Abstract
Decellularized extracellular matrices (dECMs) have demonstrated excellent utility as bioscaffolds in recapitulating the complex biochemical microenvironment, however, their use as bioinks in 3D bioprinting to generate functional biomimetic tissues has been limited by their printability and lack of tunable physical properties. Here, we describe a method to produce photocrosslinkable tissue-specific dECM bioinks for fabricating patient-specific tissues with high control over complex microarchitecture and mechanical properties using a digital light processing (DLP)-based scanningless and continuous 3D bioprinter. We demonstrated that tissue-matched dECM bioinks provided a conducive environment for maintaining high viability and maturation of human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes and hepatocytes. Microscale patterning also guided spontaneous cellular reorganization into predesigned striated heart and lobular liver structures through biophysical cues. Our methodology enables a light-based approach to rapidly bioprint dECM bioinks with accurate tissue-scale design to engineer physiologically-relevant functional human tissues for applications in biology, regenerative medicine, and diagnostics.
Keywords: 3D bioprinting, decellularized extracellular matrix, tissue-specific bioinks, human induced-pluripotent stem cells, biomimetic tissues
Graphical Abstract
Introduction
With the convergence of tissue engineering and 3D bioprinting platforms, these technologies have driven significant advancements towards the creation of more physiologically relevant tissue and organ substitutes in regenerative medicine. In particular, 3D bioprinting systems offer a unique approach for the controlled assembly of cells and biomaterials as well as providing well-defined microstructural topographies and biomechanical properties to recapitulate key features of the native microenvironment[1–3]. Attaining this level of detail during microfabrication is critical since multiple cell populations in vivo are arranged into highly ordered geometries and are in constant communication with their surrounding matrix to form a tissue-specific function[4].
While the fabrication of various tissues has been demonstrated via 3D bioprinting[5–10], a major challenge is the limited selection of naturally-derived bioinks that mimic the distinct complex biochemical composition of individual tissues to provide the necessary cues to improve and maintain cell phenotype, viability, function, and maturation in vitro[11–13]. In the advent of decellularization technologies, this strategy offers a promising tool to be able to preserve the innate biochemical constituents and ultrastructure of the native extracellular matrix (ECM)[14–16]. Furthermore, decellularized extracellular matrix (dECM) has been widely demonstrated as bio-instructive scaffolds to direct and modulate cellular responses including proliferation and differentiation in addition to promoting in vivo tissue repair and regeneration[14,17]. It has also been recognized that cells cultured on dECM derived from their tissue of origin improved expansion and differentiation potential[18–21]. Though the exact mechanism is not clear, it is hypothesized that the unique compositional makeup of the ECM in different tissues provides a favorable environment for tissue-matched cell types, thus leading to the concept of applying a tissue-specific approach in tissue engineering[22,23]. Until recently, the application of dECM as novel bioinks has been adopted for extrusion-based 3D bioprinting systems. Pati et al. first demonstrated the successful application of dECM bioinks to construct cell-laden porous adipose, cartilage, and heart tissue analogues using a nozzle-based bioprinter[24]. In this case, the bioprinted tissues supported high viability and enhanced the tissue-specific gene expression of human adipose-derived stem cells (ADSCs), human turbinate mesenchymal stem cells (hTMSCs), and rat myoblast cells when cultured in their respective tissue-matched dECM bioinks compared to collagen I controls, which further highlights the advantages of tissuespecific ECM[24].
Although promising, dECM hydrogels on their own are inherently mechanically weak with poor structural fidelity. To circumvent these issues, polycaprolactone (PCL) frameworks printed in between dECM bioinks have been used to provide physical support when constructing 3D structures, however, PCL is not readily degradable or remodeled and limits the translation of these tissues for clinical applications[25]. The addition of different molecular weight polyethylene glycol (PEG)-based crosslinkers as well as thiolated gelatin and hyaluronic acid into solubilized dECM have also been used to improve the viscosity of the bioinks and enable better control over the mechanical properties of the final printed tissue[26]. For instance, a twostep crosslinking mechanism that relies on the initial spontaneous crosslinking of the thiol groups with PEG acrylate groups to form a soft extrudable hydrogel followed by UV photopolymerization of the thiol and PEG alkyne groups was employed to stabilize the construct[26]. Similarly, the incorporation of vitamin B2 photoinitiator into heart dECM bioinks has been used to provide tunable mechanical properties through a combination of covalent UVA photocrosslinking and subsequent thermal gelation[27]. Nonetheless while structural supports and crosslinkers are an improvement for dECM bioinks in extrusion bioprinting, the reported printing resolution remains no less than 100 μm and these approaches are relatively slow involving multiple steps which limits their scalability[11,28]. To this end, there is a critical need for new techniques to 3D bioprint dECM bioinks that enable the creation of tailorable robust microgeometries critical for recapitulating tissue-scale organization and resolution coupled with high-speed fabrication. Furthermore, applying bioprinted dECM to create tunable patientspecific tissues would also represent significant progress in the development of next-generation biomimetic human tissue platforms for future applications in drug screening and as models for biological studies.
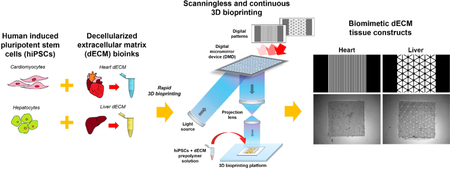
To address these challenges, we developed a novel direct method to produce highly tunable tissue-specific dECM-based constructs possessing biomimetic microarchitectures by using a custom digital light processing (DLP)-based scanningless and continuous 3D bioprinter (Fig. 1)[5]. Specifically, our light-based 3D bioprinting process enabled the simultaneous tuning of mechanical properties and the generation of high resolution complex microscale geometries in mere seconds. We applied this approach to create biomimetically patterned cell-laden 3D dECM heart and liver tissue constructs to guide cellular organization and provide a complex biochemical microenvironment for promoting the maturation of human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes (hiPSC-CMs) and hiPSC-hepatocytes (hiPSC-Heps), respectively. The work presented combines the DLP-based bioprinting of photocrosslinkable dECM bioinks with hiPSC-derived cells as a proof-of-concept for realizing the potential of engineering personalized human tissue platforms. In addition, the methods described can be readily adapted to incorporate dECM from other tissues for customizing existing photocrosslinkable hydrogels and serves as a foundation to broaden the library of available tissue-specific bioinks for the production of complex physiologically-relevant tissues and organs.
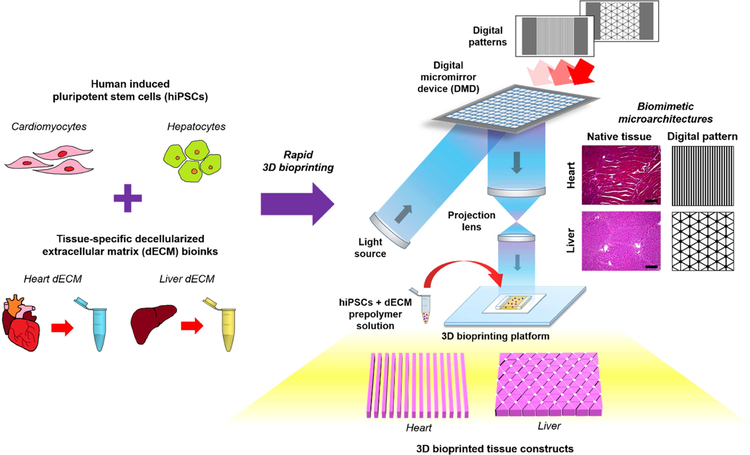
Figure 1: Overview of the rapid 3D bioprinting process to fabricate dECM tissue constructs with tissue-specific hiPSCs.
Human iPSCs were differentiated into hiPSC-CMs and hiPSC-Heps then subsequently combined with their respective tissue-specific photocrosslinkable dECM bioinks. Digital patterns recapitulating key histological features of each tissue were designed and inputted into a custom-built rapid DLP-based 3D bioprinter to create tissue constructs possessing microscale biomimetic microarchitecture in the order of seconds. This approach can be readily extended to create a wide range of tissue-specific photocrosslinkable dECM bioinks to build personalized human tissue platforms for broad applications in tissue engineering and regenerative medicine.
Materials and Methods
Tissue procurement
Heart and liver tissues were freshly harvested from three-month-old healthy Yorkshire pigs weighing approximately 40–45 kg supplied by S&S Farms (Ramona, CA), an approved animal vendor by the Institutional Animal Care and Use Committee (IACUC) at the University of California San Diego (UCSD). The pigs were euthanized by administering a lethal overdose of pentobarbital and the tissues were immediately harvested, transported to the lab on ice, and rinsed of residual blood and clots. To prepare the tissues for decellularization, only the left ventricle was collected from the heart and the hepatic vein and arteries were removed from the liver. All tissues were then minced into 0.5 cm3 pieces, portioned into tubs, and stored in 1X phosphate buffered saline (PBS, Cat. # 10010023, Gibco™) supplemented with 1% antibioticantimycotic (ABAM, Cat. # 15240062, Thermofisher Scientific) at −80°C until further use. All steps from tissue harvest to storage were performed within 2 h to ensure preservation of tissue quality and at least four of each tissue was pooled to minimize potential batch-to-batch variability.
Preparation of decellularized tissues
All decellularization steps were performed in a 37°C benchtop incubator shaker set at 120 rpm and all solutions were supplemented with 1% ABAM and 0.01 mM phenylmethylsulfonyl fluoride (PMSF, Cat. # P7626, Sigma-Aldrich), with the exception of trypsin-EDTA (Cat#: 25200056, Gibco™) and Sorensen’s Digest Buffer solution.
Decellularized heart left ventricle (HdECM) was performed as follows. Tissues were first frozen in hypotonic solution at −80°C then thawed and agitated in the incubator shaker for 2 h. Fresh hypotonic solution was replaced and the freeze-thaw process was repeated for a total of four cycles then rinsed with 1X PBS three times for 30 min each. The tissues were then digested in 0.25% trypsin-EDTA for 2 h and rinsed with 1X PBS three times for 30 min each. Next, tissues were incubated in 4% (w/v) sodium deoxycholate (SDC) (Cat. # D6750, Sigma-Aldrich) solution in 1X PBS for 24 h followed by three 30 min 1x PBS rinses and washed in 0.5% (w/v) sodium dodecyl sulfate (SDS) (Cat#: 436143, Sigma-Aldrich) solution in 1X PBS for 4 h. Tissues were rinsed again with 1X PBS three times 30 min each and digested with DNase (0.66 Units/mL, Cat#: D4513, Sigma-Aldrich) in Sorensen’s Digest Buffer solution for 24 h. Finally, the tissues were rinsed for 24 h with deionized water (DI) water and stored in 70% ethanol at 4°C. Decellularized liver (LdECM) was carried out by performing three rounds of freeze-thaw cycles involving freezing of the tissue in hypotonic solution, thawing, and then agitation for 2 h. Tissues were then rinsed three times with 1X PBS for 30 min each and washed in 1% (w/v) SDS solution for 48 h. Following this DI water was used to thoroughly rinse the tissues for 24 h prior to storing them in 70% ethanol at 4°C.
Histological and immunohistochemical analysis of decellularized tissues
Decellularized tissue samples were processed for histological analysis to assess the tissue architecture and removal of cellular content. Samples were fixed in 4% (w/v) paraformaldehyde (PFA) (Cat#: 161–20145, Wako) overnight and rinsed three times with 1X PBS prior to paraffin embedding. Next, sample blocks were sectioned into 5 μm thickness, stained with hematoxylin and eosin (H&E), and imaged with a Keyence BZ-9000 microscope with a multicolor CCD camera.
Immunohistochemical staining was performed to visualize the presence of extracellular matrix constituents post decellularization. Unfixed decellularized tissue samples were soaked in 30% sucrose in 1X PBS for 24 h, cryoembedded with Tissue-Tek ® O.C.T. compound (Sakura® FineTek USA Inc.), and sectioned using a cryostat at 10 μm thickness. To prepare the samples for immunostaining, the slides were brought to room temperature and incubated with blocking solution consisting of 10% (v/v) goat serum (Cat#: S-1000, Vector Laboratories) diluted in 1X PBS with 0.2% (v/v) Tween 20 (Cat. # TCI-T0543, Spectrum Chemicals) for 1 h at room temperature. Sections were then stained with the following antibodies purchased from Abcam diluted in blocking solution: monoclonal mouse anti-collagen I (1:2000, Cat. # ab90395), polyclonal rabbit anti-collagen IV (1:100, Cat. # ab6586), polyclonal rabbit anti-laminin (1:100, Cat. # ab11575), and monoclonal mouse anti-fibronectin (1:100, Cat #: ab6328). Incubation of anti-collagen I was performed for 1 h at room temperature. Anti-collagen IV was incubated at 37°C for 1 h. Anti-laminin and anti-fibronectin were incubated at 4°C overnight. Following primary antibody staining, samples were rinsed three times with 1X PBS and stained with the following secondary antibodies diluted in blocking solution for 1 h at room temperature: CF647 donkey anti-mouse IgG (H+L) (1:200, Cat. # 20046, Biotium) and CF647 donkey anti-rabbit IgG (H+L) (1:200, Cat. # 20047, Biotium). Samples were rinsed three times with 1X PBS and mounted with Fluorshield Mounting medium (Abcam) prior to imaging with a Leica DMI 6000B microscope. Native tissue controls and no primary controls were included to confirm the specificity of all antibodies.
Scanning electron microscopy
To assess the ultrastructure, decellularized tissues were prepared for scanning electron microscopy (SEM) imaging using established protocols[29]. Briefly, samples were fixed for 1 h at room temperature then overnight in 2.5% glutaraldehyde (Cat. # G5882, Sigma-Aldrich). Gradual dehydration was performed using ethanol followed by chemical drying in hexamethyldisilazane (Cat. # H4875, Sigma-Aldrich) overnight, and then sputtered coated with iridium for 7 s prior to imaging with a Zeiss Sigma 500 scanning electron microscope.
Quantification of dsDNA, glycosaminoglycan, and collagen content
Decellularized tissue samples as well as their respective native tissue controls were frozen and lyophilized for 48 h. For dsDNA and glycosaminoglycan (GAG) content measurement, 50 mg of dried powdered sample was placed in a 1.5 mL eppendorf tube and digested with 1 mL of papain solution (Cat. # P3125, Sigma-Aldrich) comprised of 0.1 mg/mL papain in 0.2 M sodium phosphate buffer solution (pH 6.4) at 65°C for 20 h with periodic vortexing. After digestion, the samples were centrifuged at 10,000 rpm for 10 min and the supernatant was collected. Residual dsDNA content was measured using the Quant-iT™ PicoGreen dsDNA (ThermoFisher Scientific) and the GAG content with the Blyscan™ Glycosaminoglycan Kit (Biocolor) following the manufacturer’s instructions. To quantify the collagen content, 10 mg of dried powdered sample was placed in a 1.5 mL polypropylene tube and homogenized with 100 μL of DI water for 30 s using a vortex. Next, 100 μL of 12 M HCl (Millipore Sigma) was added to each tube and hydrolyzed at 120°C for 3 h in a heat block. The samples were centrifuged at 10,000 rpm for 10 min and the supernatant was collected for collagen measurement using the Hydroxyproline Assay Kit (Sigma-Aldrich) according the manufacturer’s protocol.
Preparation of decellularized extracellular matrix (dECM) solutions
Lyophilized HdECM and LdECM as well as collagen I controls (Cat. # C9879, Sigma-Aldrich) were processed into solution by first mincing them finely with scissors and cryomilled into a fine powder to ensure uniform digestion. Briefly, the minced samples were transferred into a cryomilling chamber containing three 10 mm stainless steel milling balls. The chambers were precooled in liquid nitrogen for 3 min then placed into a Retsch™ MM400 mixer mill and milled at 30 Hz for 2 min. Next, 10 mg/mL of tissue powder was digested for 24 h at room temperature in pepsin digest solution consisting of 1 mg/mL pepsin (Cat. # P7012, Sigma-Aldrich) dissolved in 0.1 M HCl. The resulting solution was neutralized by dropwise addition of NaOH, frozen at 80°C, and lyophilized for 24 h. Finally, the lyophilized solutions were cryomilled again under the same conditions mentioned prior and stored at room temperature until further use.
Synthesis and preparation of GelMA prepolymer, LAP photoinitiator, and dECM bioinks
GelMA was synthesized using previously established protocols[5,9,30]. Briefly, a solution of 10% (w/v) porcine gelatin type A (Cat. # G2500, Sigma-Aldrich) in 10X PBS was stirred in a round bottom flask heated to 60°C until dissolved. Next, methacrylic anhydride (Cat. # 276685, Sigma-Aldrich) was added dropwise to a final concentration of 8% (v/v) then stirred for 3 h at 60°C and diluted with an equal volume of prewarmed 1X PBS. The resulting GelMA solution was dialyzed against distilled water using a dialysis tube (13.5 kDa, Cat. # 888–10990, Spectrum Laboratories) for 7 days at 45°C. Once dialyzed the GelMA solution was frozen and the lyophilized product was stored at −80°C until further use.
The photoinitiator, lithium phenyl-2,4,6 trimethylbenzoylphosphinate (LAP), was synthesized using previously reported protocols[31,32]. Under continuous stirring at room temperature under argon, 3.2 g (0.018 mol) of dimethyl phenylphosphonite (Cat. # 2946–61-4, Acros Organics) was added dropwise to an equimolar amount of 3 g (0.018 mol) of dimethyl phenylphosphinite (Cat. # 149470, Sigma-Aldrich) and allowed to react for 18 h. Afterwards, a 4-fold excess of 6.1 g lithium bromide (Cat. # 213225, Sigma-Aldrich) dissolved in 100 mL of 2butanone (Cat. # M209–4, ThermoFisher Scientific) was added to the mixture and then heated to 50°C. After 10 min a solid precipitate formed and cooled to room temperature overnight. The unreacted lithium bromide was removed from the precipitate via washing with 2-butanone followed by filtration for a total of three times. The resulting white LAP powder solid precipitate was ground with a mortar and pestle and stored at room temperature under argon until needed.
Preparation for each of the photocrosslinkable dECM bioink formulations were conducted as follows. Note that formulations were optimized and chosen for each specific tissue in this study to produce mechanically soft hydrogels suitable for cell encapsulation in the heart and liver tissue constructs. For the heart tissue constructs, stock solutions of 10% (w/v) GelMA + 0.5% (w/v) LAP and 100 mg/mL HdECM solution in 1X PBS were made separately and mixed in a 1:1 ratio immediately prior to bioprinting to yield a final solution containing 5% (w/v) GelMA + 5% (w/v) HdECM + 0.25% (w/v) LAP. Similarly, for the liver tissue constructs the dECM prepolymer solutions were prepared in the same way yielding a final solution concentration containing 5% (w/v) GelMA + 5% (w/v) LdECM + 0.25% (w/v) LAP. Collagen I controls for each formulation were prepared in the same manner by replacing the dECM solution with collagen I solution.
Characterization of swelling ratio
Printed acellular dECM constructs as well as their respective collagen I controls were characterized for long term stability by measuring the swelling ratio. For swelling ratio measurements, triplicate samples were immersed into Ringer’s buffer solution at 37°C and images of the hydrated cross-sectional area (Awet) were taken at 24 h, 72 h, 7 day, 21 day, and 28 day time points with a Leica DMI 6000-B microscope. A separate set of triplicate samples for each group was air dried overnight and used to measure the cross sectional dry area (Adry). The swelling ratio for each time point was calculated by normalizing the average hydrated cross-sectional area to the average dry cross-sectional area (Awet/Adry). The average cross-sectional area of each construct was measured using ImageJ software.
Mechanical testing
The compressive Young’s modulus of printed acellular dECM constructs and respective collagen I controls were measured using a CellScale MicroSquisher (Waterloo, Canada) microscale mechanical testing apparatus. Briefly, samples were equilibrated at 37°C overnight and prior to each measurement the samples were preconditioned for two cycles at 0.8 μm/s strain rate to remove the effects of hysteresis. For data collection, compression was performed at 10% strain with 30 s compression followed by 2 s hold and 10 s recovery. The Young’s modulus was calculated using a custom MATLAB code by measuring the slope of the force versus displacement curve.
Human iPSC-derived cardiomyocyte differentiation and culture
Human iPSCs (hiPSCs) were produced using previously established methods[5,33]. Cells sourced from human adult dermal fibroblasts (HDF, Cat. # 106K-05a, Cell Applications Inc.) and human perinatal foreskin fibroblasts (BJ, Cat. # CRL-2522, ATCC) were cultured in complete medium comprised of DMEM (Cat. # MT10013CV, Corning) supplemented with 10% fetal bovine serum (Cat. # 101, Tissue Culture Biologicals) and 1% ABAM (Cat. # 45000–616, Corning). Cells were incubated at 37°C with 5% CO2 and passaged every 3–5 days at a ratio of 1:6 using 0.25% trypsin-EDTA (Cat. # MT25053CI, Corning). Reprogramming was performed by plating the cells at 2 × 105 cells per 6-well plate and incubated for 48 h. Using the CytoTune™-iPS 2.0 Sendai Reprogramming Kit (Cat. # A16517, Life Technologies), the fibroblasts were reprogrammed via delivery of the four factors (i.e c-Myc, Sox2, Klf4, and Oct4) in accordance with the manufacturer’s protocol. After reprogramming, hiPSCs were transferred to Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix (Cat. # 356230, Corning) coated wells, cultured in xeno-free and feeder-free Essential 8™ medium (Cat. # A1517001, Life Technologies), and passaged every 3–4 days at a ratio of 1:8 using Versene (Cat. # 15040066, Life Technologies).
Cardiac differentiation was performed using the PSC Cardiomyocytes Differentiation Kit (Cat. # A25042SA, Life Technologies) and for all steps cells were cultured at 37°C with 5% CO2 under normoxia with medium exchange every two days (Supplementary Figure 1). Prior to differentiation, hiPSCs were dissociated by incubation with Accutase (Cat. # AT104, STEMCELL™ Technologies) and seeded into 12-well plates coated with hESC-qualified Matrigel substrate (Cat. # 354277, Corning) and allowed to reach 80–90% confluence after 3 days of culture. Differentiation was induced by incubating in cardiomyocyte differentiation medium A in the first 48 h and subsequent incubation in cardiomyocyte differentiation medium B for an additional 48 h. Next, the cells were cultured in cardiomyocyte maintenance medium for 8 days. Upon observing spontaneous contraction at 8–9 days of differentiation, at day 12 the cardiomyocytes were purified by incubating in RPMI1640 medium without glucose (Cat. # 11879020, Life Technologies) supplemented with 4 mM lactate (Cat. # 129–02666, Wako Pure Chemical) for an additional 6 days. Following purification, cells were cultured in HEPES buffered RPMI1640 medium (Cat. # 22400089, Life Technologies) supplemented with 2% (v/v) B27 (Cat. # 17504044, Thermo Fisher Scientific) for 7 to 10 days. To prepare the cells for bioprinting, hiPSC-cardiomyocytes (hiPSC-CMs) (Supplementary Figure 2a) were dissociated with 0.25% trypsin-EDTA for 5 min, pelleted at 300 xg for 3 min with the supernatant removed, and the cells were placed on ice.
Human iPSC-derived hepatocyte culture
Human iPSC-derived hepatocytes (hiPSC-Heps), iCell® Hepatocytes 2.0 (Cat. # PHC100–020-001, Cellular Dynamics International), were purchased and used according to the manufacturer’s instructions. In general, the cells were thawed and plated into a 6-well plate coated with collagen I and 2D-plating medium was exchanged daily for the first 4 days post plating, after which maintenance medium was used and exchanged every other day (Supplementary Figure 2b). At day 7 post plating, the cells were dissociated with Accutase for 3 min at 37°C, counted with a hemacytometer, and pelleted by centrifugation at 200 xg for 3 min. The supernatant was carefully removed and the cells were placed on ice for subsequent bioprinting.
3D bioprinting system and fabrication of dECM tissue constructs
An in-house rapid digital light processing (DLP)-based 3D bioprinting system (Fig. 1) was used to fabricate the tissue constructs in this study. The rapid 3D bioprinter consists of: i) a UV light source (365 nm, Hamamatsu); ii) a computer for sliced image-flow generation and system synchronization; iii) a digital micromirror device (DMD, Texas Instruments) for optical pattern generation; iv) a set of projection optics; and v) a stage for sample position control (Newport). In particular, the DMD chip (composed of approximately two million micro-mirrors) modulates the uniform UV light and projects an optical pattern dictated by user-defined images onto the photo-crosslinkable solution. The linear stage, controlled by custom computer software, allows for simultaneous and continuous control of the projected image and the focal plane. All digital patterns were designed in Adobe Photoshop and loaded as PNG files into the custom 3D bioprinting software. More specifically, the bioprinted heart and liver tissue construct dimensions measured 3 mm (L) x 3 mm (W) x 250 μm (H) as shown in Fig. 4b. For the heart tissue constructs a striated pattern consisting of parallel lines 60 μm in width separated by 60 μm spacing was designed. For the liver tissue constructs, a lobular pattern with small circular regions representing the portal triad and central vein was designed such that each hexagonal lobule was 1 mm in diameter and the sinusoidal regions were 35 μm in width.
Figure 4. Bioprinting of biomimetically patterned heart and liver dECM tissue constructs.
(a) Designed biomimetic digital patterns and (b) their respective acellular 3D bioprinted heart and liver tissue constructs. Scale bar, 1 mm. Microstructural features of the printed (c) acellular and (d) cellularized heart and liver tissue constructs. Scale bar, 200 μm. (e,f) Macroscopic images of the bioprinted dECM tissue constructs. (g,h) To evaluate the long term stability of acellular bioprinted dECM tissue constructs and their respective collagen I controls, samples were immersed in Ringer’s physiological solution at 37°C and the swelling ratio was measured over 28 days. All data are expressed as mean ± standard deviation. (n=3).
Setup of the bioprinting platform for fabricating the heart and liver tissue constructs consisted of two 250 μm thick polydimethylsiloxane (PDMS) spacers positioned between a methacrylated coverslip and a PDMS base fixed onto a glass slide. The prepolymer solution was then carefully pipetted between the PDMS spacers and the entire glass slide was placed onto the motion controller stage followed by light exposure to produce the biomimetic patterned construct. For the cellularized heart and liver tissue constructs, hiPSC-cardiomyocytes and hiPSC-hepatocytes were pelleted and resuspended in the dECM bioink or collagen I bioink prepolymer solutions at a final concentration of 50 million cells/mL and 30 million cells/mL, respectively, prior to loading into the bioprinting platform. Immediately after printing, the tissue constructs were placed in a 24-well plate and rinsed first with prewarmed 1X PBS followed by medium to remove residual prepolymer solution. Fresh medium was then added to each of the wells and the tissue constructs were incubated at 37°C and 5% CO2 for the entire culture period and medium was exchanged every other day. For the heart tissue constructs, RPMI640 medium with ROCK inhibitor and 20% (v/v) fetal bovine serum (Cat. # 10438026, Gibco) was used to culture for the first 48 h post printing after which RPMI1640 only medium was used for subsequent culture. Meanwhile, hepatocyte maintenance medium was used to culture the liver tissue constructs. All bioprinting steps were performed within 2 h of cell dissociation to ensure optimum viability is maintained.
Cell viability assessment
Cell viability was assessed in the bioprinted dECM tissue constructs and corresponding collagen I control at 24 h, 72 h, and 7 day time points. Live/Dead™ Viability/Cytotoxicity kit (Cat. # L3224, Thermo Fisher Scientific) was used to visualize cell viability in the tissue constructs by staining with 2 uM calcein AM and 4 uM ethidium homodimer-1 in 1X PBS for 30 min at 37°C. Fluorescent images were taken using the Leica DMI 6000-B microscope. CellTiter-Glo® 3D cell viability assay (Cat. # G9681, Promega) was used to quantify the cell viability across the culture period. Briefly, duplicate samples were placed in a 24-well plate and equal volumes of media and CellTiter-Glo® 3D Reagent was added. The samples were then shaken at 160 rpm for 1 h at room temperature to ensure effective extraction of ATP from the tissue constructs. Next, 200 μL of each sample was transferred into a white opaque-walled 96-well plate and the ATP content was calculated based on an ATP standard curve generated using ATP disodium salt (Cat. # P1132, Promega) according to the manufacturer’s protocol. Luminescence was measured with a Tecan Infinite® M200 PRO microplate reader.
Immunohistochemical staining of cell-specific markers
Immunohistochemical staining was used to visualize for the presence of cell-specific marker expression of the hiPSCs after 7 days of culture. Briefly, the bioprinted dECM tissue constructs and respective collagen I controls were rinsed with 1X PBS and fixed in 4% PFA for 15 min at room temperature, followed by rinsing three times with 1X PBS to remove residual PFA. The samples were then incubated in blocking solution, comprised of 2% (w/v) bovine serum albumin (BSA, Cat. #700–100P, Gemini Bio-Products) with 0.1% (v/v) Triton X-100 in 1X PBS, for 1 h at room temperature. Next, primary antibodies were diluted in blocking solution and incubated overnight at 4°C as follows: 1) heart tissue constructs: α-actinin (1:100, Cat. # ab9465, Abcam); and 2) liver tissue constructs: E-cadherin (E-cad, 1:100, Cat. # ab1416, Abcam), albumin (Alb, 1:100, Cat. # ab137885, Abcam). The samples were rinsed three times with 1X PBS and incubated with secondary antibodies diluted in blocking solution for 1 h at room temperature as follows: 1) heart tissue constructs: phalloidin (actin) CF633 conjugate (1:200, Cat. # 00046, Biotium), CF488A donkey anti-mouse IgG (H+L) (1:200, Cat. # 20014, Biotium; and 2) liver tissue constructs: CF488A donkey anti-mouse IgG (H+L) (1:200, Cat. # 20014, Biotium), CF647 donkey anti-rabbit IgG (H+L) (1:200, Cat. # 20047, Biotium). For all secondary solutions Hoescht 33258 pentahydrate (bis-benzimide) was added to stain for nuclei. After incubation, samples were rinsed three times in 1X PBS and immersed in 1X PBS supplemented with 0.05% (v/v) sodium azide (Cat. # 14314–09, Alfa Aesar) prior to taking fluorescent images with an Olympus FV1000 microscope.
Quantitative RT-PCR analysis
Quantitative reverse transcription-polymer chain reaction (RT-PCR) was performed at 7 days for all dECM tissue constructs and corresponding collagen I controls to assess the expression level of cell-specific genes for each tissue group. RNA was isolated from the samples by pooling at least 10 samples from each group by adding ice cold TRIzol reagent (Ambion Life Technologies) and vigorously pipetting to fully lyse the cells. For 2D hiPSC controls, TRIzol reagent was added to each well, incubated for 5 min, and pipetted multiple times. Total RNA was isolated and purified using the Direct-zol RNA MiniPrep Kit (Zymo Research) in accordance with the manufacturer’s instructions. RNA concentration was measured using the NanoQuant™ Plate and read with a Tecan Infinite® M200 PRO microplate reader. The extracted RNA was then stored at −80°C until further required.
cDNA synthesis was performed on 100 ng of input RNA from each sample group using the PhotoScript® first strand cDNA synthesis kit (Cat. # E6300S, New England Biolabs) following the manufacturer’s protocol. Real-time RT-PCR was conducted using the KAPA SYBR9® Fast Universal kit (Cat. # KK4602, KAPA Biosystems) and detected using a StepOne™ Real-Time PCR System (Thermo Fisher Scientific), along with human specificprimers (Integrated DNA Technologies) for the heart (i.e. NKX2.5, MLC-2v, TNNT,) and liver (i.e. AFP, TTR, ALB) (Supplementary Table 1). No-RT and no template controls were included on every plate. Data was analyzed using the comparative Ct method with normalization to the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and reported as relative to their respective 2D cultured controls without dECM or collagen I.
Statistics
All data are expressed as a mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism version 6.0 Software by one-way ANOVA with Tukey’s post-hoc comparison or two-tailed parametric paired t-test. Significant differences were considered when p<0.05.
Results
Characterization of dECM
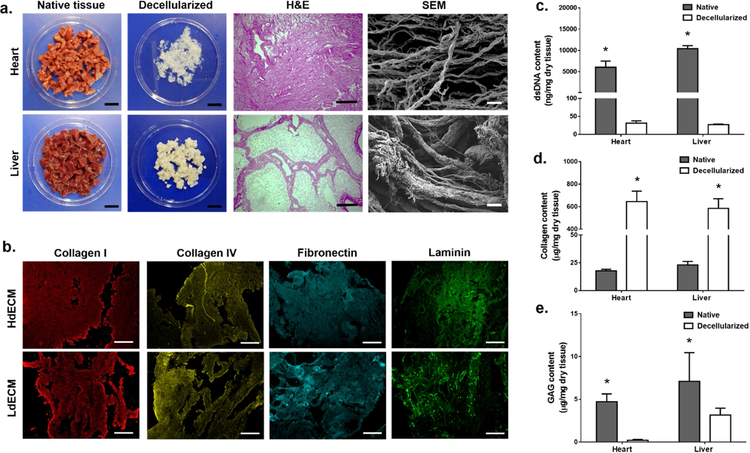
Tissue-specific dECM from porcine heart left ventricle (HdECM) and liver (LdECM) were prepared using optimized decellularization protocols to yield a white intact fibrous biomaterial (Fig. 2a). For each of the dECM tissues, the decellularization conditions sufficiently removed all cellular components while being gentle enough to preserve the native ultrastructure of the tissues as shown in the hematoxylin and eosin (H&E) stained sections (Fig. 2a). Namely, the distinct microarchitectures of each tissue were clearly visible showing the dense striated matrix of the heart as well as the lobular and sinusoidal regions of the liver (Fig. 2a). Upon closer examination under SEM, higher ordered architectures of collagen in all dECM tissues can be seen with networks of thin and thick fibrillar structures (Fig. 2a), which confirms minimal structural damage as collagen is the predominant component of the ECM[34].
Figure 2: Tissue decellularization process and characterization of ECM components.
(a) Porcine heart left ventricle and liver were decellularized using optimized protocols to yield a fibrous extracellular matrix. Scale bar, 2 cm. Histological images of hematoxylin and eosin (H&E) stained sections of the decellularized tissues showing the removal of cellular content and preservation of native microarchitecture. Scale bar, 200 μm. Scanning electron microscopy images of the ultrastructure confirming the presence of thick and thin collagen fibrils post decellularization in all tissues. Scale bar, 10 μm. (b) Representative immunohistochemical stained sections of HdECM and LdECM tissues for the presence of key extracellular matrix components pseudo colored: red (collagen I), yellow (collagen IV), cyan (fibronectin), green (laminin). Scale bar, 200 μm. (c) Residual dsDNA content quantification. (d) Collagen content quantification. (e) Glycosaminoglycan (GAG) content quantification. All data are expressed as mean ± standard deviation. * = significant within each tissue group (p< 0.05). (n=3).
The retention of key ECM constituents for each of the dECM tissues was qualitatively verified by immunohistochemistry. Positive staining for collagen I, collagen IV, fibronectin, and laminin was detected for HdECM and LdECM (Fig. 2b). Quantitative measurements for double-stranded DNA (dsDNA) content in the native and dECM tissues demonstrated a significant reduction in residual dsDNA for all dECMs to less than the 50 ng/mg dry tissue, which is the criteria for successful decellularization (Fig. 2c)[15]. The measured collagen content showed a significant enrichment of collagen in all dECM tissues relative to their native tissue controls due to the removal of cellular contents and purification of the ECM constituents post-decellularization (Fig. 2d). Although there was an observed significant reduction in glycosaminoglycan (GAG) content for HdECM and LdECM compared to their respective native tissue controls (Fig. 2e), this occurrence can be attributed to the detergent based decellularization treatments which has been shown to remove GAGs with prolonged washing[35]. In general, the decellularization protocols for each tissue were able to preserve the structural integrity of the native ECM as well as retention of major ECM components.
Photocrosslinkable dECM bioinks with tailorable microgeometry and mechanical properties
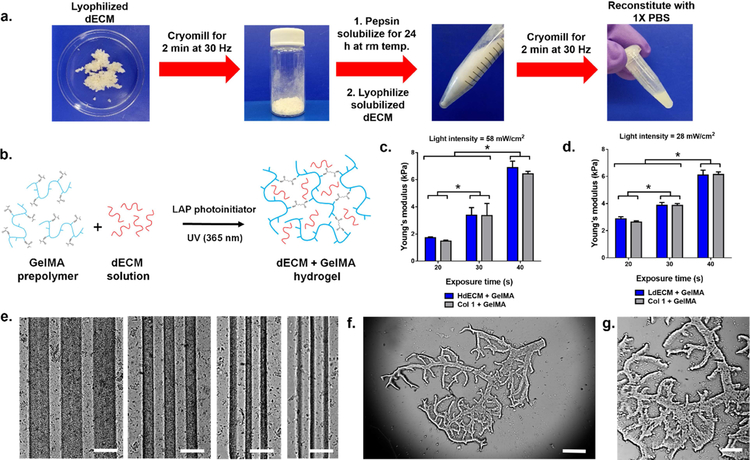
To process the tissue-specific dECMs into a photocrosslinkable bioink compatible with DLP-based 3D bioprinting, the dECMs were first prepared into a solubilized form via a combination of mechanical processing and enzymatic digestion (Fig. 3a). This was carried out by lyophilizing the dECM and cryomilling into a fine powder to ensure uniform digestion prior to treatment with pepsin. During this time, samples were digested at room temperature for 24 h to minimize the loss of collagen molecular integrity in the dECMs[34]. The resulting dECM solution was then lyophilized and cryomilled once more to yield a fine powder that could be readily reconstituted with 1X PBS to desired concentrations. Collagen I for each tissue group was prepared in the same fashion to serve as controls. This multistep process used to form the dECM into a homogenous solution was critical for physically reducing the dECM particle size to ensure high resolution of the final printed construct. To create mechanically soft 3D printed hydrogels for encapsulating cells in the heart and liver tissue constructs, HdECM or LdECM solutions were combined with photocrosslinkable gelatin methacrylate (GelMA) (Fig. 3b) at ratio of 1:1 to produce a bioink composed of 5% (w/v) of dECM and 5% (w/v) GelMA with the addition of 0.25% (w/v) lithium phenyl-2,4,6 trimethylbenzoylphosphinate (LAP) as the photoinitiator. All dECM bioink formulations remained liquid at room temperature as a homogeneous solution and were easily loaded into the printing chamber for subsequent 3D bioprinting.
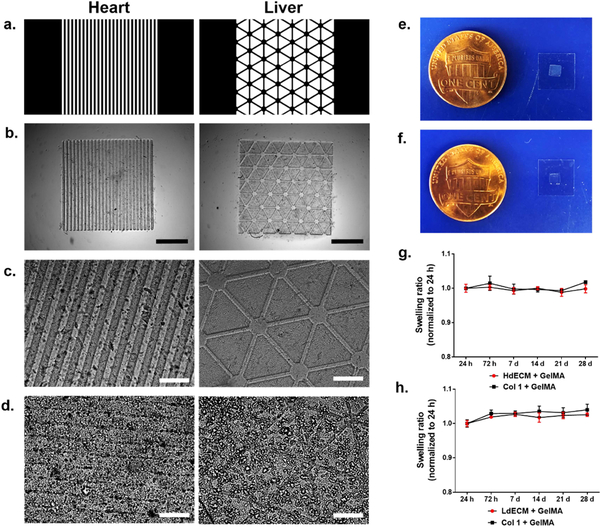
Figure 3: Preparation of photocrosslinkable dECM bioinks and characterization of printed dECM constructs.
(a) Lyophilized decellularized tissues were cryomilled and pepsin solubilized followed by a second lyophilization and cryomilling step to yield a fine dECM powder that can be reconstituted with 1X PBS. (b) Schematic of the crosslinking mechanism to form a printed dECM hydrogel construct for mechanically soft heart and liver tissue constructs through the incorporation of GelMA (c,d) Plots of the compressive Young’s modulus as a function of printing exposure time. All data are expressed as mean ± standard deviation. * = significant between exposure times (p< 0.05). (n=4). (e) Resolution of printed dECM bioinks ranging in 250, 125, 60, and 30 μm width lines (left to right). Scale bar, 250 μm. (f,g) Example of a printed hierarchal vascular network with detailed features of complex branched structures. Scale bar, 500 μm and 250 μm, respectively.
We next investigated the ability to modulate the mechanical properties of our dECM constructs since matching the matrix stiffness to that of native tissues is a crucial parameter for directing stem cell differentiation and behavior[36,37]. The effects of exposure time on the modulus of the printed dECM constructs was evaluated to demonstrate the range of mechanical properties achievable with each dECM bioink formulation while maintaining structural integrity (Fig. 3c,d). This was accomplished by precisely controlling the crosslinking density via free-radical polymerization of the GelMA component in our dECM bioinks through UV irradiation. In this case, the light intensity was optimized for each dECM bioink formulation to generate well-defined structures and was kept constant under each printing exposure time condition. In general, increasing the exposure time resulted in a successively higher compressive Young’s modulus for both dECM bioinks tested. For the HdECM and LdECM bioink formulations, mechanically soft constructs ranging in approximately 1.5–6.5 kPa and 2.5–6.0 kPa, respectively, were measured after exposure to UV light for 20, 30, and 40 s. No significant differences in modulus was measured between each of the dECM bioinks and their collagen I bioink controls for each printing condition tested. Furthermore, when comparing between each exposure time for both the HdECM and LdECM constructs the increase in mechanical stiffness were significantly different. These results validated that by using our DLP-based bioprinter, the compressive modulus of the resulting dECM hydrogel construct can be easily tuned by varying the exposure time variable without altering the bioink composition in each formulation. To demonstrate the resolution achieved with the dECM bioinks a series of parallel lines spaced 125 μm apart with widths of 250, 125, 60 and 30 μm were successfully printed (Fig. 3e). The dECM lines were well-defined with clear separation between adjacent lines and no visible deformities post printing. The versatility of the DLP-based 3D bioprinting system to fabricate complex 2.5D dECM-based biomimetic microarchitectures in a single print was also shown. In this case, hierarchal vascular networks (Fig. 3f,g) of varying widths ranging from 30 to 180 μm show the generation of intricate bifurcated geometries at physiologically relevant length scales.
Bioprinting of biomimetic dECM tissue constructs
Due to the high resolution and tunable modulus achieved by printing with the dECM bioink formulations, we assessed their application for the production of cellularized miniature biomimetic dECM tissue constructs. A set of two different biomimetic patterns capturing the key histological features of the native heart and liver were designed at tissue-length scales (Fig. 4a). In particular, equidistant parallel lines 60 μm in width and spacing was chosen for encouraging aligned myocardial tissue formation. Hexagonal lobular structures 1 mm in diameter separated by sinusoidal regions were designed to recapitulate the unit structure of the liver.
The bioprinting exposure times for each of the heart and liver dECM tissue constructs were chosen based on the minimum time required to attain high feature resolution and structural stability. In this case, the modulus for the heart tissues were 3.4 ± 0.9 kPa for the HdECM construct and 3.4 ± 0.6 kPa for the collagen I construct control. Similarly, the modulus of the liver tissues was 2.9 ± 0.1 kPa for the LdECM construct and 2.6 ± 0.1 kPa for the collagen construct control. In both cases, the soft mechanical properties were important to ensure that spreading of the encapsulated cells and remodeling could occur in the bioprinted tissue construct.
The printed miniature acellular dECM constructs (Fig. 4b,c) for each tissue type closely resembled their assigned digital patterns as shown in Fig. 4a with well-defined micron feature sizes matched to their intended dimensions. When hiPSC-CMs and hiPSC-Heps were encapsulated at high densities of 50 and 30 million cells/mL, respectively, the microarchitecture of the printed cellularized constructs after 24 h in culture could be visibly seen (Fig. 4d). Macroscopically, the printed dECM constructs maintained their shape and were easily handled by manipulation of the methacrylated coverslip (Fig. 4e,f). The equilibrium swelling ratio for each of the acellular printed dECM constructs was also measured over 28 days to evaluate the physical stability under prolonged incubation at 37°C (Fig. 4h–j). At all subsequent time points, the HdECM and LdECM constructs remained at an approximate swelling ratio of one relative to 24 h post printing with no significant changes in swelling over time and were structurally robust. Collagen I construct controls for each tissue also exhibited the same behavior and were not significantly different at each time point in relation to their respective dECM constructs.
dECM tissue constructs maintain high viability of hiPSC-derived cells and patterned microarchitectures guide cellular organization
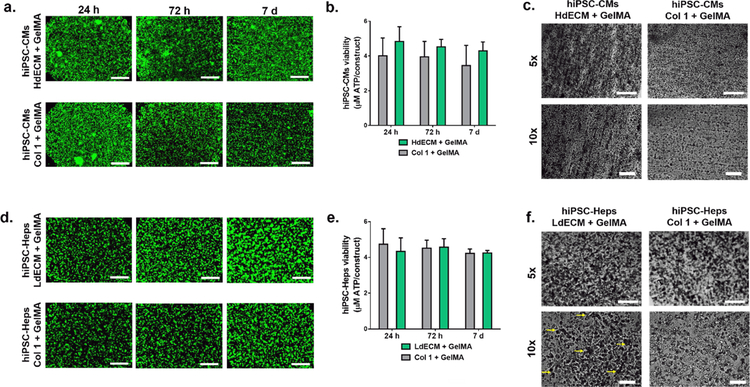
The viability of hiPSC-derived cells cultured in each of their tissue-specific dECM constructs was evaluated using Live/Dead™ staining and CellTiter-Glo ® 3D cell viability assay at time points of 24 h, 72 h, and 7 days (Fig. 5). In this case, the printed striated and lobular patterns were maintained across the entire culture period for both the HdECM and LdECM constructs as well as their respective collagen controls.
Figure 5: Viability of tissue-specific hiPSCs-derived cells cultured in the bioprinted dECM tissue constructs.
(a) Representative Live/Dead™ images of the heart dECM and collagen I tissue constructs. Scale bar, 500 μm. (n=3). (b) CellTiter-Glo® 3D cell viability assay was used to measure the metabolic activity of the heart dECM tissue constructs relative to collagen I controls. All data are expressed as mean ± standard deviation, (n=3). (c) Representative images of heart dECM tissue constructs at 7 days with striated patterns and collagen I controls. Scale bar, 500 μm (5x) and 100 μm (10x). (d) Representative Live/Dead™ images of the liver dECM and collagen I tissue constructs. Scale bar, 500 μm. (n=3). (e) CellTiter-Glo® 3D cell viability assay showing measured metabolic activity of the liver dECM tissue constructs relative to collagen I controls. All data are expressed as mean ± standard deviation, (n=3). (f) Liver dECM tissue constructs at 7 days showing preservation of printed lobular patterns relative to collagen I controls. Yellow arrows point to vertices of the hexagonal pattern. Scale bar, 500 μm (5x) and 100 μm (10x).
Interestingly, in the heart tissue constructs hiPSC-CMs parallel to the printed lines were observed after 24 h in the HdECM constructs and collagen, which gradually became more organized by 7 days. Live/Dead™ fluorescent images also showed high cell viability with minimal death in both types of constructs across all time points (Fig. 5a). This was consistent with the metabolic activity, as measured by the CellTiter-Glo® 3D cell viability assay, showing constant levels across the culture period for both the HdECM constructs and collagen I controls with no significant differences detected among the two groups and between each time point (Fig. 5b). However, after 7 days in culture more densely packed cells in areas parallel to the printed lines was observed in the HdECM constructs compared to a thinner network of aligned cells in the collagen controls (Fig. 5c). Furthermore, in the absence of external stimuli cardiomyocyte beating was recovered by 72 h in both constructs and synchronous contraction parallel to the printed lines was present in more regions of the HdECM construct (Supplementary Movie S1) at 7 days compared to the collagen I controls.
In the liver tissue constructs, Live/Dead™ staining also confirmed that high cellular viability was maintained for both the LdECM and collagen I controls over 7 days (Fig. 5d), as well as exhibiting similar levels of metabolic activity with no significant changes between time points (Fig. 5e). Qualitatively greater cellular aggregation was observed by 7 days for the hiPSC-Heps cultured in the LdECM constructs compared to the collagen I controls. Notably, high magnifications of the LdECM construct at 7 days revealed relatively larger hiPSC-Hep cluster formation throughout the scaffold with regions of cellular aggregates conforming to the printed hexagonal patterns as compared to the collagen I controls (Fig. 5f).
dECM tissue constructs provide a conducive environment to support the tissue-specific maturation of hiPSC-derived cells
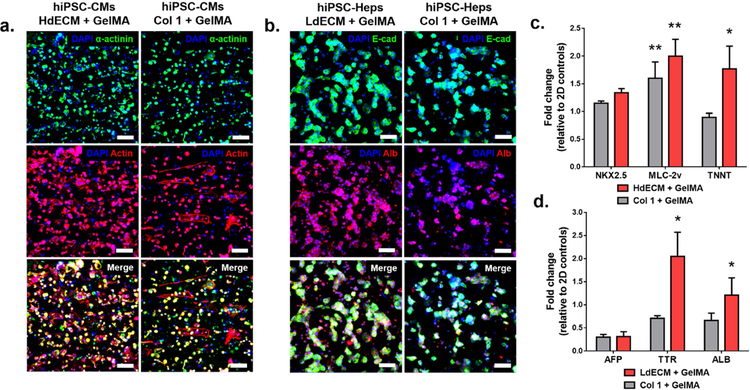
To determine the effects of dECM on promoting tissue-specific phenotype expression and cellular morphology of hiPSC-derived cells, immunostaining of key markers was used for qualitative assessment. Furthermore, we evaluated cell maturation using quantitative RT-PCR on the relative expression levels of early and late specific genes for each tissue type normalized to their individual 2D controls cultured without dECM or collagen I. Both assays were conducted on the dECM tissues and their respective collagen I controls after 7 days in culture for the heart and tissue constructs.
For the heart tissues, fluorescent images of hiPSC-CMs revealed more areas of positive staining for both α-actinin and actin in comparison to collagen I controls (Fig. 6a). Cellular localization along the direction of the patterned lines was also more visible in the HdECM constructs with greater co-staining of actin filaments and sarcomeric α-actinin markers. The hiPSC-CMs in the dECM construct within these parallel regions also revealed a denser cellular network. These qualitative findings were consistent with the measured gene expression showing an upregulation of the early transcription factor NK2 homeobox 5 (NKX2.5) relative to the 2D baseline controls (Fig. 6d), with higher expression in the HdECM constructs although not significant. In addition, compared to the 2D baseline control, both the HdECM and collagen I constructs exhibited a significant increase in expression for myosin regulatory light chain 2 (MLC-2v), which is involved in force generation for the movement of actin filaments. Furthermore, troponin T (TNNT), a mature cardiac marker for regulating muscle contraction, was significantly upregulated in the HdECM constructs compared to the collagen I constructs and 2D baseline controls.
Figure 6: Phenotype and gene expression profile of tissue-specific markers of hiPSCs cultured in the bioprinted dECM tissue constructs after 7 days.
Representative confocal images of immunohistochemical staining of the bioprinted (a) heart (α-actinin, actin) and (b) liver (E-cadherin (E-cad), albumin (ALB)) tissue constructs and their respective collagen I controls. All samples were counterstained with DAPI nucleic dye. Scale bar, 50 μm. (n=4). Gene expression of early and late tissue-specific markers for the bioprinted (c) heart and (d) liver tissue constructs relative to collagen I controls. All values were normalized to 2D controls and GAPDH served as the housekeeping gene. All data are expressed as mean ± standard deviation. * = significant to collagen I and 2D controls; ** = significant to 2D controls, (p< 0.05). (n=3).
For the liver tissues, visibly larger hiPSC-Hep aggregates and multicellular spheroids were present throughout the LdECM constructs compared to the collagen I controls in addition to more areas of positive staining of both E-cadherin (E-cad) and albumin (Alb) in the LdECM constructs (Fig. 6b). Gene expression analysis also revealed similar levels of alpha-fetoprotein (AFP) for both the LdECM and collagen I constructs showing a reduction in expression compared to the 2D controls, thus indicating a maturing state of the cells. Consistent with reduced AFP expression, the expression of transthyretin (TTR), which is a plasma protein synthesized and secreted by functional hepatocytes, was significantly upregulated in the dECM constructs relative to collagen I and 2D controls. Furthermore, concomitant to the observed strong albumin staining the expression of albumin (ALB) was also significantly higher in the LdECM constructs relative to collagen I and 2D controls.
Discussion
Over the years, dECM has become an attractive bioscaffold by providing complex structural and biochemical cues mimicking the native niche microenvironment to maintain cell phenotype and tissue-specific differentiation[14,15,17]. While solubilized dECM has enabled the formation of hydrogels[38], the translation of these materials into tissue-specific bioinks for 3D bioprinting applications is still in its infancy. A primary concern with dECM bioinks is their inherent low viscosity which consequently compromises the shape fidelity, resolution, and mechanical properties in extrusion-based 3D bioprinting systems[28]. Current efforts to develop techniques to circumvent these challenges have been mainly achieved through the use of synthetic polymer frameworks and post-print crosslinking[24,26,27]. However, a strategy that implements a one-step direct method to rapidly bioprint dECM bioinks into high resolution microgeometries that recapitulate detailed tissue-scale features with well-defined mechanical properties would greatly advance the development of next-generation functional dECM tissue constructs for tissue engineering and regenerative medicine.
In this work, we present a novel approach to fabricate microscale biomimetic tissue constructs using photocrosslinkable dECM bioinks with a digital light processing (DLP)-based scanningless and continuous 3D bioprinter. Compared to conventional extrusion-based bioprinters, our DLP-based bioprinting platform enables the fabrication of fine microarchitectures through the rapid projection of assigned digital patterns onto a photopolymerizable bioink. Moreover, smooth microscale features can be produced in seconds, relative to minutes or hours in raster-like bioprinting modalities, along with high flexibility in generating structures of complex designs[5,9,39]. Here, we aimed to evaluate the efficacy of our printing approach to produce biomimetic human heart and liver tissue constructs using hiPSCsderived cells with tissue-matched dECM bioinks and assess their viability as well as maturation.
Our method of decellularization produced collagen I rich heart and liver dECM with retained GAGs, cell adhesive matrix proteins including fibronectin and laminin, and basement membrane collagen IV that are critical in mediating cellular behavior[40,41]. Upon successful decellularization of each tissue, we processed the dECMs into a fine soluble powder using a combination of pepsin solubilization and a two-step cryomilling and lyophilization procedure to ensure dECM bioink homogeneity for high resolution printing. The powdered dECM can be stored as a dry off-the-shelf product that can be readily reconstituted with other bioink materials at desired concentrations. To impart photocrosslinking capability, we then combined the processed dECM with GelMA to produce bioink formulations for mechanically soft constructs. This simple method of processing the dECM can be easily adapted to create a wide variety of dECM bioinks derived from other tissue sources to incorporate native biochemical constituents for tissue-specific microenvironments.
The ability to fabricate complex shapes with high resolution as well as the modulation of mechanical properties is necessary for the generation of physiologically-relevant tissues. Using our bioprinting platform, we were capable of printing dECM-based structures as fine as 30 μm and producing complex hierarchal branched geometries in mere seconds, which represents a significant improvement in printing resolution (i.e. <100 μm) and orders of magnitude faster in fabrication speed compared to extrusion-based systems[11,28]. Moreover, the resulting vascular structure exhibited high resolution pattern fidelity with fine micron scale features that cannot be readily achieved with current methods using line-by-line or drop-by-drop bioprinting systems[11,28]. Together these features enable the scalability of our DLP-based 3D bioprinting process to reproducibly generate dECM-based constructs without compromising resolution and design complexity. Having control over mechanical properties during fabrication would also be advantageous since light penetration depth for post-print photocrosslinking would be limited in larger or thicker constructs. In our case, we showed that varying the light exposure time during printing for each of the heart dECM (HdECM) and liver dECM (LdECM) bioink formulations produced a range of compressive moduli. Furthermore, the observed mechanical properties in the soft LdECM and HdECM bioink formulations fell within the spectrum of normal liver[42] (i.e. <6 kPa) stiffness and the temporal stiffness values of the developing heart[43] (i.e. 2–9 kPa), respectively. Although we demonstrated that the modulus can be tuned through exposure time, other parameters including light intensity as well as bioink concentration and formulation can also be easily varied to control the mechanical properties of the final printed construct. These results showcase the flexibility of our rapid DLP-based 3D bioprinting platform to simultaneously produce smooth well-defined microarchitectures and tailorable mechanical properties during fabrication for any chosen design.
Current studies have revealed that engineered microscale topographical geometries play an important role in influencing cell function, migration, differentiation, and organization[44,45]. This phenomenon is mediated through contact guidance via the interaction of cells and biophysical signals from their surroundings[46]. In this context, we designed two individual patterns for the bioprinting of cellularized biomimetic heart and liver tissue constructs. These digital patterns were created to recapitulate the basic unit structure of each tissue, specifically the striated heart and lobular liver structure with tissue-scale feature sizes. The printed acellular tissue structures were robust with high pattern definition and exhibited long-term stability over 28 days with no observed changes in shape fidelity. For 3D bioprinted tissues, attaining physiologically-relevant cellular densities is a desired goal since appropriate cell-cell contact is necessary, particularly for maintaining the viability and function of cardiomyocyte synchronous beating and hepatocyte spheroid formation[47–49]. Here, we demonstrated that patterned microarchitectures were preserved post bioprinting with the incorporation of high cellular densities of 50 and 30 million cells/mL encapsulated in the heart and liver tissue constructs, respectively. In addition, the high cell viability observed in both the HdECM and LdECM tissue constructs confirmed that the tissue-specific dECM bioinks provided a favorable environment to support tissue-matched hiPSC-derived culture. Interestingly, denser hiPSC-CM alignment along the printed parallel lines was observed over time in the HdECM along with greater regions of synchronous beating by 7 days compared to collagen I controls. Similarly, larger hiPSC-Hep aggregation was visible in the LdECM constructs with more regions conforming to the patterned lobular structure in the LdECM constructs at 7 days relative to collagen I constructs. These findings suggest that hiPSC-derived cells cultured in their native dECM provided a stimulating environment to promote a maturing state while the printed patterns served as guide during tissue development. In particular, cells naturally sense and respond to their biophysical surroundings and this observation can be attributed to microstructural guidance effects on cellular arrangement and function[50,51]. As such, our bioprinting approach opens the possibility of designing highordered dECM-based microstructures at multiple length scales that reflect the detailed geometries of various tissues to provide physically instructive cues for promoting tissue-specific morphology.
Since the recent discovery of iPSCs, their use as a model for studying development and biological mechanisms has grown due to their unlimited self-renewal and pluripotent differentiation capability[52]. In particular, iPSC is an attractive cell source due to their patient-specificity and avoidance of potential ethical issues[52,53]. It has also been recognized that providing a conducive in vitro 3D microenvironment to improve the differentiation and maturation of hiPSC-derived cells is critical since they are generally immature relative to primary cells[54,55]. For the bioprinted heart constructs, we found that hiPSC-CMs in the HdECM constructs exhibited improved expression of key cardiac-specific markers as compared to the collagen I controls, owing to the more positive co-staining of α-actinin and actin as well as more visible elongation of the cells. Furthermore, this was coupled by the upregulation of the early cardiac transcription factor NK2 homeobox 5 (Nkx2.5) as well as a significantly higher expression of both the mature markers myosin regulatory light chain 2 (MLC-2v) and troponin T (TNNT) in the HdECM constructs relative to the collagen I and 2D controls. In the liver tissue constructs, a similar phenomenon was also revealed in which hiPSC-Heps cultured in the LdECM constructs had visibly larger spheroids, which corresponds to improved hepatocyte function, as well as more positive staining for both albumin and E-cadherin. The improved maturation state of these hiPSC-Heps was further confirmed by the decrease in alpha fetal protein (AFP) expression relative to 2D controls and significantly increased expression of both late markers, transthyretin (TTR) and albumin (ALB), compared to the collagen I constructs. Together these findings suggest that the complex constituents of the ECM incorporated into the bioinks served as a favorable environment to promote the maturation of tissue-matched hiPSC-derived cells in vitro. This is in agreement with other studies demonstrating that single component ECM substrates such as collagen or 2D cultures with limited biochemical diversity lack the necessary conditions to promote hiPSC-derived cell maturation[56,57]. Overall, these results provide a promising basis for future work to further investigate the functionality of hiPSC-derived cells within these dECM constructs to generate a robust and mature cell population for drug testing and disease modeling applications.
By interfacing dECM bioinks with DLP-based 3D bioprinting, we presented a novel approach for the rapid construction of biomimetic human tissues possessing tissue-specific biochemical constituents, microscale microarchitecture, and tailorable modulus. Moreover, we demonstrated that our photocrosslinkable dECM bioinks supported high cell viability and improved the maturation of tissue-matched hiPSC-derived cardiomyocytes and hepatocytes in a tissue-specific manner compared to collagen I controls. The methods described in this work can be adapted to develop a wide range of tissue-specific dECM bioinks for the rational design of biomimetic tissues having tissue-like cellular density with high resolution topographies to guide cellular organization and well-controlled mechanical properties. Together, these dECM-based liver and heart tissue constructs can open the door for future studies on the long-term stability and functional maturation of hiPSC-derived cells in complex biomimetic tissue systems. In future directions, our scanningless and continuous 3D bioprinting platform can also be used to incorporate multiple cells types to create heterogeneous dECM-based tissue constructs as well as the possibility to rapidly generate miniature tissue microarrays for high throughput operations. We envision that our bioprinted dECM-based tissues can serve as potential physiologically-relevant living human tissue platforms and offer a new venue for researchers to study biological disease mechanisms, develop personalized medicine, as well as for diagnostic drug screening applications.
Supplementary Material
Acknowledgements
The authors would like to thank the staff at Moores Cancer Center Histology Core for performing the H&E staining of the samples, and Patricia Pizarro at the Center for the Future of Surgery for providing the porcine liver tissues used in this study. This work was supported by National Institutes of Health (R21HD090662, R01EB021857, NS047101) and National Science Foundation (CMMI-1547005 and CMMI-1644967). Scholarship funding for Dr. Claire Yu was provided by the Natural Sciences and Engineering Research Council (NSERC) Postdoctoral Fellowship Scholarship of Canada.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests
The authors declare no competing financial interests.
Data Availability
The raw/processed data required to reproduce these findings can be shared by the authors upon request.
References
- [1].V Murphy S, Atala A, 3D bioprinting of tissues and organs, Nat. Biotechnol 32 (2014) 773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- [2].Zhu W, Ma X, Gou M, Mei D, Zhang K, Chen S, 3D printing of functional biomaterials for tissue engineering, Curr. Opin. Biotechnol 40 (2016) 103–112. doi: 10.1016/J.COPBIO.2016.03.014. [DOI] [PubMed] [Google Scholar]
- [3].Lee JM, Yeong WY, Design and Printing Strategies in 3D Bioprinting of CellHydrogels: A Review, Adv. Healthc. Mater 5 (2016) 2856–2865. doi: 10.1002/adhm.201600435. [DOI] [PubMed] [Google Scholar]
- [4].Rosso F, Giordano A, Barbarisi M, Barbarisi A, From Cell-ECM interactions to tissue engineering, J. Cell. Physiol 199 (2004) 174–180. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]
- [5].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc. Natl. Acad. Sci 113 (2016) 2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA, Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips, Sci. Rep 6 (2016) 34845. doi: 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park S-J, Kotikian A, Nesmith AP, Campbell PH, Vlassak JJ, Lewis JA, Parker KK, Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing., Nat. Mater 16 (2017) 303–308. doi: 10.1038/nmat4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Laronda MM, Rutz AL, Xiao S, Whelan KA, Duncan FE, Roth EW, Woodruff TK, Shah RN, A bioprosthetic ovary created using 3D printed microporous scaffolds restores ovarian function in sterilized mice, Nat. Commun 8 (2017) 15261. doi: 10.1038/ncomms15261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Zhang K, Chen S, Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture, Biomaterials. 124 (2017) 106–115. doi: 10.1016/J.BIOMATERIALS.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, A 3D bioprinting system to produce human-scale tissue constructs with structural integrity, Nat. Biotechnol 34 (2016) 312–319. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- [11].Garreta E, Oria R, Tarantino C, Pla-Roca M, Prado P, Fernández-Avilés F, Campistol JM, Samitier J, Montserrat N, Tissue engineering by decellularization and 3D bioprinting, Mater. Today. 20 (2017) 166–178. doi: 10.1016/J.MATTOD.2016.12.005. [DOI] [Google Scholar]
- [12].Kyle S, Jessop ZM, Tarassoli SP, Al-Sabah A, Whitaker IS, Assessing printability of bioinks, in: 3D Bioprinting Reconstr. Surg, Elsevier, 2018: pp. 173–189. doi: 10.1016/B978-0-08-101103-4.00027-2. [DOI] [Google Scholar]
- [13].Chimene D, Lennox KK, Kaunas RR, Gaharwar AK, Advanced Bioinks for 3D Printing: A Materials Science Perspective, Ann. Biomed. Eng 44 (2016) 2090–2102. doi: 10.1007/s10439-016-1638-y. [DOI] [PubMed] [Google Scholar]
- [14].Badylak SF, The extracellular matrix as a biologic scaffold material, Biomaterials. 28 (2007) 3587–3593. doi: 10.1016/J.BIOMATERIALS.2007.04.043. [DOI] [PubMed] [Google Scholar]
- [15].Crapo PM, Gilbert TW, Badylak SF, An overview of tissue and whole organ decellularization processes, Biomaterials. 32 (2011) 3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nakayama KH, Batchelder CA, Lee CI, Tarantal AF, Decellularized rhesus monkey kidney as a three-dimensional scaffold for renal tissue engineering., Tissue Eng. Part A. 16 (2010) 2207–16. doi: 10.1089/ten.TEA.2009.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Badylak SF, Freytes DO, Gilbert TW, Extracellular matrix as a biological scaffold material: Structure and function, Acta Biomater. 5 (2009) 1–13. doi: 10.1016/J.ACTBIO.2008.09.013. [DOI] [PubMed] [Google Scholar]
- [18].Zhang Y, He Y, Bharadwaj S, Hammam N, Carnagey K, Myers R, Atala A, Van Dyke M, Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype, Biomaterials. 30 (2009) 4021–4028. doi: 10.1016/j.biomaterials.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kornmuller A, Brown CFC, Yu C, Flynn LE, Fabrication of Extracellular Matrixderived Foams and Microcarriers as Tissue-specific Cell Culture and Delivery Platforms., J. Vis. Exp (2017) 55436. doi: 10.3791/55436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yu C, Kornmuller A, Brown C, Hoare T, Flynn LE, Decellularized adipose tissue microcarriers as a dynamic culture platform for human adipose-derived stem/stromal cell expansion, Biomaterials. 120 (2017) 66–80. doi: 10.1016/J.BIOMATERIALS.2016.12.017. [DOI] [PubMed] [Google Scholar]
- [21].Yu C, Bianco J, Brown C, Fuetterer L, Watkins JF, Samani A, Flynn LE, Porous decellularized adipose tissue foams for soft tissue regeneration, Biomaterials. 34 (2013) 3290–3302. doi: 10.1016/J.BIOMATERIALS.2013.01.056. [DOI] [PubMed] [Google Scholar]
- [22].Chan BP, Leong KW, Scaffolding in tissue engineering: general approaches and tissuespecific considerations., Eur. Spine J 17 Suppl 4 (2008) 467–79. doi: 10.1007/s00586008-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sachs PC, Mollica PA, Bruno RD, Tissue specific microenvironments: a key tool for tissue engineering and regenerative medicine., J. Biol. Eng 11 (2017) 34. doi: 10.1186/s13036-017-0077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pati F, Jang J, Ha D-H, Won Kim S, Rhie J-W, Shim J-H, Kim D-H, Cho D-W, Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink, Nat. Commun 5 (2014) 3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mondal D, Griffith M, Venkatraman SS, Polycaprolactone-based biomaterials for tissue engineering and drug delivery: Current scenario and challenges, Int. J. Polym. Mater. Polym. Biomater 65 (2016) 255–265. doi: 10.1080/00914037.2015.1103241. [DOI] [Google Scholar]
- [26].Skardal A, Devarasetty M, Kang H-W, Mead I, Bishop C, Shupe T, Lee SJ, Jackson J, Yoo J, Soker S, Atala A, A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs, Acta Biomater. 25 (2015) 24–34. doi: 10.1016/J.ACTBIO.2015.07.030. [DOI] [PubMed] [Google Scholar]
- [27].Jang J, Kim TG, Kim BS, Kim S-W, Kwon S-M, Cho D-W, Tailoring mechanical properties of decellularized extracellular matrix bioink by vitamin B2-induced photocrosslinking, Acta Biomater. 33 (2016) 88–95. doi: 10.1016/j.actbio.2016.01.013. [DOI] [PubMed] [Google Scholar]
- [28].Panwar A, Tan LP, Current status of bioinks for micro-extrusion-based 3D bioprinting, Molecules. 21 (2016) 685. doi: 10.3390/molecules21060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Flynn LE, The use of decellularized adipose tissue to provide an inductive microenvironment for the adipogenic differentiation of human adipose-derived stem cells., Biomaterials. 31 (2010) 4715–24. doi: 10.1016/j.biomaterials.2010.02.046. [DOI] [PubMed] [Google Scholar]
- [30].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Cellladen microengineered gelatin methacrylate hydrogels, Biomaterials. 31 (2010) 5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhu W, Li J, Leong YJ, Rozen I, Qu X, Dong R, Wu Z, Gao W, Chung PH, Wang J, Chen S, 3D-Printed Artificial Microfish, Adv. Mater 27 (2015) 4411–4417. doi: 10.1002/adma.201501372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS, Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility, Biomaterials. 30 (2009) 6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hribar KC, Finlay D, Ma X, Qu X, Ondeck MG, Chung PH, Zanella F, Engler AJ, Sheikh F, Vuori K, Chen SC, Nonlinear 3D projection printing of concave hydrogel microstructures for long-term multicellular spheroid and embryoid body culture., Lab Chip. 15 (2015) 2412–8. doi: 10.1039/c5lc00159e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lin YK, Liu DC, Effects of pepsin digestion at different temperatures and times on properties of telopeptide-poor collagen from bird feet, Food Chem. 94 (2006) 621–625. doi: 10.1016/J.FOODCHEM.2004.12.007. [DOI] [Google Scholar]
- [35].Farrokhi A, Pakyari M, Nabai L, Pourghadiri A, Hartwell R, Jalili RB, Ghahary A, Evaluation of detergent-free and detergent-based methods for decellularization of murine skin, Tissue Eng. Part A. 24 (2018) 955–967. doi: 10.1089/ten.TEA.2017.0273. [DOI] [PubMed] [Google Scholar]
- [36].Ghasemi-Mobarakeh L, Prabhakaran MP, Tian L, Shamirzaei-Jeshvaghani E, Dehghani L, Ramakrishna S, Structural properties of scaffolds: Crucial parameters towards stem cells differentiation., World J. Stem Cells. 7 (2015) 728–44. doi: 10.4252/wjsc.v7.i4.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li D, Zhou J, Chowdhury F, Cheng J, Wang N, Wang F, Role of mechanical factors in fate decisions of stem cells., Regen. Med 6 (2011) 229–40. doi: 10.2217/rme.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Saldin LT, Cramer MC, Velankar SS, White LJ, Badylak SF, Extracellular matrix hydrogels from decellularized tissues: Structure and function, Acta Biomater. 49 (2017) 1–15. doi: 10.1016/J.ACTBIO.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang AP, Qu X, Soman P, Hribar KC, Lee JW, Chen S, He S, Rapid fabrication of complex 3D extracellular microenvironments by dynamic optical projection stereolithography., Adv. Mater 24 (2012) 4266–70. doi: 10.1002/adma.201202024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yurchenco PD, Basement membranes: Cell scaffoldings and signaling platforms, Cold Spring Harb. Perspect. Biol 3 (2011) 1–27. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hynes RO, Integrins: Versatility, modulation, and signaling in cell adhesion, Cell. 69 (1992) 11–25. doi: 10.1016/0092-8674(92)90115-S. [DOI] [PubMed] [Google Scholar]
- [42].Mueller S, Sandrin L, Liver stiffness: a novel parameter for the diagnosis of liver disease., Hepat. Med 2 (2010) 49–67. http://www.ncbi.nlm.nih.gov/pubmed/24367208 (accessed April 25, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Young JL, Engler AJ, Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro, Biomaterials. 32 (2011) 1002–1009. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A, Engineering microscale topographies to control the cell-substrate interface., Biomaterials. 33 (2012) 5230–46. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE, Geometric control of cell life and death., Science. 276 (1997) 1425–8. http://www.ncbi.nlm.nih.gov/pubmed/9162012 (accessed April 25, 2018). [DOI] [PubMed] [Google Scholar]
- [46].Rørth P, Whence Directionality: Guidance Mechanisms in Solitary and Collective Cell Migration, Dev. Cell 20 (2011) 9–18. doi: 10.1016/j.devcel.2010.12.014. [DOI] [PubMed] [Google Scholar]
- [47].Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, Vitale L, Pelleri MC, Tassani S, Piva F, Perez-Amodio S, Strippoli P, Canaider S, An estimation of the number of cells in the human body, Ann. Hum. Biol 40 (2013) 463–471. doi: 10.3109/03014460.2013.807878. [DOI] [PubMed] [Google Scholar]
- [48].Radisic M, Euloth M, Yang L, Langer R, Freed LE, Vunjak-Novakovic G, Highdensity seeding of myocyte cells for cardiac tissue engineering, Biotechnol. Bioeng 82 (2003) 403–414. doi: 10.1002/bit.10594. [DOI] [PubMed] [Google Scholar]
- [49].Dvir-Ginzberg M, Gamlieli-Bonshtein I, Agbaria R, Cohen S, Liver Tissue Engineering within Alginate Scaffolds: Effects of Cell-Seeding Density on Hepatocyte Viability, Morphology, and Function, Tissue Eng. 9 (2003) 757–766. doi: 10.1089/107632703768247430. [DOI] [PubMed] [Google Scholar]
- [50].Walters BD, Stegemann JP, Strategies for directing the structure and function of threedimensional collagen biomaterials across length scales, Acta Biomater. 10 (2014) 1488–1501. doi: 10.1016/J.ACTBIO.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Harley BAC, Kim H-D, Zaman MH, Yannas IV, Lauffenburger DA, Gibson LJ, Microarchitecture of Three-Dimensional Scaffolds Influences Cell Migration Behavior via Junction Interactions, Biophys. J 95 (2008) 4013–4024. doi: 10.1529/BIOPHYSJ.107.122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Medvedev SP, Shevchenko AI, Zakian SM, Induced Pluripotent Stem Cells: Problems and Advantages when Applying them in Regenerative Medicine., Acta Naturae. 2 (2010) 18–28. http://www.ncbi.nlm.nih.gov/pubmed/22649638 (accessed April 27, 2018). [PMC free article] [PubMed] [Google Scholar]
- [53].Volarevic V, Markovic BS, Gazdic M, Volarevic A, Jovicic N, Arsenijevic N, Armstrong L, Djonov V, Lako M, Stojkovic M, Ethical and Safety Issues of Stem CellBased Therapy., Int. J. Med. Sci 15 (2018) 36–45. doi: 10.7150/ijms.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yang X, Pabon L, Murry CE, Engineering Adolescence: Maturation of Human Pluripotent Stem Cell-Derived Cardiomyocytes, Circ. Res 114 (2014) 511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Meseguer-Ripolles J, Khetani SR, Blanco JG, Iredale M, Hay DC, Pluripotent Stem Cell-Derived Human Tissue: Platforms to Evaluate Drug Metabolism and Safety., AAPS J. 20 (2017) 20. doi: 10.1208/s12248-017-0171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang B, Jakus AE, Baptista PM, Soker S, Soto-Gutierrez A, Abecassis MM, Shah RN, Wertheim JA, Functional Maturation of Induced Pluripotent Stem Cell Hepatocytes in Extracellular Matrix-A Comparative Analysis of Bioartificial Liver Microenvironments., Stem Cells Transl. Med 5 (2016) 1257–67. doi: 10.5966/sctm.20150235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fong AH, Romero-López M, Heylman CM, Keating M, Tran D, Sobrino A, Tran AQ, Pham HH, Fimbres C, Gershon PD, Botvinick EL, George SC, Hughes CCW, Three-Dimensional Adult Cardiac Extracellular Matrix Promotes Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes, Tissue Eng. Part A. 22 (2016) 1016–1025. doi: 10.1089/ten.tea.2016.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.