017).
Footnotes
Competing interests
The authors declare competing interests: see Web version for details.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
RELATED LINKS
Anamorelin: https://en.wikipedia.org/wiki/Anamorelin
ATC codes: https://www.whocc.no/atc_ddd_index/
Channelopathy: https://en.wikipedia.org/wiki/Channelopathy
ChemIDplus: https://chem.nlm.nih.gov/chemidplus/
ClinicalTrials.gov: https://clinicaltrials.gov/
Cochrane Collaboration: http://www.cochrane.org/
DrugCentral: http://drugcentral.org/
Drug Target Ontology: http://drugtargetontology.org/
Edgar Allan Poe quotes: https://quoteinvestigator.com/2017/07/12/many-books/
Evidence of absence: https://en.wikipedia.org/wiki/Evidence_of_absence
Harmonizome: http://amp.pharm.mssm.edu/Harmonizome/
Illuminating the Druggable Genome: https://commonfund.nih.gov/idg
IMPC (International Mouse Phenotyping Consortium): http://www.mousephenotype.org/
IMPC information about Adgrd1: http://www.mousephenotype.org/data/genes/MGI:3041203
IMPC information about Alpk3: http://www.mousephenotype.org/data/genes/MGI:2151224
Integrity | Available from Clarivate Analytics at: https://clarivate.com/products/integrity/
L1000: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL20573
MIDAS Platform | IQVIA analytics platform for industryleading sales and medical data available at: https://www.iqvia.com/solutions/commercialization/geographies/midas
Monarch Initiative: http://www.monarchinitiative.org/
NIH Common Fund: https://commonfund.nih.gov/
NIH ExPORTER: https://exporter.nih.gov/
NIH RePORTER: https://projectreporter.nih.gov/reporter.cfm
Pharos: https://pharos.nih.gov
PubMed: https://pubmed.gov
STRING: https://string-db.org/
The Cancer Genome Atlas: https://cancergenome.nih.gov/
Target Central Resource Database: http://juniper.health.unm.edu/tcrd/
William Gibson Wikiquotes: https://en.wikiquote.org/wiki/William_Gibson
References
- 1.Knowles J & Gromo G Target selection in drug discovery. Nat. Rev. Drug Discov. 2, 63–69 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Edwards AM et al. Too many roads not taken.Nature 470, 163–165 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Alberts B, Kirschner MW, Tilghman S &Varmus H Rescuing US biomedical research from its systemic flaws. Proc. Natl Acad. Sci. USA 111, 5773–5777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S et al. PubChem Substance and Compound databases. Nucleic Acids Res. 44, D1202–D1213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaulton A et al. The ChEMBL database in 2017.Nucleic Acids Res. 45, D945–D954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomczak K, Czerwin´ska P & Wiznerowicz M The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol 19, A68–A77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munafò MR et al. A manifesto for reproducible science. Nat. Hum. Behav. 1, 0021 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickerson RS Confirmation bias: a ubiquitous phenomenon in many guises. Rev. Gen. Psychol. 2, 175–220 (1998). [Google Scholar]
- 9.Santos R et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ursu O et al. DrugCentral: online drug compendium.Nucleic Acids Res. 45, D932–D939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amberger J, Bocchini CA, Scott AF &Hamosh A McKusick’s Online Mendelian Inheritance in Man (OMIM). Nucleic Acids Res. 37, D793–D796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M et al. Gene ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pletscher-Frankild S, Pallejà A, Tsafou K,Binder JX & Jensen LJ Diseases: text mining and data integration of disease-gene associations. Methods 74, 83–89 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Kiermer V Antibodypedia. Nat. Methods 5, 860–861(2008). [Google Scholar]
- 15.Consortium UniProt. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadatos G et al. SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 44, D1220–1228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouillard AD et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajduk PJ, Huth JR & Tse C Predicting protein druggability. Drug Discov. Today 10, 1675–1682 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Hopkins AL & Groom CR The druggable genome.Nat. Rev. Drug Discov 1, 727–730 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Surade S & Blundell TL Structural biology and drug discovery of difficult targets: the limits of ligandability. Chem. Biol. 19, 42–50 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Kubinyi H Drug research: myths, hype and reality.Nat. Rev. Drug Discov 2, 665–668 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Yang X et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805–817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mestres J, Gregori-Puigjané E, Valverde S & Solé RV Data completeness—the Achilles heel of drug-target networks. Nat. Biotechnol. 26, 983–984 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Schreiber SL et al. Advancing biological understanding and therapeutics discovery with smallmolecule probes. Cell 161, 1252–1265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin CP, Brady LS, Insel TR & Collins FS NIH molecular libraries initiative. Science 306, 1138–1139 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Southan C et al. The IUPHAR/BPS guide to pharmacology in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 44, D1054–D1068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waring MJ et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 14, 475–486 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Hunter S et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–312 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruger FA, Gaulton A, Nowotka M & Overington JP PPDMs-a resource for mapping small molecule bioactivities from ChEMBL to Pfam-A protein domains. Bioinformatics 31, 776–778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campillos M, Kuhn M, Gavin A-C, Jensen LJ & Bork P Drug target identification using side-effect similarity. Science 321, 263–266 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Keiser MJ et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang X & Dixit VM Drugging the undruggables: exploring the ubiquitin system for drug development. Cell Res. 26, 484–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai AC & Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakamoto KM et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. 98, 8554–8559 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gadd MS et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mungall CJ et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 45, D712–D722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickinson ME et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlén M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Kim M-S et al. A draft map of the human proteome. Nature 509, 575–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenat DB & Feigenbaum EA On the thresholds of knowledge. Artif. Intell 47, 185–250 (1991). [Google Scholar]
- 45.Fishilevich S et al. Genic insights from integrated human proteomics in GeneCards. Database 2016, baw030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smirnov DA et al. Genetic variation in radiationinduced cell death. Genome Res. 22, 332–339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrison JL & Knight ZA Linking smell to metabolism and aging. Science 358, 718–719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kliewer SA, Lehmann JM & Willson TM Orphan nuclear receptors: shifting endocrinology into reverse. Science 284, 757–760 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Willson TM, Jones SA, Moore JT & Kliewer SA Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med. Res. Rev. 21, 513–522 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Moore LB et al. Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 275, 15122–15127 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Pellicciari R et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45, 3569–3572 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Hambruch E, Kinzel O & Kremoser C On the pharmacology of farnesoid X receptor agonists: give me an ‘A’, like in ‘acid’. Nucl. Recep. Res 3, 101207 (2016). [Google Scholar]
- 53.Wacker D, Stevens RC & Roth BL How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth BL, Irwin JJ & Shoichet BK Discovery of new GPCR ligands to illuminate new biology. Nat. Chem. Biol. 13, 1143–1151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roth BL, Sheffler DJ & Kroeze WK Magic shotguns versus magic bullets: selectively nonselective drugs for mood disorders and schizophrenia. Nat. Rev. Drug Discov. 3, 353–359 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Hernandez PA et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat. Genet. 34, 70–74 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Sternini C Receptors and transmission in the braingut axis: potential for novel therapies. III. Mu-opioid receptors in the enteric nervous system. Am. J. Physiol. Gastrointest. Liver Physiol 281, G8–15 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Sternini C Taste receptors in the gastrointestinal tract. IV. Functional implications of bitter taste receptors in gastrointestinal chemosensing. Am. J. Physiol. Gastrointest. Liver Physiol 292, G457–461 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Rockman HA, Koch WJ & Lefkowitz RJ Seventransmembrane-spanning receptors and heart function. Nature 415, 206–212 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Elphick GF et al. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 306, 1380–1383 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Roth BL & Kroeze WK Integrated approaches for genome-wide interrogation of the druggable non-olfactory G protein-coupled receptor superfamily. J. Biol. Chem. 290, 19471–19477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elkins JM et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat. Biotechnol. 34, 95–103 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Lin X et al. Life beyond kinases: structure-based discovery of sorafenib as nanomolar antagonist of 5-HT receptors. J. Med. Chem. 55, 5749–5759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X-P et al. Allosteric ligands for the pharmacologically dark receptors GPR68 and GPR65. Nature 527, 477–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan JD et al. The anthelmintic praziquantel is a human serotoninergic G-protein-coupled receptor ligand. Nat. Commun. 8, 1910 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roth BL Drugs and valvular heart disease. N. Engl. J. Med. 356, 6–9 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Kroeze WK et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lansu K et al. In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat. Chem. Biol. 13, 529–536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pafilis E et al. The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text. PLoS ONE 8, e65390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okajima D, Kudo G & Yokota H Antidepressantlike behavior in brain-specific angiogenesis inhibitor 2-deficient mice. J. Physiol. Sci. 61, 47–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katsu T et al. The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci. Lett. 353, 53–56 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Wei J & Hemmings GP Lack of a genetic association between the frizzled-3 gene and schizophrenia in a British population. Neurosci. Lett. 366, 336–338 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Jeong SH, Joo EJ, Ahn YM, Lee KY & Kim YS Investigation of genetic association between human Frizzled homolog 3 gene (FZD3) and schizophrenia: results in a Korean population and evidence from meta-analysis. Psychiatry Res. 143, 1–11 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Wu P, Nielsen TE & Clausen MH Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discov. Today 21, 5–10 (2016). [DOI] [PubMed] [Google Scholar]
- 75.Zawistowski JS et al. Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P-TEFb complex. Cancer Discov. 7, 302–321 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kullmann DM The neuronal channelopathies. Brain 125, 1177–1195 (2002). [DOI] [PubMed] [Google Scholar]
- 77.Gloyn AL et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 52, 568–572 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Marbán E Cardiac channelopathies. Nature 415, 213–218 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Berman RM et al. Antidepressant effects of ketamine in depressed patients. Biol. Psychiatry 47, 351–354 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Kirby T Ketamine for depression: the highs and lows. Lancet Psychiatry 2, 783–784 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Zanos P et al. NMDAR inhibition-independent antidepressant actions of ketamine metabolites. Nature 533, 481–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pedersen SF, Klausen TK & Nilius B The identification of a volume-regulated anion channel: an amazing Odyssey. Acta Physiol. 213, 868–881 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Niemeyer BA Changing calcium: CRAC channel (STIM and Orai) expression, splicing, and posttranslational modifiers. Am. J. Physiol. Cell Physiol. 310, C701–709 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Dauner K, Lissmann J, Jeridi S, Frings S & Möhrlen F Expression patterns of anoctamin 1 and anoctamin 2 chloride channels in the mammalian nose. Cell Tissue Res. 347, 327–341 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Pandey AK, Lu L, Wang X, Homayouni R & Williams RW Functionally enigmatic genes: a case study of the brain ignorome. PLoS ONE 9, e88889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfeffer C & Olsen BR Editorial: Journal of negative results in biomedicine. J. Negat. Results Biomed. 1, 2 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Groth P, Gibson A & Velterop J The anatomy of a nanopublication. Inf. Serv. Use 30, 51–56 (2010). [Google Scholar]
- 88.Agarwal P & Searls DB Can literature analysis identify innovation drivers in drug discovery? Nat. Rev. Drug Discov. 8, 865–878 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nguyen D-T et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 45, D995–D1002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wishart DS et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Griffith M et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koscielny G et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 45, D985–D994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin Y et al. Drug target ontology to classify and integrate drug discovery data. J. Biomed. Semant. 8, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maggon K Best-selling human medicines 2002– 2004. Drug Discov. Today 10, 739–742 (2005). [DOI] [PubMed] [Google Scholar]
- 96.Stebbins S The world’s 15 top selling drugs. 24/7 Wall St http://247wallst.com/special-report/2016/04/26/top-selling-drugs-in-the-world/ (2016).
- 97.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB & Gloriam DE Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug Discov 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shih H-P, Zhang X & Aronov AM Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat. Rev. Drug Discov. 17, 19–33 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Tartaglia LA et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 83, 1263–1271 (1995). [DOI] [PubMed] [Google Scholar]
- 100.Xie J et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–92 (1998). [DOI] [PubMed] [Google Scholar]
- 101.Lee MJ et al. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science 279, 1552–1555 (1998). [DOI] [PubMed] [Google Scholar]
- 102.Sakurai T et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G proteincoupled receptors that regulate feeding behavior. Cell 92, 573–585 (1998). [DOI] [PubMed] [Google Scholar]
- 103.Abifadel M et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34, 154–156 (2003). [DOI] [PubMed] [Google Scholar]
- 104.Kojima M et al. Ghrelin is a growth-hormonereleasing acylated peptide from stomach. Nature 402, 656–660 (1999). [DOI] [PubMed] [Google Scholar]
- 105.Temel JS et al. Anamorelin in patients with nonsmall-cell lung cancer and cachexia (ROMANA 1 and ROMANA 2): results from two randomised, doubleblind, phase 3 trials. Lancet Oncol. 17, 519–531 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.