Abstract
Misfolding and amyloid formation of transthyretin (TTR) is implicated in numerous degenerative diseases. TTR misfolding is greatly accelerated under acidic conditions, and thus most of the mechanistic studies of TTR amyloid formation have been conducted at various acidic pH values (2 – 5). In this study, we report the effect of pH on TTR misfolding pathways and amyloid structures. Our combined solution and solid-state NMR studies revealed that TTR amyloid formation can proceed via at least two distinct misfolding pathways depending on the acidic conditions. Under mildly acidic conditions (pH 4.4), tetrameric native TTR appears to dissociate to monomers that maintain most of the native-like β-sheet structures. The amyloidogenic protein undergoes a conformational transition to largely unfolded states at more acidic conditions (pH 2.4), leading to amyloid with distinct molecular structures. Aggregation kinetics is also highly dependent upon the acidic conditions. TTR quickly forms moderately ordered amyloids at pH 4.4, while the aggregation kinetics is dramatically reduced at a lower pH of 2.4. The effect of the pathogenic mutations on aggregation kinetics is also markedly different under the two different acidic conditions. Pathogenic TTR variants (V30M and L55P) aggregate more aggressively than WT TTR at pH 4.4. In contrast, the single-point mutations do not affect the aggregation kinetics at the more acidic condition of pH 2.4. Given that the pathogenic mutations lead to more aggressive forms of TTR amyloidoses, the mildly acidic condition might be more suitable for mechanistic studies of TTR misfolding and aggregation.
Keywords: Transthyretin, misfolding, amyloid formation, solid-state NMR
1. Introduction
Transthyretin (TTR) is a natively folded tetrameric protein that is secreted into the blood and cerebrospinal fluid (CSF) by the liver and choroid plexus, respectively. TTR acts as a carrier of retinol-binding protein in the blood and thyroid hormone (T4) in the CSF. Amyloid formation of wild-type (WT) TTR is associated with senile systemic amyloidosis (SSA) affecting as much as 25% of the population over age 80. More than 100 single-point mutations are known to accelerate amyloid formation, leading to earlier onset of TTR amyloidoses such as familial amyloidotic polyneuropathy (FAP), familial amyloid cardiomyopathy (FAC), [1] and rarely central nervous system selective amyloidosis (CNSA). [2,3] Previous biophysical studies showed that TTR amyloidoses are initiated by the dissociation of the tetramers to monomers followed by a conformational transition of the monomer to a partially unfolded intermediate, which self-assembles into amyloid.[4–7]
The conformational transition of the globular protein to amyloidogenic intermediates is facilitated under partly denaturing acidic conditions. [8–11] It has been hypothesized that TTR misfolding and aggregation might be triggered in the acidic environments in vivo such as endosomes and lysosomes. [12] Although it is still unknown whether TTR is exposed to the acidic environments in vivo, acid-induced TTR misfolding studies in vitro at pH 4 – 5 have provided valuable insights into the molecular mechanism of TTR amyloidogenesis, [4–7,13] leading to the development of therapeutic agents such as tafamidis approved in Europe and Japan.
Recently, more acidic conditions (pH 2 – 3) were also used to investigate TTR amyloid formation process, providing additional insights into aggregation kinetics, assembly-disassembly properties of TTR protofibrils, and structural features of intermediate aggregates. [14–16] The biophysical studies at various acidic conditions enhanced our understanding of molecular basis of TTR amyloid formation. It is, however, unclear whether the TTR aggregations proceed via a single misfolding pathway at the acidic pHs or multiple pathways depending on the pH values.
In the present study, we systematically investigated the effect of pH on the TTR misfolding and amyloid formation pathways using solution and solid-state NMR spectroscopy. Our previous biophysical studies revealed that tetrameric TTR dissociates into aggregation-prone monomers that contain extensive native-like β-sheet conformations at the mildly acidic pH of 4.4. [17] In addition, the amyloidogenic TTR monomers were shown to form moderately ordered amyloid. Here we report that TTR undergoes a conformational transition to more unfolded states at pH 2.4, and slowly forms amyloid with distinct molecular structures. Wild-type and mutant forms of TTR also have markedly different aggregation properties at the different pH values. At the mildly acidic pH of 4.4, clinically more aggressive mutations (V30M and L55P) accelerate TTR aggregations, while the aggregation kinetics was not altered by the pathogenic mutations at pH 2.4. These results indicate that the TTR misfolding and aggregation process observed at pH 4.4 might be more relevant to that observed in vivo.
2. Materials and methods
2.1. Protein Expression.
Wild-type and two mutant forms of TTR (V30M and L55P) were expressed in an E. coli expression system, as previously described. [4] The overexpressed proteins were purified by using an anion exchange Q column and a Superdex 75 gel filtration column (GE Healthcare, Pittsburg, PA) running on an ÄKTA FPLC system. Uniformly 15N-labeled protein was prepared using 15N-NH4Cl as a sole source of nitrogen in M9 minimal medium. The protein concentration was measured and calculated using an extinction coefficient of 77,600 M-l cm−1 at 280 nm.
For selectively 13CO- and 13Cα-labeled samples, the M9 media supplemented with unlabeled amino acids (100 mg per liter culture) and 13CO- and 13Cα-labeled amino acids (50 mg/L) were used for protein expression. The selective labeling of the specific amino acid was confirmed by 13C solid-state NMR spectra of the native state of TTR.
2.2. Amyloid Formation.
Amyloid samples at pH 4.4 were obtained by mixing the protein (0.2 mg/mL in 10 mM PBS buffer) with 200 mM acetate buffer (100 mM KCl, 1 mM EDTA, pH 4.4). In order to trigger TTR aggregation at a low pH of 2.4, 1 – 2 μL of concentrated H3PO4 was added to the protein solution (10 mM PBS buffer) to lower the pH directly to 2.4.
2.3. NMR Spectroscopy.
1H/15N heteronuclear single-quantum coherence (HSQC) NMR spectra were recorded at 15 °C on a 500 MHz Varian Inova spectrometer equipped with a z-axis gradient triple resonance probe. Solid-state NMR spectra were recorded using Bruker 830 MHz spectrometers equipped with a 3.2 mm MAS probe. Two-dimensional 13C-13C correlation NMR spectra were collected using a proton-driven spin diffusion (PDSD) mixing scheme [18] at spinning frequencies of 11 – 12 kHz. The spinning speed was set close to the Δωiso = 2ωr rotational resonance (RR) conditions [19,20] for efficient polarization transfer. The 90° pulse-lengths for 1H and 13C were 3.0 and 2.5 μs, respectively, and the two-pulse phase-modulated (TPPM) decoupling scheme was employed with a radio-frequency field strength of 85 kHz. For the 2D PDSD spectra, complex data points of 1024 × 145 were collected for amyloid states with an acquisition delay of 2 sec, and 48 – 64 FIDs were accumulated for each t1 data point. The cross-peaks observed in all of the TTR amyloid formed at pH 4.4 were assigned in our previous solid-state NMR study. [17]
2.4. Circular dichroism (CD) Spectroscopy.
The CD spectra were recorded by scanning from 260 nm to 190 nm on a Jasco J-810 spectropolarimeter (Easton, MD) using a 1 mm path length Suprasil quartz cell at pH 2.4 – 7.0. The protein samples (0.1 mg/ml) were pre-equilibrated at 15 °C for 30 min before acquisitions.
2.5. Transmission electron microscopy (TEM).
Insoluble TTR aggregates were spun down at 13,000 g for 1 hour and washed twice with deionized water to remove remaining soluble tetramers and soluble aggregates. The TTR amyloid samples were negatively stained with 2 % uranyl acetate. Aliquots of 5 uL TTR samples were applied to 200 Formvar-coated copper grids, and the TTR amyloids were visualized using Philips CM12 transmission electron microscope at 80 kV.
2.6. Aggregation essay.
Under the mildly acidic condition (pH 4.4), TTR precipitates as large aggregates that can be quantified by measuring optical density at 400 nm. The turbidity of the protein solution was recorded over a period of 10 hrs to monitor aggregation kinetics at pH 4.4. At a more acidic pH of 2.4, the protein sample forms transparent gel-like aggregates, which cannot be quantified by the turbidity measurements. Dynamic light scattering (DLS) was used to probe structural changes from the initial unfolded states to β-structured amyloid over a period of 24 hrs.
3. Results
3.1. Misfolding pathways
Amyloid formation of globular proteins involves a global/local unfolding of natively folded proteins to intermediates that self-assemble into amyloid adopting a cross-β structure. Partial denaturation of globular proteins has been used to promote misfolding and amyloid formation to investigate molecular mechanism of the aggregation process in vitro. [21–25] Previous studies of the acid-mediated denaturation pathway of TTR showed that mild acidifications with optimum pH of 4.4 trigger rate-limiting tetramer dissociation and subsequent monomer unfolding that is required for TTR amyloid formation. [4–6] Recently, stronger acidic conditions (pH 2 – 3) have also been used to induce TTR amyloid formation. [14–16] It is, however, unclear whether the TTR aggregations proceed via a similar misfolding mechanism or distinct molecular mechanisms at different acidic conditions.
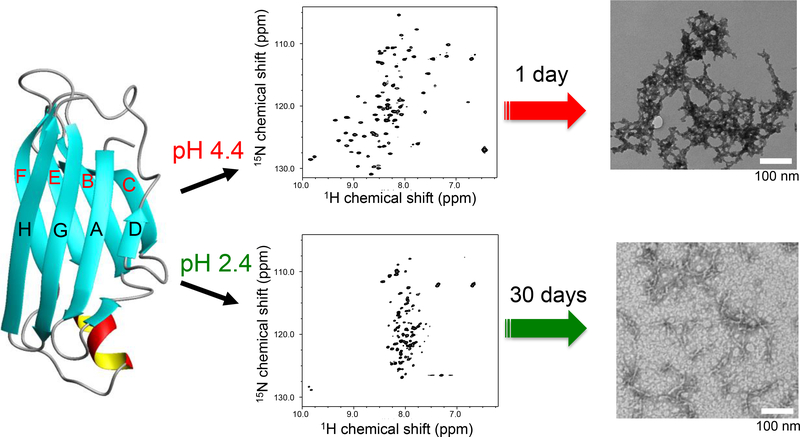
In order to investigate the misfolding pathways at the acidic conditions in more detail, two dimensional 1H/15N HSQC NMR spectra were recorded at the two different pH values of 4.4 and 2.4 (Figure 1). Under the mildly acidic condition (pH 4.4), the NMR resonances in the HSQC spectrum are well-dispersed, indicating that TTR still adopts a folded conformation at the amyloidogenic pH. In contrast to pH 4.4, the NMR cross-peaks appear in completely different positions at pH 2.4 (Figure S1), suggesting that TTR undergoes a drastic conformational change at the more acidic condition. The clustered NMR resonances in the 1H dimension are typically observed for unfolded states of proteins, indicating that TTR becomes largely unfolded at pH 2.4.
Figure 1.
A schematic diagram for the two distinct misfolding and amyloid formation pathways of TTR at the two different acidic conditions (pH 2.4 and 4.4) along with 2D 1H/15N HSQC NMR spectra and TEM images. A very low protein concentration of 0.15 mg/ml was used for the 2D 1H/15N HSQC NMR experiments to ensure TTR exist as monomeric forms. The TEM images were obtained from the TTR samples (0.2 mg/ml and 10 mg/ml) incubated for 1 day and one month at pH 4.4 and 2.4, respectively. The different protein concentrations and incubation times were used because of the different aggregation kinetics. At pH 2.4, a much higher protein concentration was required to obtain amyloid precipitates due to the much slower aggregation kinetics than those at pH 4.4.
The distinct structural features of the monomeric intermediate states observed in the HSQC NMR spectra were confirmed with CD spectroscopy (Figure S2). The native TTR predominantly adopts β-sheet conformations consisting of the two β-sheets (CBEF and DAGH β-sheets, Figure 1). The CD spectrum collected at pH 7.0 shows a minimum between 210 and 220 nm indicative of typical β-sheet conformations, indicating that TTR adopts β-sheet conformations at the physiological pH. It is interesting to note that the CD spectrum recorded at the amyloidogenic pH 4.4 is almost identical to that observed at the native condition (pH 7.0). The CD spectrum recorded at the more acidic pH of 2.4 displays a marked difference from that obtained at pH 4.4. The minimum intensity at 190 nm clearly indicates that TTR undergoes an unfolding transition to more disordered states at pH 2.4.
3.2. Structural features of amyloids.
TTR amyloids incubated at the two different conditions (pH 4.4 and 2.4) were examined with TEM and thioflavin T (ThT) fluorescence measurements. Under the mildly acidic condition of pH 4.4, TTR forms moderately ordered aggregates (amyloid-4, Figure 1). On the other hand, small filamentous aggregates were observed at more acidic condition (amyloid-2, Figure 1), as was previously observed. [26] The ThT fluorescence intensities upon binding to the two amyloids were also consistent with the TEM images (Figure S3). A more enhanced ThT fluorescence for the amyloid formed at pH 2.4 suggests that the filamentous aggregates might have larger surfaces where the small molecule can bind than the TTR aggregates obtained at pH 4.4.
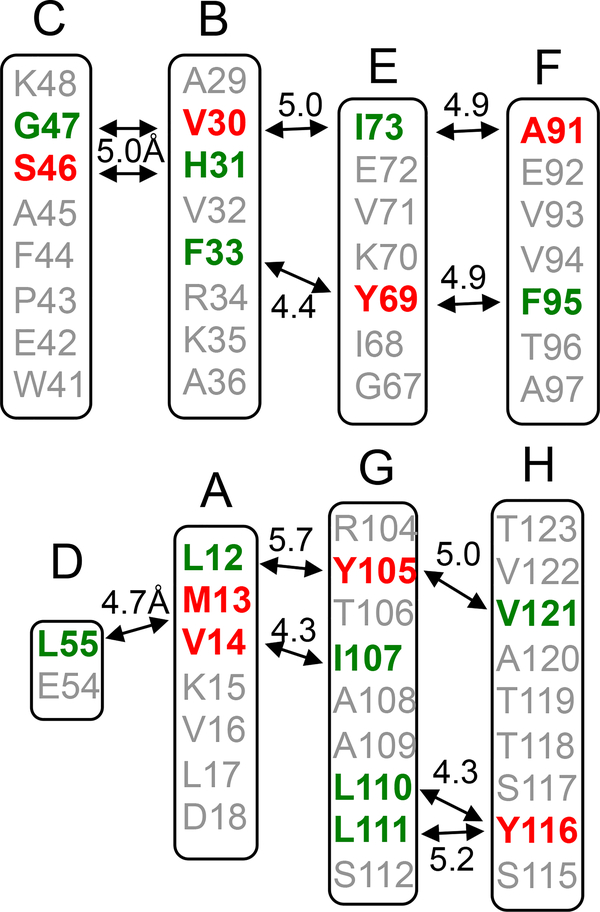
We employed solid-state NMR to compare molecular structures of the two amyloids in more detail. Two-dimensional (2D) 13C-13C correlation NMR spectra were acquired to compare overall structural features of the two amyloid states (Figure S4). The 2D NMR spectra for the two amyloid states are overlapped well, suggesting that the two amyloid forms have overall similar structures. Our previous structural studies showed that TTR amyloid formed at pH 4.4 has extensive native-like β-sheet structures. [17] Thus, we investigated native-like β-structures of the two amyloid states using selective 13CO/13Cα-labeling schemes (Figure 2), which was shown to be of great use in investigating native-like β-structures of amyloid states of TTR. [17] For example, selective 13CO-Leu and 13Cα-Met labeling will produce one dipolar-coupled 13CO-13Cα spin pair at distances of 4–6 Å in the DA strands (Figure 2). Solid-state NMR experiments that detect spin pairs with a separation of up to 6 Å can then be used to investigate any structural changes in the β-structure.
Figure 2.
Selective labeling schemes for the solid-state NMR experiments. The 13CO (green) and 13Cα (red) carbons of the amino acids are labeled if the internuclear distance in the native TTR tetramer is between 4 and 6 Å.
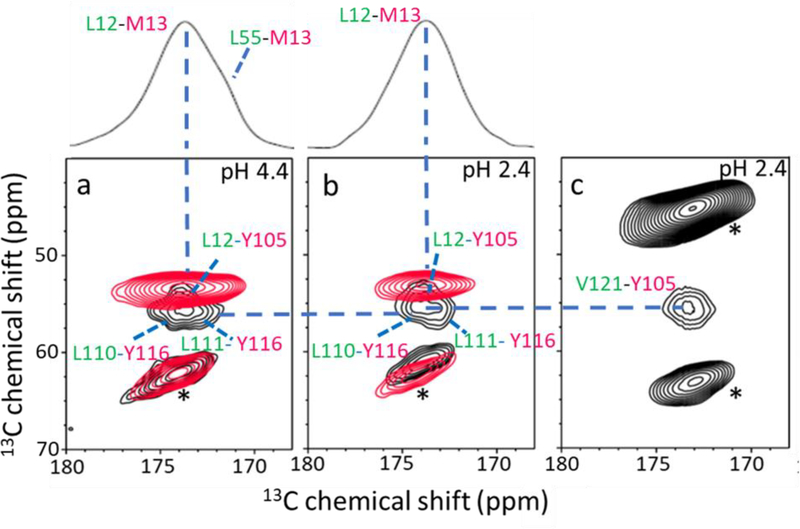
Figure 3 shows 2D 13C-13C correlation NMR spectra obtained with a proton-driven spin diffusion (PDSD) mixing scheme for the 13CO-Leu and 13Cα-Met (L/M) labeled TTR amyloids (red in Figure 3a and 3b). In the amyloid state of TTR formed at pH 4.4 (amyloid-4), two cross-peaks for the L55-M13 and L12-M13 spin pairs were detected, suggesting the presence of native-like DA β-structure in the amyloid state (red in Figure 3a and Figure S5). However, the cross-peak for L55-M13 was not observed for the amyloid obtained at pH 2.4 (amyloid-2, red in Figure 3b), indicating a structural change in the DA structure at the lower pH. The solid-state NMR experiments were then used to probe the AGH β-structure in the amyloid states (black in Figure 3a and 3b). In the overlaid PDSD spectra for the L/Y (black) and L/M (red) spin pairs, the two cross-peaks for the L110-Y116 and L111-Y116 (black in Figure 3a) were observed in amyloid-4. The same cross-peaks for the L/Y spin pairs were also detected in the amyloid-2 (black in Figure 3b). In addition, one cross-peak for L12-Y105 in AG β-structure was detected in the amyloid-2 (Figure 3c). The cross-peak for the for L12-Y105 spin pair was also observed in amyloid-4 state in our previous solid-state NMR studies, [17] suggesting that the AGH β-structure is maintained in the two amyloid states.
Figure 3.
(a) Overlaid 2D PDSD spectra of the amyloid-4 with 13CO-Leu/13Cα-Met (red; labeled on DA), and 13CO-Leu/13Cα-Tyr (black; labeled on AGH) TTR. (b) Overlaid 2D PDSD spectra of the amyloid-2 with the same labeling schemes in a. (c) 2D PDSD spectrum of the amyloid-2 with 13CO-Val/13Cα-Tyr labeled on GH strands. The 1D slice in Figure 3a and 3b was drawn for the L/M spin pairs. The NMR spectra were obtained with a mixing time of 500 ms at a 1H frequency of 830 MHz. The L/Y and L/M spin pairs in the amyloid-4 state were unambiguously assigned in our previous solid-state NMR studies. [17] In order to confirm that the cross-peaks originate from the spin pairs within the TTR monomer, the 2D PDSD experiments were conducted on the mixture of singly 13CO- and 13Cα- labeled TTRs (1:1 ratio, for example a mixture of 13CO-Leu-TTR and 13Cα-Tyr-TTR) and no cross-peak was observed from the mixture. * denotes spinning sidebands.
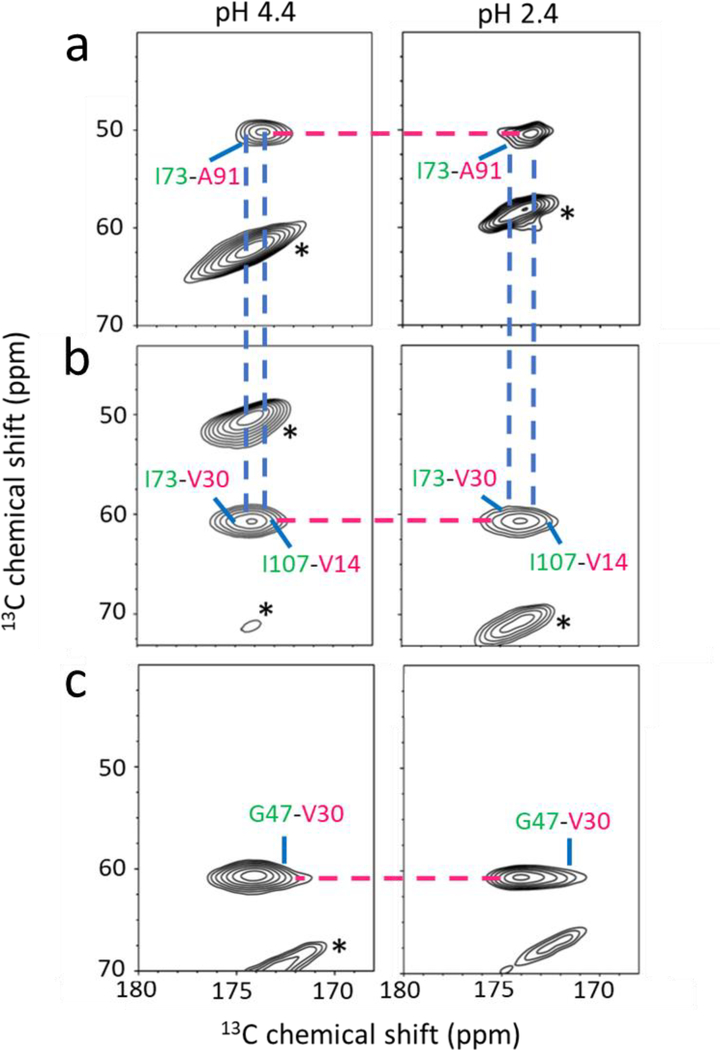
The solid-state NMR experiments were extended to probe native-like CBEF β-structures in the amyloid states using the 13CO and 13Cα labeling schemes (Figure 2). The 2D PDSD solid-state NMR spectra for various labeling schemes were compared for the two amyloid states. Our previous solid-state NMR experiments with the amino acid selective labeling schemes in the CBEF β-structure showed that the native-like CBEF β-structure is maintained in TTR amyloid formed at pH 4.4. [17] For example, the cross-peaks for I73-V30 and I73-A91 spin pairs in the BEF β-structure (left in Figure 4a and 4b) were observed in the 2D PDSD spectra for the amyloid-4. The 2D PDSD experiments with additional labeling schemes in CB β-structure also showed that the CB β-structure is maintained in the amyloid-4 on the basis of the cross-peaks for the H31-S46 (Figure S6) and G47-V30 (left in Figure 4c) spin pairs in the CB β-structure (Figure 2).
Figure 4.
2D PDSD spectra of the two amyloid states for the labeling schemes shown in Figure 2. (a) 13CO-Ile/13Cα-Ala for the EF strands. (b) 13CO-Ile/13Cα-Val for the BE strands. (c) 13CO-Gly/13Cα-Val for the CB strands. The spin pairs in the amyloid-4 state were unambiguously assigned in our previous solid-state NMR studies. [17]
The 2D PDSD spectra for the same labeling schemes were acquired for the amyloid formed at pH 2.4 and compared to those of the other amyloid state (right in Figure 4 and Figure S6). The NMR spectra for the I/A and I/V spin pairs in the BEF β-structure are almost identical (Figure 4a and 4b), suggesting the presence of the native-like BEF β-structure in the amyloid-2. The native-like structure is further evidenced by the two cross-peaks for the F95-Y69 and F33-Y69 spin pairs in the BEF β-structure (Figure 2 and S7). In addition, similar PDSD spectra for the labeling schemes of CB β-structure (G47-V30 and H31-S46 in Figure 4c and Figure S6, respectively) were observed for both amyloid states. The extensive 2D solid-state NMR experiments reveal that both TTR amyloid states contain native-like CBEF β-structures. On the other hand, the DAGH β-structure is maintained in the amyloid-4, while only AGH β-structure is preserved in the amyloid-2.
3.3. The effect of the pathogenic mutations on aggregation kinetics.
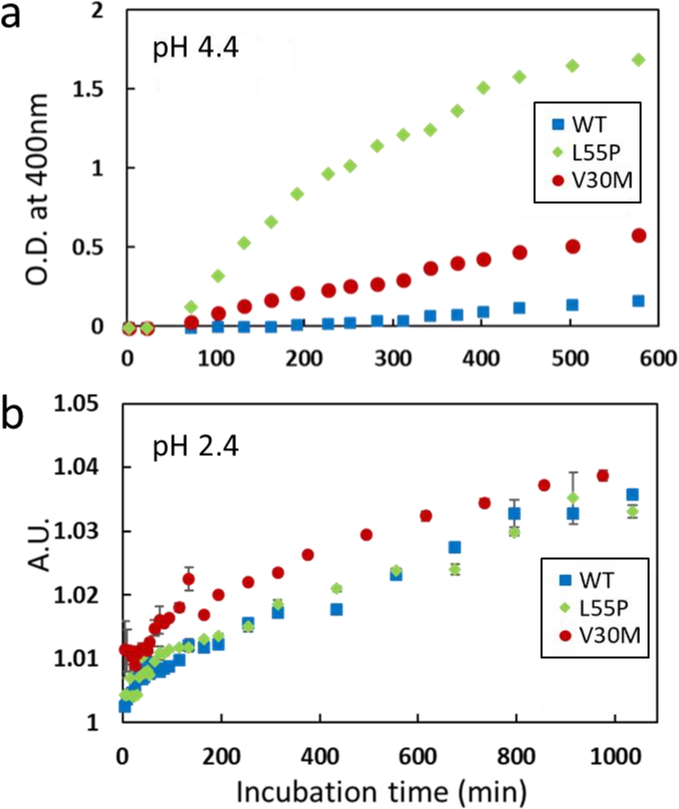
A variety of pathogenic mutations are known to destabilize native TTR tetramers, facilitating dissociation to monomers, which is the rate-limiting step in TTR amyloid formation process. Aggregation kinetics of WT and two mutant forms of TTR (V30M and L55P) was monitored and compared at the two acidic conditions of pH 2.4 and 4.4 (Figure 5). Turbidity was monitored by measuring optical density at 400 nm for the aggregation kinetics at pH 4.4 (Figure 5a). The kinetics studies showed that the pathogenic mutant TTR aggregates much faster than WT TTR. Aggregation properties of the WT and mutant TTRs were also examined at the more acidic pH value of 2.4 (Figure S8 and 5b). Dynamic light scattering was used to probe aggregation kinetics since TTR forms transparent gel-like aggregates at pH 2.4, rather than white precipitates formed at pH 4.4. Interestingly, WT and mutant forms of TTR exhibit similar aggregation behaviors at pH 2.4, suggesting that the pathogenic mutations do not affect the aggregation property of TTR at the more acidic condition.
Figure 5.
Aggregation kinetics of the WT and mutant forms of TTR measured at pH 4.4 (a) and 2.4 (b). Optical density at 400 nm was monitored at pH 4.4, and dynamic light scattering was used to probe aggregation at pH 2.4. The scattering intensity at 100 μs in the scattering profile shown in Figure S8 was plotted and compared for the WT and mutant forms of TTR. Errors in the experimental measurements were mostly smaller than the size of the markers.
4. Discussion
Acid-denaturation is required to induce local and/or global unfolding of TTR to aggregation-prone monomeric intermediate states. Various acidic conditions (pH 2 – 5) have, therefore, been used to investigate molecular mechanism of TTR misfolding and amyloid formation. Initial biophysical studies showed that amyloids derived by a variety of polypeptides shares common structural features, suggesting the presence of similar misfolding mechanisms. However, recent high-resolution structural studies suggested that amyloids formed by even a single protein can adopt diverse molecular structures depending on experimental conditions, [27–33] implying that TTR may have multiple misfolding pathways depending on the pH values.
Previous biochemical studies of TTR aggregation showed that mildly acidic pH of 4 – 5 are the optimum conditions to promote TTR misfolding and amyloid formation. [7] Further acidification (below pH 3) significantly slowed down the aggregation kinetics and no visible precipitates were observed at the same incubation conditions (protein concentration and temperature).[4] Thus mechanistic studies of TTR amyloid formation have been predominantly carried out under the mildly acidic conditions of pH 4 – 5 based on the hypothesis that TTR may be exposed to acidic environments such as lysosomes in vivo. Recently, more acidic pH of 2 – 3 has been used to induce TTR unfolding and subsequent amyloid formation for the mechanistic studies of TTR aggregation. In this study, we investigated TTR misfolding and aggregation pathways under the two acidic conditions to explore whether TTR aggregates via distinct misfolding pathways or similar pathways with common misfolding intermediates.
It was previously shown that TTR forms moderately ordered amyloid through native-like misfolding intermediate states with CBEF and DAGH β-structures at pH 4.4. [17] Our current solution and solid-state NMR studies suggested a structural change in the DA β-structure under the more acidic condition of pH 2.4, which might lead to distinct amyloid with different molecular conformations. In addition, effect of the pathogenic mutations on the TTR aggregation kinetics is markedly different under the different acidic conditions. A majority of the pathogenic mutations were shown to destabilize the native tetrameric TTR structure, facilitating dissociation of the tetramer to monomers that is the rate-limiting step in the TTR amyloid formation process. [5,34,35] It was demonstrated that V30M and L55P TTR tetramers exhibit less thermodynamic stabilities than WT tetramer. The L55P mutant tetramer was also shown to dissociate into monomers much faster than V30M and WT TTR. The thermodynamic and kinetic stabilities of WT and mutant TTRs are well correlated with the aggregation kinetics measured at pH 4.4 (Figure 5a). On the contrary, the pathogenic TTR variants (V30M and L55P) exhibit similar aggregation behavior at pH 2.4. Previous biophysical studies showed that the two single-point mutations selectively affect the DA β-structure, accelerating misfolding and aggregation. [36–38] Under the more acidic condition of pH 2.4, the TTR misfolding intermediate state is largely unfolded (Figure 1), and the DA β-structure is already disrupted even in the WT TTR, as shown in Figure 3b. Thus the effect of the pathogenic mutations on structural changes might be negligible at pH 2.4, leading to the similar aggregation kinetics (Figure 5b).
It is also interesting to note that the amyloid state of TTR formed at pH 2.4 contains extensive native-like β-structures (CBEF and AGH β-sheet), although monomeric intermediate states are largely unfolded at the acidic condition, as shown by NMR and CD spectra (Figure 1 and Figure S1). These results indicate that only partially unfolded TTR conformers in the dynamic conformational ensemble of unfolded states can form amyloid. Finally, the reduced aggregation kinetics at pH 2.4 might originate from the larger entropic penalty for the conformational transition from more dynamic misfolding intermediates to β-structured aggregates at pH 2.4, compared to that from more structured intermediates to amyloid at pH 4.4.
5. Conclusion
We investigated distinct misfolding pathways of transthyretin (TTR) under different aggregation conditions, which have been used for previous biochemical studies of TTR misfolding and aggregation. Our combined solution and solid-state NMR studies revealed that TTR amyloid formation can proceed via at least two distinct misfolding pathways depending on the aggregation conditions. In addition, we showed that the effect of the pathogenic mutations on aggregation kinetics is also markedly different under the two different conditions. The experimental results provided useful information as to the suitable aggregation condition for TTR misfolding and amyloid formation. Our experimental data suggested that mildly acidic conditions (pH 4.4) are physiologically more relevant for TTR misfolding studies than more acidic conditions (pH 2.4), which would be of great use for future mechanistic studies of TTR misfolding and aggregation.
Supplementary Material
Highlights.
Transthyretin can aggregate via two distinct misfolding pathways, leading to distinct amyloid structures.
Under mildly acidic conditions (pH 4.4), TTR quickly forms moderately ordered amyloid and pathogenic mutations (L55P and V30M) accelerate aggregation.
At a more acidic condition of pH 2.4, TTR becomes more unfolded and slowly forms a different type of amyloid, and the pathogenic mutations do not affect TTR aggregation propensity.
Acknowledgements
This work was supported by the NIH grant (NS084138 and NS097490, K.H.L.). The solid-state NMR spectra were acquired at the National High Magnetic Field Laboratory, which is supported by NSF Cooperative Agreement No. DMR-1157490 and the State of Florida.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online. Overlaid 2D PDSD NMR spectra for the amyloid-4 state were included in Figure S9 to confirm the reproducibility of the NMR spectra.
Conflict of interest
We hereby declare that the authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jacobson DR, Pastore RD, Yaghoubian R, Kane I, Gallo G, Buck FS, Buxbaum JN, Variant-sequence transthyretin (isoleucine 122) in late-onset cardiac amyloidosis in black Americans, N. Engl. J. Med, 336 (1997), 466–473. [DOI] [PubMed] [Google Scholar]
- [2].Connors LH, Lim A, Prokaeva T, Roskens VA, Costello CE, Tabulation of human transthyretin (TTR) variants, 2003, Amyloid, 10 (2003), 160–184. [DOI] [PubMed] [Google Scholar]
- [3].Saraiva MJ, Transthyretin mutations in health and disease, Hum. Mutat, 5 (1995), 191–196. [DOI] [PubMed] [Google Scholar]
- [4].Lai ZH, Colon W, Kelly JW, The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid, Biochemistry, 35 (1996), 6470–6482. [DOI] [PubMed] [Google Scholar]
- [5].Lashuel HA, Lai ZH, Kelly JW, Characterization of the transthyretin acid denaturation pathways by analytical ultracentrifugation: Implications for wild-type, V30M, and L55P amyloid fibril formation, Biochemistry, 37 (1998), 17851–17864. [DOI] [PubMed] [Google Scholar]
- [6].Quintas A, Vaz DC, Cardoso I, Saraiva MJ, Brito RM, Tetramer dissociation and monomer partial unfolding precedes protofibril formation in amyloidogenic transthyretin variants, J. Biol. Chem, 276 (2001), 27207–27213. [DOI] [PubMed] [Google Scholar]
- [7].Colon W, Kelly JW, Partial Denaturation of Transthyretin Is Sufficient for Amyloid Fibril Formation Invitro, Biochemistry, 31 (1992), 8654–8660. [DOI] [PubMed] [Google Scholar]
- [8].Kelly JW, The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways, Curr. Opin. Struct. Biol, 8 (1998), 101–106. [DOI] [PubMed] [Google Scholar]
- [9].Booth DR, Sunde M, Bellotti V, Robinson CV, Hutchinson WL, Fraser PE, Hawkins PN, Dobson CM, Radford SE, Blake CCF, Pepys MB, Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis, Nature, 385 (1997), 787–793. [DOI] [PubMed] [Google Scholar]
- [10].Knowles TP, Vendruscolo M, Dobson CM, The amyloid state and its association with protein misfolding diseases, Nat. Rev. Mol. Cell Biol, 15 (2014), 384–396. [DOI] [PubMed] [Google Scholar]
- [11].Bourgault S, Solomon JP, Reixach N, Kelly JW, Sulfated glycosaminoglycans accelerate transthyretin amyloidogenesis by quaternary structural conversion, Biochemistry, 50 (2011), 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Connelly S, Choi S, Johnson SM, Kelly JW, Wilson IA, Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses, Curr. Opin. Struct. Biol, 20 (2010), 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nakanishi T, Yoshioka M, Moriuchi K, Yamamoto D, Tsuji M, Takubo T, S-sulfonation of transthyretin is an important trigger step in the formation of transthyretin-related amyloid fibril, Biochim. Biophys. Acta, 1804 (2010), 1449–1456. [DOI] [PubMed] [Google Scholar]
- [14].Pires RH, Karsai A, Saraiva MJ, Damas AM, Kellermayer MS, Distinct annular oligomers captured along the assembly and disassembly pathways of transthyretin amyloid protofibrils, PLoS One, 7 (2012), e44992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Faria TQ, Almeida ZL, Cruz PF, Jesus CS, Castanheira P, Brito RM, A look into amyloid formation by transthyretin: aggregation pathway and a novel kinetic model, Phys. Chem. Chem. Phys, 17 (2015), 7255–7263. [DOI] [PubMed] [Google Scholar]
- [16].Groenning M, Campos RI, Hirschberg D, Hammarstrom P, Vestergaard B, Considerably Unfolded Transthyretin Monomers Preceed and Exchange with Dynamically Structured Amyloid Protofibrils, Sci. Rep, 5 (2015), 11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lim KH, Dasari AK, Hung I, Gan Z, Kelly JW, Wright PE, Wemmer DE, Solid-State NMR Studies Reveal Native-like beta-Sheet Structures in Transthyretin Amyloid, Biochemistry, 55 (2016), 5272–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Szeverenyi NM, Sullivan MJ, Maciel GE, Observation of spin exchange by two-dimensional fourier transform 13C cross polarization-magic-angle spinning, J. Magn. Reson, 47 (1982), 462–475. [Google Scholar]
- [19].Raleigh DP, Levitt MH, Griffin RG, Rotational resonance in solid state NMR, Chem. Phys. Lett, 147 (1988), 71–76. [Google Scholar]
- [20].Seidel K, Lange A, Becker S, Hughes CE, Heise H, Baldus M, Protein solid-state NMR resonance assignments from (13C,13C) correlation spectroscopy, Phys. Chem. Chem. Phys, 6 (2004), 5090–5093. [Google Scholar]
- [21].Dobson CM, Protein folding and misfolding, Nature, 426 (2003), 884–890. [DOI] [PubMed] [Google Scholar]
- [22].Tycko R, Molecular structure of amyloid fibrils: insights from solid-state NMR, Q. Rev. Biophys, 39 (2006), 1–55. [DOI] [PubMed] [Google Scholar]
- [23].Sunde M, Blake CC, From the globular to the fibrous state: protein structure and structural conversion in amyloid formation, Q. Rev. Biophys, 31 (1998), 1–39. [DOI] [PubMed] [Google Scholar]
- [24].Kelly JW, The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways, Curr. Opin. Struct. Biol, 8 (1998), 101–106. [DOI] [PubMed] [Google Scholar]
- [25].Uversky VN, Fink AL, Conformational constraints for amyloid fibrillation: the importance of being unfolded, Biochim. Biophys. Acta, 1698 (2004), 131–153. [DOI] [PubMed] [Google Scholar]
- [26].Carulla N, Caddy GL, Hall DR, Zurdo J, Gairi M, Feliz M, Giralt E, Robinson CV, Dobson CM, Molecular recycling within amyloid fibrils, Nature, 436 (2005), 554–558. [DOI] [PubMed] [Google Scholar]
- [27].Miller Y, Ma B, Nussinov R, Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in a rugged energy landscape, Chem. Rev, 110 (2010), 4820–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Greenwald J, Riek R, Biology of amyloid: structure, function, and regulation, Structure, 18 (2010), 1244–60. [DOI] [PubMed] [Google Scholar]
- [29].Kodali R, Wetzel R, Polymorphism in the intermediates and products of amyloid assembly, Curr. Opin. Struct. Biol, 17 (2007), 48–57. [DOI] [PubMed] [Google Scholar]
- [30].Paravastu AK, Leapman RD, Yau WM, Tycko R, Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils, Proc. Natl. Aca. Sci. USA, 105 (2008), 18349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang L, Schubert D, Sawaya MR, Eisenberg D, Riek R, Multidimensional structure-activity relationship of a protein in its aggregated states, Angew. Chem. Int. Ed, 49 (2010), 3904–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tycko R, Amyloid polymorphism: structural basis and neurobiological relevance, Neuron, 86 (2015), 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pedersen JS, Andersen CB, Otzen DE, Amyloid structure--one but not the same: the many levels of fibrillar polymorphism, FEBS J, 277 (2010), 4591–4601. [DOI] [PubMed] [Google Scholar]
- [34].Lashuel HA, Wurth C, Woo L, Kelly JW, The most pathogenic transthyretin variant, L55P, forms amyloid fibrils under acidic conditions and protofilaments under physiological conditions, Biochemistry, 38 (1999), 13560–73. [DOI] [PubMed] [Google Scholar]
- [35].Hammarstrom P, Jiang X, Hurshman AR, Powers ET, Kelly JW, Sequence-dependent denaturation energetics: A major determinant in amyloid disease diversity, Proc. Natl. Aca. Sci. USA, 99 Suppl 4 (2002), 16427–16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lim KH, Dasari AKR, Ma R, Hung I, Gan Z, Kelly JW, Fitzgerald MC, Pathogenic Mutations Induce Partial Structural Changes in the Native beta-Sheet Structure of Transthyretin and Accelerate Aggregation, Biochemistry, 56 (2017), 4808–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yang M, Lei M, Huo S, Why is Leu55-->Pro55 transthyretin variant the most amyloidogenic: insights from molecular dynamics simulations of transthyretin monomers, Protein. Sci, 12 (2003), 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hornberg A, Olofsson A, Eneqvist T, Lundgren E, Sauer-Eriksson AE, The beta-strand D of transthyretin trapped in two discrete conformations, Biochim. Biophys. Acta, 1700 (2004), 93–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.