Abstract
Chromatin-associated proteins play critical roles in many cellular processes, including gene expression, epigenetic regulation, DNA repair, recombination and replication. Especially, epigenetic landscape, shaped by a variety of chromatin-binding proteins, is dynamic and regulated in a context-dependent manner. In situ chromatin binding assay is a powerful but simple tool to investigate how proteins, such as epigenetic components, associate with chromatin. This approach relies on the fact that chromatin bound proteins are more resistant to detergent extraction. Here we describe a protocol for the in situ chromatin binding assay used in Schizosaccaromyces pombe.
Keywords: Epigenetics, in situ chromatin binding, H3, CENP-A, GFP, Fission yeast, Schizosaccaromyces pombe
1. Introduction
Eukaryotic DNA is packaged into chromatin by wrapping around histone proteins. The organization of chromatin is tightly regulated and can be divided into two major domains: heterochromatin and euchromatin. Whereas heterochromatin is a tightly packed form of chromatin containing few genes, euchromatin is a more open chromatin region where genes are actively transcribed. In addition to histones, a variety of proteins are also associated with chromatin and play important roles in a wide range of cellular activities, such as DNA replication, transcriptional regulation, chromatin remodeling, cell cycle progression, aging, tumorigenesis, development and differentiation. One major group of proteins associated with chromatin is epigenetic regulators. These proteins mediate epigenetic modifications on chromatin, such as histone modification, DNA methylation, and histone variants, which exert effects on cellular processes without changing genetic sequences. Epigenetic regulators are usually associated with chromatin in a dynamic manner. The epigenetic hallmark for heterochromatin is histone 3 lysine 9 methylation (H3K9me), conserved from fission yeast to human. H3K9me is essential for heterochromatin structure and function. In fission yeast, the modification is catalyzed by histone methyltrasferase Clr4 (a human Suv39 homolog) and recognized by the conserved HP1 homolog, Swi6 [1]. The CLRC complex, which is composed of Clr4, Rik1, Cul4, Dos1/Raf1, Dos2/Raf2, and Lid2, is recruited to heterochromatin during S phase, and promotes heterochromatin assembly [2–6].
A distinct chromatin structure in all eukaryotes is the centromere that provides foundation for kinetochore assembly and is crucial for proper chromosome segregation during mitosis and meiosis. In most eukaryotes including fission yeast and humans, centromere is epigenetically defined by CENP-A, a centromere-specific H3 variant [4,7–9]. CENP-A loading to centromeres is cell cycle-dependent, and is mediated by multiple CENP-A loading factors. Analyzing the chromatin association of specific proteins is thus critical for elucidating the epigenetic mechanisms used to govern chromatin structures, such as heterochromatin and centromeres.
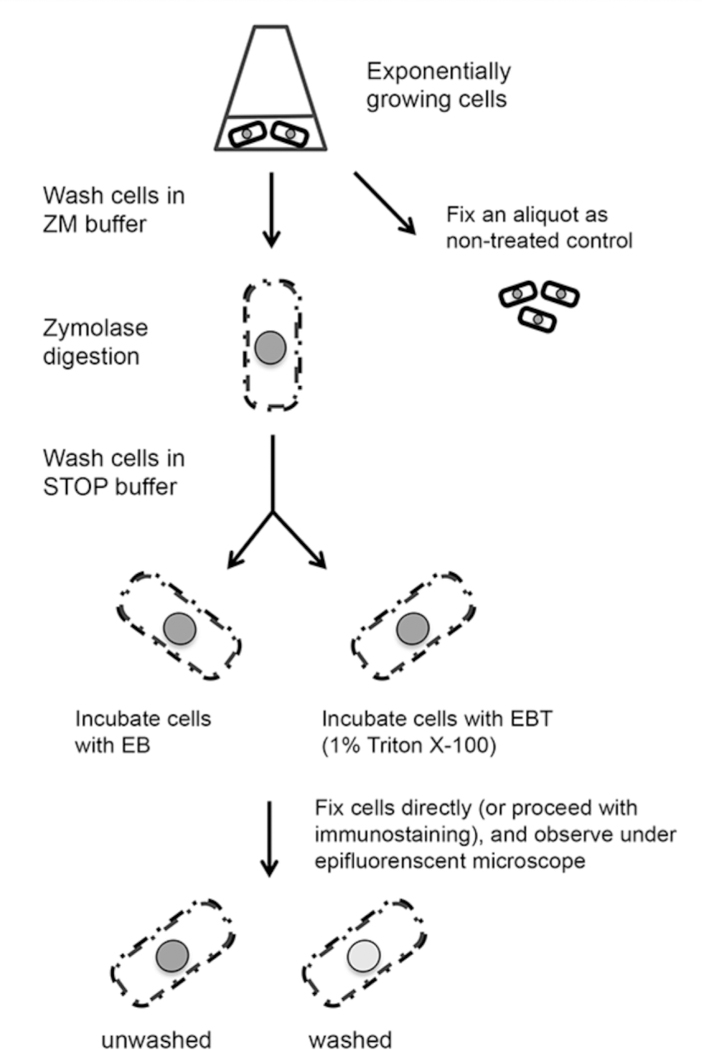
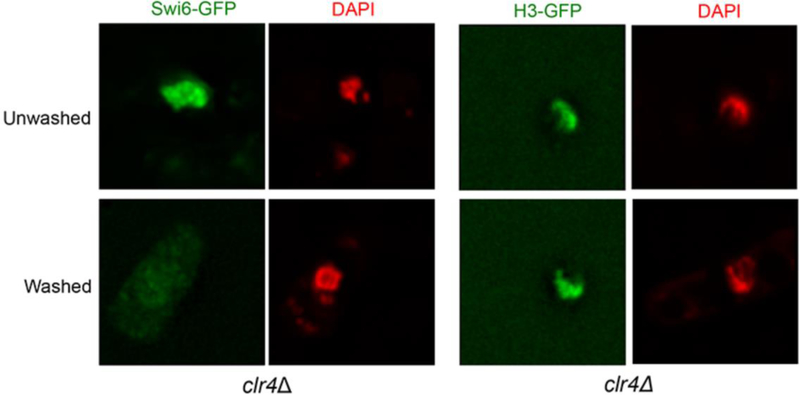
Chromatin immunoprecipitation (ChIP) is a widely used method to study the binding of proteins to chromatin, and has paved the way for better understanding of chromatin and epigenetic regulation. ChIP uses formaldehyde to create chemical crosslinks between proteins and DNA. The chromatin is mechanically sheared and subsequently precipitated by an antibody specific to the protein of interest. DNA fragments co-precipitated with the protein are analyzed by Southern hybridization or PCR [10–13]. However, the outcome of ChIP depends on the efficiency of crosslink, variation of immunoprecipitation and quality of antibodies. In addition, ChIP can only determine the protein binding ability to chromatin on average from a pool of cells. In situ chromatin binding assay has been developed to study the protein-chromatin interaction at the single-cell level in fission yeast. It originally was used to analyze the binding of replication and transcription factors to chromatin [14]. We have adapted it for use to study histone variants and histone modification complexes, such as centromere and heterochromatin regulators [6,15]. The method starts with partial digestion of fission yeast cell wall using zymolyase, followed by detergent extraction (washing with Triton X-100). As a result, soluble nucleoplasmic proteins and non-chromatin bound proteins are washed away, while proteins associated with chromatin remain, which can be detected using either a GFP (or GFP variants) tag, or indirect immunofluorescence (Fig. 1). Importantly, partial digestion of cell wall by zymolyase allows cells to maintain their structure. As an example, this technique has been used to examine the chromatin association of the human HP1 homolog, Swi6, in a clr4 mutant background. Swi6-GFP can be washed away upon detergent extraction in clr4Δ cells, demonstrating that Swi6 has little association with chromatin in the absence of H3K9me. On the other hand, the association of histone H3-GFP with chromatin is independent of Clr4 activity and therefore in clr4Δ cells the nuclear localization of H3 is retained after the same detergent treatment (Fig. 2).
Figure 1.
Schematic flow diagram for in situ chromatin binding assay.
Figure 2. In situ chromatin-binding assay for clr4Δ cells expressing either Swi6-GFP or H3-GFP.
Swi6-GFP is diffused in the nucleus in clr4Δ cells before washing with Triton X‐100 (top panel). The signal can be readily removed upon washing with the detergent (bottom panel). In contrast, the H3-GFP signal in clr4Δ cells is retained in the nucleus before (top panel) or after (bottom panel) Triton X‐100 extraction, indicating that H3-GFP is stably bound with the chromatin. Cells are counterstained with DAPI (red) to visualize the nucleus.
This in situ approach enables examination of protein distribution at the single-cell level while cell structure is largely maintained. In addition, since this method allows analysis of large cell population at single-cell level, cell cycle synchronization is not necessary, and comparative analyses between different mutants, time courses, or temperature conditions in fission yeast are simple and easy. When the protein of interest is directly tagged with a fluorescent tag, this method is most straightforward. But indirect immunofluorescence can also be used to detect proteins of interest. The described protocol and buffer conditions are optimized for histone variants and histone modifying complexes. However, this approach can be adapted to study other chromatin binding proteins. Note that the detergent extraction conditions have to be determined empirically for specific proteins, and it also is important to have proper control samples in order to demonstrate the washing is effective but not excessive.
2. Materials
ZM buffer: 50 mM sodium citrate pH 5.6, 1.2 M sorbitol, 0.5mM MgAc.
STOP buffer: 0.1 M MES pH 6.4, 1.2 M sorbitol, 1 mM EDTA, 0.5 mM MgAc.
Extraction buffer (EB): 20 mM PIPES-KOH, pH 6.8, 0.4 M sorbitol, 150 mM KAc, 2 mM MgAc. Store at –20°C.
EBT buffer: 20 mM PIPES-KOH, pH 6.8, 0.4 M sorbitol, 150 mM KAc, 2 mM MgAc, 10% Triton X-100.
20,000 U/g Zymolyase 20-T (Amsbio, cat. no. 120491–1), store at 4°C.
10% Triton X-100 (in EBT buffer).
2% Sodium dodecyl sulfate (SDS) (optional).
1 M Dithiothreitol (DTT), store at –20°C.
100% Methanol.
100% Acetone.
Protease inhibitor cocktail 100X (Sigma, cat.no. P8215), store at −20°C.
Fixation solution: 3.7% formaldehyde, 10% methanol.
Mounting medium: Vectashield anti-fade mounting media with DAPI, (Vector, cat. no. H-1200), store at 4°C.
Microscope slides, precleaned Plain Microscope Slides, (Fisherbrand, cat. no. 12–550-A3).
Poly-L-lysine–coated cover slips. Soak 13-mm cover slips (ThermoFisher Scientific cat. No. 174950) in 0.1% poly-L-lysine (Sigma, cat. no. P 8920) for 10 min. Drain, dry, rinse in deionized water, and dry.
VWR micro cover glass, cat. no. 48366–227.
PEM buffer: 100 mM PIPES, 1mM EGTA, 1mM MgSO4, adjust the pH to 6.9.
PEMSorb buffer: PEM, 1.2M Sorbitol.
PBSBAL buffer: PBS, 1% BSA, 100mM Lysine hydrochloride, and 0.01% NaN3, pH 6.9.
- Media:
- YES media:
For 1 Liter Final concentration Yeast extract 5g 0.5% w/v Glucose 30g 3% w/v adenine 225 mg histidine 225 mg leucine 225 mg uracil 225 mg lysine 225 mg - PMG media:
KH Phthallate 3g 14.7 mM Na2HPO4 2.2g 15.5 mM L-glutamic acid, monosodium salt 3.75g Glucose 20g 2% w/v Salt stock (50x) 20mL Vitamin stock (1000x) 1mL Mineral stock (10,000x) 0.1mL adenine 225 mg histidine 225 mg leucine 225 mg uracil 225 mg lysine 225 mg - 50× Salt stock
For 1 Liter Final concentration MgCl2.6H20 53.3g 0.26 M CaCl2.2H20 0.735g 4.99 mM KCl 50g 0.67 M Na2SO4 2g 14.1 mM - 1000× Vitamin stock
For 1 Liter Final concentration pantothenic acid 1g 4.2 mM nicotinic acid 10g 81.2 mM inositol 10g 55.5 mM biotin 10mg 40.8 μM - 10,000× Mineral stock
For 1 Liter Final concentration boric acid 5g 80.9 mM MnSO4 4g 23.7 mM ZnSO4.7H2O 4g 13.9 mM FeCl2.6H2O 2g 7.4 mM molybdic acid 0.4g 2.47 mM KI 1g 6.02 mM CuSO4.5H2O 0.4g 1.6 mM citric acid 10g 47.6 mM
3. Methods
3.1. in situ Chromatin Binding Assay using GFP-tagged proteins.
Inoculate the fission yeast strain into 10 mL YES or PMG media, and grow overnight at permissive temperature. Culture should be in log phase when cells are collected for assay. In general, 10 mL of culture at OD595nm = 0.2 (~108 cells in total) is enough for one to three assays (See Notes 1~4).
Spin down an aliquot of cells by at 3000 × g for 5 minutes. Fix cells by resuspending the pellet in fixation solution (See Note 5). These cells will be used as untreated control, and be examined in step 9.
Collect the remaining cells by centrifugation at 3000 × g for 5 min. Resuspend the cells in 1 mL ice-cold ZM buffer containing 10 mM DTT (See Note 6). Transfer to a 1.5-mL Eppendorf tube.
Spin cells down at 3000 × g for 5 min, and then resuspend in 0.5 mL ZM buffer containing 10 mM DTT and 1mg zymolyase 20-T (See Note 7). Incubate cells on a rocker platform at 32°C for 30 min. To test for adequate digestion, a few microliters of cells can be mixed with an equal volume of 2% SDS on a slide and examined under phase contrast microscopy. Cells should go phase dark. (Aim for >90% cells going phase dark.) If digestion is not adequate with the previous described procedures, add more zymolyase (at 0.2mg interval) or extend incubation time (check every 5 minutes).
When adequate digestion has been achieved, wash cells 3 times with ice-cold STOP buffer. (All subsequent centrifugations from here to step 8 are at 1000 × g for 1 min using an Eppendorf centrifuge at 4°C).
Resuspend cells in 1 mL ice-cold EB, and spin down.
Resuspend cells in 0.9 mL extraction buffer containing protease inhibitors. Transfer 0.45 mL of the cell suspension to a new 1.5-mL Eppendorf tube containing 50 μL EBT buffer. Mix and transfer to 20°C water bath for 7 min (See Note 8). Remaining cells will be used as unwashed control.
Spin both washed and unwashed cells down, and resuspend them in 20~50 μL ice-cold fixation solution. For proteins that are not directly tagged with GFP (or other fluorescent proteins), proceed to step 1 in Subheading 3.2.
To examine the cells by epifluorescence microscopy, spread a thin film of cells on microscope slide. Apply 1~2 μL Vectashield mounting solution and a cover slip to the sample. The cover slip can be sealed with nail varnish, but this is not necessary if the slides are to be viewed immediately (See Note 9). When obtaining images of cells, first check the cells under phase contrast to make sure cells within the field have been properly digested, then take a green fluorescent protein (GFP) image, and finally a DAPI image (See Note 10).
3.2. in situ Chromatin Binding Assay Using Indirect Immunofluorescence Staining.
Spin down cells prepared at step 8 in Subheading 3.1 at 1000 × g for 3 min and remove the fixation solution. Resuspend cells in 0.5mL PEMSorb buffer, and wash three times with PEM.
Resuspend cells with 50μL PEM by vortexing, and make sure that there aren’t any cell clumps. Spread about 10μL of the cell suspension onto a poly-L-lysine–coated 13-mm cover slip. Drain excessive liquid with kimwipes.
Transfer coverslips to a petri dish, wash in PBS for 5 mins.
Replace PBS with PBSBAL buffer, and incubate for 1 hour at room temperature (See Note 11).
Drain off PBSBAL buffer, and add 10μL primary antibody (diluted in PBSBAL) to coverslip. Incubate for 1 hour at room temperature in a humid chamber.
Wash three times in PBSBAL (5 min each).
Apply 20μL secondary antibody to the cover slip (conjugated with fluorescent dye). Incubate in dark humid container for at least 1 h at room temperature (See Note 12).
Wash three times in PBS (5 min each).
Drain the cover slip well. Mount in Vectashield anti-fade mounting media with DAPI. Seal the cover slip with nail varnish (See Note 13). Obtain images of cells taking first a fluorescent image, and then a DAPI image for each field of cells.
Acknowledgements
We thank Dr. Qianhua Dong, Hyoju Ban, and David Aristizabal Corrales for reading the manuscript. This work was supported by National Institutes of Health grant R01GM106037 (to F.L.) and NSF grant MCB-1330557 (to F.L.). F. L. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts.
Footnotes
Notes
Cells grow relatively healthier in rich (YES) medium than in minimum medium (PMG). However, for GFP-tagged proteins expressed on a non-integrated plasmid (such as pREP1), minimum medium lacking specific nutrient (for example, no leucine in the case of pREP1) should be used to make sure that the plasmid is properly maintained. We found that it is a good practice to culture cells in two-steps for cells carrying plasmid such as pREP1: Step 1) inoculate a single colony into 2 mL YES medium, and grow overnight. Collect cells by centrifugation at 3000 × g for 3 minutes, followed by washing once with sterile water. pREP1 plasmid harbors nmt1 promoter, which is repressed in the presence of thiamine. It is important to remove YES medium as much as possible because YES contains low level of thiamine. Step 2) inoculate cells from step 1 into 20 mL PMG without leucine, and culture at permissive temperature for 18~28 hours.
The optimal induction time for nmt1 promoter should be determined empirically for specific GFP-tagged proteins. Before processing cells for in situ chromatin binding assay, take an aliquot and examine under epifluorescent microscope to make sure that the GFP signals are visible and proteins are properly expressed.
Proteins analyzed should have a GFP (or other fluorescent) tag; alternatively, proteins can be detected via indirect immunofluorescence as described.
The Mg2+-containing EB is suitable for analysis of Swi6, CENP-ACnp1 and H3 proteins in log phase cells. For other proteins, modifications of the buffer condition may be necessary by the end user.
Alternatively, cells can be fixed by resuspending in 2 mL ice-cold 100 % methanol for 10 mins. Spin cells down and then resuspend cells in 1 mL ice-cold 100% acetone. The best fixation procedure has to be optimized for specific protein examined.
DTT should add freshly to the ZM buffer prior each use. To avoid repeated thaw and freeze, store small aliquots (no more than 1mL) of 1M DTT in −20°C.
Zymolyase in ZM buffer recommended here reaches final concentration of 2mg/ml, or 20U per 108 cells. Cells that were not adequately digested during the zymolyase digestion step appear bright under phase contrast microscope when examined by 2% SDS.
The exact time for washing with the detergent (1% Triton X-100) should be decided for individual proteins analyzed. We found that 7 minutes at 20°C is sufficient for washing away of Swi6 and CENP-ACnp1. It is highly recommended to use proper control groups to determine the optimal washing time, and to run control groups in parallel with each experiment. If it is suspected that the protein of interest may be hold in the nucleus by structures other than chromatin, such as nuclear matrix, it is recommended to include micrococcal nuclease digestion as an additional control to determine whether the retention of protein after detergent washing is chromatin-dependent. Method for micrococcal nuclease digestion control has been previously described [14].
When cover slips are sealed with nail varnish, slides can be stored at 4°C for up to 7 days. When ready to analyze, warm the slides in room temperature until the condensation is gone.
Other than epifluorenscent microscope, flow cytometry analysis can be employed to further analyze the fluorescent signal within the cell population. Detailed methods have been previously described [14].
This step is used to block nonspecific binding of the epitope. The appropriate time can be determined based on different antibodies. Coverslip can be rocked gently to speed up the process.
Spin the secondary antibody before use to get rid of any fluorescent precipitate.
After nail varnish is completely dried, slides can be stored at −20°C.
References
- 1.Allshire RC, Ekwall K (2015) Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe. Cold Spring Harbor perspectives in biology 7 (7):a018770. doi: 10.1101/cshperspect.a018770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buscaino A, White SA, Houston DR, Lejeune E, Simmer F, de Lima Alves F, Diyora PT, Urano T, Bayne EH, Rappsilber J, Allshire RC (2012) Raf1 Is a DCAF for the Rik1 DDB1-like protein and has separable roles in siRNA generation and chromatin modification. PLoS genetics 8 (2):e1002499 10.1371/journal.pgen.1002499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez M, He H, Sun S, Li C, Li F (2013) Cell cycle-dependent deposition of CENP-A requires the Dos1/2-Cdc20 complex. Proceedings of the National Academy of Sciences of the United States of America 110 (2):606–611. 10.1073/pnas.1214874110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez M, Li F (2012) DNA replication, RNAi and epigenetic inheritance. Epigenetics 7 (1):14–19. 10.4161/epi.7.1.18545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ (2005) Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Current biology : CB 15 (16):1448–1457. 10.1016/j.cub.2005.07.021 [DOI] [PubMed] [Google Scholar]
- 6.Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ (2008) Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell 135 (2):272–283. 10.1016/j.cell.2008.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He H, Gonzalez M, Zhang F, Li F (2014) DNA replication components as regulators of epigenetic inheritance--lesson from fission yeast centromere. Protein & cell 5 (6):411–419. 10.1007/s13238-014-0049-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 (5532):1098–1102. 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Li F (2016) Are all repeats created equal? Understanding DNA repeats at an individual level. Curr Genet 10.1007/s00294-016-0619-x [DOI] [PMC free article] [PubMed]
- 10.Dedon PC, Soults JA, Allis CD, Gorovsky MA (1991) A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem 197 (1):83–90 [DOI] [PubMed] [Google Scholar]
- 11.Collas P (2010) The current state of chromatin immunoprecipitation. Mol Biotechnol 45 (1):87–100. 10.1007/s12033-009-9239-8 [DOI] [PubMed] [Google Scholar]
- 12.Bawa-Khalfe T (2016) Isolation of In Vivo SUMOylated Chromatin-Bound Proteins. Methods Mol Biol 1475:205–216. 10.1007/978-1-4939-6358-4_15 [DOI] [PubMed] [Google Scholar]
- 13.Ricke RM, Bielinsky AK (2005) Easy detection of chromatin binding proteins by the Histone Association Assay. Biol Proced Online 7:60–69. 10.1251/bpo106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kearsey SE, Montgomery S, Labib K, Lindner K (2000) Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J 19 (7):1681–1690. 10.1093/emboj/19.7.1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez M, He H, Dong Q, Sun S, Li F (2014) Ectopic centromere nucleation by CENP--A in fission yeast. Genetics 198 (4):1433–1446. 10.1534/genetics.114.171173 [DOI] [PMC free article] [PubMed] [Google Scholar]