Abstract
Background:
The response to first-line, platinum-based treatment of muscle-invasive bladder cancer has not improved in 3 decades.
Objective:
To identify genes that influence cisplatin resistance in bladder cancer.
Design, setting, and participants:
We performed a whole-genome CRISPR screen in a bladder cancer cell line to identify genes that mediate resistance to cisplatin.
Outcome measurements and statistical analysis:
Targeted validation was performed in two bladder cancer cell lines. The top gene candidate was validated in a publicly available bladder cancer dataset.
Results and limitations:
From the CRISPR screen, we identified MSH2 as the most significantly enriched gene and mismatch repair as the most significantly enriched pathway that promoted resistance to cisplatin. Bladder cancer cells with knockdown of MSH2 showed a reduction in cisplatin-mediated apoptosis. MSH2 loss did not impact the sensitivity to other chemotherapies, including the cisplatin analog oxaliplatin. Bladder tumors with low MSH2 protein levels, quantified using reverse-phase protein array, showed poorer survival when treated with cisplatin- or carboplatin-based therapy; these results require future validation using immunohistochemistry. Additionally, results are retrospective from patients with primarily high-grade tumors; thus, validation in a controlled clinical trial is needed.
Conclusions:
We generated in vitro evidence that bladder cancer cell lines depleted of MSH2 are more resistant to cisplatin. We additionally found an association between low MSH2 in bladder tumors and poorer patient survival when treated with platinum-based chemotherapy. If successfully validated prospectively, MSH2 protein level could assist in the selection of patients for chemotherapy.
Patient summary:
We report the first evidence that MSH2 protein level may contribute to chemotherapy resistance observed in muscle-invasive bladder cancer. MSH2 has potential as a biomarker predictive of response to platinum-based therapy.
Keywords: MSH2, Mismatch repair, CRISPR screen, Bladder cancer, Cisplatin, Patient stratification, Chemotherapy
1. Introduction
Bladder cancer is the ninth most common cancer type, with an estimated 430 000 new cases and 165 000 deaths worldwide in 2012 [1]. In addition to radical cystectomy, patients with muscle-invasive bladder cancer (MIBC) are typically treated with one of two chemotherapy regimens, methotrexate, vinblastine, doxorubicin, and cisplatin (MVAC) or gemcitabine and cisplatin (GC). The response rate to these cisplatin-based chemotherapies remains 50% in MIBC [2]. Identifying the molecular mechanisms that determine patient response to these treatments has a direct clinical impact, including as a biomarker to stratify patients according to response and potentially as an avenue to explore novel therapeutic targets.
Cancer cells that lack components of the mismatch repair (MMR) pathway, such as MLH1 and MSH2, are resistant to cisplatin in vitro [3,4]. Ovarian tumors with low MLH1 expression associate with poorer survival in platinum-treated patients [5–7]. Similarly, a study of 17 ovarian cancer patients treated with cisplatin-based chemotherapy found that patients with a poor response had significantly lower MSH2 protein levels [8]. Interestingly, low MMR protein expression does not always correlate with resistance to platinum. In contrast to ovarian cancer, non-small cell lung cancer (NSCLC) patients with low MSH2 had improved survival when treated with platinum [9,10]. While the MMR pathway has not been associated with platinum resistance in bladder cancer, a subset of bladder cancers have reduced or absent protein expression of MSH2 as determined by immunohistochemistry (IHC) [11–15].
Here, we take an unbiased approach to investigate mediators of cisplatin resistance by performing, to the best of our knowledge, the first genome-wide CRISPR-Cas9 cisplatin resistance screen in bladder cancer cells. Our screen results show that cells with loss of MSH2 are more resistant to cisplatin. We validated this finding by showing that bladder cancer cell lines with knockdown of MSH2 had a reduction in cisplatin-mediated apoptosis. Consistent with our in vitro results, we found that MIBC tumors with low levels of MSH2 protein expression had poorer survival when treated with platinum-based chemotherapy compared with those with higher MSH2 expression. MSH2 levels did not associate with survival in patients who did not receive a pharmacologic or radiation treatment, suggesting that the association with survival is specific to platinum treatment.
2. Patients and methods
2.1. Cell culture, shRNA knockdown, and drug treatments
MGHU4 and 253J bladder cancer cell lines are representative of alterations found in MIBC [16,17]. Cells were cultured in Minimum Essential Medium media (Gibco) supplemented with 10% fetal bovine serum (Sigma-Aldrich). PLKO.1 TRC shRNAs were transduced using psPAX2 (Addgene plasmid #12260) and pMD2.G (Addgene plasmid #12259; see Supplementary Table 1 for shRNAs) [18]. Transduced cells were selected with 2–5 μg/ml puromycin (Sigma-Aldrich). Cisplatin, gemcitabine (Sigma-Aldrich), methotrexate, doxorubicin, vinblastine, and oxaliplatin (ApexBio) were solubilized in water or dimethyl sulfoxide. For dose response experiments, cell viability was measured using the CellTiter-Glo luminescent assay (Promega).
2.2. Performing the CRISPR resistance screen
To generate sgRNA lentivirus, 12 μg of the human GeCKO (Genome-Scale CRISPR Knock-Out) lentiviral A library was transfected into 30 million HEK293T cells for 24 h [19,20]. Viral supernatant was harvested, and MGHU4 cells were transduced at a calculated multiplicity of infection of 0.3 followed by selection with 2 μg/ml puromycin. Two million MGHU4 cells were plated in quadruplicate for each condition. Cells were treated with 3 μM cisplatin or vehicle for 30 h. Treatment media were removed and cells were allowed to grow to confluency prior to harvesting genomic DNA (Machery-Nagel).
2.3. Sequencing of sgRNA
The sgRNA sequences of each sample were polymerase chain reaction (PCR) amplified from 4 μg of genomic DNA using primers containing adaptor and barcoding sequences. DNA fragments were size selected using agarose gel and sequenced using a 1 × 125 bp run on the HiSeq2500 (Illumina). Reads generated from each sample were aligned to the indexed sgRNA sequences with the Bowtie2 sequence aligner using the “very-sensitive-local” option [21]. The counts of sgRNA were summarized using htseq [22]; sgRNA counts across all samples were compiled, and differential sgRNA abundance was calculated using DESeq2 [23]. To map the sgRNA results to the gene level (approximately three gRNAs per gene), we calculated the mean fold change and combined individual sgRNA p values from DESeq2 using Fisher’s method followed by multiple hypothesis testing correction using the Benjamini-Hochberg procedure [24]. The top resistant gene thresholds were set at a q value of <0.01 and log2 fold change of >2 or <−2 in the cisplatin-treated group compared with the vehicle control group.
2.4. Screen enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID), which uses a modified Fisher’s exact test followed by Benjamini-Hochberg multiple hypothesis testing correction [24], was used to determine the enriched gene ontology (GO) term biological processes [25]. In addition to GO term enrichment, Gene Set Enrichment Analysis (GSEA), which uses the Kolmogorov-Smirnov statistic, was used to determine the enrichment of the KEGG MMR pathway across the entire ranked gene list [26,27].
2.5. Western blots
For each sample, 25 μg of protein was loaded onto 10% SDS-PAGE and transferred to nitrocellulose membranes (BioRad) followed by a Western blot (see Supplementary Table 2 for antibodies). Membranes were imaged using the Odyssey Fc Imaging System (LI-COR) following application of ECL (Millipore Sigma).
2.6. Caspase activation
Cells were cotreated with the indicated concentration of cisplatin and 4 μM CellEvent Caspase-3/7 Green Detection Reagent (Thermo Fisher Scientific) according to manufacturer instructions. Phase contrast and green fluorescence were imaged every 3 h following initial treatment using the Incucyte Zoom system (Essen Bioscience) with a 4× objective. The displayed results are cumulative green cell counts normalized to total cell confluency. We compared the induction in cisplatin-treated MSH2 knockdown cells with that in treated nontargeting cells using a repeated-measure one-way analysis of variance (ANOVA; independent variable is knockdown).
2.7. Quantitative reverse transcriptase PCR
Total RNA was extracted using the NucleoSpin RNA kit (Machery-Nagel). Synthesis of cDNA was performed with the iScript cDNA Synthesis Kit (BioRad). The SsoFast Evagreen supermix (BioRad) was used for quantitative reverse transcriptase PCR assays (see Supplementary Table 3 for primer sequences). Target genes were normalized to mean GAPDH expression and a two-way ANOVA was performed, where the two factors were knockdown and treatment.
2.8. Analysis of bladder tumors
All human MIBC data were from The Cancer Genome Atlas (TCGA) [16]. Clinical, treatment, and survival data were downloaded using the TCGAbiolinks R package [28]. One hundred patients were treated with platinum-based (cisplatin or carboplatin) chemotherapy, 12 of whom were in the neoadjuvant setting, but molecular characterization was performed prior to any chemotherapy treatment [16]. Level 4, batch-normalized reverse-phase protein array (RPPA) was from the study by Li et al [29] and MSH2 mRNA (RNA Seq V2 RSEM) expression was from the studies by Gao et al [30] and Cerami et al [31]. TCGA bladder cancer patients receiving cystectomies were identified using data from the Broad GDAC Firehose [32]. We used neoantigens published by Robertson et al [16]. Overall, 340 TCGA patients had RPPA and clinical data (Supplementary Table 4). Overall survival was analyzed using the survival, survminer, and ggplot2 R packages [33–35]. Patient characteristics were compared using a Student t test or Fisher’s exact test. The primary endpoint is overall survival, which was defined as the time from diagnosis to death from any cause. Patients were censored at the point of their last follow-up (last known time the patient was alive). For all survival analyses, each arm was censored after the at-risk population had <10 patients. Statistical differences between two patient groups were evaluated with the log-rank test.
3. Results
3.1. Whole-genome CRISPR screen identifies mediators of cisplatin resistance
We executed an unbiased CRISPR screen (65 383 sgRNAs; approximately three sgRNAs per gene [19,20]) to identify sgRNA constructs that increased in the population of cisplatin-treated (3 μM for 30 h) compared with vehicle-treated cells (Fig. 1A). We identified 9757 sgRNA constructs that were differentially abundant between the two groups (Fig. 1B and 1C, and Supplementary Table 5). The top two gene candidates are MSH2 and MLH1 (Fig. 1D and Supplementary Table 6).
Fig. 1.
A whole-genome CRISPR screen to identify mediators of cisplatin resistance in a bladder cancer cell line. (A) Experimental outline of the screen and analysis. (B) A heatmap displaying median-centered counts for 9757 differentially abundant sgRNAs. (c) Counts from the screen for the top 10 resistant sgRNAs. (D) A volcano plot displaying the log2 fold change and adjusted p value for all sgRNAs identified in the screen. A threshold of adjusted p < 0.05 is indicated in the plot.
3.2. MMR pathway mediates cisplatin sensitivity
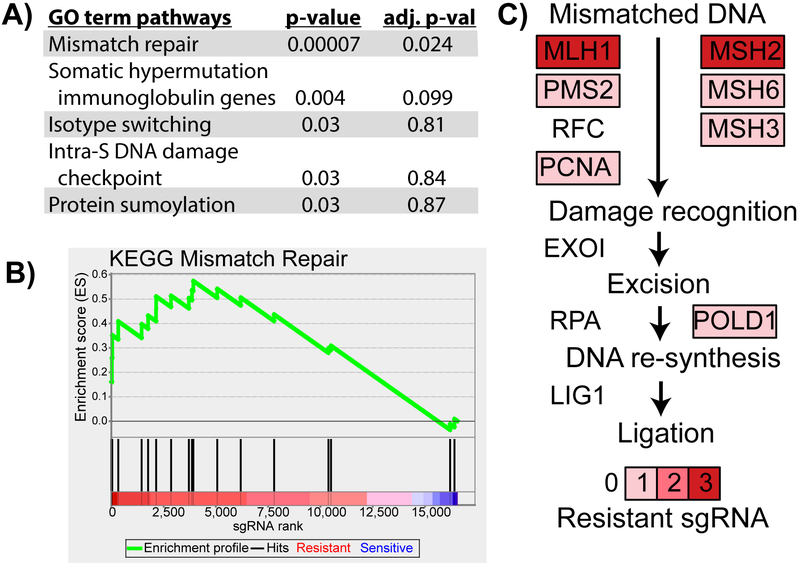
We performed pathway-level analysis to identify processes that mediate cisplatin resistance. GO term enrichment analysis [25] on the top 48 significantly resistant genes identified MMR as the top hit (Fig. 2A). To confirm this result, we tested MMR using GSEA across the 16 564 genes in our screen (ranked on mean fold change) and confirmed MMR pathway significance (Fig. 2B) [26]. MSH2 and MLH1 each had three significantly resistant sgRNAs, and several other genes in the MMR pathway had a single resistant sgRNA (Fig. 2C) [27].
Fig. 2.
sgRNA constructs targeting genes of the mismatch repair pathway are enriched in the cisplatin-treated MGHU4 cells. The mean log2 fold change and combined p value were calculated using the three sgRNAs targeting each gene in the screen. (A) Top GO term biological process results from DAVID enrichment analysis of the 48 genes in the screen with a combined q value of <0.01 and log2 mean fold change of >2 of the cisplatin- versus vehicle-treated group. Enrichment was determined by a modified Fisher’s exact test and corrected for multiple hypothesis testing (Benjamini-Hochberg). (B) GSEA results of the KEGG mismatch repair pathway on the ranked list of genes by mean log2 fold change (p < 0.05, Kolmogorov-Smirnov test). (C) Components and genes of the mismatch repair pathway are depicted with the number of significantly resistant sgRNAs from our synthetic lethal screen targeting each gene (out of three). adj. p-val = adjusted p value; DAVID = Database for Annotation, Visualization and Integrated Discovery; GO = gene ontology; GSEA = Gene Set Enrichment Analysis.
3.3. Bladder cancer cell lines with knockdown of MSH2 are resistant to cisplatin
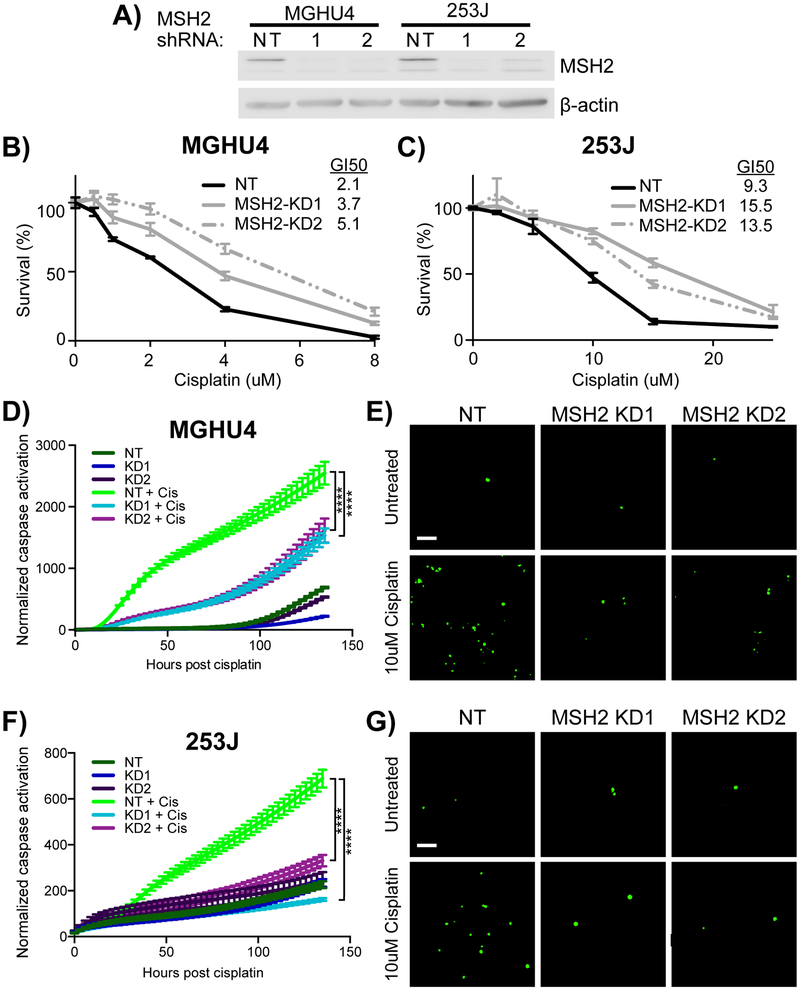
We moved forward with MSH2 because it was the top ranked and most statistically significant hit in our screen (Fig. 1D). Additionally, five studies found a subset of bladder cancers with lower expression of MSH2 [11–15]. Strong shRNA knockdowns of MSH2 resulted in 45–142% increases in resistance to cisplatin in MGHU4 and 253J bladder cancer cell lines (Fig. 3A–C). These cells had reduced caspase activation compared with nontargeting control, cisplatin-treated cells (Fig. 3D–G). These data show that the loss of MSH2 in cisplatin-treated bladder cancer cells increases survival and reduces apoptosis.
Fig. 3.
Knockdown of MSH2 increases cisplatin resistance in bladder cancer cell lines. Two shRNA constructs targeting MSH2 and one nontargeting shRNA control were transduced into MGHU4 and 253J bladder cancer cell lines. (A) Western blot showing the level of knockdown of MSH2. (B) MGHU4 and (C) 253J cell lines were treated with the indicated doses of cisplatin for 48 h, and cell viability was measured using an ATP-based assay (mean ± SEM of three independent experiments). The GI50 (μM) of cell line was calculated with using a four-parameter nonlinear regression fit using least squares. (D) MGHU4 and (F) 253J cells were treated with 10 and 15 μM cisplatin, respectively, or vehicle. Cumulative caspase activation (green cells) was measured and normalized to cell confluency. Representative images of (E) MGHU4 and (G) 253J cells are shown at the 24 h time point. Statistical significance of Figures 3D and 3F were determined using a repeated-measure one-way ANOVA (independent variable is knockdown). Data represents the mean ± SEM of a representative experiment (of three experiments). ANOVA = analysis of variance; SEM = standard error of the mean. **** p < 0.0001.
To investigate the role of the DNA-damage response, we treated MGHU4 and 253J MSH2 knockdown cells with cisplatin for 48 h, followed by measurements of genes involved in DNA-damage response (Supplementary Fig. 1). Cisplatin treatment in cells transduced with nontargeting shRNA led to an accumulation of p-ATM and p53. We observed reductions in the accumulation p-ATM and stabilization of p53 in MSH2 knockdown cells. Cisplatin increased the transcriptional response of BAX, MDM2, p21, and PUMA in nontargeting cells. However, in MSH2 knockdowns, induction of MDM2 is reduced, and in the 253J cells, induction of BAX and p21 was also attenuated. While we observed a stronger DNA-damage response in MSH2 knockdown cells than was reported in past studies, there was a reduction in some of these factors [3,36,37].
3.4. Cells with low MSH2 are equally sensitive to oxaliplatin and other chemotherapies
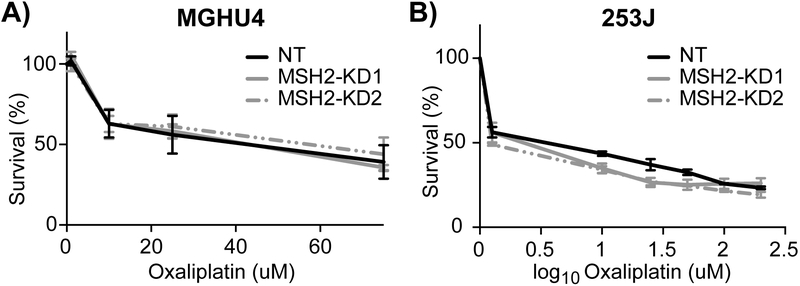
Previous work has shown that the loss of MMR does not affect sensitivity to the cisplatin analog oxaliplatin [38]. We tested oxaliplatin response, as well as the other constituent components of the MVAC and GC regimens, in bladder cancer cells. Cells with reduced MSH2 were equally sensitive to all of the tested chemotherapies compared with control (Fig. 4 and Supplementary Fig. 2). These results indicate that MSH2 loss mediates resistance to cisplatin specifically, without affecting the other drugs in the standard MVAC and GC treatments or the cisplatin analog oxaliplatin.
Fig. 4.
MSH2 knockdown bladder cancer cell lines are equally sensitive to oxaliplatin. (A) MGHU4 and (B) 253J bladder cancer cell lines were treated with the indicated doses of oxaliplatin for 48 h, and cell viability was measured using an ATP-based assay.
3.5. MSH2 protein levels do not correlate with clinicopathologic features in bladder cancer
Studies have shown conflicting evidence that bladder cancers with reduced MSH2 protein expression associate with stage, grade, and survival [11–15]. To investigate the clinical impact of MSH2, we analyzed 340 bladder cancer patients from TCGA with both molecular and clinical data [16]. MSH2 and MSH6 are tightly linked as they are components of the MutSα MMR complex; their strong correlation strengthens our confidence in the TCGA RPPA data (Supplementary Fig. 3A, ρ = 0.66). We found a weak correlation between MSH2 mRNA and protein levels, indicating that mRNA levels are a poor surrogate for MSH2 protein levels in bladder cancer (Supplementary Fig. 3B, ρ = 0.29). Surprisingly, low MSH2 protein levels did not have an impact on the number of neoantigens, stage, or grade, suggesting that MSH2 is independent of these features (Supplementary Fig. 3C–E) [16].
3.6. MSH2 protein levels correlate with survival in platinum-treated bladder cancer patients
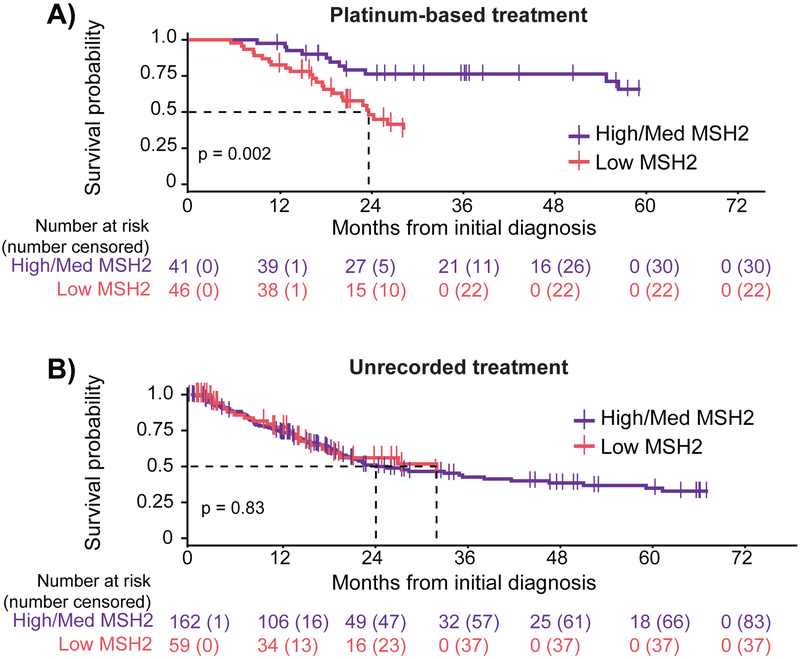
We investigated whether MSH2 protein levels correlate with survival in platinum-treated (cisplatin or carboplatin) patients. We divided TCGA bladder cancer patients into three equal groups based on their MSH2 protein expression (Supplementary Fig. 4A), and following the work of prior biomarker studies [39,40], we compared the low MSH2 group with the combined medium/high groups. We found that for patients treated with platinum-based therapy, individuals with low MSH2 had significantly poorer survival compared with patients with medium or high protein levels (Supplementary Fig. 4B and Fig. 5A, p = 0.002, log-rank test). We next investigated low MSH2 protein levels in patients without a recorded pharmacologic or radiation treatment, and found that in these patients, MSH2 protein level did not have any association with survival (Supplementary Fig. 4C and Fig. 5B, p = 0.83, log-rank test). For these patients, we found no discernable differences in clinical characteristics (Supplementary Tables 7 and 8). These results suggest that low MSH2 protein identifies patients less responsive to platinum therapy and is not simply prognostic of overall survival.
Fig. 5.
Platinum-treated bladder cancer patients with low levels of MSH2 protein correlate with poorer survival. Survival of patients was compared with MSH2 RPPA level in 340 bladder cancer patients. Overall survival is plotted for bladder cancer patients with a (A) platinum-based treatment or (B) nonpharmacologic or radiation treatment. Survival of patients with low MSH2 (red) is compared with those with medium or high levels of mSh2 (purple). The statistical difference in survival was calculated using a log-rank test. med = medium; RPPA = reverse-phase protein array.
4. Discussion
No clinically actionable biomarker of resistance to first-line chemotherapy in MIBC is currently available. The MMR pathway has been shown to impact the response to cisplatin-based chemotherapy in multiple cancers [3–6,8–10]; however, clinical responses vary across cancer type. For platinum-treated patients, low MSH2 correlates with poor survival in ovarian cancer [8] but improved survival in NSCLC [9,10]. Here, we report the first association between low MSH2 protein and poorer survival in bladder cancer patients treated with platinum-based therapy, and this biomarker is not simply prognostic of patient survival.
Consistent with in vitro studies in other cancer types [3,36,37], we observed a reduction in the DNA-damage response when cells had low levels of MSH2. It is unclear whether this reduction alone accounts for the cisplatin resistance that we observed. Future studies are needed to address additional mechanisms of response and also to determine whether the DNA-damage response can serve as a therapeutic target to address MMR-mediated cisplatin resistance. Microsatellite instability has been linked to both MMR and chemotherapy response in other cancer types; yet, it is infrequent in bladder tumors [12,13,41] and thus cannot be used as an effective bladder cancer biomarker. It has previously been shown that a subset of bladder tumors have low to no expression of MSH2 [11–15]. While patients expressing low levels of MSH2 protein had an impaired response to platinum-based chemotherapy, they do not have microsatellite instability or an increase in neoantigens (Supplementary Fig. 3C). Future work will investigate the potential separation of MMR functions in regard to DNA repair activity versus the response to cisplatin.
The disease control rate of oxaliplatin-based therapy in patients who have failed cisplatin- or carboplatin-based chemotherapy is 36% [42]. We found that oxaliplatin and the noncisplatin components of the MVAC and GC chemotherapy regimens were equally effective in bladder cancer cell lines irrespective of MSH2 expression. Therefore, an unanswered question is whether bladder tumors with low MSH2 levels would respond well to MVAC and GC regimens with oxaliplatin being substituted for cisplatin.
Current protein diagnostics rely on IHC [43]. For this reason, it will be necessary to validate our findings using IHC on patient samples. Our results are based on retrospective patient data that were not collected as a standardized clinical trial [16]. The collected samples covered a broad range of patient characteristics in relation to clinicopathologic features. Similarly, some of these patients were treated with MVAC, while others received GC; 12% of platinum-treated patients were provided neoadjuvant treatment. The lack of standardized patient selection presents confounding factors that are better controlled within a clinical trial. However, using the patient and tumor characteristics available to us, we did not find notable differences by MSH2 expression that would influence our results (Supplementary Tables 7 and 8).
5. Conclusions
In this study, we found that MSH2 contributes to in vitro resistance to cisplatin in bladder cancer cell lines. We showed that patients with low levels of MSH2 had poorer survival when treated with platinum-based chemotherapy. Future studies will be necessary to validate MSH2 protein levels as a prospective biomarker of platinum-based therapeutic resistance in bladder cancer.
Supplementary Material
Acknowledgments:
We would like to thank Robert T. Jones, Kelly D. Sullivan, and Rani K. Powers for insightful discussions. We would also like to thank the Functional Genomics Facility for their genomic library expertise.
Funding/Support and role of the sponsor: This work was supported by the Boettcher Foundation (J.C.C.), T32GM007635 (A.G.), the Front Range Cancer Challenge (A.G.), and in part by the Genomics, Functional Genomics, and the Biostatistics & Bioinformatics Shared Resource of the University of Colorado Cancer Center (P30CA046934).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A whole-genome screen identified MSH2 loss as a mediator of cisplatin resistance in bladder cancer cell lines. In vitro results showed that MSH2 depletion reduced cisplatin-mediated apoptosis. Bladder tumors with low MSH2 protein are resistant to platinum-based therapy.
Financial disclosures: James C. Costello certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
References
- [1].Mahdavifar N, Ghoncheh M, Pakzad R, Momenimovahed Z, Salehiniya H. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer Prev APJCP 2016;17:381–6. [DOI] [PubMed] [Google Scholar]
- [2].von der Maase H, Hansen SW, Roberts JT, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: Results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol 2000;18:3068–77. [DOI] [PubMed] [Google Scholar]
- [3].Gong JG, Costanzo A, Yang HQ, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999;399:806–9. [DOI] [PubMed] [Google Scholar]
- [4].Brown R, Hirst GL, Gallagher WM, et al. hMLH1 expression and cellular responses of ovarian tumour cells to treatment with cytotoxic anticancer agents. Oncogene 1997;15:45–52. [DOI] [PubMed] [Google Scholar]
- [5].Samimi G, Fink D, Varki NM, et al. Analysis of MLH1 and MSH2 expression in ovarian cancer before and after platinum drug-based chemotherapy. Clin Cancer Res 2000;6:1415–21. [PubMed] [Google Scholar]
- [6].Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res 2004;10:4420–6. [DOI] [PubMed] [Google Scholar]
- [7].Ding X, Mohd AB, Huang Z, et al. MLH1 expression sensitises ovarian cancer cells to cell death mediated by XIAP inhibition. Br J Cancer 2009;101:269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ercoli A, Ferrandina G, Raspaglio G, et al. hMSH2 and GTBP expression in advanced stage epithelial ovarian cancer. Br J Cancer 1999;80:1665–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kamal NS, Soria J-C, Mendiboure J, et al. MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clin Cancer Res 2010;16:1206–15. [DOI] [PubMed] [Google Scholar]
- [10].Levallet G, Dubois F, Fouret P, et al. MSH2/BRCA1 expression as a DNA-repair signature predicting survival in early-stage lung cancer patients from the IFCT-0002 phase 3 trial. Oncotarget 2017;8:4313–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mylona E, Zarogiannos A, Nomikos A, et al. Prognostic value of microsatellite instability determined by immunohistochemical staining of hMSH2 and hMSH6 in urothelial carcinoma of the bladder. APMIS 2008;116:59–65. [DOI] [PubMed] [Google Scholar]
- [12].Roupret M, Fromont G, Azzouzi A-R, et al. Microsatellite instability as predictor of survival in patients with invasive upper urinary tract transitional cell carcinoma. Urology 2005;65:1233–7. [DOI] [PubMed] [Google Scholar]
- [13].Catto JWF, Xinarianos G, Burton JL, Meuth M, Hamdy FC. Differential expression of hMLH1 and hMSH2 is related to bladder cancer grade, stage and prognosis but not microsatellite instability. Int J Cancer 2003;105:484–90. [DOI] [PubMed] [Google Scholar]
- [14].Kassem HS, Varley JM, Hamam SM, Margison GP. Immunohistochemical analysis of expression and allelotype of mismatch repair genes (hMLH1 and hMSH2) in bladder cancer. Br J Cancer 2001;84:321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jin T-X, Furihata M, Yamasaki I, et al. Human mismatch repair gene (hMSH2) product expression in relation to recurrence of transitional cell carcinoma of the urinary bladder. Cancer 1999;85:478–84. [PubMed] [Google Scholar]
- [16].Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017;171:540–56. e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nickerson ML, Witte N, Im KM, et al. Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene 2017;36:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moffat J, Grueneberg DA, Yang X, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 2006;124:1283–98. [DOI] [PubMed] [Google Scholar]
- [19].Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 1995;57:289–300. [Google Scholar]
- [25].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [26].Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016;44: D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li J, Lu Y, Akbani R, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013;10:1046–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Broad Institute of MIT and Harvard. Broad Institute TCGA Genome Data Analysis Center (2016): Firehose stddata_2016_01_28 run. http://ezid.cdlib.org/id/doi:10.7908/C11G0KM9 [Google Scholar]
- [33].Therneau TM, Grambsch PM. Modeling survival data: extending the Cox model. New York, NY: Springer-Verlag; 2000. [Google Scholar]
- [34].Kassambara A, Kosinski M, Biecek P, Fabian S. survminer, CRAN, v0.4.3; 2018. https://cran.r-project.org/web/packages/survminer/index.html [Google Scholar]
- [35].Wickham H ggplot2: Elegant graphics for data analysis. New York, NY: Springer; 2016. [Google Scholar]
- [36].Luo Y, Lin F-T, Lin W-C. ATM-mediated stabilization of hMutL DNA mismatch repair proteins augments p53 activation during DNA damage. Mol Cell Biol 2004;24:6430–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yoshida K, Ozaki T, Furuya K, et al. ATM-dependent nuclear accumulation of IKK-alpha plays an important role in the regulation of p73-mediated apoptosis in response to cisplatin. Oncogene 2008;27:1183–8. [DOI] [PubMed] [Google Scholar]
- [38].Fink D, Nebel S, Aebi S, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Res 1996;56:4881–6. [PubMed] [Google Scholar]
- [39].Font A, Taron M, Gago JL, et al. BRCA1 mRNA expression and outcome to neoadjuvant cisplatin-based chemotherapy in bladder cancer. Ann Oncol 2011;22:139–44. [DOI] [PubMed] [Google Scholar]
- [40].Hoffmann A-C, Wild P, Leicht C, et al. MDR1 and ERCC1 expression predict outcome of patients with locally advanced bladder cancer receiving adjuvant chemotherapy. Neoplasia 2010;12:628–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342–50. [DOI] [PubMed] [Google Scholar]
- [42].Srinivas S, Harshman LC. A phase II study of docetaxel and oxaliplatin for second-line treatment of urothelial carcinoma. Chemotherapy 2009;55:321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Boellner S, Becker K-F. Reverse phase protein arrays—quantitative assessment of multiple biomarkers in biopsies for clinical use. Microarrays 2015;4:98–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.